Abstract
This study is based on the evidence that immunization of macaques with human CD4+ T cells elicits prevention of simian immunodeficiency virus (SIV) infection. We hypothesized that heat-shock protein 70 (HSP70) isolated from CD4+ T cells may act as a chaperone and carry the protective host proteins. Two moieties of HSP70 were affinity-purified from human CD4+ T cells; an ADP preparation with HSP70-bound proteins (ADP-HSP) and an ATP control preparation. Immunization of rhesus macaques with these preparations showed significant inhibition of SIVmac251 infectivity ex vivo in CD4+ T cells only with the ADP-HSP (P = 0·01). Proteomic analysis identified three cytoskeletal elements, cofilin, profilin and γ-actin, exclusively in the ADP-HSP preparation. Investigation of the mechanism of prevention of SIV replication suggests that antibodies to the cytoskeletal proteins may inhibit actin depolymerization and facilitate viral degradation by the innate antiviral APOBEC3G. As cytoskeletal proteins are critical in the formation of virological and immunological synapses, finding specific antibodies and anti-SIV/human immunodeficiency virus (HIV) factors suggests a novel insight into HIV-1 immunopathogenesis.
Keywords: AIDS, T cells, vaccination, viral
Introduction
The goal of a preventive vaccine against human immunodeficiency virus (HIV)-1 has not so far been realized and the HIV-1 epidemic in many developing countries has not been contained. An early finding that simian immunodeficiency virus (SIV) infection can be prevented in 85% of rhesus macaques immunized with SIV grown in human CD4+ T-cells lines1–6 was not pursued after the demonstration that uninfected human CD4+ T cells could also induce significant prevention of SIV infection in macaques.7–9 This protection was ascribed to multifactorial immune responses against xenogeneic human CD4+ cellular antigens, especially human leucocyte antigens (HLAs), but the mechanism of protection has not been established.10 The present investigation was based on the hypothesis that the protective CD4+ T-cell antigen might be carried by HSP70 chaperones. HSP70 plays an important role in innate and adaptive immunity11 and may facilitate loading and processing of antigenic epitopes into major histocompatibility complex (MHC) class II.12 It is also capable of cross-presentation of peptides by the HLA class I pathway.13 HSP70 is also a potent stimulator of CC chemokines14 and cytokine production.15 It may function as an intrinsic adjuvant by virtue of binding the C-C motif of CCR5, CD40 and Toll-like receptor 4 (TLR4).14,16–18 Furthermore, HSP70 stimulates CD4+ T cells and dendritic cells (DCs) to produce apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G), which inhibits HIV-1 or SIV.19
The rationale for the protocol design was based on the structure and chaperone function of HSP70. This consists of a 44-kDa ATPase, an 18-kDa substrate-binding domain and a 10-kDa C-terminal fragment. The C-terminal substrate-binding domain of the Escherichia coli HSP70 homologue (termed DnaK) consists of a β-sandwich and an α-helical subdomain. The upper sheet of the β-sandwich forms the substrate-binding site with loops L1-2 and L3-4 forming the sides of a channel, which is the primary site of interaction with the substrate. Any protein or peptide bound in the channel can be eluted by ATP, whereas ADP treatment retains it. This led to the hypothesis that isolating HSP70 from human CD4+ T cells using an ADP column may retain any protective proteins, whereas these will be eluted from the HSP70 prepared on an ATP column. Proteomic analysis of these two preparations may identify the putative protein retained by the ADP-HSP70 preparation. Thus, the hypothesis was examined that a preparation of HSP70 isolated from human CD4+ T cells may carry proteins or peptides that protect macaques from SIV infection. ADP and ATP column preparations of HSP70 from a human CD4+ T-cell line (C8166) were adsorbed to alum and used to immunize rhesus macaques. Significant inhibition of SIVmac251 resulted when macaques were immunized with the ADP-HSP70 moiety, which would retain proteins derived from the host CD4+ T cells, unlike that with ATP-HSP70. This was correlated with a significant increase in the intracellular innate anti-HIV-1 APOBEC3G (A3G) factor in CD4+ T cells in the ADP-HSP70, compared with the ATP-HSP70 moiety. To identify the putative proteins, two-dimensional electrophoresis, followed by mass spectrometry of the resulting spots, revealed cofilin, profilin and γ-actin in the ADP-HSP70 preparation. These three cytoskeletal elements stimulated up-regulation of APOBEC3G expression in CD4+ T cells from ADP-HSP-immunized macaques and showed an increase in serum immunoglobulin G (IgG) antibodies. These findings suggest a potentially novel strategy of immunization against HIV-1 infection.
Materials and methods
Extraction of human HSP70 from C8166 cells
C8166 cells (ECACC, Porton Down, UK) were grown in RPMI-1640 medium containing 10% fetal calf serum (FCS) and supplemented with 2 mm l-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin (Sigma-Aldrich Ltd, Poole, UK). HSP70 was extracted from a 10-ml packed cell volume by Dounce homogenization in 40 ml of hypotonic buffer [10 mm NaHCO3 and 0·5 mm phenylmethylsulphonyl fluoride (PMSF), pH 7·1]. After centrifugation at 100 000 g, the supernatant was buffer-exchanged into 20 mm Tris-acetate, 20 mm NaCl, 15 mmβ-mercaptoethanol, 3 mm MgCl2 and 0·5 mm PMSF, pH 7·5 (buffer D). The sample was applied to a 5-ml ADP-agarose column (Sigma-Aldrich) equilibrated with buffer D. The column was washed with buffer D containing 0·5 m NaCl and then buffer D alone, until no more protein could be detected by Bradford protein assay (BioRad, Hemel Hempstead, UK). The column was then incubated at room temperature for 30 min in buffer D containing 3 mm ADP (Sigma-Aldrich) and bound proteins were eluted with the same buffer. The same procedure was performed for the HSP70 preparation with an ATP-agarose column and ATP elution.20 The preparations were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) on 12% gels, which were either silver-stained with Silver Quest® (Invitrogen, Paisley, UK) or stained with Colloidal Coomassie Blue (Sigma-Aldrich) (Fig. 1a). The preparations were then concentrated by centrifugal filtration using a molecular weight cut-off of 10 kDa (Millipore, UK Ltd, Watford, UK) and dialysed against sterile saline for use in immunogens.
