Abstract
The human chemokine receptor CRAM (chemokine receptor on activated macrophages), encoded by the gene CCRL2, is a new candidate for the atypical chemokine receptor family that includes the receptors DARC, D6 and chemocentryx chemokine receptor (CCX-CKR). CRAM is maturation-stage-dependently expressed on human B lymphocytes and its surface expression is up-regulated upon short-term CCL5 exposure. Here, we demonstrate that the homeostatic chemokine CCL19 is a specific ligand for CRAM. In radioactive labelling studies CCL19 bound to CRAM-expressing cells with an affinity similar to the described binding of its other receptor CCR7. In contrast to the known CCL19/CCR7 ligand/receptor pair, CRAM stimulation by CCL19 did not result in typical chemokine-receptor-dependent cellular activation like calcium mobilization or migration. Instead, we demonstrate that CRAM is constitutively recycling via clathrin-coated pits and able to internalize CCL19 as well as anti-CRAM antibodies. As this absence of classical chemokine receptor responses and the recycling and internalization features are characteristic for non-classical chemokine receptors, we suggest that CRAM is the newest member of this group. As CCL19 is known to be critically involved in lymphocyte and dendritic cell trafficking, CCL19-binding competition by CRAM might be involved in modulating these processes.
Keywords: B cells, chemokine receptor, chemotaxis, signal transduction
Introduction
Chemokines and their receptors play a major part in homing of leucocytes and recruiting them to sites of inflammation.1 There are approximately 50 different human chemokines and 21 identified receptors expressed on specific subpopulations of leucocytes. Most chemokine receptors bind different chemokines with variable affinities.2 Classically, chemokine receptors react to ligand binding by triggering an intracellular signalling cascade mediated by G-protein activation. This leads to receptor phosphorylation by a G-protein-coupled receptor kinase, which results in uncoupling of the receptor from G-proteins and finally desensitization through β-arrestin binding to the phosphorylated receptor.3,4 This in turn promotes receptor internalization by linking it to the clathrin coat together with adaptor proteins.5,6
The complex system of chemokine receptors is supplemented and modulated by the group of so-called atypical or non-classical chemokine receptors [including D6, duffy antigen receptor for chemokines (DARC), chemocentryx chemokine receptor (CCX-CKR)], which are believed to be non-signalling or chemokine-regulating receptors, thereby modulating local chemokine concentrations (reviewed by Haraldsen and Rot7). Despite some common features like the lack of signalling and the modified DRYLAIV motif, the members of this group are heterogeneous in their behaviour. For example, D6 is a receptor expressed on endothelium and some leucocytes and is able to bind, scavenge and degrade most pro-inflammatory CC-chemokines. This activity is mediated by a constitutive internalization process.8 CCX-CKR is restricted to the homeostatic chemokines CCL19, CCL21 and CXCL13 and specializes in their degradation.9 On the other hand, DARC is expressed on erythrocytes and vascular endothelium and does not scavenge chemokines per se but acts as a buffer for both CXC and CC pro-inflammatory chemokines. It also effectively mediates transcytosis of its ligands through endothelial layers.10,11
The human gene CCRL2 encodes a putative chemokine receptor first described in 1998 and produces two splice variants known as CRAM-A (chemokine receptor on activated macrophages) and the 36 base pair shorter, more common variant, CRAM-B (also known as human chemokine receptor; HCR12). CRAM shares over 40% sequence homology with known chemokine receptors such as CCR1, -2, -3 and -5, but bears a mutation in the highly conserved DRYLAIV motif (Table 1), with a glutamine (Q) at position 127 instead of aspartic acid (D), replacing an acidic residue with a neutral one and rendering coupling to the Gi protein highly unlikely. Until recently, no competitive ligand of CRAM had been found, although considerable data on its expression in tissues and haematopoietic cell types are available.13
Table 1.
Comparison of amino acid sequences of the conserved DRYLAIV motif between chemokine receptor on activated macrophages (CRAM) and other atypical chemokine receptors
| Receptor | % Receptor sequence identity (amino acids) | DRYLAIV |
|---|---|---|
| CRAM (human) | 100 | QRYLVFL |
| CRAM (mouse) | 51·6 | QGYRVFS |
| DARC (human) | – | AEALAIL |
| D6 (human) | 3·2 | DKYLEIV |
| CCX-CKR (human) | 28·3 | DRYVAVT |
| U51 (HHV6) | – | ERINHFC |
| U12(HHV6) | – | IRYKTLK |
| ORF74 (HHV8) | 18·9 | VRYLLVA |
Overall identity is indicated in the second column and sequence of the highly conserved second intracellular loop is in the third column, with conserved amino acids in bold in reference to the classical sequence.
Extensive work has also been carried out on the murine homologue to CRAM, L-CCR (lipopolysaccharide inducible C-C chemokine receptor related gene), which shares only 51% sequence identity with the human gene, a very low value of identity compared with other chemokine receptor orthologues.14–16 It has been reported that L-CCR binds chemokines such as CCL2, -5, -7 and -8, but this remains controversial and has never been confirmed (discussed by Mantovani et al.17). Only recently, murine and human CRAM have been found to bind to human chemerin, an adipokine previously described as an agonist for the G-protein-coupled receptor CMKLR1 (also known as ChemR23).15,18
Binding of chemerin was not followed by classical receptor activation and it was postulated that CRAM concentrates and presents its ligands without degrading them. We have previously described CRAM as being widely expressed on B cells in a maturation-stage-dependent manner, with the highest expression on pro-B and pre-B cells and CRAM expression as being altered in the presence of CCL5.19 We did not observe any classical chemokine receptor responses such as migration or calcium mobilization upon CCL5 stimulation of CRAM-expressing cells. Instead, the surface expression was up-regulated, in a similar manner to D6,20 supporting the theory that the receptor could belong to the group of atypical receptors as already suggested by others.17
In this report, we further characterized CRAM ligand binding and internalization on transfectants and a pre-B cell line with a high endogenous level of CRAM expression. We show that CRAM is a receptor for the homeostatic chemokine CCL19 (ELC, MIP-3β). This binding is also not followed by classical chemokine activation of cells, pointing towards an atypical status. Furthermore, CRAM recycles constitutively, and can internalize CCL19 within a few minutes, suggesting an implication in the regulation of immune responses and homing processes.
