Abstract
To explore whether bacterial secreted 4-hydroxy-2-alkylquinolines (HAQs) can regulate host innate immune responses, we used the extracts of bacterial culture supernatants from a wild-type (PA14) and two mutants of Pseudomonas aeruginosa that have defects in making HAQs. Surprisingly, the extract of supernatants from the P. aeruginosa pqsA mutant that does not make HAQs showed strong stimulating activity for the production of innate cytokines such as tumour necrosis factor-α and interleukin-6 in the J774A.1 mouse monocyte/macrophage cell line, whereas the extract from the wild-type did not. The addition of 4-hydroxy-2-heptylquinoline (HHQ) or 2-heptyl-3,4-dihydroxyquinoline (PQS, Pseudomonas quinolone signal) to mammalian cell culture media abolished this stimulating activity of the extracts of supernatants from the pqsA mutant on the expression of innate cytokines in J774A.1 cells and in the primary bronchoalveolar lavage cells from C57BL/6 mice, suggesting that HHQ and PQS can suppress the host innate immune responses. The pqsA mutant showed reduced dissemination in the lung tissue compared with the wild-type strain in a mouse in vivo intranasal infection model, suggesting that HHQ and PQS may play a role in the pathogenicity of P. aeruginosa. HHQ and PQS reduced the nuclear factor-κB (NF-κB) binding to its binding sites and the expression of NF-κB target genes, and PQS delayed inhibitor of κB degradation, indicating that the effect of HHQ and PQS was mediated through the NF-κB pathway. Our results suggest that HHQ and PQS produced by P. aeruginosa actively suppress host innate immune responses.
Keywords: 4-hydroxy-2-alkylquinolines, immune evasion, innate cytokine, innate immunity, macrophage, nuclear factor-κB, quorum sensing
Introduction
During coevolution with their hosts, pathogens have developed the means to evade the innate or adaptive immune systems to survive.1 The bacterium Pseudomonas aeruginosa has been studied as a model organism for investigating host–pathogen interactions. Pseudomonas aeruginosa is an opportunistic pathogen that causes diseases in patients with impaired host defences and is often responsible for life-threatening nosocomial infections among immunocompromised individuals.2,3 It is also the main morbidity- and mortality-causing agent in people suffering from cystic fibrosis.
Pseudomonas aeruginosa uses cell-to-cell communication, also known as quorum sensing, to modulate gene expression in phenotypes such as biofilm development, swarming motility and the production of an arsenal of extracellular virulence factors that are capable of causing extensive tissue damage, bloodstream invasion, and consequently the promotion of systemic dissemination.4 Quorum sensing provides bacteria with the means to co-ordinately regulate the gene expression of a large array of target genes in a cell population density-dependent manner via the exchange of small signalling molecules.5,6
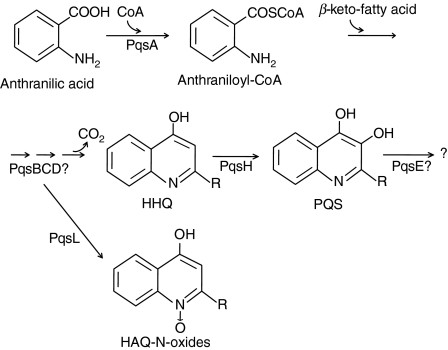
One class of quorum-sensing molecules produced by P. aeruginosa is the N-acylhomoserine lactone family including N-(3-oxo-dodecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) and N-butanoyl-homoserine lactone (C4-HSL). The synthesis of N-acylhomoserine lactone is regulated by las and rhl quorum-sensing systems, consisting of LasRI and RhlRI, respectively.7–9 A second class of molecules with quorum-sensing signals is 4-hydroxy-2-alkylquinolines (HAQs), including derivatives of 4-hydroxy-2-heptylquinoline (HHQ) and the corresponding dihydroxylated derivatives, such as 2-heptyl-3,4-dihydroxyquinoline (PQS, pseudomonas quinolone signal).10–13 The biosynthesis of HAQs in P. aeruginosa (Fig. 1) requires genes encoded by the pqsABCDE and phnAB operons; HHQ is synthesized by PqsABCD from anthranilic acid14,15 and PQS is synthesized by hydroxylation of HHQ by PqsH which is a putative monoxygenase.11,12 Both HHQ and PQS act as co-inducing ligands of MvfR (multiple virulence factor regulator), the transcriptional regulator controlling pqsABCDE and phnAB.15–17
Figure 1.
Biosynthetic pathway of 4-hydroxy-2-alkylquinolines (HAQs) in Pseudomonas aeruginosa. Anthranilate is activated by PqsA to form anthraniloyl-CoA, and this is condensed with β-keto-fatty acid to produce 4-hydroxy-2-heptylquinoline (HHQ). The condensation is postulated to be catalysed by PqsB, PqsC and PqsD. Alternatively, the intermediates in this pathway can be converted to HAQ-N-oxides by PqsL. HHQ is hydroxylated by PqsH to produce 2-heptyl-3,4-dihydroxyquinoline (PQS, Pseudomonas quinolone signal).
The quorum-sensing signals of P. aeruginosa play a role during infection by regulating the expression of many virulence factors and by inducing inflammation.4–6,18 Although the roles of the N-acylhomoserine lactones in the modulation of immune responses have been well studied,4–6,18 the effects of HAQs on the modulation of immune responses are poorly understood. Mutations in a number of genes required for HAQ biosynthesis result in significantly reduced virulence in both plant and animal infection models, suggesting that HAQs are important for the virulence in the host.10,16,19–21 It is known that PQS is produced in the lungs of cystic fibrosis patients infected with P. aeruginosa,22 but it is not clear whether PQS is required for virulence.11,16,23–26 In a previous study using human peripheral blood mononuclear cells (PBMC), Hooi et al.20 have shown that PQS has an immune modulating activity that is different from that of 3-oxo-C12-HSL in cell proliferation and in the production of interleukin-2 (IL-2) and tumour necrosis factor-α (TNF-α). However, the functions of HAQ in the innate immune responses are still poorly understood.
