Abstract
Mutations of mitochondrial DNA are associated with a wide spectrum of disorders, primarily affecting the central nervous system and muscle function. The specific consequences of mitochondrial DNA mutations for neuronal pathophysiology are not understood. In order to explore the impact of mitochondrial mutations on neuronal biochemistry and physiology, we have used fluorescence imaging techniques to examine changes in mitochondrial function in neurons differentiated from mouse embryonic stem-cell cybrids containing mitochondrial DNA polymorphic variants or mutations. Surprisingly, in neurons carrying a severe mutation in respiratory complex I (<10% residual complex I activity) the mitochondrial membrane potential was significantly increased, but collapsed in response to oligomycin, suggesting that the mitochondrial membrane potential was maintained by the F1Fo ATPase operating in ‘reverse’ mode. In cells with a mutation in complex IV causing ∼40% residual complex IV activity, the mitochondrial membrane potential was not significantly different from controls. The rate of generation of mitochondrial reactive oxygen species, measured using hydroethidium and signals from the mitochondrially targeted hydroethidine, was increased in neurons with both the complex I and complex IV mutations. Glutathione was depleted, suggesting significant oxidative stress in neurons with a complex I deficiency, but not in those with a complex IV defect. In the neurons with complex I deficiency but not the complex IV defect, neuronal death was increased and was attenuated by reactive oxygen species scavengers. Thus, in neurons with a severe mutation of complex I, the maintenance of a high potential by F1Fo ATPase activity combined with an impaired respiratory chain causes oxidative stress which promotes cell death.
Keywords: mitochondrial diseases, oxidative stress, calcium signalling, neuronal degeneration, neuronal loss
Introduction
The most prominent and disabling features in patients with mitochondrial disease are often due to neuronal dysfunction and neurodegeneration (Taylor and Turnbull, 2005). The clinical presentation is extremely variable, ranging from focal deficits such as optic neuropathy to more global disease such as dementia and ataxia (McFarland et al., 2007). Only a limited number of neuropathological studies are available and these show widespread evidence of neuronal loss and dysfunction (Oldfors et al., 1990; Sparaco et al., 2003; Betts et al., 2004). Currently, however, we have limited information on the mechanisms involved in these patterns of neuronal dysfunction and neurodegeneration. Understanding these pathological mechanisms is crucial if we are to develop strategies to treat or prevent the development of symptoms in patients with mitochondrial disease.
Mitochondrial disease may result from defects of either the nuclear or mitochondrial genome and there is an increasing recognition of the importance of these disorders. For example, mitochondrial DNA (mtDNA) disorders affect a minimum of 1 in 10 000 adults in the UK (Schaefer et al., 2008). Other reports suggest that the burden of disease may be much more common. A study of more than 3000 sequential umbilical-cord blood samples, taken to investigate the birth prevalence of pathogenic mtDNA mutations, showed that one mitochondrial tRNA mutation, the m.3243A>G MTTL1 gene mutation, was present in the general population at a frequency of 0.14% (Elliott et al., 2008). The m.1555A>G mitochondrial RNA mutation associated with aminoglycoside deafness is present in 1 in 200 individuals from two separate populations (Bitner-Glindzicz et al., 2009; Vandebona et al., 2009). Furthermore, strikingly similar mitochondrial abnormalities and mitochondrial respiratory chain deficient cells are also present in patients with Alzheimer’s disease (Bender et al., 2008), Parkinson’s disease and in normal brain ageing (Bender et al., 2006; Kraytsberg et al., 2006). Therefore establishing the pathophysiological mechanisms underlying mtDNA disease may have important implications for other common forms of neurodegeneration.
There are many different possible approaches to understanding the mechanisms involved in mitochondrial neurodegeneration, ranging from post-mortem studies in patients (Oldfors et al., 1990) to the creation of animal models of human disease (Inoue et al., 2000; Fan et al., 2008; Tyynismaa and Suomalainen, 2009). The latter approach has many attractions but for mtDNA mutations it is proving challenging, at least in part because of difficulty in manipulating the mitochondrial genome. In addition, there is now good evidence that many pathogenic mtDNA mutations are not transmitted (Stewart et al., 2008), thus limiting the availability of mice that can be studied. The approach we have taken is to use cybrid technology to generate embryonic stem cells that contain pathogenic mtDNA mutations (Kirby et al., 2009). These embryonic stem cells can then be differentiated into neurons and glia, and their biochemical and physiological properties investigated. These cell lines also offer a major advantage in terms of cell imaging, which enables a comprehensive analysis of the impact of mtDNA mutations specifically on neuronal and glial metabolism and physiology.