Materials and methods
Cell culture and transfection
All cell line media (Invitrogen, Carlsbad, CA) were supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin. The human pre-B-cell line Nalm6 [American Type Culture Collection (ATCC), Manassas, VA], was grown in RPMI-1640, and the human B-cell chronic lymphatic leukaemia cell line Mec1 (ATCC), was grown in Iscove’s modified Dulbecco’s medium in a humidified atmosphere (5% CO2) at 37°. The Chinese hamster ovary CHO-K1 and GAG-deficient CHO-pgsA-745 cells (ATCC) were maintained in Ham’s F12 medium. Stable transfectants, generated with Polyfect® (Qiagen, Hilden, Germany) according to the provider’s instructions, were additionally provided with 800 μg/ml G418.
The polymerase chain reaction (PCR) products of CRAM-B (GenBank/EMBL accession number U97123) were cloned in pcDEF3 as described previously.19 The complementary DNA clone for human CRAM-B N-terminally tagged with a haemagglutinin (HA) triplicate in pcDNA3.1 was obtained from the Missouri S&T cDNA Resource Center (http://www.cdna.org).
Receptor binding and ligand internalization assays with radiolabelled chemokines
The CRAM-B or mock-transfected CHO-K1 and CHO-pgsA-745 adherent cell cultures were detached using a 10 mm ethylenediaminetetraacetic acid (EDTA) solution and then washed in RPMI-1640 and resuspended in binding medium [RPMI 1640, 0·1% bovine serum albumin (BSA), and 20 mm HEPES (pH 7·4)] on ice. Assays in triplicate contained 125 pm of the radiolabelled chemokine CCL19 (specific activity, 2200 Ci/mm; Perkin Elmer, Waltham, MA), and diluted concentrations of unlabelled competitor chemokines (R&D Systems, Minneapolis, MN). After 2-hr incubations on ice, cells were separated from the unbound chemokine by microcentrifugation through a phthalate oil cushion [1·5 parts dibutylphthalate to 1 part bis(2-ethylhexyl)phthalate] and bound radioactivity was counted with a γ-counter. Data were analysed and fitted to a sigmoidal dose–response curve with variable slope model using Prism 0.1.53 software (GraphPad, La Jolla, CA). Expression of CRAM receptor was tested by flow cytometry before the binding experiments.
Internalization assays were performed as in Comerford et al.9 The cells prepared as described for binding assays were incubated for 2 hr with the labelled chemokine, washed, resuspended in binding buffer and subsequently shifted to 37° for 10 min. After centrifugation, cell pellets were washed with ice-cold phosphate-buffered saline (PBS) or acid washed (PBS pH 2·2 with 1 m HCl) for 2 min. Acid treatment was interrupted by addition of binding buffer and cells were immediately centrifuged through a phthalate oil cushion as described above for measurements of cell-associated counts/min.
Chemotaxis and calcium mobilization measurements
For chemotaxis assays, a cell suspension (in RPMI-1640, 0·5% BSA) was loaded to the top chamber of transwell culture inserts (8-μm pore; Costar, Lowell, MA). Cells transmigrated for 2 hr in response to a chemokine stimulus in the bottom chamber. Duplicates were assayed for each condition. Cells in the lower chamber were counted with a flow cytometer at high flow for 40 seconds.
For calcium measurements, cells were incubated for 30 min in 2 μm fluo-3-AM (Invitrogen Molecular Probes, Carlsbad, CA) in PBS. After two PBS-washes, cells were resuspended in PBS with 0·1% BSA. Cells were diluted 1 : 10 in RPMI-1640, 1·5 mm CaCl2/0·1% BSA, and fluorescence kinetics were cytometrically determined up to 120 seconds after addition of the chemokine.
Western blot
Cells were starved in fetal calf serum-free medium for 2 hr and stimulated with 50 nm CCL19 or 10 nm stromal-cell-derived factor-1 for the indicated times at 37°. Protein lysates were prepared with 100 μl lysis buffer. Lysis buffer was 20 mm Tris–HCl pH 8·0, 150 mm KCl, 1 mm EDTA, 0·2 mm Na3VO4, 1% Triton X, 0·5 mm phenylmethylsulphonyl fluoride, protein inhibitor cocktail (complete®; Roche Applied Science, Basel, Switzerland). Equal amounts of protein were separated by 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Western blot analysis was performed using the appropriate antibodies recognizing the phosphorylated form of the proteins or the total proteins after stripping the membrane using Re-Blot plus solution (Millipore, Billerica, MA). The antibodies for p44/42 mitogen-activated protein kinase (MAPK) were provided by Cell Signaling Technology (Beverly, MA). Immunoreactive bands were visualized using horseradish peroxidase-conjugated secondary antibody and the enhanced chemiluminescence system (GE Amersham, Fairfield, CT).
Flow cytometry
Murine monoclonal antibodies against CRAM (anti-HCR/CRAM-A/B, clone 152254 or clone 152211 in the indicated cases, recognizing both splice variants) were produced by R&D Systems. Anti-HA (clone 12CA5) and secondary antibody (R-phycoerythrin-labelled rabbit anti-mouse immunoglobulin G) were from Roche and DakoCytomation (Glostrup, Denmark), respectively. Biotinylated anti-CCL19 antibody was from R&D Systems and fluorescein isothiocyanate (FITC)–conjugated avidin was from Sigma-Aldrich (St Louis, MO). The binding of CCL19 to Nalm6 cells was evaluated using 106 cells incubated for 30 min with the indicated concentrations of CCL19 in binding medium, then washed once and incubated with CCL19 antibody (0.1 μg/ml) in the same medium for 30 min at 4°, washed again and finally incubated in the same conditions with FITC-avidin (100 μg/ml). Samples were acquired using a FacsCalibur™ flow cytometer (Beckton Dickinson Immunocytometry Systems, Heidelberg, Germany) and analysed using FlowJo software (Tree Star, Inc., Ashland, OR), the binding experiment was curve fitted as described for radiolabelled ligand.