The induction of innate immune responses is mediated by transcription factor nuclear factor-κB (NF-κB), which also plays an essential role in host inflammatory responses.27,28 The NF-κB can be induced in different cell types by certain microbial components and can translocate into the nucleus upon cellular stimulation, where it binds target genes and regulates their transcription.28,29 In an unstimulated state, the nuclear translocation of NF-κB is inhibited by the inhibitor of κB (IκB) which is regulated by the IκB kinase (IKK) complex. Upon stimulation, the IKK complex phosphorylates and marks IκB for degradation and subsequent ubiquitination. This phosphorylation allows NF-κB to be released from IκB and to translocate to the nucleus.
In the present study, we explored the regulatory function of HAQs in the innate immune responses using a wild-type (PA14) and two mutants of P. aeruginosa and synthetic HHQ and PQS. The results showed that HHQ and PQS strongly suppressed innate immune responses in vitro and ex vivo, and that this suppressive activity was mediated by the NF-κB signalling pathway.
Materials and methods
Experimental reagents
Both HHQ and PQS were chemically synthesized as previously described.30,31 They were then dissolved in methanol in a concentration of 1 mg/ml (stock solution).
Generation of the pqsA and pqsH mutants
The P. aeruginosa strain PA1432 and two pqs isogenic mutants (pqsA and pqsH) were used in this study. The pqsA insertion mutant was provided from the non-redundant PA14 transposon library,33 and the pqsH in-frame deletion mutant was created by splicing by overlap extension (i.e. SOEing)34 using an allelic exchange vector, pEX18T,35 and four oligonucleotide primers (pqsH-N1: 5′-TGAATTCGATCTTCTGATAGG-3′, pqsH-UC: 5′-GAACAGGATCAGCGTCATCGACATCAGCATCG-3′, pqsH-DN: 5′-CGATGCTGATGTCGATGACGCTGATCCTGTT C-3′, pqsH-C1: 5′-ATCAAGCGTCATGGATCCATC-3′) to amplify the 5′ and 3′ segments of the pqsH gene. The phenotypes of the pqs mutants were verified by measuring the production of PQS-related compounds by liquid chromatography/mass spectrometry analysis following high performance liquid chromatography of the mutant extracts, as described previously.13 The PQS congeners can be identified by their mass (m/z value), and we could identify two major peaks for PQS (m/z = 260) and HHQ (m/z = 244) in the wild-type PA14 strain, which were absent in the pqsA mutant, whereas only the PQS peak was absent in the pqsH mutant (Fig. S1). Furthermore, episomal copy of each gene complemented the deficiency of pyocyanin production in the mutants (Fig. S2).
Preparation of extracts from P. aeruginosa culture supernatants
The wild-type and two pqs mutants were grown overnight in Luria–Bertani (LB) broth (1% tryptone, 0·5% yeast extract, and 1% NaCl). Each overnight culture was subcultured to an optical density at 600 nm (OD600) of 0·02 in 200 ml of the LB broth. Aeration was performed for 12 hr in 1-l baffled flasks at 37° and 1·4 g. These cultures were then centrifuged at 8000 g for 15 min. Supernatant acidification, extraction and evaporation took place as described previously.13 Finally, the extracts were dissolved in 0·5% dimethylsulphoxide (final concentration).
Cell culture and cytokine measurement
A mouse monocyte/macrophage cell line J774A.1 was purchased from the Korean Cell Line Bank (Seoul, Korea) and maintained in RPMI-1640 supplemented with 5% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and penicillin/streptomycin. For measuring the expression of innate cytokines, 3 × 105 J774A.1 cells were plated onto a 24-well culture plate and treated with ethyl acetate extracts from P. aeruginosa PA14 or two mutants in the presence or absence of purified PQS or HHQ for 48 hr. The amount of TNF-α and IL-6 was measured by an enzyme-linked immunosorbent assay (ELISA).
Cell viability assay
J774A.1 cells (2 × 104) were plated onto 96-well plates. After overnight culture, the cells were treated with various concentrations of extracts from P. aeruginosa culture supernatants or with various concentrations of HHQ or PQS. After 48 hr, cell viability was measured by MTS assay using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Immune responses in the bronchoalveolar lavage cells
The cells in the bronchoalveolar lavage (BAL) fluid from nine C57BL/6 mice (age 6–8 weeks) at each experiment were harvested in phosphate-buffered saline (PBS), and pelleted by centrifugation. A total of 4·2 × 104 BAL cells were cultured in vitro in 150 μl media in the 96 wells with extracts of culture supernatants from PA14 or the pqsA mutant in the presence or absence of 0·28 μg/ml of HHQ or PQS for 48 hr. The expression of TNF-α and IL-6 in the culture supernatants was measured by ELISA.
In vivo lung infection model
The PA14 and pqsA mutant strains were inoculated in 4 ml liquid LB media and cultured in the shaking incubator at 37° for 8 hr. The bacteria were then harvested, washed with PBS, and resuspended in PBS. The bacteria [1 × 105 colony-forming units (CFU)] in 40 μl PBS were injected into the lungs of anaesthetized C57BL/6 mice intranasally. After 12 hr the mice were killed, and lung lavage was performed three times with 1 ml PBS. The lungs were recovered, resuspended in 2 ml PBS, and homogenized using a PT-1200E homogenizer (Kinematica AG, Littau, Switzerland). Lung homogenates (100 μl) were plated onto Difco Cetrimide agar plate (Difco, Livonia, MI, USA) and incubated at 37° for 16 hr, and the CFUs were counted. The experiments with live mice were approved by the Sogang University Institutional Animal Care and Use Committee.