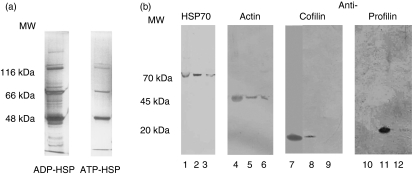
Figure 1.
(a) Separation and characterization of ADP-heat-shock protein (HSP) and ATP-HSP proteins using sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining. (b) Western blot analysis of ADP-HSP and ATP-HSP proteins. Controls: lane 1, C8166 cell lysate; lane 4, non-muscle actin; lane 7, recombinant cofilin; lane 11, recombinant profilin; ADP-HSP: lanes 2, 5, 8 and 12; ATP-HSP: lanes 3, 6, 9 and 10. MW, molecular weight.
Characterization of proteins and peptides
2D gel electrophoresis and mass spectrometry
The extracted ADP-HSP and ATP-HSP preparations were concentrated and loaded onto 24-cm immobiline DryStrip first-dimension IPG strips, pH 3–11, non-linear (GE Healthcare, Little Chalfont, UK) by passive rehydration. Isoelectric focusing was performed (500 V for 1 hr, 1000 V for 1 hr and 8000 V for 8·5 hr). The focused strips were incubated for 15 min sequentially in SDS equilibration buffer (50 mm Tris–HCl, pH 8·8, 6 m urea, 30% (v/v) glycerol, 2% SDS and 0·002% bromophenol blue), firstly with 1% (w/v) dithiothreitol (DTT) and then with 2·5% (w/v) iodoacetamide. Proteins were then separated in the second dimension on 12·5% SDS-PAGE gels (Ettan DALT 6 separation unit; GE Healthcare). Gels were fixed and stained overnight using Sypro Ruby total protein stain (Invitrogen). A total of 40 spots were analysed (34 from ADP-HSP; six from the ATP-HSP 2D gels). Spots were excised from the gel using the Investigator ProPic automated spot picker (Perkin Elmer, Waltham, MA) and digested with trypsin using the ProGest automated digestion unit (Perkin Elmer). Peptides were analysed using a 4700 MALDI-Tof/Tof mass spectrometer (Applied Biosystems, Foster City, CA) to give a peptide mass fingerprint and peptide sequence information, which was searched against the mass spectrometry database (MSDB) using the Mascot search engine from Matrix Science (London, UK) to identify the protein.
Peptide elution
Material bound to ADP-HSP was eluted by incubation of 100 μg of ADP-HSP or ATP-HSP (negative control) with 10 mm ATP plus 4 mm MgCl2 for 30 min at room temperature. Peptides were then separated using 5-kDa centrifugal filters. The released proteins were subjected to proteomic analysis.
Western blot analysis
ADP-HSP and ATP-HSP preparations were separated on 12% SDS-PAGE gels and western blotting was carried out following transfer onto nitrocellulose filters. Blots were probed with antibodies specific for human HSP70 and heat shock protein cognate-1 (HSC70) (Stressgen, Cambridge, UK), anti-human cofilin, profilin and non-muscle actin antibodies (Bioquote, York, UK and Sigma-Aldrich) (Fig. 1a,b).
Immunization
Nine adult rhesus macaques were housed according to the UK Home Office guidelines as set out in the Animals (Scientific Procedures) Act 1996. The animals were sedated with ketamine hydrochloride (10 mg/kg) and dopram (10 mg/kg). The dialysed HSP preparations were adsorbed to alum and administered intramuscularly (i.m.) (× 3) at monthly intervals into the buttocks and biceps (100 μg of ADP-HSP, n = 5; and 100 μg of ATP-HSP, n = 4). Venous blood samples were taken before and 4 weeks after each immunization. At the end of the study the animals were killed using an anaesthetic overdose.
Serum antibody responses
An enzyme-linked immunosorbent assay (ELISA) was used for antibodies to the three cytoskeletal proteins identified by proteomic analysis and for HSP70. Recombinant human HSP70 was provided by Dr Paul Lehner (University of Cambridge, Cambridge, UK), and cofilin, profilin and non-muscle actin were purchased from Bioquote. γ- and β-actin were purchased from GenWay Biotech Inc. (San Diego, CA) A Luminex assay (BioRad, Watford, UK) was used for antibodies to human HLA class I and class II.
Dose-dependent inhibition of SIV infectivity of CD4+ T cells from macaques immunized with ATP-HSP and ADP-HSP
Infectivity studies were carried out using CD4+ T cells enriched from the peripheral blood mononuclear cells (PBMCs) of macaques, by negative selection on magnetic cell-sorting columns (Miltenyi Biotec, Bisley, UK). More than 95% of the cells that did not bind to the column were CD4+. The CD4+ T cells were activated with 10 μg/ml phytohaemagglutinin (PHA) (Sigma-Aldrich) in RPMI and 10% FCS (Biosera, Ringer, UK), penicillin at 100 U/ml, streptomycin at 100 μg/ml, 2 mm l-glutamine (Sigma-Aldrich) and 20 IU interleukin-2 (IL-2; Schiaparelli Biosystoems BV, Woerden, the Netherlands) in culture medium for 3 days at 37° in 5% CO2. Activated CD4+ T cells were infected over 2 hr with serial dilutions of SIVmac251 [30 000 counts per minute (cpm)/100 μl reverse transcriptase (RT) activity; obtained from the NIBSC, Potters Bar, UK], giving multiplicity of infection (MOI) values of 10–104. The cells were then washed three times and plated out in a 96-well plate at 2 × 105 cells/well. Every 2 days, 100 μl of the culture supernatant was replaced with 100 μl of medium supplemented with IL-2 and on day 9 the RT activity was assayed.21 CD4+ T cells were studied from macaques immunized with the ADP-HSP70 and ATP-HSP70 material, before and after immunization.
Serum neutralizing activity
The SIV inoculum was adjusted to contain 2000 TCID50/ml in medium (TCID50 being the 50% tissue culture infective dose). Aliquots of 60 ug were incubated with 60 ul of a serial dilution of immunized and control monkey sera for 1 hr at 37°22. Then 100 μl of the mixture was transferred to 96-well flat-bottom plates containing 10 × 104 C8166 cells in 100 μl medium/well. The cultures were incubated in CO2 at 37° for 3 days and washed (five times) with medium, and 150 μl of uninfected C8166 cells (1 × 104 cells) were added, with the final volume adjusted to 200 μl/well. The cultures were incubated for three more days and on day 6 post-infected 100 μl of the supernatant was assayed for SIV p27 using the Zeptometrix kit (Zeptometrix, Buffalo, NY).