Small interfering RNA knockdown
Expression of clathrin heavy chain or caveolin 1 was knocked down in Nalm6 cells using small interfering RNA (siRNA; sc-35067 and sc-29241, respectively; Santa Cruz Biotechnologies, Santa Cruz, CA) by electroporation at 230 V and 800 μFd. Briefly, 2 × 106 cells were suspended in phenol-red-free RPMI-1640 with 25 mm HEPES buffer, incubated in 2 mm electroporation cuvettes with 500 nm siRNA for 20 min at room temperature, then submitted to electroporation. After another 5-min incubation period on ice, the cells were transferred to a six-well plate with 2 ml full medium. Internalization assays and evaluation of knockdown efficiency by Western blot were performed 72 hr after transfection.
Receptor internalization
For internalization experiments, Nalm6 cells or CRAM-B-transfected CHO-K1 cells were pre-treated with the inhibitors filipin (5 μg/ml), nystatin (50 μg/ml) or hypertonic sucrose (0·45 m) (all Sigma Aldrich) for 1 hr at 37° where indicated. Depletion of K+ was achieved by a 2-hr incubation in K+-free medium (100 mm NaCl, 1.2 mm CaCl2, 0.5 mm MgCl2, 20 mm HEPES) at 37° after a 5 min hypotonic shock treatment in medium diluted 1 : 1 with water. Subsequently, cells were stained with the primary anti-CRAM or anti-HA antibody at 4° for 1 hr, washed and either shifted to 37° for the indicated time-points or kept at 4° as a control in the presence of chemokines where indicated, before staining with secondary antibody. Alternatively, cells could be restained with the primary antibody before staining with secondary antibody as described in Bonecchi et al.21 The per cent of receptor internalization was calculated as the geometric mean channel fluorescence values of cells incubated for the indicated times at 37° relative to cells kept at 4°.
High content live cell imaging of internalization
The CRAM-B transfected CHO cells were grown on micro-slide eight-well microscopy chambers overnight (Ibitreat; Ibidi, Martinsried, Germany). For the visualization of the cytoplasm and as a cell mask, cells were stained with 2 μm CellTracker™ Orange (Molecular Probes, Invitrogen) for 30 min at 37°. Subsequent staining was with anti-CRAM (5 μg/ml) and secondary antibodies (Alexa488-conjugated anti-mouse; Molecular Probes, Invitrogen) at 4°. Imaging of receptor internalization was performed with an Olympus Scan^R High-Content-Screening-Station. For Alexa488, the excitation filter used was 494/20 nm combined with an ET-Multiband Filter Set for DAPI/FITC/Texas Red (Chroma, Rockingham, VT) for emission and mirror. For Cell Tracker Orange the U-MWIG3 TRITC filter set (Olympus, Tokyo, Japan) was used for both excitation and emission. The microscope was equipped with a climate chamber (Evotec Technologies, Hamburg, Germany) heated to 37°. Images of the cells were taken every 3 min from 25 different positions over a total time period of 30 min and later statistically analysed using the Olympus Scan^R Analysis software (v. 1.06 beta). The Cell Tracker signal was used to delineate the cells, and a combination of circularity and area was used to identify single cells within these images. Of these, the Alexa488-positive cells (i.e. CRAM-positive) were then gated for further analysis. The total amount of fluorescence within each cell, corrected for area, was measured in the outer cytoplasmic region and this value was compared with the amount of fluorescence present in the inner cytoplasm and nucleus. These measurements were carried out for each time-point.
Confocal imaging
For immunofluorescent staining, Nalm6 on Bross adhesion slides were fixed in 4% paraformaldehyde, washed, permeabilized with 0·2% Triton-X, and stained with anti-CRAM (5 μg/ml) and secondary antibodies (Alexa488-conjugated anti-mouse immunoglobulin; Molecular Probes, Invitrogen). The cells were then stained with one of the following antibodies: 1 μg/ml anti-β-arrestin 1/2/3 (Santa Cruz technologies) or 1 μg/ml anti-Caveolin (BD Transduction, Heidelberg, Germany), a secondary antibody (10 μg/ml Alexa647-conjugated anti-rabbit immunoglobulin) and 100 μm DAPI (Sigma Aldrich). Slides were mounted in ProLong Antifade (Molecular Probes) and analysed using a confocal microsope (Leica TCS SP2 AOBS spectral confocal microscope).
Results
CCL19 is a novel ligand of CRAM
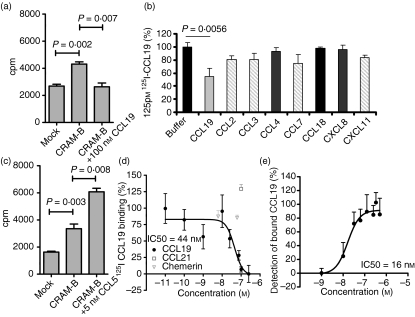
Recently, when screening for CRAM ligands based on functional responses, we demonstrated a CCL5-induced increase of CRAM surface expression accompanied by stress fibre formation and extracellular signal-regulated kinase activation.19 Further binding screening of possible ligands allowed identification of CCL19 as a competitive ligand for CRAM-B (Fig. 1a). Although competition with other unlabelled chemokines revealed CCL2, CCL3, CCL7 and CXCL11, but not CCL18, CCL4 and CXCL8, as potential other low-affinity ligands to CRAM (Fig. 1b), this could not be confirmed by additional flow cytometry detection of CCL19-Fc in the presence of increasing concentrations of other chemokines from 0.1 to 200 nm (data not shown). Competition with CCL5 did not reduce binding but repeatedly resulted in increased cell-associated radioactivity (Fig. 1c). Although we have previously described active receptor up-regulation after CCL5 treatment, this can be ruled out here as a cause of this increased binding because binding experiments are performed exclusively at 4°. The observed effect is most likely the result of chemokine aggregations as often described for CCL5.22 We further investigated CCL19 binding to CRAM-B by a dose–effect experiment on CHO-K1 CRAM-B cells and obtained a 50% inhibitory concentration (IC50) of 44 nm (Fig. 1d). We confirmed CCL19 binding to CRAM-Bhigh but CCR7low cells (the human pre-B-cell line Nalm6) by flow cytometry (Fig. 1e), obtaining an IC50 in the same range of affinity. Incidentally, by the time we were performing this assay, Zabel et al.15 had reported that murine and human chemerin were ligands for CRAM and its murine orthologue, hence we included human chemerin as well as CCL21, the second ligand for CCR7, the CCL19 signalling receptor, in our competition assay against 125I-labelled CCL19. Unexpectedly, none of the tested concentrations of either ligand (CCL21 from 0.1 to 100 nm, chemerin from 6.25 pm to 62.5 nm) was able to compete with 125I-labelled CCL19 binding to the cells [respective highest concentrations shown in (Fig. 1d)].