The biochemical consequences of mtDNA defects have been explored on several different cell types, but there have been relatively few studies on neurons (Wong et al., 2002), the main site of the neuropathology and subsequent clinical features. In the present study, we have performed an analysis of the mitochondrial metabolism of neurons containing mtDNA mutations that generate either a mild defect of complex IV or a severe defect of complex I (Kirby et al., 2009). Our studies show very different biochemical phenotypes and responses between the two cell lines and both are different from control cell lines. These studies give an important insight into potential mechanisms and approaches to treatment of neurodegeneration due to mtDNA mutations.
Materials and methods
Cell lines used in this study were described in detail by Kirby et al. (2009). All cybrids were derived from ES-1 (CC9.3.1). Control cell lines were the parental embryonic stem-cell line ES-I and a cybrid (Cy1-I) with a polymorphic variant (m.9821Adel) in the mitochondrial tRNA gene for arginine (MTTR). The polymorphic cell line provides an important control for the process of derivation of the cybrids. Cybrids with altered electron transport chain function were Cy2-I and Cy3-I. Cy2-I harbours the m.6589 T>C mutation in one of the three mtDNA genes encoding complex IV subunits (MTCO1) and causes a mild complex IV deficit (∼40% residual complex IV activity). Cy3-I has two mutations in mtDNA genes encoding different complex I subunits (m.13887Cins in MTND6 and m.12273G>A in MTND5) and causes a severe complex I defect (<10% residual complex I activity).
Differentiation into neurons
Parental embryonic stem cells and cybrids were differentiated into neurons using the 4+/4– method (Bain et al., 1995) as described by Kirby et al. (2009) which also gives full details of markers of differentiation. Cultures were maintained on poly-d-lysine/laminin coated coverslips. Studies were performed on differentiated neurons 7–9 days post plating.
Imaging cytosolic free calcium concentration, mitochondrial membrane potential, reactive oxygen species generation and glutathione concentration
Cells were loaded for 30 min at room temperature with 5 µM fura-2 AM (Molecular Probes, Eugene, OR) and 0.005% Pluronic in a 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-buffered salt solution composed of (mM): 156 NaCl, 3 KCl, 2MgSO4, 1.25 KH2PO4, 2 CaCl2, 10 glucose and 10 HEPES, pH adjusted to 7.35 with NaOH. For simultaneous measurement of cytosolic free calcium concentration ([Ca2+]c), and mitochondrial membrane potential (Δψm), rhodamine 123 (10 µM, Molecular Probes, Eugene, OR) was added into the cultures during the last 15 min of the fura-2 loading period. For measurements of Δψm, cells were loaded with 25 nM tetramethylrhodamine methylester for 30 min at room temperature and the dye was present at the same concentration in all solutions throughout the experiment. In these experiments tetramethylrhodamine methylester is used in the ‘redistribution mode’ (Duchen et al., 2003) to assess Δψm, and therefore a reduction in mitochondrial localized tetramethylrhodamine methylester fluorescence represents mitochondrial depolarization.
Fluorescence measurements of fura-2 and rhodamine 123 loaded cells were made using an epifluorescence inverted microscope equipped with a 20 × fluorite objective. [Ca2+]c and Δψm were monitored in single cells using excitation light provided by a Xenon arc lamp, the beam passing through a monochromator (Cairn research, Kent, UK) to select wavelengths sequentially at 340, 380 and 490 nm (Cairn Research, Kent, UK) with bandwidths of 10 nm. Emitted fluorescence light was reflected through a 515 nm long-pass filter to a frame transfer cooled charge-coupled device camera (Hamamatsu Orca ER). All imaging data were collected and analysed using software iQ (Andor, Belfast, UK). The fluorescence data were acquired at intervals of 5–10 s. The fura-2 data have not been calibrated in terms of [Ca2+]c because of the uncertainty arising from the use of different calibration techniques. Accumulation of rhodamine 123 in polarized mitochondria quenches the fluorescent signal, in response to depolarization the fluorescence signal is dequenched; an increase in rhodamine 123 signal therefore signals mitochondrial depolarization. Wherever possible, we have normalized the signals between resting level (set to 0) and a maximal signal generated in response to the uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) (1 μM; set to 100%) (Abramov and Duchen, 2008).