APOBEC3G mRNA assay of PBMCs from macaques immunized with ATP-HSP and ADP-HSP
RNA was isolated from 2 × 106 PBMCs using the Total RNA Isolation Kit (Promega, Southampton, UK) and cDNA was generated from RNA by reverse transcription–polymerase chain reaction (RT-PCR) using the Reverse Transcription System (Promega). Rhesus macaque A3G mRNA expression was determined by real-time PCR using previously described macaque A3G primers and method.19 Real-time PCR amplification within samples was normalized using GAPDH amplification and data analysis was performed as described before.19 The same method was used to assay A3G mRNA expression in PBMCs stimulated with optimal concentrations of cofilin (10 μg/ml), profilin (0·3 μg/ml) and β/γ non-muscle actin (0·15 μg/ml) purchased from Genway Biotech Inc.
Statistics
The data were analysed by the paired Student’s t-test, with the pre- and post-immunization data being compared in the same macaques or two means being compared between different animal groups.
Results
The effects of ADP-HSP and ATP-HSP on SIV infectivity of CD4+ T cells ex vivo following immunization of macaques
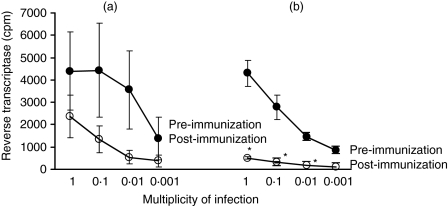
The rationale for studying immunization with the ADP-HSP70 is that it may bind a protective protein, which would have been eluted in the ATP-HSP70 preparation. Macaques immunized with the ADP-HSP material in alum (× 3), in which proteins or peptides bound to the HSP70 may be retained, showed a significant decrease in infectivity when the pre- and post-immunization CD4+ T cells were compared with all except the last dilution of SIV (P = 0·01, Fig. 2b). Although immunization with ATP-HSP in alum showed a greater ex vivo decrease in SIV infectivity for the post- than pre-immunization CD4+ T cells, this was statistically not significant (Fig. 2a). SIV infectivity of CD4+ T cells from macaques immunized with ADP-HSP70 was also significantly lower compared with those from ATP-HSP70-immunized macaques at SIV concentrations of 1 (P = 0·020) and 0·1 MOI (P = 0·036) (Fig. 2a,b). An intriguing observation was the difference in sensitivity to SIV infection in the pre-immune CD4+ T cells between the ATP-HSP70 and ADP-HSP70 preparations, but there was no statistically significant difference between them (P = 0·976). These results are consistent with the hypothesis that ADP-HSP70 may include proteins or peptides acquired from the human CD4+ T cells, which elicited a protective anti-SIV response.
Figure 2.
Dose-dependent simian immunodeficiency virus (SIV) infectivity of CD4+ T cells before and after immunization of macaques (n = 9) with heat-shock protein 70 (HSP70) isolated from a human CD4+ T-cell line (C8166) by (a) ATP (n = 4) compared with (b) ADP (n = 5) column separation. *P = 0·01 pre- versus post-immunization.
Identification of the ADP- and ATP-HSP bound material
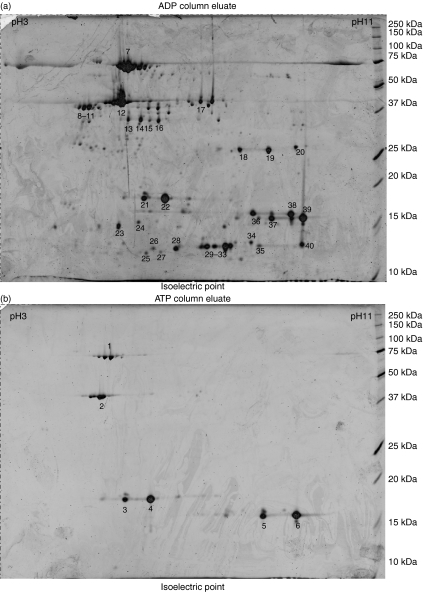
Having established that the putative protective antigens might be associated with ADP-HSP70, the critical point was to identify the protein. Proteomic analysis was carried out on material separated by 2D gel electrophoresis (Fig. 3a,b). Of the six spots analysed from the ATP-HSP preparation, spot 1 was identified as human DnaK, spot 2 as β-actin and spots 3–6 as nucleoside diphosphate kinase (NDPK). Thirty-four spots were analysed from the ADP-HSP preparation; NDPK was identified in 12 spots (22–30, 32–34, 37 and 39), DnaK in spots 7 and 19, HSC-1 in three spots (8–10), HSP70–2 in spot 11 and HSC54 in spots 18 and 20. In addition, pyridoxal kinase, adenosine kinase and protein kinase C interacting protein 1 were identified (spots 15–16, 17 and 31, respectively).
Figure 3.
Two-dimensional electrophoresis (DE) gel map of human heat-shock protein 70 (HSP70)-associated proteins eluted from (a) ADP-agarose with 3 mm ADP and (b) ATP-agarose with 3 mm ATP. The proteins were separated using a 24-cm immobiline non-linear strip (pH 3–11) followed by 12·5% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were stained with Sypro Ruby and 40 representative spots were selected for mass spectrometry.
Additional spots were observed in the ADP-HSP and identified as the cytoskeletal proteins cofilin (spots 36, 38), profilin (spots 35 and 40) and γ-actin (spot 12), along with two spots identified as hypothetical proteins of the actin family (spots 13 and 14) (Fig. 3 and Table 1). Thus, further studies were pursued with these proteins, as they were limited to the ADP-HSP70 preparations and may account for protection in SIV infection. Western blot analysis of the ADP-HSP and ATP-HSP proteins with antibodies to the three cytoskeletal proteins confirmed cofilin, profilin and β/γ-actin in the ADP-HSP preparation (Fig. 1b), but β-actin was also found in the ATP-HSP preparation, as indicated by the proteomic analysis. Proteomic analysis of proteins released from the ADP-HSP moiety following incubation with 10 mm ATP identified plexin B and protein kinase c inhibiting protein 1 (data not shown).
Table 1.