NF-κB activity assay
J774A.1 cells were pre-treated with HHQ or PQS for 2 hr, and treated additionally with the extracts from pqsA (or PA14) culture supernatants for 30 min. The cells were harvested, washed with PBS, and resuspended in Buffer A [10 mm HEPES, pH 7·9, 10 mm KCl, 0·1 mm dithiothreitol (DTT), 0·5 mm phenylmethylsulphonyl fluoride (PMSF)]. For preparation of nuclear extracts, cells in Buffer A were gently mixed by vortex and kept on ice for 15 min. Then 10% nonidet P-40 (NP-40) was added to the mixture and strongly mixed by vortex for 10 seconds. Pellets were obtained by centrifugation. The pellets were extracted by vortex in Buffer B [20 mm HEPES, pH 7·9, 0·4 m NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 1 mm DTT, 1 mm PMSF, 10% NP-40) for 10 min. The mixture was then centrifuged and the supernatants were used for nuclear extracts. The binding of NF-κB to its responsive elements was measured by the electrophoretic mobility shift assay (EMSA). Nuclear extracts (10 μg) were incubated with 1 μg poly dI : dC (Sigma Aldrich, St Louis, MO, USA) for 30 min at room temperature in the binding buffer (10 mm Tris–HCl, pH 7·5, 50 mm NaCl, 1 mm DTT, 1 mm EDTA, 5% glycerol) and then the 32P-labelled oligomers were added to the mixture and incubated for 15 min on ice. The sequence of the oligomer for NF-κB consensus binding sites is 5′-AGTTGAGGGGACTTTC CCAGGC-3′ and that of the p65 mutant oligomer is 5′-AGTTGAGGCGACTTTCCCAGGC-3′. The mixtures were resolved by polyacrylamide gel electrophoresis and subjected to autoradiography. In the competition assay, 50–100 times more non-labelled oligomers were added. In the supershift assay, 2 μg anti-NF-κB p65 (sc-8008X; Santa Cruz, CA, USA), anti-NF-κB p50 (sc-8414; Santa Cruz), or anti-NF-κB c-Rel (sc-71X; Santa Cruz) antibody was added to the binding mixture and incubated for a further 15 min on ice.
Western blotting
J774A.1 cells were treated with extracts of pqsA supernatant in the presence or absence of HHQ or PQS for 5–120 min. The whole cell lysates from 2 × 106 cells were electrophoresed on a sodium dodecyl sulphate–polyacrylamide gel and transferred to a polyvinylidene difluoride membrane for 1 hr. The membranes were blocked for 1 hr in Tris-buffered saline with 0·05% Tween-20 (TBST) and 5% skim milk. Anti-IκB-α (#9242, Cell Signaling, Danvers, MA, USA) or anti-β-actin (sc-47778; Santa Cruz) antibodies were diluted 1 : 1000 in TBST, and incubated with the membrane overnight at 4°. Blots were washed with TBST three times for 10 min and incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (A90-116P; Bethyl, Montgomery, TX, USA) or goat anti-goat immunoglobulin G (A50-110P; Bethyl) antibodies for 1 hr at room temperature. Proteins were detected by incubation with enhanced chemiluminescence reagent and exposure to X-ray film.
RNA isolation and reverse transcription-polymerase chain reaction
Total RNA was isolated from J774A.1 cells treated with extracts from P. aeruginosa or with various concentrations of HHQ or PQS for 90 min using TRIzol reagent (Invitrogen). Complementary DNA was synthesized using SuperScript II reverse transcriptase and oligo-dT (Invitrogen) according to the manufacturer’s protocol. Polymerase chain reactions were carried out with Taq polymerase (Takara, Shiga, Japan) using the primers listed in Table 1.
Table 1.
Primers used for reverse transcription–polymerase chain reaction
| Genes | Sequences (5′→3′) | |
|---|---|---|
| IL12β | F: TGGTTTGCCATCGTTTTGCTG | R: ACAGGTGAGGTTCACTGTTTCT |
| MCP3 | F: GCTGCTTTCAGCATCCAAGTG | R: CCAGGGACACCGACTACTG |
| MIP2 | F: GCGCCCAGACAGAAGTCATAG | R: AGCCTTGCCTTTGTTCAGTATC |
| B7h | F: TAAAGTGTCCCTGTTTTGTGTCC | R: ATTGCACCGACTTCAGTCTCT |
| IP10 | F: CCAAGTGCTGCCGTCATTTTC | R: GGCTCGCAGGGATGATTTCAA |
| IRF1 | F: ATGCCAATCACTCGAATGCG | R: TTGTATCGGCCTGTGTGAATG |
| Teck | F: TTACCAGCACAGGATCAAATGG | R: CGGAAGTAGAATCTCACAGCAC |
| IL1β | F: GCAACTGTTCCTGAACTCAACT | R: ATCTTTTGGGGTCCGTCAACT |
| HPRT | F: GTTGGATACAGGCCAGACTTTGTTG | R: GAGGGTAGGCTGGCCTATAGGCT |
F, forward; R, reverse.
Statistical analysis
Results were expressed as mean ± standard deviation (SD). Differences between groups were determined by a Student’s t-test or analysis of variance (anova) when the data passed four sets of normality tests (Shapiro–Wilk, Kolmogorov–Smirnov, Cramer–von Mises, and Anderson–Darling), or Kruskal–Wallis analysis when the data did not pass the normality tests using sas9.1 software (SAS Institute Inc., Cary, NC, USA).
Results
Secreted molecules from P. aeruginosa can induce innate cytokines
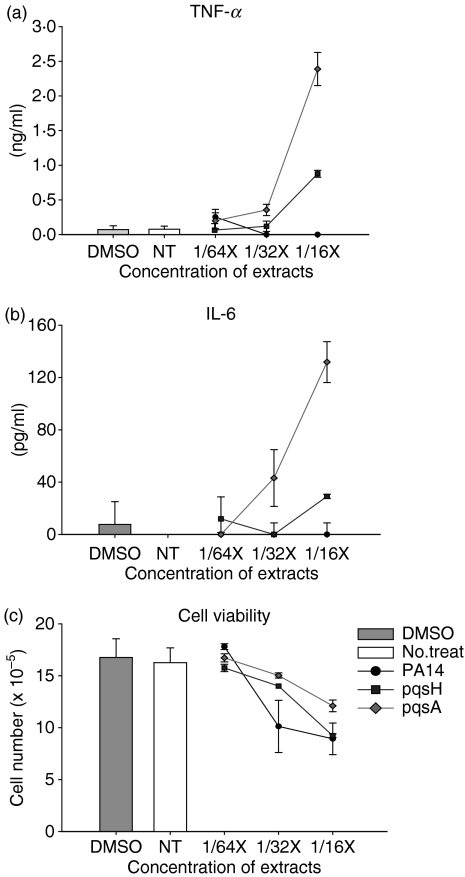
To initially investigate whether released HAQs can influence the host immune system, we extracted with ethyl acetate a series of culture supernatants from wild-type P. aeruginosa, PA14, and two mutants, pqsA and pqsH, defective in the biosynthesis of HAQs10 (Fig. 1). We used a mouse monocyte/macrophage cell line J774A.1 as a model system to examine the effect of the secreted molecules on the expression of innate cytokines. We treated J774A.1 cells with these extracts for 2 days and measured the expression of innate cytokines in the culture supernatants from J774A.1 cells using an ELISA. Surprisingly, the extracts from the pqsA mutant, at the dilution of 1/16×, strongly induced TNF-α and IL-6 expression in J774A.1 cells, and extracts from the pqsH mutant induced an intermediate level of cytokines, whereas the wild-type did not (Fig. 2a,b). This result suggests that the induction of host immune responses may be suppressed by molecules produced by wild-type P. aeruginosa, and partly suppressed by a pqsH mutant, but are not suppressed by a pqsA mutant. The pqsA mutant is defective in the production of HAQs such as HHQ and PQS, and the pqsH mutant is defective in the production of PQS, it therefore follows that HHQ and PQS are good candidates to be responsible for the suppression activity.
