Abstract
We investigated the phenotypic basis for genetically determined differences in susceptibility and resistance to Chlamydia muridarum pulmonary infection using BALB/c and C57BL/6 mice. Following C. muridarum intranasal inoculation, the intensity of infection was very different between BALB/c and C57BL/6 beginning as early as 3 days post-infection. Intrapulmonary cytokine patterns also differed at early time-points (days 2 and 4) between these two strains of mice. The early recruitment of neutrophils to lung tissue was greater in BALB/c than in C57BL/6 mice and correlated with a higher number of inclusion forming units (IFU) of C. muridarum. At day 12 post-infection, BALB/c mice continued to demonstrate a greater burden of infection, significantly higher lung cytokine levels for tumour necrosis factor-α and interleukin-17 (IL-17) and a significantly lower level for interferon-γ than did C57BL/6 mice. In vitro, bone-marrow-derived dendritic cells (BMDCs) from BALB/c mice underwent less functional maturation in response to C. muridarum infection than did BMDCs from C57BL/6 mice. The BMDCs of BALB/c mice expressed lower levels of activation markers (CD80, CD86, CD40 and major histocompatibility complex class II) and secreted less IL-12 and more IL-23 than BMDCs from C57BL/6 mice. Overall, the data demonstrate that the differences exhibited by BALB/c and C57BL/6 mice following C. muridarum pulmonary infection are associated with differences in early innate cytokine and cellular responses that are correlated with late differences in T helper type 17 versus type 1 adaptive immune responses.
Keywords: adaptive immunity, Chlamydia muridarum, innate immunity, rodent, susceptibility
Introduction
The Chlamydiae is a phylum of intracellular bacteria with unique phylogenetics. Chlamydia trachomatis is the major human aetiological agent responsible for several diseases of importance to public health including trachoma and a variety of sexually transmitted diseases which have marked risks for blindness and infertility.1 There has been long-standing interest in developing an effective vaccine against C. trachomatis infection, but progress has been limited in part because of an incomplete understanding of the nature of protective immunity, infection-induced immunopathology and its underlying molecular and cellular basis.2–6
Chlamydia muridarum is a mouse-adapted strain whose genome exhibits high similarity to the C. trachomatis genome in terms of gene content and chromosomal synteny.7 Infection of mice with C. muridarum has proved to be surprisingly useful in suggesting models for the immunobiology of C. trachomatis infection in humans.8–10 Research regarding the immunology of murine C. muridarum infection has revealed that the resolution of primary and secondary infection is highly dependent on T helper type 1 (Th1) –cell-mediated immune responses.10–12 The Th1-type cytokines such as interleukin-12 (IL-12) and interferon-γ (IFN-γ) play critical roles in controlling and resolving C. muridarum infection.13–18 Adaptive immune responses such as the Th1 response are built upon early innate cellular and cytokine responses, which shape the developmental and migratory pathway exhibited by effector and memory T cells.19,20 Recruiting effector cells to sites of infection and amplifying and regulating inflammation are also among the major events orchestrated by the innate immune system. The role of dendritic cells (DCs), natural killer cells, macrophages and neutrophils during the innate phase after C. muridarum infection remains largely unknown.21–25 In particular DCs are likely to be particularly important because they are the major professional antigen-presenting cells crucial for initiation of the adaptive T-cell immune response.5,26
Toll-like receptors (TLRs) and other classes of pathogen-associated molecular pattern (PAMP) sensors are the fundamental components of the innate immune response that trigger early recruitment of inflammatory cells.25,27,28 The roles of PAMPs in the host inflammatory response to C. muridarum infection are still under active study and limited evidence shows that TLR-2 may be an important mediator in the innate immune response to C. muridarum infection, playing a role in both early production of inflammatory cytokine mediators and in the development of chronic inflammatory pathology.28Chlamydia muridarum molecules that induce PAMP sensor activation remain undefined although plasmid encoded proteins may be associated with activation of TLR-2.29
Heterogeneity in host susceptibility to C. muridarum infection in both human and murine systems has been well-documented.10,30–32 In particular, epidemiological observations suggest that the intensity of the early inflammatory response to C. trachomatis infection is correlated with the risk of development of late sequelae.33 Furthermore, it has been suggested that approximately 40% of the heterogeneity in response to C. trachomatis infection has genetic underpinnings. The mechanisms underlying genetic variation in the susceptibility to primary C. muridarum infection have been best studied using inbred mouse strains.9,10,32 Yang et al.10 demonstrated that the faster clearance of C. muridarum in C57BL/6 mice than in BALB/c mice was related to Th1 cell cytokine IFN-γ production and consequent adaptive T-cell immunity, and was inversely related to early IL-10 production. However, that report did not study the possible connection between variations in innate and adaptive immunity in relation to host susceptibility to C. muridarum infection; nor did it evaluate the potential role for the newest T helper lineage member – Th17 cells – in C. muridarum immunobiology. We chose the murine model of respiratory C. muridarum infection to explore the phenotypic basis for the genetically determined differences that lead to differential outcomes of C. muridarum infection in these two inbred strains of mice. We measured microbial growth kinetics, intrapulmonary local cytokine production and inflammatory cell recruitment at both early and late stages of infection. We also evaluated in vitro DC responses following C. muridarum infection using bone-marrow-derived DCs (BMDCs) collected from these two strains of mice. The findings provide a molecular and cellular explanation for the genetically determined difference in how BALB/c and C57BL/6 mice handle C. muridarum infection.
Materials and methods
Reagents and antibodies
All antibodies used for fluorescence-activated cell sorting (FACS) were from BD Pharmingen (Mississauga, ON, Canada), except fluorescein isothiocyanate (FITC)-conjugated anti-mouse F4/80 from Cedarlane Laboratories (Belington, ON, Canada). Chemokine keratinocyte-derived chemokine (KC/CXCL1), and macrophage inflammatory protein 2 (MIP-2/CXCL2) and mouse IL-23 immunoassay were purchased from R&D Systems (Minneapolis, MN); other cytokines for enzyme-linked immunosorbent assay (ELISA) were purchased from BD Pharmingen. Iscove’s modified Dulbecco’s medium (IMDM), minimum essential medium (MEM), gentamicin, 2-methyl-diethyltryptamine (2-ME) and sodium pyruvate were purchased from Sigma (St Louis, MO); fetal calf serum (FCS) and l-glutamine were purchased from Gibco (Grand Island, NY) and murine granulocyte–macrophage colony-stimulating factor (GM-CSF) was purchased from R&D Systems.