Characterization of proteins fractionated by two-dimensional polyacrylamide gel electrophoresis (PAGE) and identified by MALDI-Tof/Tof mass spectrometry
| Spot no. | Protein name | Accession number | Theoretical pI | Theoretical MW | Protein score | Sequence coverage (%) | Number of matched peptides | Precursor ion mass | Peptide sequence | Ion score |
|---|---|---|---|---|---|---|---|---|---|---|
| ATP 1 | DnaK-Hu | A27077 | 5·37 | 70 854·2 | 565 | 45·66 | 26 | 1981·9978 | TVTWAWTVPAYFNDSQR | 129 |
| 2 | β-Actin | Q96HG5 | 5·78 | 40 477·2 | 608 | 58·9 | 16 | 1790·8918 | SYELPDGQVITGNER | 122 |
| 3 | NDPK | A33386 | 5·83 | 17 137·7 | 354 | 50·65 | 11 | 1344·7634 | TFIAIKPDGVQR | 84 |
| 4 | NDPK | A33386 | 5·83 | 17 137·7 | 344 | 70·39 | 12 | 1149·6415 | DRPFFAGLVK | 79 |
| 5 | NDPK | 1NUEA | 8·55 | 17 155·9 | 368 | 74·8 | 12 | 1344·7634 | TFIAIKPDGVQR | 100 |
| 6 | NDPK | 1NUEA | 8·55 | 17 155·9 | 305 | 78·14 | 12 | 1175·6571 | DRPFFPGLVK | 50 |
| ADP 7 | DnaK-Hu | A27077 | 5·37 | 70 854·2 | 397 | 48·29 | 26 | 2774·3267 | QTQFTTYSDNA QPGVLIQVYEGER | 34 |
| 8 | HSC-1 | Q961S6 | 5·36 | 64 562·1 | 273 | 26·92 | 15 | 1081·5676 | LLQDFFNGK | 54 |
| 9 | HSC-1 | Q961S6 | 5·36 | 64 562·1 | 331 | 23·67 | 14 | 1253·6161 | FEELNADLFR | 72 |
| 10 | HSC-1 | Q961S6 | 5·36 | 64 562·1 | 335 | 33·73 | 16 | 1253·6161 | FEELNADLFR | 72 |
| 11 | HSP70-2 | AAD21815 | 5·48 | 69 982 | 182 | 24·02 | 15 | 1109·5789 | LLQDFFNGR | 29 |
| 12 | γ-Actin | P63261 | 5·55 | 40 194·1 | 431 | 54·7 | 17 | 1516·7026 | QEYDESGPSIVHR | 94 |
| 13 | HP (actin family) | Q8WVW5 | 5·78 | 40 477·2 | 367 | 48·7 | 12 | 1790·8916 | SYELPDGQTIVIGNER | 90 |
| 14 | HP (actin family) | Q8WVW5 | 5·78 | 40 477·2 | 210 | 22·8 | 6 | 1790·8916 | SYELPDGQTIVIGNER | 83 |
| 15 | Pyridoxal kinase | BAA95540 | 5·75 | 35 079·9 | 115 | 29·5 | 10 | – | – | |
| 16 | Pyridoxal kinase | BAA95540 | 5·75 | 35 079·9 | 390 | 48·29 | 16 | 2282·289 | VVPLADIITPNQFEAELLSGR | 73 |
| 17 | Adenosine kinase | G02049 | 6·09 | 37 488 | 370 | 32·6 | 14 | 1171·6582 | AGHYAASIIIR | 80 |
| 18 | HSC-54 | BAB18615 | 5·62 | 53 484 | 446 | 37·93 | 19 | 1981·9978 | TVTNAVVTVPAYFNDSQR | 88 |
| 19 | DnaK-Hu | A27077 | 5·37 | 70 854·2 | 356 | 30·03 | 20 | 1961·9976 | TVTNAVVTVPAYFNDSQR | 70 |
| 20 | HSC-54 | BAB18615 | 5·62 | 53 484·4 | 308 | 33·26 | 15 | 1961·9978 | TVTNAVVTVPAYFNDSQR | 42 |
| 21 | HSnm23 | CAA35621 | 7·07 | 20 398·3 | 226 | 63·33 | 12 | 1344·7634 | TFIAIKPDGVOR | 45 |
| 22 | NDPK | A33386 | 5·83 | 17 137·6 | 162 | 70·39 | 12 | 1149·6415 | DRPFFAGLVK | 11 |
| 23 | NDPK | A33386 | 5·83 | 17 137·6 | 119 | 56·57 | 8 | 1149·6415 | DRPFFAGLVK | 18 |
| 24 | NDPK | A33386 | 5·83 | 17 137·6 | 151 | 56·77 | 9 | 1149·6415 | DRPFFAGLVK | 17 |
| 26 | NDPK | 1NSKR | 8·87 | 17 157·9 | 70 | 50·33 | 6 | 1175·6571 | DRPFFPGLVK | 5 |
| 28 | NDPK | AAC05177 | 8·76 | 15 519 | 116 | 54·0 | 7 | 1175·6571 | DRPFFPGLVK | 29 |
| 29 | NDPK | AAC05177 | 8·76 | 15 519 | 84 | 42·34 | 6 | 1175·6571 | DRPFFPGLVK | 17 |
| 30 | NDPK | 1NSKR | 8·87 | 17 157·9 | 136 | 62·5 | 8 | 1175·6571 | DRPFFPGLVK | 37 |
| 31 | PKC-1 inhibitor | HINT_HUMAN | 6·46 | 13 662 | 97 | 54·76 | 5 | 1416·7594 | AQVARPGGDTIFGK | 19 |
| 32 | NDPK | QNSKR | 8·55 | 1715·9 | 158 | 62·91 | 8 | 1801·9113 | VMLGETNPADSKPGTR | 43 |
| 33 | NDPK | AAC05177 | 8·76 | 15 519 | 142 | 54·01 | 7 | 1175·6571 | DRPFFPGLVK | 41 |
| 34 | NDPK | AAC05177 | 8·76 | 15 519 | 131 | 67·88 | 8 | 1175·6571 | DRPFFPGLVK | 30 |
| 35 | Profilin | 1AWIA | 8·46 | 14 842·8 | 163 | 65·94 | 9 | 1470·7587 | SSFYVNGLTLGGQK | 50 |
| 36 | Cofilin | S12632 | 8·22 | 18 490·7 | 226 | 52·41 | 8 | 2166·0964 | EILVGDGQTVDDPYATFVK | 68 |
| 37 | NDPK | 1NUEA | 8·55 | 17 155·9 | 186 | 64·23 | 11 | 1344·7634 | TFIAIKPDGVQR | 42 |
| 38 | Cofilin | S12632 | 8·22 | 18 490·7 | 147 | 46·98 | 7 | 1990·0684 | KEDLVFIFWAPESAPLK | 38 |
| 39 | NDPK | 1NSKR | 8·87 | 17 157·9 | 143 | 77·63 | 12 | −1175·6571 | DRPFFPGLVK | 12 |
| 40 | Profilin | A28622 | 8·44 | 15 044·6 | 154 | 92·02 | 11 | 1470·7587 | DRPFFPGLVK | 18 |
The protein name and accession number represent the candidate proteins identified with the highest significant score from the mass spectrometry database (MSDB) by the Mascot search engine. The protein score is defined as −10 × log(P), where P is the probability that the observed match is a random event and protein scores greater than 64 are significant (P < 0·05). Non-significant scores are not shown. The theoretical isoelectric point (pI) and molecular weight (MW, Daltons) for each identified protein are shown. Where tandem mass spectrometry (MSMS) was performed, the precursor ion mass and resulting amino acid sequences are shown together with the corresponding ion score. Only the highest ion score and its corresponding peptide are shown.