For measurement of mitochondrial reactive oxygen species production, cells were pre-incubated with mitochondrially targeted hydroethidine (5 µM, Molecular Probes, Eugene, OR) for 10 min at room temperature. For measurement of cytosolic reactive oxygen species production, dihydroethidium (2 μM) was present in the solution during the experiment. No preincubation (‘loading’) was used for dihydroethidium to limit the intracellular accumulation of oxidized products.
Tetramethylrhodamine methylester measurements were made using a Zeiss 510 UV-VIS confocal laser scanning microscope equipped with a META detection system and a 40× oil immersion objective. Illumination intensity was kept to a minimum to avoid phototoxicity and the pinhole set to give an optical slice of ∼2 μm. Tetramethylrhodamine methylester was excited using the 543 nm laser line and fluorescence measured using a 560 nm long-pass filter. For hydroethidine and mitochondrially targeted hydroethidine measurements a ratio of the oxidized/reduced form was measured: the 543 nm laser line and 560 nm long-pass filter were used to excite the oxidized form (ethidium) while excitation 351 nm and measurement at 405–470 nm was used to measure changes in the reduced form (hydroethidium). All data presented were obtained from at least five coverslips and two to three different cell preparations.
Glutathione measurements
To measure glutathione, cells were incubated with 50 µM monochlorobimane (MCB) (Molecular Probes, Eugene, OR). As MCB reacts with glutathione in a reaction catalyzed by gluthatione-s-trasferase, generating a fluorescent adduct, cells were incubated with the dye in HEPES-buffered salt solution at room temperature for 40 min, or until a steady state had been reached before images were acquired for quantitation (Keelan et al., 2001). The cells were then washed with HEPES-buffered salt solution and images of the fluorescence of the MCB-glutathione adduct were acquired using the cooled charge-coupled device imaging system as described using excitation at 380 nm and emission at >400 nm.
Cell-death experiments
Cells were stained simultaneously with 20 µM propidium iodide, which is excluded from viable cells but exhibits a red fluorescence following a loss of membrane integrity, and 4.5 µM Hoechst 33342 (Molecular Probes, Eugene, OR), which stains all nuclei and yields blue fluorescence to UV illumination, to count the total number of cells. Using phase contrast optics, a bright field image allowed identification of neurons, which show a quite distinct morphology compared with the flatter glial cells and also lie in a different focal plane, above the glial layer. A total number of 600–800 neurons or glial cells were counted in 20–25 fields of each coverslip. Each experiment was repeated five or more times, using separate cultures.
Statistical analysis
Statistical analysis was performed with the aid of Origin 8 (Microcal Software Inc., Northampton, MA, USA) software. Means expressed ± SEM.