Chlamydia
Chlamydia muridarum mouse pneumonitis strain Nigg was grown in HeLa 229 cells in MEM containing 10% FCS and 0·5 mg/ml cyclohexamide. The C. muridarum live elementary bodies (EBs) were purified from infected HeLa cells by discontinuous density gradient centrifugation as previously described34 and stored in sucrose–phosphate–glutamic acid buffer (SPG) at −80°. The infectivity of purified EBs was determined by infecting HeLa monolayers for 24 hr, followed by immunostaining and enumeration of inclusions.
Generation and purification of BMDCs
The protocol for generation and purification of BMDCs used in this study has been described previously.35 Briefly, bone marrow cells were flushed from the femurs of mice and cultured in 20 ml IMDM supplemented with 10% FCS, 4 mm l-glutamine, 50 μg/ml gentamicin, 0·5 mm 2-ME, 10 ng/ml GM-CSF and 5% IL-4 culture supernatants of hybridoma X63. After 7–8 days of culture, the non-adherent cells were harvested and purified using anti-CD11c magnetic beads (Miltenyi Biotech Ltd, Auburn, CA). Routinely the purity of CD11c+ cells was 95 ± 5% (data not shown) as determined by FACS.
Murine pulmonary infection
Eight-week-old female C57BL/6 and BALB/c mice were purchased from Charles River (St Constant, QC, Canada) and housed in a pathogen-free animal facility. All experimental procedures were in accordance with the guidelines approved by the animal care committee of the University of British Columbia. Animals were inoculated intranasally with live C. muridarum EBs as previously described.36 The mice were monitored daily and their body weights were measured. At the indicated time-points, animals were killed by cervical dislocation and the lungs were aseptically harvested for weight, IFU titration, cytokine determination and processing the phenotyping cells using FACS analysis. Susceptibility to C. muridarum infection was assessed by changes in body weight, lung weight and lung bacterial burden.
Quantification of bacterial burden in lung
Confluent monolayers of HeLa 229 cells were prepared in flat-bottom 96-well plates after an overnight culture, and then pre-treated with 100 μl per well Hanks’ buffered salt solution (Gibco) containing 30 μg/ml diethylaminoethyl-dextran (Pharmacia Fine Chemicals, Uppsala, Sweden) at room temperature for 20 min followed by washing with SPG. Lung tissues were mechanically homogenized with tissue grinders in 5 ml cold phosphate-buffered saline (PBS) per lung followed by centrifugation at 1000 g for 10 min at 4° to remove coarse tissue debris. The fresh clarified tissue suspensions were serially diluted in SPG and immediately inoculated 100 μl per well onto HeLa monolayers. Left lung homogenates were divided into aliquots and stored at −80° for cytokine assays. After incubation at 37° in 5% CO2 for 2 hr, the supernatants were removed from HeLa cells and 100 μl per well MEM containing 1 μg/ml cycloheximide (Sigma) was added. The plates were placed in a cell incubator for 24 hr, after which cells were fixed. Immunostaining was carried out using an in-house prepared mouse anti-EB serum as primary antibody and horseradish peroxidase anti-mouse immunoglobulin G as secondary antibody. The colour was developed using 1 × 3,3-diaminobenzidine tetrahydrochloride (DAB)/metal concentrate in stable peroxide substrate buffer (Pierce, Rockford, IL). The C. muridarum inclusions were counted under a microscope at × 200 magnifications.
Analysis of cytokine and chemokine by ELISA
Purified BMDCs generated from C57BL/6 or BALB/c mice were plated into U-bottom 96-well plates at 2 × 105 cells in 200 μl of complete IMDM per well and pretreated with or without live C. muridarum EBs at a multiplicity of infection of 3 for 48 hr. Spleens were harvested from C57BL/6 or BALB/c mice individually at day 12 after C. muridarum intranasal infection and 2 × 106/ml of single-cell suspensions were cocultured with or without 1 × 105/ml of heat-killed C. muridarum EBs (HK EBs) in RPMI-1640 medium containing 10% FCS, 4 mm l-glutamine, 50 μg/ml gentamicin and 0·5 mm 2-ME in 24-well plates for 72 hr. Concanavalin A (ConA, 2 μg/ml) was used as a positive control. The culture supernatants were collected and stored at − 80° until they were tested. Lung homogenates were obtained as described above, thawed then vortexed and spun down before they were tested. Cytokines [IL-12, IL-10, IL-6, IL-17, IFN-γ and tumour necrosis factor-α (TNF-α)] in culture supernatants or lung homogenates were determined by ELISA as previously described.36 Paired anti-mouse IL-12 (p40/p70) (C15.6, C17.8), IL-10 (JESS-2A5, SXC-1), IL-6 (MP5-20F3, MP5-32C11), IL-17 (TC11-18H10.1, TC11-8H4), IFN-γ and TNF-α monoclonal antibodies for ELISA were used. The chemokines KC/CXCL1 and MIP-2/CXCL2 in total lung homogenates were measured using commercially available ELISAs. Production of IL-23 was tested using Quantikine®(R&D Systems, Minneapolis, MN) mouse IL-23 immunoassay according to the manufacturer’s instructions.
FACS analysis
For lung inflammatory cell enumeration, groups of four mice were inoculated intranasally with PBS or 8000 IFU live-EBs resuspended in 40 μl PBS. Animals were killed on day 2 post-infection, and their lungs were perfused through the right ventricle of the heart and harvested for total cell isolation. Lung tissues of each mouse were individually minced with scissors to a fine slurry in 0·5 ml PBS containing 2% FCS and subjected to collagenase digestion at room temperature for 20 min in 5 ml of 1 mg/ml collagenase (Sigma) followed by the addition of 200 μl 0·5 m ethylenediaminetetraacetic acid (EDTA, Gibco) per lung. After incubation at room temperature for 5 min, undigested fragments were dispersed by drawing the solution up and down through the bore of a 10-ml syringe with an 18-gauge needle and were further homogenized using a 40-μm nylon strainer and the plunger of a 1-ml syringe. Red blood cells were lysed by incubating with 0·5 m ammonium chloride. The suspensions of lung cells were centrifuged at 500 g at 4° for 5 min and washed twice with FACS buffer (PBS containing 2% FCS and 0·1% sodium azide) before staining. For phenotypic analysis of BMDCs, cells were incubated with media alone or live C. muridarum EBs at multiplicity of infection of 3 for 24 hr and collected. After washing with FACS buffer twice, cells were ready for staining. Cell staining was performed as previously described.35 Analysis was carried out on FACS Caliber (Becton Dickinson, San Jose, CA) using CellQuest software (BD Bioscience, Mississauga, ON, Canada) or FlowJo software (Tree star, Ashland, OR); 2 × 105 lung cells and 1 × 104 BMDCs were counted and 1·5 × 104 lung cells (dots) were shown.