Figure 1.
CCL19 is a ligand to chemokine receptor on activated macrophages (CRAM). (a) 125I-labelled CCL19 association to 106 CHO-K1 mock-transfected or CRAM-B-transfected cells with or without addition of 100 nm cold CCL19 was assessed by γ-counter. (b) Screening of several chemokines for competitive binding was carried out using 125 pm125I-labelled CCL19 and adding 25 nm CCL19 (dark grey) as a control or 100 nm for other chemokines. Potential low-affinity binding is indicated by light grey columns, no competition is shown in black, results shown are from one experiment performed in triplicate and are expressed in % of the maximal binding. (c) 125I-labelled CCL5 association to 106 CHO-K1 mock-transfected or CRAM-B-transfected cells with or without addition of 5 nm of cold CCL5 was assessed by γ-counter. (d) Competitive binding assay, with data expressed in percentage of maximum bound iodinated chemokine, determined in the absence of cold competitor. Total displacement was defined by 0·25 μm CCL19; data shown represent one representative experiment out of three, each point represents mean values and SD of triplicates and was curve fitted using a sigmoidal dose–response curve (GraphPad Prism Software) to evaluate the 50% inhibitory concentration (IC50). (e) Direct interaction of CCL19 at the indicated concentrations to Nalm6 cells was revealed by flow cytometry after addition of CCL19-biotin antibody and strepatvidin-fluorescein isothiocyanate. Data represent triplicate determinations of geometric mean fluorescence intensity from two independent evaluations and are normalized to maximal binding obtained with 100 nm CCL19.
CCL19 binding to CRAM on pre-B ALL cells does not induce classical chemokine receptor signalling
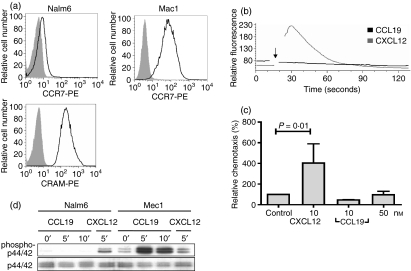
We went on to characterize CCL19–CRAM interactions in more detail. The pre-B-cell line Nalm6 endogenously expresses high levels of both CRAM-A and -B, whereas expression of CCR7, the other CCL19 binding receptor, is only weakly or not detectable by fluorescence-activated cell sorting (FACS) stain and messenger RNA level23 (Fig. 2a, upper left panel). As a comparison, high CCR7 levels of the human chronic lymphatic leukaemia cell line Mec1 are shown. Primary pre-B cells have already been described as CCR7low and unresponsive to CCL19 in chemotaxis and calcium assays,24 but the role of CRAM has never been investigated. CCL19 stimulation of Nalm6 cells did not lead to calcium signalling (Fig. 2b). In addition, CCL19 did not induce migration in chemotaxis assays (Fig. 2c) or MAPK phosphorylation (Fig. 2d). Nalm6 cells were perfectly capable of MAPK phosphorylation in response to CXCL12. On the other hand, Mec1 cells responded strongly to CCL19 stimulation (Fig. 2d), confirming the quality of the chemokine used and also undermining the influence of the low levels of CCR7 in Nalm6 cells. Taken together, these results confirm the previously described ‘silent’ behaviour of CRAM.15,19
Figure 2.
CCL19 does not induce any functional responses in cells expressing high levels of chemokine receptor on activated macrophages (CRAM). (a) CCR7 expresssion in Nalm6 and Mec1 cells (upper panels) and CRAM expression (lower panel). Histograms show protein expression (black line) and corresponding isotype (tinted histogram). (b) Nalm6 cells were stimulated with different CCL19 concentrations (ranging from 5 to 50 nm) and calcium transient releases were measured. For clarity, only stimulation with 20 nm CCL19 is shown. The arrow shows the time at which chemokines were added. Calcium transient releases were determined with flow cytometry using fluo-3-AM. For comparison, results after stimulation with 20 nm CXCL12 are shown. (c) Chemotaxis of Nalm6 cells towards different concentrations of CCL19 (10 and 50 nm) and CXCL12 (10 nm) as a positive control. Results show the percentage of migrating cells compared with an unstimulated control and the mean and range of double samples of two independent experiments. (d) Absence of p44/42 mitogen-activated protein kinase phosphorylation of Nalm6 cells in response to 50 nm CCL19 and presence in response to 10 nm CXCL12 for the indicated times are shown. Detection was achieved with antibodies against phospho-p44/42 (upper panel) and total p44/42. Successful phosphorylation is shown for Mec1 cells under the same experimental conditions.
CRAM internalizes CCL19 upon binding
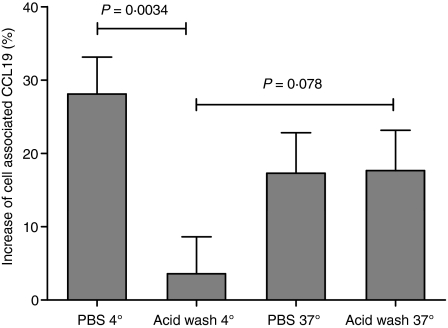
To investigate the fate of CCL19 after binding to CRAM-B, an internalization assay was performed in CHO-K1 cells transfected with CRAM-B (Fig. 3). We measured an increased cell-association of 125I-labelled CCL19 compared with mock-transfected cells treated in the same conditions (PBS as a control or acid wash to remove surface association; both conditions were measured at 4° to prevent internalization or 37° to allow internalization). When kept at 4°, CRAM-B-transfected cell-associated radioactivity was significantly reduced by acid wash to a similar level as in mock-transfected cells treated in the same conditions, confirming that nearly all receptor-specific chemokine binding can be removed by the experimental procedure. When incubated at 37°, a large part of the radioactivity associated with CRAM-B-transfected cells became resistant to acid wash, with levels similar to those obtained by washing with PBS only. This clear result demonstrates that CRAM-B-transfected cells are capable of internalizing the chemokine CCL19.
Figure 3.