Results
Effect of mtDNA mutations on mitochondrial membrane potential
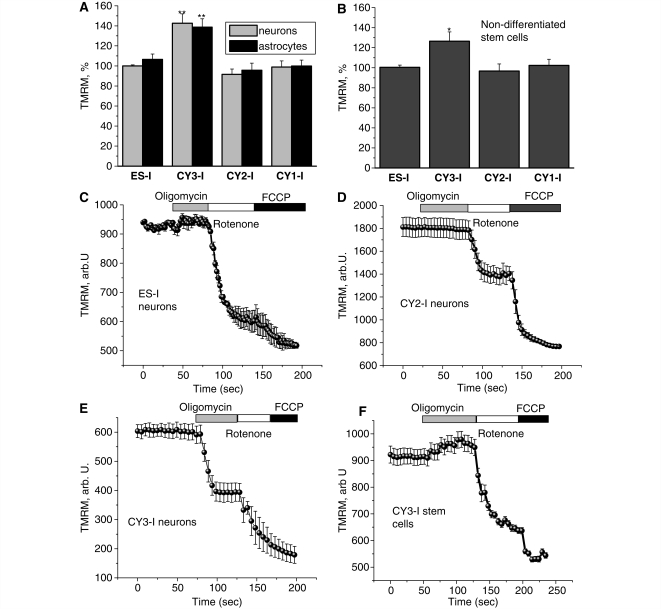
Mutations of mtDNA affecting subunits of mitochondrial complexes impair the efficiency of respiration and as a result might be expected to alter the Δψm. However, in the severe mutant cells Cy3-l (Fig. 1A and B), both stem cells and differentiated cells (neurons and glial-like cells) showed a significantly increased tetramethylrhodamine methylester signal (suggesting an increase in Δψm). Relative to the parental cell line ES-I (taken as 100%) the measurements of tetramethylrhodamine methylester signal intensity were 144 ± 8% for neurons (n = 112), 140 ± 7% for glial cells (n = 89; P < 0.001 for both cell types). In the stem cells there was also an increased Δψm (127 ± 8%) (n = 44 cells; P < 0.05) compared with control cell lines, but this was significantly lower than for differentiated cells, suggesting a different possible mechanism. In Cy1-I and Cy2-I cells, values of tetramethylrhodamine methylester fluorescence were not significantly different from the control ES-I cells either as differentiated cells or as undifferentiated stem cells (Fig. 1A and B).
Figure 1.
Characteristics of mitochondrial membrane potential (Δψm) in cells with mitochondrial mutations. (A–B) Neurons and astrocytes with severe mutation in complex I (CY3-I) showed a significant increase (P < 0.001) in Δψm compared with control cells. The mitochondrial potential in cells with a mutation in complex IV (CY2-I) showed no significant difference from control. Non-differentiated CY3-I cells also exhibited a 26% increase (P < 0.05) in tetramethylrhodamine methylester (TMRM) fluorescence (i.e. an increased Δψm) compared with controls. In control and CY2-I neurons (C–D), oligomycin did not affect Δψm; rotenone induced a partial depolarization; FCCP induced complete depolarization. In CY3-I neurons (E), oligomycin caused a mitochondrial depolarization. In the CY3-I stem cells (F) the Δψm was not maintained by reverse electron flow since oligomycin did not cause mitochondrial depolarization. *P < 0.05; **P < 0.001.
Mechanism of maintenance of mitochondrial membrane potential in cells with severe complex I deficiency
To investigate how a mutation that severely impairs complex I activity can not only maintain Δψm but also be associated with a value greater than seen in control cells, we explored the roles of different mitochondrial mechanisms in the maintenance of membrane potential. In cells with normal oxidative phosphorylation, Δψm is maintained by the proton pumping activity of the respiratory chain. However if oxidative phosphorylation is impaired, the F1Fo-ATP synthase (complex V) may reverse, hydrolyse ATP and pump protons across the inner membrane, so maintaining Δψm (e.g. McKenzie et al., 2007). Application of oligomycin (2 μg/ml), the inhibitor of the F1Fo-ATP synthase, caused a profound decrease in Δψm in CY3-I neurons, with a decrease in the tetramethylrhodamine methylester signal by 58 ± 5%, (n = 82; Fig. 1E). Oligomycin either increased or did not affect Δψm in the other cell lines (Fig. 1C and D). Thus, in CY3-I cells, in response to the impaired activity of the respiratory chain, the F1Fo complex switched to ATP consumption mode which maintained Δψm.
Despite carrying the same mtDNA mutation, the response of the undifferentiated CY3-I stem cells to oligomycin was different compared with the differentiated neuronal CY3-I cells (Fig. 1F). In the undifferentiated cells, application of oligomycin increased tetramethylrhodamine methylester fluorescence by 7.4 ± 0.4% (n = 99), in contrast to the depolarization seen in the differentiated cells. Tetramethylrhodamine methylester fluorescence was then significantly reduced by the subsequent addition of rotenone (by 38.2 ± 4.1%), indicating a fall in Δψm. A relatively large further decrease in signal, in response to FCCP, then suggested that complex II activity must also be relatively active as a donor of electrons in these cells. The effects of oligomycin, rotenone and FCCP in non-differentiated cells CY2-I (n = 76), ES-I (n = 41) and CY1-I (n = 67) were equivalent to effects in the differentiated neuron and glial cells.