The following monoclonal antibodies were used for cell staining: phycoerythrin (PE)-conjugated anti-mouse CD11c (HL3); PE-conjugated anti-mouse CD49b (DX5); peridinin chlorophyll protein-Cy™5.5-conjugated anti-mouse (BD Pharmingen, Missisauga, ON, Canada) CD11b (M1/70); FITC-conjugated anti-mouse CD80 (16-10A1); FITC-conjugated anti-mouse CD86 (GL1); FITC-conjugated anti-mouse I-A/I-E (2G9); FITC-conjugated anti-mouse CD40 (HM40-3); FITC-conjugated rat anti-mouse Gr-1 (RB6-8C5); FITC-conjugated anti-mouse CD3 (17A2); FITC-conjugated anti-mouse F4/80 (CL8940F); and FITC-conjugated anti-mouse TLR-2 (6C2).
Statistical analysis
Statistical analysis was conducted using two-tailed unpaired Student’s t-test for ELISA, FACS analysis, IFU, body weight and lung weight comparisons of each time-point. Repeated measures analysis of variance (RM anova) was used for the changes in body weight over time between the two groups. Differences at the P < 0·05 level were considered statistically significant.
Results
Susceptibility to C. muridarum infection and disease in BALB/c versus C57BL/6 mice
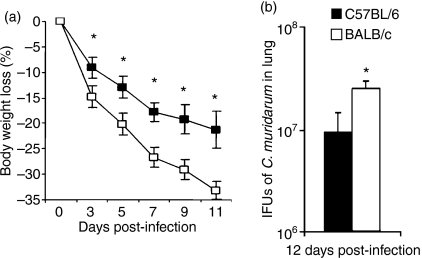
Each group of animals was infected in parallel with C. muridarum via intranasal inoculation. Susceptibility was assessed by monitoring weight loss and lung bacterial burden. As shown in Fig. 1, beginning at day 3 BALB/c mice lost significantly more body weight than C57BL/6 mice after infection (Fig. 1a) and at day 12 post-infection the bacterial burden in the lungs of BALB/c mice remained significantly greater than in C57BL/6 mice (182·6 ± 28·4 × 106 versus 75·2 ± 34·4 × 106 IFU) (Fig. 1b). The changes in body weight were also compared over time for the two groups by two-way RM anova; body weight loss in BALB/c was significantly greater than C57BL/6 (P < 0·001). Additionally, BALB/c mice exhibited more clinical signs of disease, as indicated by lethargy, ruffled fur and laboured breathing, than did C57BL/6 mice. The results confirm previous reports that BALB/c mice are more susceptible to C. muridarum pulmonary infection than C57BL/6 mice.
Figure 1.
Susceptibility of BALB/c mice and C57BL/6 mice following Chlamydia muridarum infection. Groups of eight mice were intranasally inoculated with 2000 inclusion-forming units (IFU) of C. muridarum and body weight changes were monitored every 2 days (a). Mice were killed at day 12 post-infection and IFU in lung tissue were titrated as described in the Materials and methods (b). The data represent the mean ± SD from eight individual mice. One of three independent trials with similar results is shown. *P < 0·05.
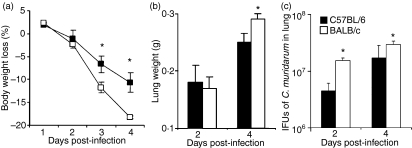
To investigate whether these differences correlate with innate immunity, we tested in separate experiments the early responses of mice following C. muridarum infection. Body weights were measured daily until day 4 post-infection. Beginning day 3 post-infection BALB/c mice showed significantly greater loss of body weight than C57BL/6 mice (Fig. 2a). Change in lung weight is an established index of disease severity in murine models following pulmonary infection.37 As shown in Fig. 2(b), lung weights exhibited no difference at day 2 post-infection but by day 4 they were significantly greater in BALB/c mice than in C57BL/6 mice, indicating more severe infection in BALB/c mice than in C57BL/6 mice. Lung titration of C. muridarum revealed significantly higher IFU in lung homogenates of BALB/c than C57BL/6 mice at both day 2 (14·2 ± 1·3 × 106 versus 4·3 ± 1·5 × 106) and day 4 (269·5 ± 36·3 × 106 versus 155·5 ± 103·5 × 106) after infection (Fig. 2c). These results show that genetic differences in susceptibility to C. muridarum lung infection are demonstrable at early stages of infection and probably reflect differences in innate response.
Figure 2.
Early stage susceptibility of BALB/c mice and C57BL/6 mice following Chlamydia muridarum infection. Groups of eight mice were intranasally inoculated with 8000 inclusion-forming units (IFU) of C. muridarum and body weight changes were monitored daily up to 4 days (a). Mice were killed at days 2 or 4 post-infection and the lungs were weighed (b). Infectivity was assessed by lung IFU titration at days 2 and 4 (c). The data represent the mean ± SD from eight individual mice. One of three independent trials with similar results is shown. *P < 0·05.
Early and late local cytokine profiles in response to C. muridarum infection
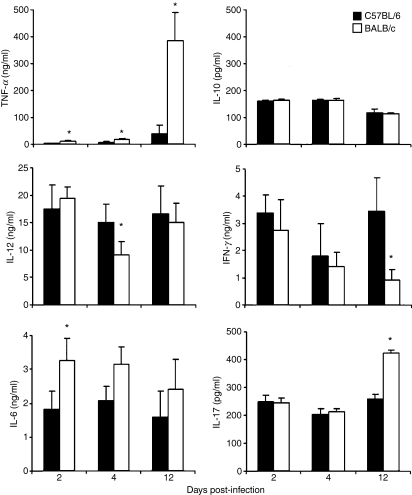
In the early stage of infection, the balance among cytokines can be very important in containing infection and in triggering T-cell and B-cell developmental pathways. Therefore, we compared the production of several important pro-inflammatory cytokines including TNF-α, IL-6, IL-10, IL-12, IL-17 and IFN-γ in the lungs at both early (days 2 and 4) and later (day 12) stages of infection. As shown in Fig. 3, BALB/c produced significantly higher levels of TNF-α in lung homogenates than C57BL/6 mice on both days 2 and 4 after C. muridarum infection as determined by ELISA. Levels of IL-6 showed a pattern similar to TNF-α although the differences were statistically significant only at day 2 post-infection. C57BL/6 mice showed significantly higher levels of IL-12 production at day 4 post-infection than BALB/c mice. There were no statistically significant differences in IFN-γ, IL-10 and IL-17 levels at the early time-points. By day 12 post-infection BALB/c mice demonstrated significantly higher TNF-α and lower IFN-γ levels than C57BL/6 mice. Notably, intrapulmonary IL-17 levels were significantly greater in BALB/c mice than in C57BL/6 mice at day 12 post-infection. These data demonstrate that different cytokine profiles occur both early and late in response to C. muridarum infection in BALB/c versus C57BL/6 mice.