Figure 2.
Stimulation of innate immune responses by the secreted molecules from pqsA mutant strain. (a,b) J774A.1 cells were treated with a vehicle control (0·5% dimethylsulphoxide) or extracts of culture supernatants from wild-type or two mutants (pqsA and pqsH) of Pseudomonas aeruginosa for 2 days. The concentrations of the extracts (1/16×, 1/64×, 1/128×) are presented as fold dilution compared with the original volume of the bacterial culture supernatants. The amount of tumour necrosis factor-α (TNF-α) and interleukin-6 (IL-6) from the supernatants from the J774A.1 culture was measured by enzyme-linked immunosorbent assay. Data represent mean ± SD (n = 3). (c) Cell viability after treatment with various concentrations of the extracts for 2 days was measure by MTS assay. Data represent mean ± SD (n = 4). All data (a–c) are representative of three independent experiments with similar results.
To check whether this effect is the result of the difference in the cytotoxicity of the extracts of bacterial culture supernatants from the wild-type and the pqsA and pqsH mutants, we performed a cytotoxicity assay. The extracts were slightly cytotoxic to J774A.1 cells at the concentration used (Fig. 2c). The cytotoxicity was not much different between cells treated with PA14 extracts and those treated with pqsA or pqsH mutant extracts at the concentrations used. However, the difference in cytokine production was much larger than the difference in cytotoxicity at higher concentrations of supernatant extracts. At the concentration of 1/16× extracts, the difference in TNF-α and IL-6 expression was more than 1000-fold between cells treated with the wild-type versus pqsA mutant culture extracts (Fig. 2). This result indicates that the cytotoxicity of the non-polar extract is not the major cause of the difference in the expression of innate cytokines.
HHQ and PQS suppress the expression of innate cytokines
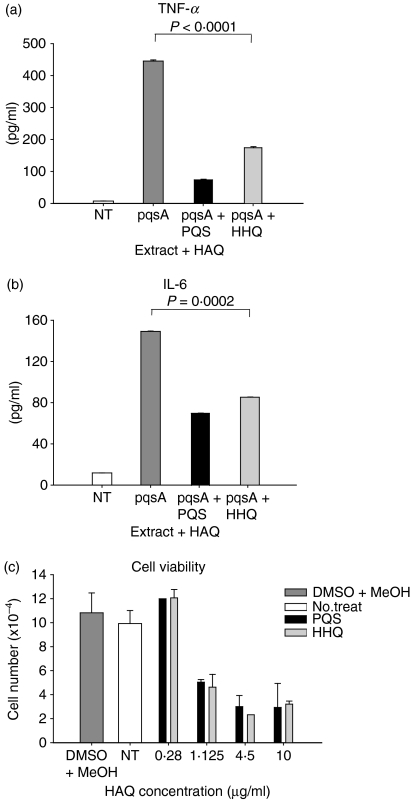
The pqsA mutant is defective in the conversion of anthranilic acid into HAQs so the result in Fig. 2 could be explained either by the fact that anthranilic acid or its derivatives induces the expression of innate cytokines because previous studies have shown that anthranilic acid accumulates in the culture supernatants of the pqsA mutant,12,21 or that HAQs suppress the expression of innate cytokines. We examined these possibilities through combined treatment of J774A.1 cells with extracts from the pqsH mutant + anthranilic acid or with extracts from the pqsA mutant + HHQ (or PQS). Expression of TNF-α and IL-6 in J774A.1 cells treated with pqsA extracts + HHQ or PQS for 2 days was significantly reduced (Fig. 3a,b), whereas in cells treated with pqsH extracts + anthranilic acid (in the range of 0·1–30 μm) there was no change (data not shown). This result suggests that HHQ and PQS suppress the expression of innate cytokines in J774A.1 cells, when induced by non-polar molecules released by the pqsA mutant strain.
Figure 3.
Suppression of innate immune responses by 4-hydroxy-2-heptylquinoline (HHQ) and 2-heptyl-3,4-dihydroxyquinoline (PQS, Pseudomonas quinolone signal). (a,b) J774A.1 cells were treated with extracts of culture supernatant from the pqsA mutant at 1/16× concentration for 24 hr in the presence or absence of 0·28 μg/ml of HHQ or PQS. The amount of tumour necrosis factor-α (TNF-α) and interleukin-6 (IL-6) from the culture supernatants was measured by enzyme-linked immunosorbent assay. Data represent mean ± SD (n = 8). Statistical differences among groups were analysed by analysis of variance. The dimethyl sulphoxide vehicle control showed no difference compared with no-treatment control in Fig. 2 so it was not included in the experiment shown here. (c) Cell viability after treatment with various concentrations of HHQ or PQS for 24 hr was measured by MTS assay. Data represent mean ± SD (n = 4). All data (a–c) are representative of three independent experiments with similar results.