Effects of substrates on the maintenance of mitochondrial membrane potential
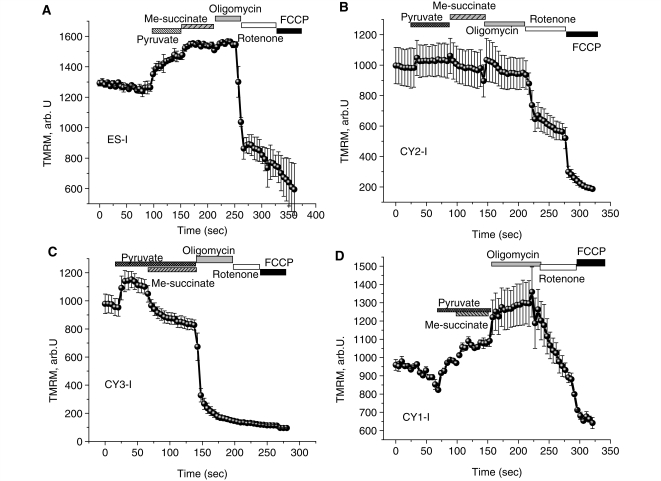
It has been shown that in some models, maintenance of the Δψm by ‘reverse’ ATPase activity can be corrected by giving additional substrate, suggesting that substrate supply may be rate limiting (Gandhi et al., 2009). Provision of additional substrate for complex I (pyruvate, 5 mM) increased Δψm in CY3-I cells (by 16.1 ± 1.1%, n = 101; P < 0.05), but methyl-succinate (5 mM) a membrane-permeable analogue of succinate, the substrate for complex II, induced a small but significant mitochondrial depolarization in CY3-I neurons (by 19.1 ± 1.1%; P < 0.05). Additional mitochondrial substrates did not change the effect of oligomycin on Δψm—application of the F1Fo-ATPase inhibitor increased Δψm in ES-I, CY1-I and CY2-I and collapsed Δψm in CY3-I neurons (Fig. 2A–D). Thus, the impaired maintenance of Δψm by the respiratory chain in CY3-I neurons is not due to lack of substrate supply.
Figure 2.
Effect of mitochondrial substrates on mechanism of maintenance of Δψm in cells with mitochondrial mutations. Application of pyruvate (5 mM) or methyl succinate (5 mM) to neurons increased Δψm, but increased substrate provision did not prevent the oligomycin induced mitochondrial depolarization in CY3-I neurons (C). In the presence of pyruvate, methyl-succinate induced further hyperpolarization of mitochondria in ES-I and CY1-I neurons (A, B, D), but a small mitochondrial depolarisation in CY3-I cells. TMRM = tetramethylrhodamine methylester.
Calcium homoeostasis
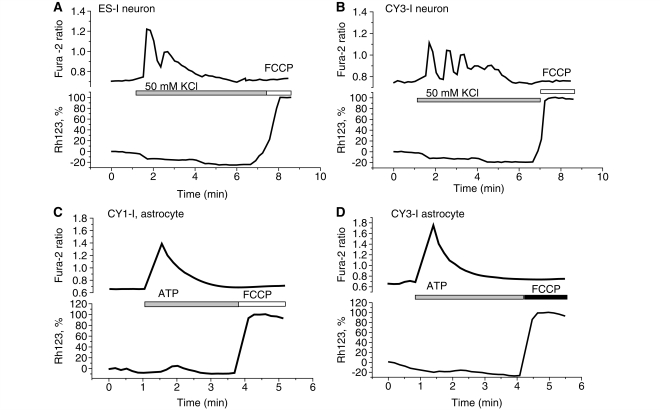
Mitochondria play a significant role as a spatial calcium buffering system in maintaining neuronal calcium homoeostasis (Duchen, 2000). Physiological stimuli do not damage mitochondrial function in healthy cells but can increase the generation of reactive oxygen species, which when associated with mitochondrial calcium overload can precipitate opening of the mitochondrial permeability transition pore. In order to investigate the influence of the mitochondrial mutations on calcium homeostasis we measured [Ca2+]c and Δψm simultaneously using fura-2 and rhodamine 123, respectively, following stimulation of cells with a variety of agonists to raise [Ca2+]c. ATP (100 μM) was used to stimulate [Ca2+]c signals in astrocytes via purinoceptors, and 50 mM KCl was used to induce depolarization of the plasma membrane and open voltage gated calcium channels, which are primarily expressed in neurons. Application of 50 mM KCl to all four cell lines produced a [Ca2+]c signal on cells of neuronal phenotype, with no significant depolarization of Δψm (Fig. 3) as characterized before in primary neurons (Keelan et al., 2001; Gandhi et al., 2009). Application of 100 μM ATP to glial cells in co-culture induced a [Ca2+]c signal with no significant difference between groups of cells in amplitude or time course (Fig. 3C–D). Raising [Ca2+]c with ATP also did not change Δψm in any of the cell lines. Thus, these mitochondrial mutations did not induce pathological changes in their mitochondrial responses to [Ca2+]c signals under these conditions.