CCL19 is internalized by chemokine receptor on activated macrophages B (CRAM-B) expressing CHO cells. CCL19 binding was assessed in cells shifted to 37° compared with 4° before a washing step in acidic buffer. Data show the relative increased association of 125I-labelled CCL19 to CRAM-B-transfected CHO-K1 when compared with mock-transfected cells in the same experimental conditions. Resistance to acid-mediated removal of bound chemokine in CRAM-B-transfected cells occurs after allowing internalization of the chemokine at 37°. The figure shows mean and SEM of three independent experiments realized in triplicate.
Receptor internalization is constitutive
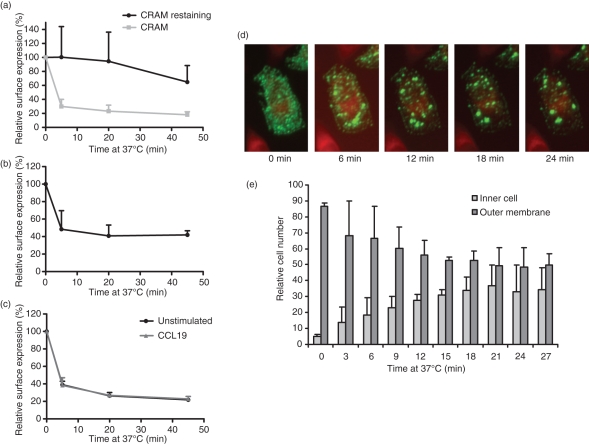
For the atypical receptors D6 and CCX-CKR, constitutive cycling to and from the cell surface has been reported, making it a characteristic of this receptor group.9,25 It was therefore a logical step to study internalization of CRAM as well. Using an antibody-feeding technique, we found that the receptor is rapidly internalized by Nalm6 cells. After only 5–10 min, surface expression of the receptor was lowered to about 30–40% of initial values (Fig. 4a). This was observed in the B-cell line Nalm6 as well as in CRAM-A-transfected and CRAM-B-transfected CHO-K1 and using different CRAM antibody clones no perceptible differences could be detected (data not shown). Relabelling of the cells with anti-CRAM antibody after the 37° incubation revealed that cell surface levels remained roughly constant. This implies that a significant fraction of internalized receptors is re-expressed on the cell membrane (Fig. 4a). To exclude the possibility that the antibody itself activates CRAM and induces the observed internalization, we reproduced the experiment in CHO cells transfected with the HA-tagged form of CRAM-B, obtaining similar results (Fig. 4b). The rate of internalization was unaltered by the presence of the chemokine CCL19 (Fig. 4c).
Figure 4.
Chemokine receptor on activated macrophages (CRAM) internalization is constitutive and independent of ligand binding. (a) Internalization of CRAM in Nalm6 cells. Cells were stained with anti-CRAM primary antibody followed by incubation at 37° for the indicated times and then either restained with the primary antibody or directly stained with the secondary antibody (t-test P values for 5, 20 and 45 min of incubation are 0.02, 0.01 and 0.008, respectively). (b) For internalization of CRAM in CHO-K1, cells were transfected with the haemagglutinin (HA) -tagged form of CRAM-B. Cells were stained with anti-HA antibody followed by incubation at 4° or 37° for the indicated times and then stained with the secondary antibody. (c) Internalization of CRAM in the presence of CCL19 was measured in Nalm6 cells stained as in (a). Chemokines were added at 25 nm during the 37° incubation step. (d) CRAM-B changes its distribution in cells when shifted to 37°. CRAM-B-transfected CHO cells were stained for CRAM (green) and cytoplasm (red) and observed with a Scan^R microscope in a climate chamber preheated to 37° during 30 min. Pictures show one representative cell over the total time period. (e) Analysis of cells treated as in (d) shows relative amounts of cells from pictures taken in 25 different positions of the slide with dominant Alexa488 Fluorescence (= CRAM-B) at their surface compared with cells with a majority of fluorescence in the cytoplasm for each time-point during the course of the experiment. Data show one representative out of three independent experiments.
We confirmed the internalization seen in FACS by observing CRAM-B-transfected CHO-K1 cells with a Scan^R High-Content-Screening station over 30 min (Fig. 4d). The first pictures of single cells clearly showed that CRAM was evenly distributed on the cell surface but later moved to the cytoplasm and towards the nucleus in granular structures. This process was very rapid and could be observed within minutes. Quantitative analysis was performed by defining and comparing number of cells with a majority of membrane-bound fluorescence (ratio of fluorescence outer/inner cell > 1) with the number of cells with higher internal fluorescence values (ratio of fluorescence outer/inner cell < 1) over the time–course (Fig. 4e). Among all cells considered, almost 90% of cells had a high cell surface expression at the beginning of the assay, and this number continually decreased to 40% after 30 min incubation. Cell numbers with a high inner fluorescence increased at the same rate from 6% to 44%.
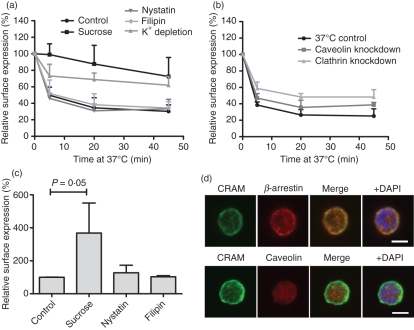
We subsequently investigated the endocytosis pathways employed by the receptor using inhibitors of the clathrin, hypertonic sucrose and K+ depletion25,26 or caveolin pathways, nystatin and filipin,27 respectively. In the pre-B-cell line Nalm6, internalization of CRAM was found to be most likely dependent on the clathrin pathway (Fig. 5a), as it could be inhibited by treatment with hypertonic sucrose and K+ depletion, whereas nystatin and filipin had no effect. We were able to confirm this finding by knocking down clathrin expression (Fig. 5b). In addition, cell surface expression was drastically increased when pre-incubating cells with sucrose but not with caveolin inhibitors (Fig. 5c). Not only does this support data on the use of the clathrin pathway, it is also in accordance with the idea of a constitutively cycling receptor that is quickly re-expressed at the cell surface. To confirm this observation, Nalm6 cells were stained with anti-CRAM antibody and anti-β-arrestin-1/2/3 antibody. β-Arrestin is a protein that directs receptors to the clathrin-coated pits. The cells were analysed in a confocal microscope, revealing consistent colocalization of CRAM and β-arrestins but not of CRAM and caveolin (Fig. 5d).