Figure 3.
Cytokine secretions in lung tissue of BALB/c mice versus C57BL/6 mice after infection with Chlamydia muridarum. Groups of eight mice for each strain were inoculated intranasally with 8000 inclusion-forming units (IFU; for days 2 and 4 infection) or 2000 IFU (for day 12 infection) of C. muridarum. At days 2, 4 and 12 post-infection, lung homogenates were prepared for lung IFU titrations and for assays of cytokine levels by enzyme-linked immunosorbent assay. The data represent the mean ± SD from eight individual mice. One of three independent trials with similar results is shown. *P < 0·05.
C. muridarum cytokine responses in BALB/c and C57BL/6 mice
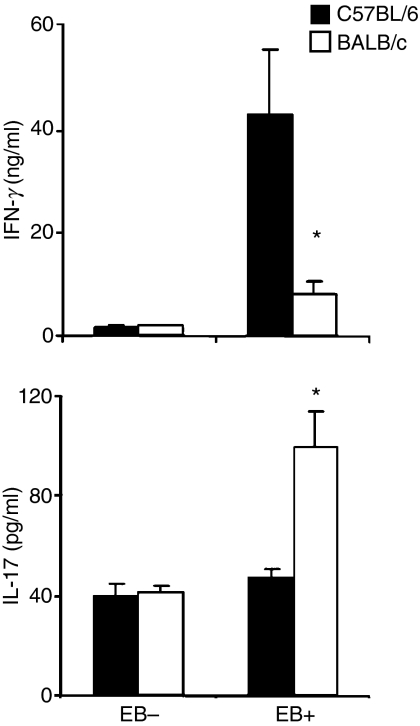
To confirm that the differences of IFN-γ and IL-17 levels at day 12 post-infection in BALB/c versus C57BL/6 mice are correlated with late adaptive Th17 and Th1 immune responses, we investigated C. muridarum cellular cytokine responses. Splenocytes were harvested at day 12 from mice infected with 2000 IFU C. muridarum and cultured in vitro with or without HK EB for 72 hr. The C. muridarum IFN-γ and IL-17 responses were measured by ELISA. As shown in Fig. 4, BALB/c mice demonstrated a significantly lower C. muridarum IFN-γ response (8·3 ± 2·4 ng/ml versus 43·3 ± 12·6 ng/ml) and higher C. muridarum IL-17 response (100·1 ± 13·9 pg/ml versus 47·6 ± 3·5 pg/ml) than C57BL/6 mice. For the positive control, ConA (0·2 μg/ml) was used (9·1 ± 1·4 ng/ml versus 15·2 ± 4·3 ng/ml for IFN-γ and 159·0 ± 19·6 pg/ml versus 112·9 ± 13·3 pg/ml for IL-17) and showed no significant difference. Higher Th17 and lower Th1 cytokine responses in splenocytes are consistent with higher IL-17 and lower IFN-γ levels directly measured in the lungs of BALB/c compared with C57BL/6 mice.
Figure 4.
Cytokine production by splenocytes in response to Chlamydia muridarum stimulation. Groups of nine mice were inoculated intranasally with 2000 inclusion-forming units (IFU) of C. muridarum and splenocytes were prepared at day 12 followed by stimulation with or without heat-killed C. muridarum elementary bodies (EBs) for 72 hr. The level of C. muridarum responsive interferon-γ (IFN-γ) and interleukin-17 (IL-17) in splenocyte culture supernatants was analysed by enzyme-linked immunosorbent assay. The data represent the mean ± SE from nine individual mice. *P < 0·05.
Early cellular recruitment and chemokine production in response to pulmonary C. muridarum infection
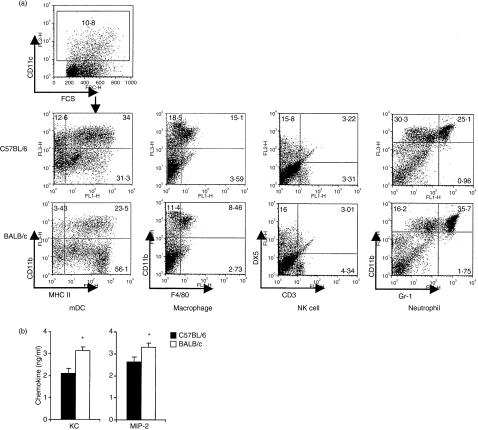
Rapid cellular recruitment and exudative responses triggered by pro-inflammatory cytokines are hallmarks of the acute inflammatory response. Accordingly, we examined the phenotype of lung cell recruitment by FACS in response to intranasal C. muridarum infection. BALB/c and C57BL/6 mice were intranasally infected with C. muridarum or with PBS alone and lung cells were analysed on day 2 by double staining of surface markers. The counts of total lung cells were 3·3 ± 1·2 × 106 cells/lung in BALB/c versus 4·2 ± 0·9 × 106 cells/lung in C57BL/6 before infection and 8·3 ± 1·3 × 106 cells/lung in BALB/c versus 8·6 ± 1·0 × 106 cells/lung in C57BL/6 after infection. Surface markers evaluated include DX5+ CD3− for NK cells, Gr1+ CD11b+ for neutrophils, CD11b+ CD11c+ major histocompatibility complex class II (MHC II)+ for myeloid DCs, and F4/80+ CD11b+ for macrophages. Compared with lung cells from PBS-inoculated mice, rapid infiltration with neutrophils, DCs and macrophages was observed in both strains of mice following infection (Table 1). Neutrophils, however, were more significantly recruited in BALB/c mice than in C57BL/6 mice in response to intranasal C. muridarum infection. Fewer DCs and macrophages were detected in BALB/c mice than in C57BL/6 mice. The recruitment of NK cells was similar between these two strains of mice (Fig. 5a, Table 1).