We then performed cytotoxicity assays with HHQ and PQS. The J774A.1 cells were treated with various concentrations of HHQ and PQS and the number of live cells was counted (Fig. 3c). Both HHQ and PQS at the concentration of 0·28 μg/ml showed no cytotoxicity compared with the vehicle control (Fig. 3c). At this concentration, production of TNF-α and IL-6 was inhibited more than 50% (Fig. 3a,b). This result indicates that the reduction of TNF-α and IL-6 shown in Fig. 3(a,b) was not the result of the cytotoxicity of HHQ and PQS. At concentrations higher than 1·12 μg/ml, the cytotoxicity of HHQ and PQS increased (Fig. 3c).
HAQs affect the pathogenicity of P. aeruginosa
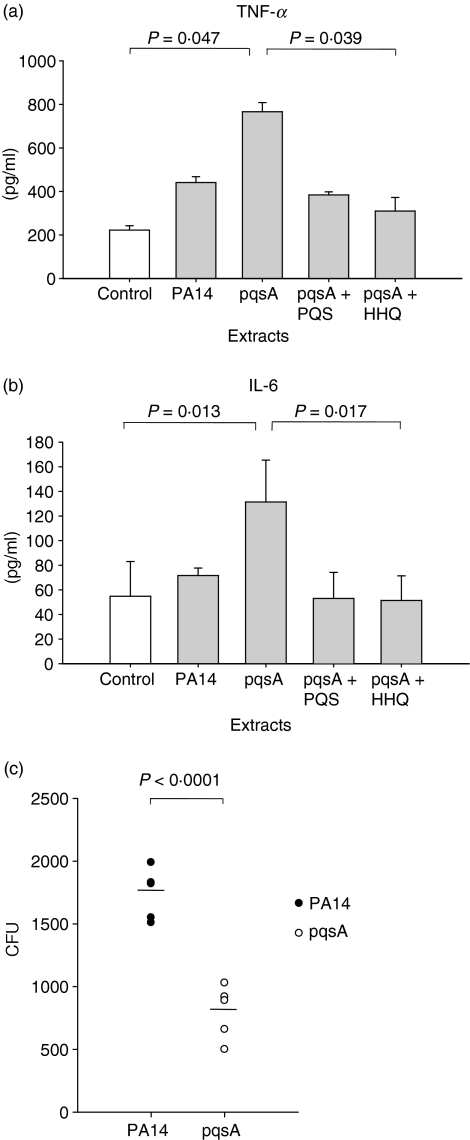
Next, we examined whether HHQ and PQS inhibit the innate immune responses in the primary cells from BAL fluid, which are more physiologically relevant. Harvested BAL cells from C57BL/6 mice were cultured in vitro for 24 hr in the presence of extracts of P. aeruginosa culture supernatants that were supplemented, or not, with HHQ or PQS, and the production of innate cytokines was measured in the culture supernatants (Fig. 4a,b). Just as with the J774A.1 cells, the extracts of pqsA mutant culture supernatants induced more TNF-α and IL-6 production by the BAL cells compared with the wild-type culture supernatants (Fig. 4a,b), suggesting that this induction also works on more physiologically relevant cells. When HHQ or PQS was added to the culture media, it effectively inhibited the expression of TNF-α and IL-6 induced by the extracts of the pqsA mutant (Fig. 4a,b), confirming that the inhibitory effect of HHQ and PQS on the expression of innate cytokines is valid for primary BAL cells as well.
Figure 4.
Immune responses in the bronchoalveolar lavage (BAL) cells (a,b) and pathogenicity in the lung (c). (a,b) The cells in the BAL fluid from C57BL/6 mice were harvested and cultured in vitro with extracts of culture supernatants from PA14 (wild-type) or a pqsA mutant in the presence or absence of 4-hydroxy-2-heptylquinoline (HHQ) or 2-heptyl-3,4-dihydroxyquinoline (PQS, Pseudomonas quinolone signal) for 48 hr. The expression of tumour necrosis factor-α (TNF-α) and interleukin-6 (IL-6) in the culture supernatants was measured by enzyme-linked immunosorbent assay. Data represent mean ± SD (n = 3). Statistical differences among groups were analysed by a Kruskal–Walis test. (c) 1 × 105 colony-forming units (CFUs) of Pseudomonas aeruginosa were injected into the lung of C57BL/6 mice intranasally (n = 5, each group). After 12 hr the mice were killed and lung lavage was performed. The lung was homogenized in 2 ml phosphate-buffered saline, and 100 μl of the lung homogenates were plated onto a Cetrimide agar (CA) plate to measure CFU. Statistical differences between groups were analysed by a Student’s t-test. All data (a–c) are representative of three independent experiments with similar results.
We further examined whether the defect in the production of HAQs influences the pathogenicity of P. aeruginosa during lung infection in an in vivo mouse model. We introduced live PA14 or the pqsA mutant strain into C57BL/6 mice intranasally. After 1 day of infection, the CFUs of tissue infiltrating P. aeruginosa were counted. The number of CFUs in the lungs infected with the pqsA mutant was reduced by about 50% compared with the wild-type-infected lungs (Fig. 4c). This result suggests that HAQs may play a role in the pathogenicity of P. aeruginosa, although it does not exclude another possibility that the pqsA strain may be generally lacking virulence.
HHQ and PQS suppress the NF-κB pathway
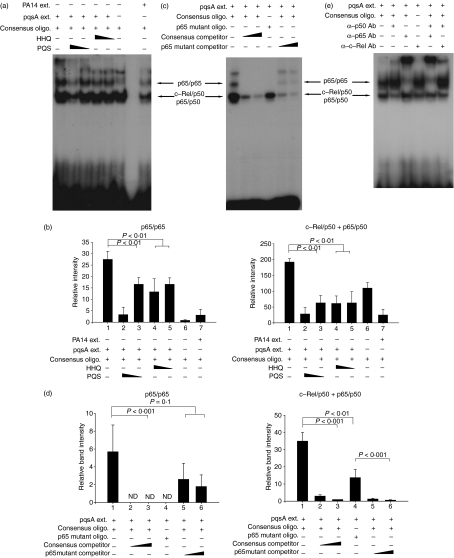
The expression of innate cytokines TNF-α and IL-6 is regulated by NF-κB27,28,36,37 so we postulated that HHQ and PQS might regulate the expression of innate cytokines by suppressing NF-κB activation. We examined the activation of NF-κB by checking how it binds to its responsive elements using the EMSA. Interestingly, the extract of the pqsA mutant induced NF-κB activation, whereas the negative control or the extract of PA14 did not (Fig. 5a). Pre-treatment with HHQ or PQS significantly inhibited the binding of NF-κB to its cognate DNA induced by extracts from the pqsA mutant (Fig. 5a). Pre-treatment of HHQ and a low dose (0·28 μg/ml) of PQS mainly reduced the binding of p65/p65 homodimer to DNA, and a high dose (1·12 μg/ml) of PQS reduced the binding of c-Rel/p50 and p65/p50 heterodimer as well as p65/p65 homodimer (Fig. 5a). Densitometric analysis with the films using ImageJ (NCBI freeware) and statistical analyses confirmed this result (Fig. 5b). Specific binding of NF-κB to its cognate DNA was confirmed by an oligonucleotide competition assay (Fig. 5c). Densitometric and statistical analyses confirmed the specific competition of NF-κB bands (Fig. 5d). Specific binding of NF-κB to its cognate DNA was also confirmed by an antibody supershift assay (Fig. 5e). Taken together, these results indicate that HHQ and PQS control the expression of innate cytokines through the NF-κB pathway.