Figure 5.
Endocytosis of chemokine receptor on activated macrophages (CRAM) is clathrin dependent in Nalm6 cells. (a) The effect of different endocytosis inhibitors on CRAM internalization was studied in Nalm6 cells. Cells were pre-incubated at 37° with the indicated inhibitors of clathrin-coated pits or caveolae-mediated pathways for 1 hr before performing an internalization assay as in Fig. 4 (t test P values at 20 min are 0·0017, 0·0019, 0·67 and 0·69 for sucrose, K+ depletion, nystatin and filipin treatments, respectively). Data shown are the mean and range of three independent experiments. (b) The effect of small interfering RNA down-regulation of clathrin and cavelin-1 was assessed in the same conditions. Data show mean and SD from three independent experiments (t-test P values are 0·0148, 0·0081, 0·0320 for clathrin knockdown at 5, 20 and 45 min, respectivelyl caveolin knockdown induced only insignificant changes in internalization rate). (c) Surface expression levels of CRAM were assessed by fluorescence activated cell sorting staining after a 1-hr incubation with different endocytosis inhibitors. (d) Immunofluorescence analysis of fixed, permeabilized Nalm6 cells stained with anti-CRAM and alexa488 coupled secondary antibody (green) shows co-localization of CRAM when stained for β-arrestin (top panels), but not when stained for caveolin (bottom panels). Both antibodies were recognized by an Alexa647 anti-rabbit secondary antibody (red). The nucleus was revealed using DAPI (blue). White bars depict 5 μm.
In summary, we have shown in this last set of experiments that CRAM is constitutively internalized via rapid internalization pathways, independently of chemokine stimulation, supporting the atypical character of CRAM.
Discussion
The four known members of the non-classical or atypical chemokine receptor family are D6, DARC, CCX-CKR and CXCR7. Their exact biological role has not yet been fully elucidated, although many possibilities have been proposed (reviewed by Comerford et al.28 and O’Hayre et al.29). It is assumed that DARC is responsible for the transportation of specific inflammatory chemokines from the basolateral surface of endothelial cells to their luminal surface.11,30 D6 is capable of a high turnover clearance of a majority of the CC chemokines. CCX-CKR has so far been described as a scavenging receptor for CCL19.9 In contrast, CXCR7 is assumed to be modulating the functioning of heterologous receptors.31
In this article, we provide evidence for CRAM to be a new member of this family with a narrow binding spectrum including CCL5 and CCL19. CCL5 was shown previously to activate cells bearing CRAM (MAPK and stress-fibre filament formation); however, binding competition has always been difficult.19 Enhancement of radiolabelled CCL5 association to CRAM-B-expressing cells when increasing cold CCL5 concentrations has been observed independently by different groups (R. Nibbs, U. Gompels personal communications), even with concentrations as low as 0.1 nm. We hypothesize that these unusual observations result from aggregation and interactions with glycosaminoglycans. These cell surface molecules have been shown to have an impact on CCL5 activity.22,32–34 Preliminary binding screening showed that the homeostatic chemokine CCL19 was able to bind to CRAM-B-expressing cells. We confirmed that CCL19 is a ligand to CRAM-B with an IC50 of 44 nm, the same range as its affinity for its classical receptor CCR7 (18 nm35); however, another atypical chemokine receptor, CCX-CKR, has been shown to be a high-affinity receptor for CCL19 with an IC50 of 6 nm.36 To further characterize CRAM-B interaction with chemokines, we screened several of them for their ability to displace 125I-labelled CCL19 binding to CRAM-B. Four chemokines were shown to displace CCL19 slightly in radiolabelled competition assays (CCL2, CCL3, CCL7 and CXCL11), suggesting that they could be low-affinity ligands for CRAM, but were not confirmed by further investigations, pointing towards a narrow binding spectrum for CRAM. Two other ligands of interest: CCL21, the other ligand for CCR7, and human chemerin, which has recently been described as a ligand for murine and human CRAM,15 were investigated against radiolabelled CCL19. Surprisingly, both ligands failed to displace CCL19 binding to the cells in concentrations as high as 100 or 62.5 nm, respectively. Chemerin has been described as a very-high-affinity ligand for murine CRAM (IC50 < 1 nm15). This discrepancy with our own observation can be explained by the fact that our assay is a heterologous assay, and chemerin could have a binding site only partially overlapping with the CCL19 binding site.
CCX-CKR and CRAM seem to have a much narrower binding spectrum than D6 and DARC, which both interact with more than 12 chemokines. The expression profiles of CCX-CKR and CRAM are different, showing a very tight and complex control of CCL19 circulation. Overall, CCX-CKR seems to have a higher affinity for CCL19 than CRAM and internalizes it more efficiently (compared with the 14% of cell-associated radioactivity preserved in CRAM-expressing cells described here, CCX-CKR expression induced the preservation of up to 60% of the chemokines linked to the cells). This difference in efficiency could of course be the result of different assay conditions; however it is also likely that CRAM could be more of a chemokine presenter (like DARC) than a scavenger (like D6).
We thoroughly investigated activation pathways that could be triggered by CCL19 binding to CRAM as chemotaxis, calcium release and p44/42 MAPK activation, which are classically involved in chemokine-induced signalling. None of these pathways were activated. This is in agreement with the unique sequence alteration in the highly conserved DRYLAIV motif in the second intracellular loop, which is usually involved in coupling to G protein for signalling chemokine receptors (Table 1). The corresponding CRAM sequence is QRYLVFL. A mutation of the aspartate to valine in receptors like CXCR2 leads to an increase of their basal activity.37 However, when also associated with a mutation of the arginin, as in the case of DARC and D6, it seems that the receptors lose their signalling capacity.