Table 1.
Lung cell recruitment was analysed by mean percentage ± SD of each cell population in total lung cells by fluorescence-activated cell sorting in C57BL/6 and BALB/c mice after 2 days intranasal infection with 8000 inclusion-fomring units of Chlamydia muridarum
| Uninfected |
Infected |
|||
|---|---|---|---|---|
| C57/BL6 | BALB/c | C57/BL6 | BALB/c | |
| Natural killer cell | 16·5 ± 0·5 | 13·7 ± 0·4 | 16·9 ± 1·9 | 17·9 ± 1·7 |
| Myeoid denritic cell | 0·1 ± 0·1 | 0·2 ± 0·01 | 2·9 ± 0·5 | 1·5 ± 0·05* |
| Neutrophil | 2·0 ± 0·03 | 3·3 ± 0·3 | 25·8 ± 2·1 | 36·8 ± 3·7* |
| Macrophage | 3·3 ± 0·5 | 1·6 ± 0·2 | 16·4 ± 1·2 | 8·4 ± 0·5* |
Three individual mice in each group. One representative experiment of two independent trials with similar results is shown. Numbers in bold represent statistically significant values when C57/BL6 and BALB/c were compared.
P < 0·05.
Figure 5.
Cell recruitment and neutrophil chemokine production in lung tissue of BALB/c and C57BL/6 mice after infection with Chlamydia muridarum. Groups of three mice were inoculated intranasally with 8000 inclusion-forming units (IFU) of C. muridarum and day 2 post-infection lungs were harvested, single cell suspensions were created and cell types were enumerated by fluorescence-activated cell sorter analysis. Two independent experiments were carried out. Dot-plot from one representative mouse shows double staining of surface markers for lung inflammatory cells (a). Chemokine secretion in lung tissue of BALB/c mice and C57BL/6 mice at day 2 after infection with C. muridarum. Lung homogenates were the same samples prepared for Fig. 3. The chemokines in lung homogenates were analysed by enzyme-linked immunosorbent assay. The data represent the mean ± SE from eight individual mice. One of three independent trials with similar results is shown. *P < 0·05 (b).
To explore potential causes for the early increased influx of neutrophils, we tested the concentrations of KC and MIP-2 chemokines in lung homogenates by ELISA. The results demonstrated that both of these neutrophil chemokines were significantly higher in BALB/c than C57BL/6 mice at day 2 after C. muridarum infection (Fig. 5b).
In vitro activation of BMDCs derived from BALB/c and C57BL/6 mice after infection with C. muridarum
We were particularly interested in DCs as critical participants in the innate response by virtue of their ability to link innate and adaptive immunity. The endocytosis of C. muridarum by BMDCs from BALB/c and C57BL/6 mice was tested and exhibited no significant difference as measured by IFU (data not shown). To evaluate the activation response of BMDCs following in vitro infection with C. muridarum from these two strains of mice, we determined the expression of cell surface molecules such as CD40, CD80, CD86, MHC II and TLR-2 on BMDCs followed by determination of secretion levels of selected cytokines. After 24-hr cultures, the cells were collected and subjected to FACS analysis; and after 48 hr of culture, the supernatants from parallel sets of cultures were collected and the cytokines in the media were tested by ELISA. The expression level of activation molecules on BMDCs was estimated from fluorescence intensity. Uninfected resting BMDCs derived from these two strains of mice displayed no difference in expression of CD40, CD80, CD86, MHC II and TLR-2 (data not shown). After C. muridarum infection, the expression of CD80, CD86, CD40 and MHC II was less increased on BMDCs derived from BALB/c mice than on those from C57BL/6 (Fig. 6a,b). Expression of TLR-2 was slightly greater on BMDCs derived from BALB/c mice than on those from C57BL/6 mice. We next evaluated the response of DCs to C. muridarum infection by comparing the cytokine secretion profiles. As shown in Fig. 6(c), C. muridarum-infected BMDCs derived from BALB/c mice secreted lower levels of IL-12 and higher levels of IL-23, IL-6, IL-10 and TNF-α than those derived from C57BL/6 mice. Remarkably these in vitro cytokine responses of infected BMDCs closely resemble those observed in vivo following pulmonary C. muridarum infection.
Figure 6.
Surface phenotype and cytokine secretion of cultured dendritic cells (DCs) derived from BALB/c mice and C57BL/6 mice. Bone-marrow-derived dendritic cells (BMDCs) were generated and purified from C57BL/6 and BALB/c mice as described in the Materials and methods. Cells were infected with live Chlamydia muridarum elementary bodies at a multiplicity of infection of 3 for 24 hr and collected for staining of costimulatory molecule expression [CD80, CD86, CD40, major histocompatibility complex class II (MHC II) and Toll-like receptor 2 (TLR2)] by fluorescence-activated cell sorting (FACS). The thin-line histograms represent unstained cells as controls and bold-line histograms represent stained cells. Numerical values depict the percentage of positive cells and fluorescence intensity of relevant surface molecules. Data are representative of three independent experiments with similar results (a). Fold increase of surface molecular expression on BMDCs after C. muridarum stimulation represent mean ± SE (n = 9) of three separate experiments (b). After 48 hr incubation with or without C. muridarum, culture supernatants were collected for enzyme-linked immunosorbent assay cytokine test. The data represent the mean ± SD of three separated experiments. *P < 0·05 (c).
Discussion
Genetic differences in susceptibility to many infectious diseases including C. muridarum have been studied using inbred mouse models.5,9,10 In this study, we have demonstrated that the genetically determined differences in susceptibility to pulmonary C. muridarum infection between BALB/c and C57BL/6 mice start at early time-points following infection during which the innate responses are occurring. The results show that the differences include greater decreases in body weight, larger increases in lung weight and accelerated growth of C. muridarum in vivo (Fig. 2) in BALB/c mice compared to C57BL/6 mice. The difference in the extent of body weight loss between BALB/c and C57BL/6 mice was first statistically observed at day 3 (Fig. 2a) with the same trend maintained through to day 12 (Fig. 1a). The greater increase in lung weight from day 2 to 4 post-infection in BALB/c mice as compared with C57BL/6 mice shown in Fig. 2(b) probably indicates more severe pathological changes in the lung and was accompanied by more severe clinical signs of respiratory disease. Study of early cellular infiltration by FACS analysis of lung cells following C. muridarum infection revealed a more intense inflammatory reaction in BALB/c than C57BL/6 mice with markedly more infiltrating neutrophils (Table 1).