Figure 5.
4-Hydroxy-2-heptylquinoline (HHQ) and 2-heptyl-3,4-dihydroxyquinoline (PQS, Pseudomonas quinolone signal) suppress the binding of nuclear factor-κB (NF-κB) to its recognition site. J774A.1 cells were pre-treated with 1·12 or 0·28 μg/ml of HHQ or PQS for 2 hr, and treated with the extracts from pqsA (or PA14) culture supernatants for 30 min. After the cells were harvested, nuclear extracts were prepared and mixed with oligomers containing the NF-κB binding sites or its mutated sequences. The binding of NF-κB to the oligomers was measured by the electrophoretic mobility shift assay. All data (a,c,e) are representative of three or four independent experiments with similar results. (b) Densitometric analysis of Fig. 5(a). Band intensity of each band in scanned films was calculated by ImageJ program and combined before statistical analysis by analysis of variance (anova). Data are presented as mean ± SD (n = 3). (d) Densitometric analysis of Fig. 5(c). Band intensity was calculated as for Fig. 5(b). Statistical analyses were performed by anova or Student’s t-test. Data are presented as mean ± SD (n = 4).
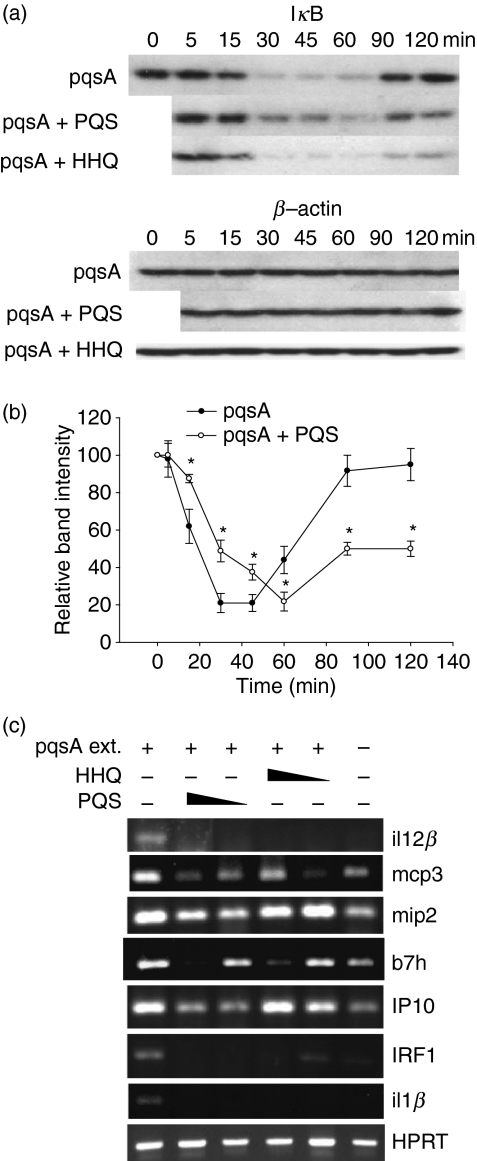
To further investigate the HHQ/PQS-mediated NF-κB modulation, we measured the amount of IκB degradation as a result of treatment with extracts of the pqsA culture supernatants. Treatment of the extract of pqsA supernatants caused rapid degradation of IκB, showing activation of NF-κB. The addition of PQS, but not HHQ, to this extract delayed the kinetics of the degradation of IκB (Fig. 6a). Densitometric and statistical analysis confirmed that PQS delayed the IκB degradation (Fig. 6b). We also examined whether the expression of NF-κB target genes is affected by the treatment of HHQ or PQS. Treatment of the extracts of pqsA induced the expression of NF-κB target genes including il12b, mcp3, mip2, b7h, IP10, IRF1 and il1b (Fig. 6c). Addition of HHQ or PQS to the extract of pqsA caused a reduction of the expression of these target genes, further confirming the inhibitory role of HHQ/PQS on the NF-κB pathway.
Figure 6.
2-Heptyl-3,4-dihydroxyquinoline (PQS, Pseudomonas quinolone signal) suppresses inhibitor of κB (IκB) degradation (a,b), and 4-hydroxy-2-heptylquinoline (HHQ) and PQS suppress the expression of its target genes (c). (a) J774A.1 cells were treated with the extract from pqsA culture supernatants in the presence or absence of 1·12 μg/ml of HHQ or PQS for various times. The cells were harvested and the IκB protein was detected by Western blotting with an anti-IκB antibody. Data are representative of five independent experiments with similar results. (b) Densitometric analysis of Fig. 6(a). Band intensity was calculated as Fig. 5(b). Statistical analyses were performed using Student’s t-test. Asterisks indicate the statistical difference (P < 0·05) between groups at the same time-point. Data are presented as mean ± SE (n = 5). (c) J774A.1 cells were pre-treated with 1·12 μg/ml or 0·28 μg/ml of HHQ or PQS for 2 hr, and treated with the extracts from pqsA (or PA14) culture supernatants for 90 min. After the cells were harvested, total RNA was isolated. Target gene expression was measure by reverse transcription–polymerase chain reaction. Data are representative of three independent experiments with similar results.