In addition to its lack of classical chemokine activation of cells, both splice variants of CRAM show a noticeable ligand-independent internalization pattern, which gives weight to its atypical status. Our results studying the internalization of CRAM indicate that this is a rapid phenomenon, with about 60% of receptors internalized after only 5 min and almost the same amount replaced at the same time, keeping cell surface levels constant. This result is in conflict with Zabel et al.’s study,15 where a similar assay was included for the murine HA-tagged receptor and only insignificant internalization was observed (reduction to about 80% of initial surface expression after 60 min). However, in light of the unusually low homology between human and murine CRAM, a direct comparison is delicate. The basal internalization of chemokine receptors in the absence of stimulation has already extensively been described for D6 and CCX-CKR. D6 internalization was described as being clathrin dependent in CHO-K1 cells, whereas CCX-CKR and DARC are said to internalize via a caveolin-dependent pathway.9,11,25 The measured internalization of CRAM involves the clathrin pathway in Nalm6 cells, as observed both with inhibitors and siRNA in flow cytometric monitoring of receptor internalization and by confocal co-localization of CRAM with β-arrestin. In all sets of experiments, no signs of caveolin influence could be observed. Addition of ligand (CCL19) did not change the internalization efficiency, although a faster re-expression would be likely.
Not only has CRAM been found to be expressed on various types of leucocytes,13,19 but also on endothelial cells and different tissues (unpublished data), which is similar to the profile seen for D6, and it has been postulated that this expression is necessary for leucocytes to leave sites of inflammation.38 Growing evidence is now pointing towards a regulating role for CRAM, one hypothesis being that it increases local concentrations of bioactive ligands by presenting them to the respective signalling receptors, based on observations made with chemerin and which fits with our work on chemokines.15 Another possible hypothesis would be that CRAM sequesters chemokines away from their signalling receptors, resulting in down-regulation of their activity. This is also in agreement with our data and supported by work from Otero Gutierrez et al.39 where it is shown that murine dendritic cell chemotaxis toward CCL19 or CCL21 via CCR7 is enhanced by neutralizing CRAM. Modulation of chemokine affinity for its receptor by another receptor has been described for CXCR7 and CXCR4 and could also be an aspect of CRAM activity.31 Finally, indirect evidence indicates that CRAM could be a transcytosis vector: in 2001, Baekkevold et al.40 showed that CCL19 is transcytosed in high endothelial venules and then recruits T cells. It was at first assumed that DARC was the transporter involved in this transcytosis, but this was excluded by in vitro experiments with transfected DARC performed by the authors. It has since been shown that CCL19 does not bind to DARC,41 making CRAM the most likely vector for these observations.
Further studies on the expression profiles as well as the fate of internalized chemokines will allow us to understand the role of CRAM in immunity and its contribution to chemokine activity. It will help to measure the relative weight of these hypotheses and the CRAM-specific role regarding CCX-CKR, which shares the binding profile and atypical status. CCL19 is a highly investigated chemokine because of its role in lymphocyte homing to lymph nodes and subsequent maturation processes, making its regulation crucial. As we have previously shown, CRAM expression is modulated throughout B-cell maturation stages, and with the addition of CRAM to the group of CCL19 binders, the characterization of CCL19 and CRAM association is an important step forward in defining B-cell biology.
Acknowledgments
The authors thank Dr Daniel F. Legler (University of Konstanz, Germany) for the gift of human CCL19-Fc and Fabian Junker for his assistance in internalization experiments.
Disclosures
The authors declare no conflict of interest. This work was supported by the Deutsche Forschungs-gemeinschaft (DFG) grant No. BU/1159/4-1 (to M.B).
References
- 1.Luster AD. Chemokines – chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–45. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A. The chemokine system: redundancy for robust outputs. Immunol Today. 1999;20:254–7. doi: 10.1016/s0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 3.Luttrell LM, Lefkowitz RJ. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci. 2002;115:455–65. doi: 10.1242/jcs.115.3.455. [DOI] [PubMed] [Google Scholar]
- 4.Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67:653–92. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson SS, Downey WE, III, Colapietro AM, Barak LS, Menard L, Caron MG. Role of beta-arrestin in mediating agonist-promoted G protein-coupled receptor internalization. Science. 1996;271:363–6. doi: 10.1126/science.271.5247.363. [DOI] [PubMed] [Google Scholar]
- 6.Goodman OB, Jr, Krupnick JG, Santini F, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–50. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 7.Haraldsen G, Rot A. Coy decoy with a new ploy: interceptor controls the levels of homeostatic chemokines. Eur J Immunol. 2006;36:1659–61. doi: 10.1002/eji.200636327. [DOI] [PubMed] [Google Scholar]
- 8.Weber M, Blair E, Simpson CV, et al. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comerford I, Milasta S, Morrow V, Milligan G, Nibbs R. The chemokine receptor CCX-CKR mediates effective scavenging of CCL19 in vitro. Eur J Immunol. 2006;36:1904–16. doi: 10.1002/eji.200535716. [DOI] [PubMed] [Google Scholar]
- 10.Kashiwazaki M, Tanaka T, Kanda H, et al. A high endothelial venule-expressing promiscuous chemokine receptor DARC can bind inflammatory, but not lymphoid, chemokines and is dispensable for lymphocyte homing under physiological conditions. Int Immunol. 2003;15:1219–27. doi: 10.1093/intimm/dxg121. [DOI] [PubMed] [Google Scholar]
- 11.Pruenster M, Mudde L, Bombosi P, et al. The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nat Immunol. 2009;10:101–8. doi: 10.1038/ni.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan P, Kyaw H, Su K, Zeng Z, Augustus M, Carter KC, Li Y. Cloning and characterization of a novel human chemokine receptor. Biochem Biophys Res Commun. 1998;243:264–8. doi: 10.1006/bbrc.1997.7981. [DOI] [PubMed] [Google Scholar]