HP, hypothetical protein; NDPK, nucleoside diphosphate kinase; HSC, heat shock protein cognate-1; PKC, protein kinase c; HSnm23, Homo sapiens NPDK like protein.
Immunogenicity of the cytoskeletal elements
Identifying the three cytoskeletal proteins raised the question of their immunogenicity. Serum IgG antibodies to the two HSP70 preparations were tested by ELISA and both showed a negligible increase in antibody titre following immunization (Table 2). However, IgG antibody titres were significantly increased to cofilin (from log2 2·8 ± 0·7 to 5·8 ± 0·7; P = 0·009) and profilin (from log2 3·6 ± 0·9 to 5·2 ± 0·6; P = 0·035) in the ADP-HSP- but not ATP-HSP-immunized macaques (Table 2). Similar analysis of serum IgG antibodies to γ- and β-actin (GenWay Biotech Inc.) showed an increase in antibodies to γ-actin in the ADP-HSP-immunized macaques (from 0·7 ± 0·2 to 3·1 ± 0·35; P = 0·078), but this failed to reach the 5% level of significance; sera from the ATP-HSP immunized macaques showed little change (Table 2). However, a significant increase in β-actin antibodies was found in sera of ATP-HSP-immunized animals (P = 0·010), and an increase was also seen with ADP-immunized animals (P = 0·059). The results suggest that the three cytoskeletal elements bound to HSP70 were immunogenic, inducing IgG-specific antibodies, which may have contributed to the post-entry prevention of SIV infection of CD4+ T cells. Serum antibodies to HLA class I and class II were examined in the Luminex assay and, with the exception of one macaque from each of the ADP-HSP and ATP-HSP groups, significant HLA antibodies were not found (data not shown). The lack of an increase in HLA antibodies and the failure to identify HLA molecules in the ADP-HSP preparation on proteomic analysis made it unlikely that HLA-induced xeno-immunity would account for the protection against SIV in the ADP-HSP-immunized macaques.
Table 2.
Serum immunoglobulin G (IgG) antibodies to heat-shock protein 70 (HSP70), cofilin and profilin pre- and post-immunization with HSP-ADP or HSP-ATP, assayed by enzyme-linked immunosorbent assay (ELISA)
| ADP-HSP |
ATP-HSP |
|||||||
|---|---|---|---|---|---|---|---|---|
| Pre | Post | t | P | Pre | Post | t | P | |
| HSP-70 | 2·2 (0·2) | 2·4 (0·5) | 0·29 | 0·783 | 2·0 (0·2) | 2·6 (0·3) | 1·47 | 0·20 |
| γ-Actin | 0·7 (0·24) | 3·1 (0·35) | 3·36 | 0·078 | 2·3 (0·58) | 3·0 (0·14) | 1·11 | 0·383 |
| β-Actin | 1·5 (0·13) | 2·7 (0·24) | 3·93 | 0·059 | 1·7 (0·36) | 3·2 (0·25) | 9·5 | 0·010 |
| Cofilin | 2·8 (0·7) | 5·8 (0·7) | 4·74 | 0·009 | 3·0 (0·4) | 3·8 (0·2) | 1·57 | 0·22 |
| Profilin | 3·6 (0·9) | 5·2 (0·6) | 3·14 | 0·035 | 3·5 (0·3) | 3·5 (0·6) | 0·00 | 1·0 |
Values are mean (± standard error of the mean) log2 titres; the t and P values were determined by paired Student’s t-test.
Evaluation of APOBEC3G mRNA in PBMCs, its relation to SIV infectivity and stimulation of the cytoskeletal proteins
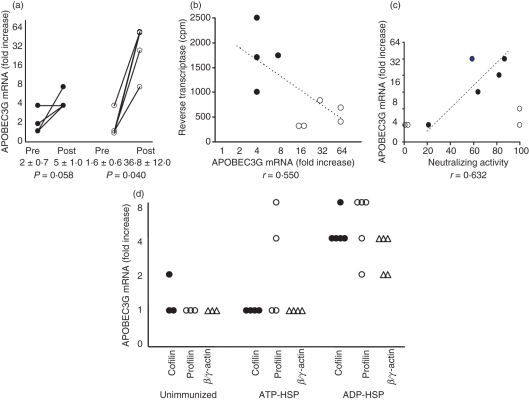
The possibility of involvement of an innate antiviral mechanism was then considered. Indeed, HSP70 up-regulates the intracellular innate antiviral APOBEC3G (A3G) in vitro with human CD4+ T cells and DCs, and in vivo in macaques (19). Evaluation of A3G mRNA in PBMCs showed a 22-fold increase in A3G mRNA (from 1·6 ± 0·6 to 36·8 ± 12·0 P = 0·040) in the ADP-HSP-immunized macaques, compared with a 2·5-fold increase (from 2·0 ± 0·7 to 5·0 ± 1·0; P = 0·058) in the ATP-HSP-immunized animals (Fig. 4a). A comparison of A3G mRNA expression with SIV infectivity showed that the raised A3G mRNA was inversely correlated with reverse transcriptase in the immunized macaques (Fig. 4b), but the 5% level of significance was not reached (r = −0·550). These results suggest that, while both preparations of HSP70 were capable of generating A3G mRNA, significantly increased expression of A3G mRNA was elicited in PBMCs from the macaques immunized with ADP-HSP than ATP-HSP (> 8-fold increase; P = 0·037) and significantly lower expression of viral replication as measured by reverse transcriptase (< 1000 cpm; P = 0·033), compared with those induced by immunization with ATP-HSP (Fig. 4b).