Discussion
In this study we demonstrated for the first time that the quorum-sensing molecules HHQ and PQS from P. aeruginosa can actively suppress the mammalian innate immune responses, and that this suppression is mediated by the NF-κB signalling pathway of the host immune cells, revealing a new aspect of host–pathogen interactions.
Suppression of the immune system with molecules secreted by micro-organisms is a common way of evading the destructive host immune responses and of surviving in the hostile host environment.1Pseudomonas aeruginosa produces many virulence factors such as hydrolytic enzymes, toxins and exopolysaccharides.38 Therefore, these molecules will probably be detected by the host immune system and induce immune responses. To adapt successfully in the host environment, P. aeruginosa needs to evade or suppress this induction of the immune system. The fact that an extract of a pqsA mutant culture induces the host immune responses more strongly than that of a wild-type culture suggests that this induction is suppressed by molecules produced only by the wild-type P. aeruginosa (Fig. 2). Using chemically synthesized HHQ and PQS, we showed that these molecules, mostly known for being involved in quorum-sensing regulation, also suppress the innate immune responses in the mouse monocyte/macrophage cell line and BAL cells (Figs 3 and 4). This suppression of the innate immune responses will benefit the survival and virulence of P. aeruginosa during infection of the host. This was also shown in an in vivo infection model, in which the pqsA mutant, which does not produce HHQ and PQS, showed reduced pathogenicity compared with the wild-type PA14 (Fig. 4c). In support of this result, Xiao et al.16 have shown that the pqsA mutant has a defect in pathogenicity in a thermally injured mouse model, whereas the wild-type and pqsH, which can produce HHQ, do not. It would therefore seem likely that HHQ and PQS may play a role in the pathogenicity of P. aeruginosa. It has been reported that PQS production was greater in strains isolated from the lungs of asymptomatic children with cystic fibrosis compared with environmental strains,39 suggesting that PQS may be important for the initial adaptation of P. aeruginosa in the lung environment. Our result that HHQ and PQS inhibit host innate immune responses provides a possible explanation for the observation made in human patients. However, there is also a possibility that molecules other than HHQ and PQS may play a role or play an additive role in the pathogenicity because the pqsA mutant also lacks other secreted compounds.
Like P. aeruginosa, several Burkholderia species also cause severe respiratory infections in human patients suffering from cystic fibrosis.40 Recently, it has been shown that HAQ derivatives are produced by some species of the Burkholderia genus,31,41 although they mostly produce HHQ and PQS analogues, which contain an unsaturated aliphatic side chain and are methylated at position 3.41 Consequently, it seems likely that the acquisition of HAQs during evolution may play beneficial roles for the survival and adaptation of these bacteria in the same ecological niches by regulating innate immune responses in the lungs.
Treatment with HHQ and PQS inhibited the production of TNF-α by the monocyte/macrophage cell line J774A.1 and by BAL cells. In our study, we used PQS at concentrations of 1·07 μm (0·28 μg/ml) and 4·3 μm (1·12 μg/ml). At this range of PQS concentrations, our result is similar to the previous study using human PBMC by Hooi et al.,20 in which they showed that PQS inhibited the production of TNF-α at low concentrations (in the range of 1–5 μm), whereas it increased the production of TNF-α at high concentrations (above 10 μm). Therefore, it seems likely that PQS has a dual effect on the production of TNF-α in the immune cells, although the reason for this dual activity is not clear. However, the concentrations of PQS above 10 μm are seemingly not adequate to reveal the effect of PQS on the immune cells because at these concentrations cell proliferation is completely blocked20 and most cells die (Fig. 3c). The experimental conditions used in our study and those of Hooi et al.20 are also different. In the present study, we used the J774A.1 mouse monocyte/macrophage cell line and mouse BAL cells rather than human PBMC, examined the effect of HHQ as well as PQS, and measured IL-6 as well as TNF-α. Hence, by using a more defined and comprehensive method, we demonstrated that HHQ and PQS can inhibit innate immune responses.
Our results showed that HHQ and PQS inhibited NF-κB activity and the expression of its target genes by preventing the binding of NF-κB to its target DNA (Figs 5 and 6). It remains to be determined how HHQ and PQS inhibit the activity of NF-κB, and whether NF-κB is a direct target of HHQ and PQS. Both HHQ and PQS contain long hydrocarbon chains so they have low water solubility. PQS has been shown to be solubilized by rhamnolipid biosurfactants, which increase the apparent solubility of hydrophobic molecules in water.42 It is speculated that rhamnolipid-solubilized HHQ and PQS can easily mix with or pass through the plasma membrane, interact with target molecules, and distort their function. It has also been shown that HHQ and PQS stimulate MvfR by directly binding to the ligand-binding domain of this LysR-type transcriptional regulator in P. aeruginosa.16 It will be interesting to examine whether this type of interaction occurs in the regulation of immune cells as well. However, our database search for an LysR-type transcriptional regulator ligand-binding domain in mammalian genomes showed no homologous domains. Therefore, HHQ and PQS probably interact with another target in mammalian cells. Recently, Kravchenko et al. reported that 3-oxo-C12-HSL, another quorum-sensing signal, selectively impairs the regulation of NF-κB signalling in activated mammalian cells,43 suggesting that modulating the NF-κB signalling pathway may be a common mechanism of quorum-sensing signals in the virulence and adaptation in the host.
Because P. aeruginosa has developed resistance to conventional antibiotics, new drugs will be needed to treat P. aeruginosa-related diseases such as cystic fibrosis and pneumonia. Quorum-sensing inhibitors have therefore been studied as possible alternative antimicrobial drugs.44 Although these studies are mostly aimed at inhibiting the biological activity of N-acylhomoserine lactones,44 recent studies have successfully targeted the biosynthesis of HAQs as a means of decreasing the virulence of the bacteria.21,45 Our results showed that HHQ and PQS suppressed the innate cytokine expression so they could shed new light on the mode of action of drugs aimed at preventing the biosynthesis of HAQs, although it is too early to speculate on clinical implications. The HAQs also have the potential to be used as an anti-inflammatory agent, being that HHQ and PQS suppress innate cytokines which are strong inducers of inflammation.