Figure 3.
Calcium homeostasis. Simultaneous measurements of [Ca2+]c and ΔΨm were made from neurons (A–B) and astrocytes (C–D) in mixed culture co-loaded with fura-2 and rhodamine 123. Traces are shown from single cells in each case. Mitochondrial mutations did not induce pathological changes in their mitochondrial responses to [Ca2+]c signals under these conditions.
Production of reactive oxygen species in mitochondria
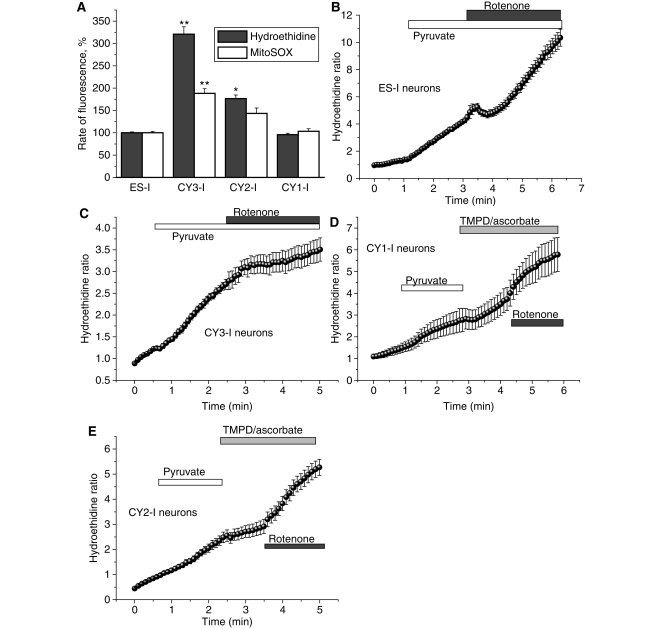
Mitochondria are widely believed to be an important source of reactive oxygen species in a number of neurodegenerative disease states. The increased generation of mitochondrial reactive oxygen species by mitochondria with an impaired respiratory chain may be a factor in cell dysfunction in the presence of mtDNA mutations (Fukui and Moraes, 2008). We have tried to differentiate between reactive oxygen species generated in the cytosol and in the mitochondrial matrix by using hydroethidine as a cytosolic indicator compared with signals from the mitochondrially targeted hydroethidine. The rate of reactive oxygen species production was assessed by measuring the ratio of the rate of increase in red fluorescence (oxidized hydroethidine or ethidium) and the rate of disappearance of the UV induced blue hydroethidine fluorescence (see ‘Materials and methods’ section). In order to isolate mitochondrial from other sources of reactive oxygen species generation, inhibitors of the nicotinamide adenine dinucleotide phosphate oxidase (apocynin, 1 mM or 0.5 µM diphenyleneiodonium) and xanthine oxidase (oxypurinol, 10 μM) were used (Abramov et al., 2007). The rates of reactive oxygen species generation both in the cytosol and in the mitochondrial matrix were not significantly different between control (ES-I) and CY1-I cells (Fig. 4A). The rate of reactive oxygen species production in CY2-I neurons was significantly higher both in the mitochondrial matrix (1.5-fold compared withwith ES-I neurons; n = 121; P < 0.05) and in the cytosol (1.8-fold increase in the rate compared with ES-I; n = 161; P < 0.05; Fig. 4A). The severe mutation in complex I (CY3-I neurons) was also associated with a significant increase in the rate of reactive oxygen species production in both the matrix (1.9-fold higher than ES-I; n = 178; P < 0.001; Fig. 4A) and the cytosol (3.3-fold higher than ES-I; n = 195; P < 0.001; Fig. 4A).