Figure 4.
The effect of immunization on A3G activity; correlation with decreased viral replication and increased viral neutralization and in vitro effect of cytoskeletal proteins on A3G activity. (a) Apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) assayed by real-time polymerase chain reaction (PCR) in peripheral blood mononuclear cells (PBMCs) before and after immunization of macaques with ATP-heat-shock protein (HSP) (•), compared with PBMCs from ADP-HSP (○). (b) Correlation between APOBEC3G mRNA and reverse transcriptase of PBMCs from post-ATP-HSP- (•) and post-ADP-HSP (○)-immunized macaques. (c) Correlation between serum neutralizing activity and APOBEC3G mRNA. (d) In vitro stimulation with cofilin (•), profilin (○) and β/γ-actin (Δ) of PBMCs from unimmunized, ATP-HSP-immunized and ADP-HSP-immunized macaques; APOBEC3G-fold increase was calculated relative to that of unstimulated cells.
The question raised by these findings was whether the increased A3G mRNA in the ADP-HSP-immunized macaques could be accounted for by the cytoskeletal proteins associated with the ADP-HSP preparation. Indeed, direct testing of the capacity of cofilin, profilin and β/γ-actin to stimulate A3G mRNA in PBMCs from immunized macaques demonstrated a significant increase in A3G mRNA expression on stimulation with cofilin (P = 0·010), profilin (P = 0·018) and β/γ-actin (P = 0·012) of cells from the ADP-HSP- but not ATP-HSP-immunized animals (Fig. 4c). However, comparing A3G mRNA expression between the ADP-HSP- and ATP-HSP-immunized macaques, only cofilin (P = 0·010) and β/γ-actin (P = 0·014) stimulated a significant increase in A3G mRNA in PBMCs from the ADP-immunized animals. The limited number of PBMCs did not permit further evaluation of the cells expressing A3G mRNA, which is found mostly in CD4+ T cells, DCs, monocytes and macrophages. The increased A3G mRNA expression elicited by the cytoskeletal proteins is consistent with ADP-HSP binding these proteins and involves post-entry inhibition of SIV infectivity of CD4+ T cells.
Serum neutralizing activity
The neutralizing activity of the sera against SIV was tested using the human CD4+ T-cell line (C8166). This showed > 50% inhibition in four of the five sera from the ADP-HSP-immunized macaques (at 1 : 8 dilution), but two of the four sera from the ATP-HSP immunized animals also yielded > 50% inhibition of SIV (Fig. 4c). However, while the former (ADP-HSP) sera showed direct correlation with A3G mRNA (r = 0·632), the latter (ATP-HSP) sera were dissociated from A3G mRNA.
Discussion
Immunization experiments in rhesus macaques with HSP70 derived from a human CD4+ T-cell line (C8166) revealed that the ADP-HSP70 preparation, which retains proteins or peptides from the CD4+ T cells, induced significant ex vivo inhibition of SIVmac251 infection in the CD4+ T cells. In contrast, immunization with the ATP-HSP70 preparation in which peptides were eluted by ATP failed to show significant inhibition of SIV infectivity of CD4+ T cells. These results raised the possibility that ADP-HSP70 may include proteins or peptides acquired from the human CD4+ T cells, which elicited a protective anti-SIV response, as was reported following immunization with SIV grown in human CD4+ T cells.1–6 The protective mechanism has not been established, but cell surface antibodies that reacted with MHC class I and II antigens, CC chemokines, CD8-suppressor factor, CCR5 antibodies, T helper type 1 (Th1) cytokines and shared epitopes between HIV and HLA have been implicated.10
Thus, a xenogeneic or allogeneic protein was an attractive possibility, as human CD4+ T cells elicited significant protection from SIV infection in macaques, as was also achieved by passive immunization with anti-HLA class II and class I antibodies. We studied the CD4+ T cell-derived proteins of the ADP-HSP70 bound material. However, proteomic analysis failed to identify HLA class I or class II proteins. A number of specific serum antibodies were tested, especially for HLA class I and II antibodies, but these largely failed to demonstrate significant correlation with ADP-HSP70 or ATP-HSP70 immunization in macaques (data not shown). Anti-cell antibodies binding to the cell surface antigens on C8166 cells were absent from these sera, but were present in the sera of animals that had been immunized with whole-cell lysates of C8166 cells, as has been demonstrated previously.7–9,23 The activity of recombinant HSP has been attributed by some to contaminants of lipopolysaccharide (LPS) and nucleotides from Escherichia coli in which the recombinant HSP70 is expressed.24 However, the HSP70 used in the present work was isolated directly from the C8166 cell line, and the procedure was carried out under sterile conditions and extensively washed, so it is unlikely that these contaminants would have been involved.
Surprisingly, the ADP-HSP material contained the cytoskeletal proteins cofilin, profilin and γ-actin, which were not detected in the ATP-HSP preparation. These or altered cytoskeletal proteins may have broken tolerance as significant IgG antibodies were found to cofilin (P = 0·009) and profilin (P = 0·035) in the ADP-HSP- but not the ATP-HSP-immunized macaques. There was no evidence of any autoimmune reaction. It is as yet not clear how the enhanced A3G expression is stimulated by the cytoskeletal proteins, but a cell surface receptor, such as TLR11, stimulated by a profilin-like molecule25 or an intracellular activation pathway will be studied. The cytoskeletal proteins induced up-regulation of A3G mRNA by stimulating PBMCs from the macaques immunized with ADP-HSP (from 1·6 ± 0·6 to 36·8 ± 12·0-fold increase; P = 0·040). Indeed, high A3G mRNA expression was correlated with low SIV infectivity of PMBCs from the ADP-HSP-immunized macaques. As CD4+ CCR5+ T cells are the primary targets of HIV-1 or SIV infection, it is noteworthy that HSP70 stimulates CCR517,18 and CD4014,16 molecules, which up-regulate A3G mRNA and protein expression.19 These results suggest that, while both preparations of HSP70 are capable of generating some A3G mRNA, the increased expression elicited by ADP-HSP may be accounted for by the additional proteins associated with the ADP-HSP moiety. These findings raise the possibility that the innate A3G antiviral factor and the anti-cofilin and profilin antibodies might account for the significant inhibition of SIV infectivity of CD4+ T cells by immunization with ADP-HSP. The mechanism of ex vivo inhibition of SIV infectivity of activated CD4+ T cells from ADP-HSP70-immunized macaques will need to be investigated. The working hypothesis, however, is based on the finding that in vivo antibodies to cofilin may have prevented depolymerization of F-actin which interferes with CD4-CCR5 colocalization and pre-entry HIV infection26,27, whereas up-regulated A3G CD4+ T cells of the ADP-HSP70-immunized macaques may have inhibited post-entry replication of SIV. Furthermore, the paradoxical possibility will be explored that immunization with cofilin increases A3G expression, while anti-cofilin antibodies may inhibit this response.