Differences in susceptibility to C. muridarum that occur at early stages in infection are most likely related to the differences in the host innate response.20 Innate immunity involves a complex network of cytokines, chemokines, plasma proteins and local cellular recruitment.19 Chemokine KC and MIP-2 are potent neutrophil chemoattractants and their concentrations in lung homogenates at day 2 were higher in BALB/c mice than in C57BL/6 (Fig. 5b). These differences in chemokine concentration may be responsible for the different neutrophil influxes between BALB/c and C57BL/6 mice (Fig. 5a and Table 1).
Many cytokines have been implicated in C. muridarum host defence and in this study we compared the cytokine profiles in lung homogenates at days 2, 4 and 12 after infection. The most remarkable difference was observed with TNF-α level. The levels of lung TNF-α were substantially higher in the BALB/c than in C57BL/6 mice at all time-points studied, especially at day 12. Interleukin-6 is another cytokine produced in significant amounts in BALB/c mice at early time-points in the lung in response to C. muridarum infection although this IL-6 response was more transient than the TNF-α response. Studies have indicated that TNF-α and IL-6 may play roles in host defence against C. muridarum infection, but it remains unclear whether they mediate protective or pathological responses.38–42 In this study, the higher TNF-α and IL-6 productions in the lung observed in BALB/c mice correlated with enhanced susceptibility to C. muridarum at both early and late stages of infection.
Interferon-γ is the cytokine that has been most thoroughly studied in C. muridarum protective immunity. Our data show that the IFN-γ production in the lungs of BALB/c and C57BL/6 mice was similar at days 2 and 4 but became statistically different at day 12, which suggests that adaptive T-cell responses are the likely source for IFN-γ during the late phases (Fig. 4). The late difference in IFN-γ secretion between the two strains of mice was temporally correlated with an observed early difference in IL-12 secretion, because the lung IL-12 level in BALB/c mice was significantly lower than in C57BL/6 mice at day 4. The lower IL-12 level in the lungs of BALB/c mice may be related to less recruitment of DCs and macrophages in early infection (Table 1) and lower secretion of IL-12 following DC infection with C. muridarum (Fig. 5).
Overall, the lung cytokine assays showed two different response profiles. One pattern observed in BALB/c mice was of high TNF-α and IL-6 levels and low IL-12 production, which was associated with more severe neutrophilic inflammation, poorer clearance of C. muridarum and later development of an adaptive Th17 immune response. The other response pattern observed in C57BL/6 mice was associated with higher IL-12 and lower TNF-α and IL-6 secretion, which was associated with less lung inflammation, more rapid clearance and the later development of Th1 IFN-γ-mediated immunity.
A novel finding in this study is the higher lung IL-17 levels in BALB/c mice than in C57BL/6 mice at day 12 after C. muridarum infection. Interleukin-17 is a recently recognized pro-inflammatory cytokine produced by Th17 cells.43 These Th17 cells appear to be critical T-cell immune effectors to extracellular pathogens at the mucosal surface and have been implicated in autoimmunity or immunopathology. The role of IL-17 in C. trachomatis immunobiology is not yet clear. As IL-6 is one of the key effector cytokines involved in triggering the Th17 cell developmental pathway, the early high levels of IL-6 in BALB/c mice may play a role in promoting the adaptive Th17 immune pathway,44 which may in turn have contributed to the stronger neutrophilic inflammatory response observed in BALB/c mice. Corresponding with the higher lung IL-17 levels at day 12 post-infection, C. muridarum-responsive IL-17 production by splenocytes from C. muridarum-infected mice was also higher in BALB/c mice than in C57BL/6 mice (Fig. 4). Importantly these differences in cytokine responses between the two strains of mice were seen only with C. muridarum EBs but not with ConA. Taken together, these data suggest that the genetic differences in innate immune responses in these two strains of mice shape the divergent T-cell adaptive immune responses and thereby determine infection kinetics and disease outcome.
Dendritic cells stand centre stage in linking innate to adaptive T-cell responses.45,46 Some studies have indicated that BMDCs from different genetic backgrounds show different levels of activation, endocytosis and cytokine production after microbial stimulation.47–50 It was therefore of great interest that when DCs from each mouse strain were infected with C. muridarum in vitro, distinct maturation patterns and cytokine secretion profiles were observed. Although no differences between resting BMDCs from the two mouse strains were observed, after in vitro C. muridarum infection the purified BMDCs derived from BALB/c mice demonstrated a smaller increase in expression of activation markers, including CD80, CD86 and MHC II, than did BMDCs from C57BL/6 mice (Fig. 6a,b). The BMDCs derived from BALB/c mice secreted lower levels of IL-12 and higher levels of IL-6, IL-10 and TNF-αin vitro compared with BMDCs from C57BL/6 mice following C. muridarum infection. Furthermore, DCs from BALB/c mice secreted more IL-23 than DCs from C57BL/6 after infection with C. muridarum (Fig. 6c). As IL-23 has an important role in the stabilization of Th17 cells, the higher IL-23 secretion from DCs may be responsible for the higher Th17 responses in BALB/c mice. The different responses of infected BMDCs may directly or indirectly alter the innate and adaptive immune responses. Darville et al.28 demonstrated that TLR-2 is an important mediator in the innate immune response to C. trachomatis infection and that it is essential for development of oviduct pathology in C. muridarum genital tract infection. Of potential interest is the finding that the TLR-2 expression on BMDCs from BALB/c mice is slightly increased compared with that on BMDCs from C57BL/6 mice after in vitro stimulation with C. muridarum. The impact of TLR-2 expression by DCs in vivo on the inflammatory response needs further study. Taken together, these findings suggest that DCs may be the target cell in which the genetically defined differences in C. muridarum immunobiology between BALB/c and C57BL/6 mice are phenotypically expressed. Overall, these findings in the murine C. muridarum model should be useful in defining new immunoepidemiological studies of human C. trachomatis infection.
Acknowledgments
This work was supported by National Institutes of Health Grant R01AI076483. We are very grateful to Dr J. Schachter for supplying C. muridarum mouse pneumonitis strain Nigg and to Dr F. Melchers (Basilea Institute, Switzerland) for the generous gift of IL-4 producing hybridoma X63.