- 13.Migeotte I, Franssen JD, Goriely S, Willems F, Parmentier M. Distribution and regulation of expression of the putative human chemokine receptor HCR in leukocyte populations. Eur J Immunol. 2002;32:494–501. doi: 10.1002/1521-4141(200202)32:2<494::AID-IMMU494>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 14.Biber K, Zuurman MW, Homan H, Boddeke HW. Expression of L-CCR in HEK 293 cells reveals functional responses to CCL2, CCL5, CCL7, and CCL8. J Leukoc Biol. 2003;74:243–51. doi: 10.1189/jlb.0802415. [DOI] [PubMed] [Google Scholar]
- 15.Zabel BA, Nakae S, Zuniga L, et al. Mast cell-expressed orphan receptor CCRL2 binds chemerin and is required for optimal induction of IgE-mediated passive cutaneous anaphylaxis. J Exp Med. 2008;205:2207–20. doi: 10.1084/jem.20080300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada T, Matsumoto M, Tatsumi Y, Kanamaru A, Akira S. A novel lipopolysaccharide inducible C-C chemokine receptor related gene in murine macrophages. FEBS Lett. 1998;425:490–4. doi: 10.1016/s0014-5793(98)00299-3. [DOI] [PubMed] [Google Scholar]
- 17.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–18. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 18.Wittamer V, Gregoire F, Robberecht P, Vassart G, Communi D, Parmentier M. The C-terminal nonapeptide of mature chemerin activates the chemerin receptor with low nanomolar potency. J Biol Chem. 2004;279:9956–62. doi: 10.1074/jbc.M313016200. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann TN, Leick M, Ewers S, Diefenbacher A, Schraufstatter I, Honczarenko M, Burger M. Human B cells express the orphan chemokine receptor CRAM-A/B in a maturation-stage-dependent and CCL5-modulated manner. Immunology. 2008;125:252–62. doi: 10.1111/j.1365-2567.2008.02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonecchi R, Borroni EM, Anselmo A, et al. Regulation of D6 chemokine scavenging activity by ligand- and Rab11-dependent surface up-regulation. Blood. 2008;112:493–503. doi: 10.1182/blood-2007-08-108316. [DOI] [PubMed] [Google Scholar]
- 21.Bonecchi R, Locati M, Galliera E, et al. Differential recognition and scavenging of native and truncated macrophage-derived chemokine (macrophage-derived chemokine/CC chemokine ligand 22) by the D6 decoy receptor. J Immunol. 2004;172:4972–6. doi: 10.4049/jimmunol.172.8.4972. [DOI] [PubMed] [Google Scholar]
- 22.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, Wells TN. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–8. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 23.Sipkins DA, Wei X, Wu JW, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–73. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honczarenko M, Glodek AM, Swierkowski M, Na IK, Silberstein LE. Developmental stage-specific shift in responsiveness to chemokines during human B-cell development. Exp Hematol. 2006;34:1093–100. doi: 10.1016/j.exphem.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Galliera E, Jala VR, Trent JO, et al. β-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–7. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- 26.Heuser JE, Anderson RG. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto Y, Ninomiya H, Miwa S, Masaki T. Cholesterol oxidation switches the internalization pathway of endothelin receptor type A from caveolae to clathrin-coated pits in Chinese hamster ovary cells. J Biol Chem. 2000;275:6439–46. doi: 10.1074/jbc.275.9.6439. [DOI] [PubMed] [Google Scholar]
- 28.Comerford I, Litchfield W, Harata-Lee Y, Nibbs RJ, McColl SR. Regulation of chemotactic networks by ‘atypical’ receptors. BioEssays. 2007;29:237–47. doi: 10.1002/bies.20537. [DOI] [PubMed] [Google Scholar]
- 29.O’Hayre M, Salanga CL, Handel TM, Allen SJ. Chemokines and cancer: migration, intracellular signalling and intercellular communication in the microenvironment. Biochem J. 2008;409:635–49. doi: 10.1042/BJ20071493. [DOI] [PubMed] [Google Scholar]
- 30.Lee JS, Frevert CW, Wurfel MM, et al. Duffy antigen facilitates movement of chemokine across the endothelium in vitro and promotes neutrophil transmigration in vitro and in vivo. J Immunol. 2003;170:5244–51. doi: 10.4049/jimmunol.170.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartmann TN, Grabovsky V, Pasvolsky R, et al. A crosstalk between intracellular CXCR7 and CXCR4 involved in rapid CXCL12-triggered integrin activation but not in chemokine-triggered motility of human T lymphocytes and CD34+ cells. J Leukoc Biol. 2008;84:1130–40. doi: 10.1189/jlb.0208088. [DOI] [PubMed] [Google Scholar]
- 32.Proudfoot AE, Handel TM, Johnson Z, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U S A. 2003;100:1885–90. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali S, Palmer AC, Banerjee B, Fritchley SJ, Kirby JA. Examination of the function of RANTES, MIP-1α, and MIP-1β following interaction with heparin-like glycosaminoglycans. J Biol Chem. 2000;275:11721–7. doi: 10.1074/jbc.275.16.11721. [DOI] [PubMed] [Google Scholar]
- 34.Skelton NJ, Aspiras F, Ogez J, Schall TJ. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry. 1995;34:5329–42. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida R, Imai T, Hieshima K, et al. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J Biol Chem. 1997;272:13803–9. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 36.Gosling J, Dairaghi DJ, Wang Y, Hanley M, Talbot D, Miao Z, Schall TJ. Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J Immunol. 2000;164:2851–6. doi: 10.4049/jimmunol.164.6.2851. [DOI] [PubMed] [Google Scholar]
- 37.Burger M, Burger JA, Hoch RC, Oades Z, Takamori H, Schraufstatter IU. Point mutation causing constitutive signaling of CXCR2 leads to transforming activity similar to Kaposi’s sarcoma herpesvirus-G protein-coupled receptor. J Immunol. 1999;163:2017–22. [PubMed] [Google Scholar]
- 38.McKimmie CS, Graham GJ. Leucocyte expression of the chemokine scavenger D6. Biochem Soc Trans. 2006;34:1002–4. doi: 10.1042/BST0341002. [DOI] [PubMed] [Google Scholar]
- 39.Otero Gutierrez K, Vecchi A, Garlanda C, Mantovani A, Sozzani S. World Intellectual Property Organization. Via Senato, 8, I-20121 Milano, Italy: LONG, Giorgio; Jacobacci & Partners S.p.A; 2008. Method for screeening drug candidates for inflammatory diseases mediated by LPS-inducible CC chemokine receptor mechanism; pp. 1–73. 028692. [Google Scholar]
- 40.Baekkevold ES, Yamanaka T, Palframan RT, et al. The CCR7 ligand elc (CCL19) is transcytosed in high endothelial venules and mediates T cell recruitment. J Exp Med. 2001;193:1105–12. doi: 10.1084/jem.193.9.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner L, Patterson AM, Ashton BA, Stone MA, Middleton J. The human Duffy antigen binds selected inflammatory but not homeostatic chemokines. Biochem Biophys Res Commun. 2004;321:306–12. doi: 10.1016/j.bbrc.2004.06.146. [DOI] [PubMed] [Google Scholar]