There is extensive evidence in the literature that cytoskeletal proteins play a key role in HIV-1–host cell interaction.28,29 The actin-binding protein cofilin depolymerizes or severs F-actin, which is required for the formation of immunological synapses.27 HIV-1 entry into host cells is dependent on actin colocalization of CD4, CCR5 or CXCR4,27 as well as envelope (Env) and group-specific antigen (Gag) polarization in lipid rafts stabilized by actin.30 Direct cellular transmission of HIV-1 occurs between HIV-infected and uninfected T cells via a virological synapse generated by the above elements and lymphocyte function-associated antigen (LFA)-1 from the uninfected cells.30,31 Interference with the cytoskeletal system by antibodies to cofilin may interfere with the depolymerization of F-actin required for the formation of the immunological synapse, colocalization of CD4 with CCR5 and HIV-1 entry into the host cell.26,27 Cofilin and profilin antibodies may also reduce transfer of HIV-1 across the virological synapse. Serum neutralizing activity from the ADP-HSP-immunized macaques showed a direct correlation with A3G mRNA (r = 0·632), but the neutralizing activity of sera from the ATP-HSP70-immunized animals was dissociated from the A3G mRNA (Fig. 4c). As noted for A3G mRNA above, HSP70 alone may elicit neutralizing activity.32 The present findings are consistent with the requirements of both A3G and serum neutralizing activity for protection against SIV. Alternatively, ADP-HSP may induce an immune response to actin-binding proteins such as cofilin, which may inhibit the actin filament function. Quite apart from the effect of actin and cofilin on the antigen-presenting cell–T-cell interaction, cytoskeletal proteins are found in HIV-1 virions with molar ratios relative to Gag of 10–15% for actin and 2–10% for cofilin. Thus antibodies to these proteins may contribute directly to inhibition of HIV-1 virions.33,34
Identification of cofilin, profilin and γ-actin in the ADP-HSP preparation, using two-dimensional gel electrophoresis, followed by mass spectrometry of the resulting spots, is consistent with the significance of these cytoskeletal proteins in the host–HIV or host–SIV interaction.26–30 Immunization of rhesus macaques with the ADP-HSP preparation elicited significant inhibition of SIV infectivity of CD4+ T cells, compared with the ATP-HSP preparation. We suggest a putative mechanism of protection in which antibodies to cofilin and profilin may inhibit depolymerization of actin and reduce transfer of HIV-1 across the virological and immunological synapses. Nuclear translocation of HIV-1 DNA will be inhibited and the viral genome may then be degraded by the activity of up-regulated A3G. However, the mechanism of prevention of HIV-1/SIV infectivity by cytoskeletal proteins is a complex emerging field, crossing virological, immunological and biological disciplines, that will require a great deal of further investigation. These findings suggest a novel strategy of preventing HIV-1 infection by utilizing epitopes within the cytoskeletal system of proteins.
Acknowledgments
We wish to thank Dr T. Whittall for the flow cytometry assays to identify CD4+ T-cell surface antibodies in the ADP-HSP preparation and Dr R. Vaughan for the Luminex assay for HLA antibodies.
Glossary
Abbreviations:
- APOBEC3G (A3G)
apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G
- DC
dendritic cell
- HSP
heat-shock protein
- RT
reverse transcriptase
- SIV
simian immunodeficiency virus
Disclosures
There are no conflicts of interest.
References
- 1.Desrosiers RC, Wyand MS, Kodama T, et al. Vaccine protection against simian immunodeficiency virus infection. Proc Natl Acad Sci U S A. 1989;86:6353–7. doi: 10.1073/pnas.86.16.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphey-Corb M, Martin LN, Davison-Fairburo B, et al. A formalin-inactivated whole SIV vaccine confers protection in macaques. Science. 1989;246:1293–7. doi: 10.1126/science.2555923. [DOI] [PubMed] [Google Scholar]
- 3.Stott EJ, Chan WL, Mills KH, Page M, Taffs F, Cranage M, Greenaway P, Kitchin P. Preliminary report: protection of cynomolgus macaques against simian immunodeficiency virus by fixed infected-cell vaccine. Lancet. 1990;336:1538–41. doi: 10.1016/0140-6736(90)93310-l. [DOI] [PubMed] [Google Scholar]
- 4.Carlson JR, McGraw TP, Keddie E, et al. Vaccine protection of rhesus macaques against simian immunodeficiency virus infection. AIDS Res Hum Retroviruses. 1990;6:1239–46. doi: 10.1089/aid.1990.6.1239. [DOI] [PubMed] [Google Scholar]
- 5.Hunsmann G, Dormont D, LeGrand R, et al. Protection of macaques against simian immunodeficiency virus infection with activated vaccines. Comparison of adjuvants, doses and challenge viruses. Vaccine. 1995;13:295–300. [PubMed] [Google Scholar]
- 6.Arthur LO, Bess JW, Jr, Urban RG, Strominger JL, Morton WR, Mann DL, Henderson LE, Benveniste RE. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. J Virol. 1995;69:3117–24. doi: 10.1128/jvi.69.5.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langlois AJ, Weinhold KJ, Matthews TJ, Greenberg ML, Bolognesi DP. The ability of certain SIV vaccines to provide reactions against normal cells. Science. 1992;255:292–3. doi: 10.1126/science.1549775. [DOI] [PubMed] [Google Scholar]
- 8.Osterhaus A, de Vries P, Heeney J. AIDS vaccine developments. Nature. 1992;255:684–6. doi: 10.1038/355684b0. [DOI] [PubMed] [Google Scholar]
- 9.Stott EJ, Almond N, West W, Kent K, Cranage MR, Rud E. Anti-cell antibody in macaques. Nature (Lond) 1991;353:393. doi: 10.1038/353393a0. [DOI] [PubMed] [Google Scholar]