Glossary
Abbreviations:
- 2-ME
2-methyl-diethyltryptamine
- BMDC
bone-marrow-derived dendritic cell
- CD
cluster of differentiation
- ConA
Concanavalin A
- EB
elementary body
- ELISA
enzyme-linked immunosorbent assay
- FACS
fluorescence-activated cell sorting
- FCS
fetal calf serum
- FITC
fluorescein isothiocyanate
- GM-CSF
murine granulocyte–macrophage colony-stimulating factor
- HK EB
heat-killed Chlamydia muridarum EB
- IFN-γ
interferon-γ
- IFU
inclusion-forming unit
- IL
interleukin
- IMDM
Iscove’s modified Dulbecco’s medium
- KC/CXCL1
keratinocyte-derived chemokine
- MEM
minimum essential medium
- MHC
major histocompatibility complex
- MIP-2/CXCL2
macrophage inflammatory protein 2
- PAMP
pathogen-associated molecular pattern
- PBS
phosphate-buffered saline
- PE
phycoerythrin
- SPG
sucrose–phosphate–glutamic acid buffer
- Th
T helper
- TLR
toll-like receptor
- TNF
tumour necrosis factor
Disclosures
The authors have no financial conflict of interest.
References
- 1.Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia. Intracellular Biology, Pathogenesis, and Immunity. Washington, DC: ASM Press; 1999. pp. 139–69. [Google Scholar]
- 2.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–61. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Yang X, Lu H, Zhong G, Brunham RC. Immunity to Chlamydia trachomatis mouse pneumonitis induced by vaccination with live organisms correlates with early granulocyte-macrophage colony-stimulating factor and interleukin-12 production and with dendritic cell-like maturation. Infect Immun. 1999;67:1606–13. doi: 10.1128/iai.67.4.1606-1613.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eko FO, He Q, Brown T, et al. A novel recombinant multisubunit vaccine against Chlamydia. J Immunol. 2004;173:3375–82. doi: 10.4049/jimmunol.173.5.3375. [DOI] [PubMed] [Google Scholar]
- 5.Su H, Messer R, Whitmire W, Fischer E, Portis JC, Caldwell HD. Vaccination against chlamydial genital tract infection after immunization with dendritic cells pulsed ex vivo with nonviable Chlamydiae. J Exp Med. 1998;188:809–18. doi: 10.1084/jem.188.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunham RC, Zhang D. Transgene as vaccine for chlamydia. Am Heart J. 1999;138:S519–22. doi: 10.1016/s0002-8703(99)70291-7. [DOI] [PubMed] [Google Scholar]
- 7.Read TD, Brunham RC, Shen C, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison RP, Caldwell HD. Immunity to murine chlamydial genital infection. Infect Immun. 2002;70:2741–51. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darville T, Andrews CW, Jr, Laffoon KK, Shymasani W, Kishen LR, Rank RG. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect Immun. 1997;65:3065–73. doi: 10.1128/iai.65.8.3065-3073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, HayGlass KT, Brunham RC. Genetically determined differences in IL-10 and IFN-gamma responses correlate with clearance of Chlamydia trachomatis mouse pneumonitis infection. J Immunol. 1996;156:4338–44. [PubMed] [Google Scholar]
- 11.Cain TK, Rank RG. Local Th1-like responses are induced by intravaginal infection of mice with the mouse pneumonitis biovar of Chlamydia trachomatis. Infect Immun. 1995;63:1784–9. doi: 10.1128/iai.63.5.1784-1789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw JH, Grund VR, Durling L, Caldwell HD. Expression of genes encoding Th1 cell-activating cytokines and lymphoid homing chemokines by chlamydia-pulsed dendritic cells correlates with protective immunizing efficacy. Infect Immun. 2001;69:4667–72. doi: 10.1128/IAI.69.7.4667-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su H, Caldwell HD. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect Immun. 1995;63:3302–8. doi: 10.1128/iai.63.9.3302-3308.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry LL, Feilzer K, Caldwell HD. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol. 1997;158:3344–52. [PubMed] [Google Scholar]
- 15.Ramsey KH, Rank RG. Resolution of chlamydial genital infection with antigen-specific T-lymphocyte lines. Infect Immun. 1991;59:925–31. doi: 10.1128/iai.59.3.925-931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H, Yang X, Takeda K, et al. Chlamydia trachomatis mouse pneumonitis lung infection in IL-18 and IL-12 knockout mice: IL-12 is dominant over IL-18 for protective immunity. Mol Med. 2000;6:604–12. [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Wang MD, Lenz S, Gao D, Kaltenboeck B. IL-12 administered during Chlamydia psittaci lung infection in mice confers immediate and long-term protection and reduces macrophage inflammatory protein-2 level and neutrophil infiltration in lung tissue. J Immunol. 1999;162:2217–26. [PubMed] [Google Scholar]
- 18.Geng Y, Berencsi K, Gyulai Z, Valyi-Nagy T, Gonczol E, Trinchieri G. Roles of interleukin-12 and gamma interferon in murine Chlamydia pneumoniae infection. Infect Immun. 2000;68:2245–53. doi: 10.1128/iai.68.4.2245-2253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Droemann D, Rupp J, Goldmann T, et al. Disparate innate immune responses to persistent and acute Chlamydia pneumoniae infection in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;175:791–7. doi: 10.1164/rccm.200607-926OC. [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 21.Bai H, Yang J, Qiu H, Wang S, Fan Y, Han X, Xie S, Yang X. Intranasal inoculation of Chlamydia trachomatis mouse pneumonitis agent induces significant neutrophil infiltration which is not efficient in controlling the infection in mice. Immunology. 2005;114:246–54. doi: 10.1111/j.1365-2567.2004.02088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jendro MC, Deutsch T, Korber B, et al. Infection of human monocyte-derived macrophages with Chlamydia trachomatis induces apoptosis of T cells: a potential mechanism for persistent infection. Infect Immun. 2000;68:6704–11. doi: 10.1128/iai.68.12.6704-6711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redecke V, Dalhoff K, Bohnet S, Braun J, Maass M. Interaction of Chlamydia pneumoniae and human alveolar macrophages: infection and inflammatory response. Am J Respir Cell Mol Biol. 1998;19:721–7. doi: 10.1165/ajrcmb.19.5.3072. [DOI] [PubMed] [Google Scholar]