In summary, this study demonstrates that HHQ and PQS, which are produced by P. aeruginosa, can regulate the expression of innate cytokines by the mouse monocyte/macrophage cell line and BAL cells, possibly facilitating bacterial adaptation and pathogenicity in the host environment. Although a detailed mechanism for this regulation awaits further studies, our study shed light on new roles of these quorum-sensing molecules in the regulation of innate cytokines and pathogenicity, which has potential therapeutic implications for human diseases such as cystic fibrosis.
Acknowledgments
We thank Eric Déziel for his critical comments on this manuscript. This work was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2006-311-C00145).
Disclosures
The authors have no potential conflicts of interest.
Supporting Information
Additional Supporting information may be found in the online version of this article:
Figure S1. LC/MS data of P. aeruginosa extracts. The production of PQS-related compounds was analyzed by LC/MS analysis following HPLC of the mutant extracts, as described in Methods. The compounds were identified by their mass (m/z value): PQS (m/z = 260) and HHQ (m/z = 244).
Figure S2. Complementation analysis of pyocyanin production. The production of pyocyanin (PYO) (blue-green color) in wild type and two pqs mutants was assessed by visual inspection. Wild type strain produced pyocyanin (PYO), whereas the pqsH mutant produced a little and the pqsA mutant no PYO. Complementation was performed using episomal copy of each gene (pUCP18 based constructs).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–34. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 2.Rello J, Diaz E. Pneumonia in the intensive care unit. Crit Care Med. 2003;31:2544–51. doi: 10.1097/01.CCM.0000089928.84326.D2. [DOI] [PubMed] [Google Scholar]
- 3.Chastre J, Fagon JY. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867–903. doi: 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt T, Givskov M. The role of quorum sensing in the pathogenicity of the cunning aggressor Pseudomonas aeruginosa. Anal Bioanal Chem. 2007;387:409–14. doi: 10.1007/s00216-006-0774-x. [DOI] [PubMed] [Google Scholar]
- 5.Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000;2:1721–31. doi: 10.1016/s1286-4579(00)01327-7. [DOI] [PubMed] [Google Scholar]
- 6.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 7.Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–9. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passador L, Cook JM, Gambello MJ, et al. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–30. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 9.Latifi A, Winson MK, Foglino M, et al. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–43. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 10.Diggle SP, Cornelis P, Williams P, et al. 4-quinolone signalling in Pseudomonas aeruginosa: old molecules, new perspectives. Int J Med Microbiol. 2006;296:83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 11.Gallagher LA, McKnight SL, Kuznetsova MS, et al. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–80. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deziel E, Lepine F, Milot S, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A. 2004;101:1339–44. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepine F, Milot S, Deziel E, et al. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom. 2004;15:862–9. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Bredenbruch F, Nimtz M, Wray V, et al. Biosynthetic pathway of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines. J Bacteriol. 2005;187:3630–5. doi: 10.1128/JB.187.11.3630-3635.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wade DS, Calfee MW, Rocha ER, et al. Regulation of pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–80. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao G, Deziel E, He J, et al. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006;62:1689–99. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- 17.Diggle SP, Matthijs S, Wright VJ, et al. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Estrada O, Zaborina O, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–7. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 19.Deziel E, Gopalan S, Tampakaki AP, et al. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- 20.Hooi DS, Bycroft BW, Chhabra SR, et al. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect Immun. 2004;72:6463–70. doi: 10.1128/IAI.72.11.6463-6470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesic B, Lepine F, Deziel E, et al. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 2007;3:1229–39. doi: 10.1371/journal.ppat.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collier DN, Anderson L, McKnight SL, et al. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett. 2002;215:41–6. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- 23.Cao H, Krishnan G, Goumnerov B, et al. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A. 2001;98:14613–8. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau GW, Ran H, Kong F, et al. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72:4275–8. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahajan-Miklos S, Tan MW, Rahme LG, et al. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 26.Rahme LG, Tan MW, Le L, et al. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1997;94:13245–50. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naumann M. Nuclear factor-κB activation and innate immune response in microbial pathogen infection. Biochem Pharmacol. 2000;60:1109–14. doi: 10.1016/s0006-2952(00)00390-7. [DOI] [PubMed] [Google Scholar]
- 28.Kopp EB, Ghosh S. NF-κB and rel proteins in innate immunity. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 29.Karin M. How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene. 1999;18:6867–74. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 30.Diggle SP, Winzer K, Chhabra SR, et al. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 31.Diggle SP, Lumjiaktase P, Dipilato F, et al. Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol. 2006;13:701–10. doi: 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Rahme LG, Stevens EJ, Wolfort SF, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 33.Liberati NT, Urbach JM, Miyata S, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–8. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horton RM, Cai ZL, Ho SN, et al. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–35. [PubMed] [Google Scholar]
- 35.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, et al. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 36.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 37.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Pollack M. The virulence of Pseudomonas aeruginosa. Rev Infect Dis. 1984;6(Suppl. 3):S617–26. doi: 10.1093/clinids/6.supplement_3.s617. [DOI] [PubMed] [Google Scholar]
- 39.Guina T, Purvine SO, Yi EC, et al. Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc Natl Acad Sci U S A. 2003;100:2771–6. doi: 10.1073/pnas.0435846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coenye T, Mahenthiralingam E, Henry D, et al. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int J Syst Evol Microbiol. 2001;51:1481–90. doi: 10.1099/00207713-51-4-1481. [DOI] [PubMed] [Google Scholar]
- 41.Vial L, Lepine F, Milot S, et al. Burkholderia pseudomallei, B. thailandensis and B. ambifaria produce 4-hydroxy-2-alkylquinoline (HAQ) analogues with a methyl group at the 3 position that is required for quorum sensing regulation. J Bacteriol. 2008;190:5339–52. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calfee MW, Shelton JG, McCubrey JA, et al. Solubility and bioactivity of the pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect Immun. 2005;73:878–82. doi: 10.1128/IAI.73.2.878-882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kravchenko VV, Kaufmann GF, Mathison JC, et al. Modulation of gene expression via disruption of NF-κB signaling by a bacterial small molecule. Science. 2008;321:259–63. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol. 2006;296:149–61. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Zaborina O, Lepine F, Xiao G, et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007;3:e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.