- 10.Lehner T, Shearer GM, Hackett CJ, Schultz A, Sharma OK. Workshop summary. Alloimmunization as a strategy for vaccine design against HIV/AIDS. AIDS Res Hum Retroviruses. 2000;16:309–13. doi: 10.1089/088922200309188. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava P. Roles of heat shock proteins in innate and adaptive immunity. Nat Rev Immunol. 2002;2:185–94. doi: 10.1038/nri749. [DOI] [PubMed] [Google Scholar]
- 12.Panjwani N, Akbari O, Garcia S, Brazil M, Stockinger B. The HSC73 molecular chaperone: involvement in MHC class II antigen presentation. J Immunol. 1999;163:1936–42. [PubMed] [Google Scholar]
- 13.Castellino F, Boucher PE, Eichelberg K, Mayhew M, Rothman JE, Houghton AN, Germain RN. Receptor-mediated uptake of antigen/heat shock protein complexes results in major histocompatibility complex class I antigen presentation via two distinct processing pathway. J Exp Med. 2000;191:1957–64. doi: 10.1084/jem.191.11.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Kelly CG, Kartunen JT, et al. CD40 is a cellular receptor mediating mycobacterial heat shock protein 70 stimulation of CC chemokines. Immunity. 2001;15:971–83. doi: 10.1016/s1074-7613(01)00242-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Kelly CG, Singh M, McGowan EG, Carrara A-S, Bergmeier LA, Lehner T. Stimulation of Th1-polarizing cytokines, C-C chemokines, maturation of dendritic cells and adjuvant function by the peptide binding fragment of heat shock protein 70. J Immunol. 2002;169:2422–9. doi: 10.4049/jimmunol.169.5.2422. [DOI] [PubMed] [Google Scholar]
- 16.Becker T, Hartl F-U, Wieland F. CD40, an extracellular receptor for binding and uptake of Hsp70-peptide. Nat Med. 2002;158:1277–85. doi: 10.1083/jcb.200208083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Floto RA, MacAry PA, Boname JM, et al. Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science. 2006;314:454–8. doi: 10.1126/science.1133515. [DOI] [PubMed] [Google Scholar]
- 18.Whittall T, Wang Y, Younson J, Kelly C, Bergmeier LA, Peters B, Singh M, Lehner T. Interaction between the CCR5 chemokine receptors and microbial HSP70. Eur J Immunol. 2006;36:2304–14. doi: 10.1002/eji.200635953. [DOI] [PubMed] [Google Scholar]
- 19.Pido-Lopez J, Whittall T, Wang Y, Bergmeier LA, Babaahmady K, Singh M, Lehner T. Stimulation of cell surface CCR5 and CD40 molecules by their ligands or by microbial HSP70 upregulates APOBEC3G expression in CD4+ T cells and dendritic cells. J Immunol. 2007;178:1671–9. doi: 10.4049/jimmunol.178.3.1671. [DOI] [PubMed] [Google Scholar]
- 20.Peng P, Menoret A, Srivastava P. Purification of immunogenic heat shock protein 70-peptide complexes by ADP-affinity chromatography. J Immunol Methods. 1997;204:13–21. doi: 10.1016/s0022-1759(97)00017-3. [DOI] [PubMed] [Google Scholar]
- 21.Babahmady K, Oehlmann W, Singh M, Lehner T. Inhibition of HIV-1 infection of human CD4+ T cells by microbial HSP70 and the peptide epitope 407–426. J Virol. 2007;81:3354–60. doi: 10.1128/JVI.02320-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol. 1999;73:8966–74. doi: 10.1128/jvi.73.11.8966-8974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmeier LA, Walker J, Tao L, Cranage M, Lehner T. Antibodies to human and non-human primate cellular and culture medium components in macaques vaccinated with the simian immunodeficiency virus. Immunology. 1994;83:213–20. [PMC free article] [PubMed] [Google Scholar]
- 24.Tsan MF, Gao B. Heat shock proteins and immune system. J Leukoc Biol. 2009;85:905–10. doi: 10.1189/jlb.0109005. [DOI] [PubMed] [Google Scholar]
- 25.Yarovinsky F, Zhang D, Andersen JF, et al. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005;308:1626–9. doi: 10.1126/science.1109893. [DOI] [PubMed] [Google Scholar]
- 26.Eibert SM, Lee K-H, Pipkorn R, Sester U, Wabnitz GH, Giese T, Meuer SC, Samstag Y. Cofilin peptide homologs interfere with immunological synapse formation and T cell activation. Proc Natl Acad Sci USA. 2004;101:1957–62. doi: 10.1073/pnas.0308282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyengar S, Hildreth J, Schwartz D. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol. 1998;72:5251–5. doi: 10.1128/jvi.72.6.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fackler OT, Krausslich H-G. Interactions of human retroviruses with the host cell cytoskeleton. Curr Opin Microbiol. 2006;9:409–15. doi: 10.1016/j.mib.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Mataresse P, Malorni W. Human immunodeficiency virus (HIV)-1 proteins and cytoskeleton: partners in viral life and host cells death. Cell Death Differ. 2005;12:932–41. doi: 10.1038/sj.cdd.4401582. [DOI] [PubMed] [Google Scholar]
- 30.Jolly C, Kashefi M, Hollinshead M, Sattentau Q. HIV cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199:283–93. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy CG, Emerman M, Hope TJ. Visualization of the intracellular behaviour of HIV in living cells. J Cell Biol. 2002;159:441–52. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babaahmady K, Bergmeier LA, Lehner T. Combining human antisera to human leukocyte antigens, HIVgp120 and 70 kDa heat shock protein results in broadly neutralizing activity to HIV-1. AIDS. 2008;22:1267–76. doi: 10.1097/QAD.0b013e328304b3a6. [DOI] [PubMed] [Google Scholar]
- 33.Chertova E, Chertov O, Coren LV, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–52. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott DE, Coren VV, Kane BP, et al. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–43. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]