- 24.Tseng CT, Rank RG. Role of NK cells in early host response to chlamydial genital infection. Infect Immun. 1998;66:5867–75. doi: 10.1128/iai.66.12.5867-5875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolard MD, Hudig D, Tabor L, Ivey JA, Simecka JW. NK cells in gamma-interferon-deficient mice suppress lung innate immunity against Mycoplasma spp. Infect Immun. 2005;73:6742–51. doi: 10.1128/IAI.73.10.6742-6751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Xing Z, Brunham RC. GM-CSF transgene-based adjuvant allows the establishment of protective mucosal immunity following vaccination with inactivated Chlamydia trachomatis. J Immunol. 2002;169:6324–31. doi: 10.4049/jimmunol.169.11.6324. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Coriolan D, Schultz K, Golenbock DT, Beasley D. Toll-like receptor 2 mediates persistent chemokine release by Chlamydia pneumoniae-infected vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2005;25:2308–14. doi: 10.1161/01.ATV.0000187468.00675.a3. [DOI] [PubMed] [Google Scholar]
- 28.Darville T, O’Neill JM, Andrews CW, Jr, Nagarajan UM, Stahl L, Ojcius DM. Toll-like receptor-2, but not toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol. 2003;171:6187–97. doi: 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- 29.O’Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol. 2007;179:4027–34. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 30.Qiu H, Wang S, Yang J, Fan Y, Joyee AG, Han X, Jiao L, Yang X. Resistance to chlamydial lung infection is dependent on major histocompatibility complex as well as non-major histocompatibility complex determinants. Immunology. 2005;116:499–506. doi: 10.1111/j.1365-2567.2005.02249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jupelli M, Guentzel MN, Meier PA, Zhong G, Murthy AK, Arulanandam BP. Endogenous IFN-gamma production is induced and required for protective immunity against pulmonary chlamydial infection in neonatal mice. J Immunol. 2008;180:4148–55. doi: 10.4049/jimmunol.180.6.4148. [DOI] [PubMed] [Google Scholar]
- 32.Miyairi I, Tatireddigari VR, Mahdi OS, Rose LA, Belland RJ, Lu L, Williams RW, Byrne GI. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J Immunol. 2007;179:1814–24. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- 33.Skwor TA, Atik B, Kandel RP, Adhikari HK, Sharma B, Dean D. Role of secreted conjunctival mucosal cytokine and chemokine proteins in different stages of trachomatous disease. PLoS Negl Trop Dis. 2008;2(e264):1–12. doi: 10.1371/journal.pntd.0000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–76. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaharik ML, Nayar T, White R, et al. Genetic profiling of dendritic cells exposed to live- or ultraviolet-irradiated Chlamydia muridarum reveals marked differences in CXC chemokine profiles. Immunology. 2007;120:160–72. doi: 10.1111/j.1365-2567.2006.02488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X, Shen C, Rey-Ladino J, Yu H, Brunham RC. Characterization of murine dendritic cell line JAWS II and primary bone marrow-derived dendritic cells in Chlamydia muridarum antigen presentation and induction of protective immunity. Infect Immun. 2008;76:2392–401. doi: 10.1128/IAI.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huppert M, Sun SH, Gleason-Jordon I, Vukovich KR. Lung weight parallels disease severity in experimental coccidioidomycosis. Infect Immun. 1976;14:1356–68. doi: 10.1128/iai.14.6.1356-1368.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Natividad A, Hanchard N, Holland MJ, et al. Genetic variation at the TNF locus and the risk of severe sequelae of ocular Chlamydia trachomatis infection in Gambians. Genes Immun. 2007;8:288–95. doi: 10.1038/sj.gene.6364384. [DOI] [PubMed] [Google Scholar]
- 39.Holtmann H, Shemer-Avni Y, Wessel K, Sarov I, Wallach D. Inhibition of growth of Chlamydia trachomatis by tumor necrosis factor is accompanied by increased prostaglandin synthesis. Infect Immun. 1990;58:3168–72. doi: 10.1128/iai.58.10.3168-3172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conway DJ, Holland MJ, Bailey RL, Campbell AE, Mahdi OS, Jennings R, Mbena E, Mabey DC. Scarring trachoma is associated with polymorphism in the tumor necrosis factor alpha (TNF-alpha) gene promoter and with elevated TNF-alpha levels in tear fluid. Infect Immun. 1997;65:1003–6. doi: 10.1128/iai.65.3.1003-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darville T, Andrews CW, Jr, Rank RG. Does inhibition of tumor necrosis factor alpha affect chlamydial genital tract infection in mice and guinea pigs? Infect Immun. 2000;68:5299–305. doi: 10.1128/iai.68.9.5299-5305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams DM, Grubbs BG, Darville T, Kelly K, Rank RG. A role for interleukin-6 in host defense against murine Chlamydia trachomatis infection. Infect Immun. 1998;66:4564–7. doi: 10.1128/iai.66.9.4564-4567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–52. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 44.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–7. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw J, Grund V, Durling L, Crane D, Caldwell HD. Dendritic cells pulsed with a recombinant chlamydial major outer membrane protein antigen elicit a CD4(+) type 2 rather than type 1 immune response that is not protective. Infect Immun. 2002;70:1097–105. doi: 10.1128/IAI.70.3.1097-1105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gervassi A, Alderson MR, Suchland R, Maisonneuve JF, Grabstein KH, Probst P. Differential regulation of inflammatory cytokine secretion by human dendritic cells upon Chlamydia trachomatis infection. Infect Immun. 2004;72:7231–9. doi: 10.1128/IAI.72.12.7231-7239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams NL, Kloeze E, Govan BL, Körner H, Ketheesan N. Burkholderia pseudomallei enhances maturation of bone marrow-derived dendritic cells. Trans R Soc Trop Med Hyg. 2008;102:S71–5. doi: 10.1016/S0035-9203(08)70019-1. [DOI] [PubMed] [Google Scholar]
- 48.Pejawar SS, Parks GD, Alexander-Miller MA. Abortive versus productive viral infection of dendritic cells with a paramyxovirus results in differential upregulation of select costimulatory molecules. J Virol. 2005;79:7544–57. doi: 10.1128/JVI.79.12.7544-7557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harada T, Miura NN, Adachi Y, Nakajima M, Yadomae T, Ohno N. Highly expressed dectin-1 on bone marrow-derived dendritic cells regulates the sensitivity to beta-glucan in DBA/2 mice. J Interferon Cytokine Res. 2008;28:477–86. doi: 10.1089/jir.2007.0101. [DOI] [PubMed] [Google Scholar]
- 50.Koike E, Takano H, Inoue K, Yanagisawa R. Accelerated differentiation of bone marrow-derived dendritic cells in atopic prone mice. Int Immunopharmacol. 2008;8:1737–43. doi: 10.1016/j.intimp.2008.08.006. [DOI] [PubMed] [Google Scholar]