Figure 4.
Mitochondrial and cytosolic reactive oxygen species production in cells with mitochondrial mutations. CY3-I neurons displayed a significantly higher basal rate of increase in mitochondrially targeted hydroethidine (Mitosox) and hydroethidine ratio, demonstrating a higher basal production of intra-mitochondrial and extra-mitochondrial reactive oxygen species compared with control (A). Histogram demonstrating percentage values of the rate of mitochondrially targeted hydroethidine or hydroethidine ratio compared with 100% for control neurons. (B–E) show increase of ΔΨm by mitochondrial substrates (pyruvate and TMPD/ascorbate) or inhibition of complex 1 with rotenone demonstrated the dependence of reactive oxygen species production on ΔΨm. *P < 0.05; **P < 0.001.
The high rate of reactive oxygen species production in CY3-I neurons may be a consequence of two features of the mitochondrial respiratory chain in these cells—a defect in complex I and a higher Δψm. To explore this we used additional substrate for complex I, 5 mM pyruvate, and induced a 2.2-fold increase of the rate of increase of the hydroethidine ratio in ES-I cells (n = 153; P < 0.001) but not in CY3-I neurons (n = 114; Fig. 4B and C). Subsequent inhibition of complex I with 5 μM rotenone induced a further increase of reactive oxygen species production in ES-I neurons (to 3.1-fold increase relative to the basal rate) but a decrease in CY3-I cells, (from a 3.3-fold increase in the basal rate in ES-I neurons to 1.5-fold increase in the rate of CY3-I cells; P < 0.05; Fig. 4B and C). In the CY3-I cells the effects of substrate and rotenone suggest that reactive oxygen species generation in CY3-I neurons is sustained by the high Δψm.
In the CY2-I cells reactive oxygen species production was increased in the presence of rotenone (to 289 ± 24% of basal rate for CY2-I). Pyruvate also increased reactive oxygen species generation in CY2-I cells (163 ± 11% of basal; n = 144) but subsequent application of Wurster’s blue (TMPD; 200 μM)/ascorbate (5 mM) inhibited reactive oxygen species production in CY2-I neurons (to 106 ± 6% of basal rate hydroethidine ratio). In the CY2-I neurons the increased reactive oxygen species production is likely to be due to defect in complex IV.
Glutathione measurements
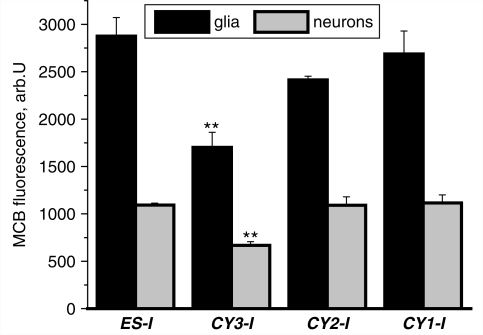
Cell viability in response to increased reactive oxygen species generation is dependent on the efficiency of cellular antioxidant systems. Glutathione provides one of the major antioxidant mechanisms in the CNS. Oxidative stress—the condition in which the rate of reactive oxygen species generation overwhelms antioxidant defence, is typically associated with glutathione depletion and leads to CNS pathology (Dringen and Hirrlinger, 2003). MCB was used to measure glutathione status in non activated neurons and glial cells from co-culture. Differences between the level of glutathione concentration in neurons and astrocytes is well established (Keelan et al., 2001; Dringen and Hirrlinger, 2003) and was observed in these differentiated stem-cell preparations, with a level of MCB fluorescence in all groups of neurons at about 40% of the signal in astrocytes in the same culture (Fig. 5). Despite the higher rate of reactive oxygen species production that we have measured in CY2-I neurons, the mutation in complex IV was not associated with a significant change in glutathione concentration in the neurons or astrocytes (Fig. 5). The severe mutation in complex I (CY3-I) was not only associated with increased reactive oxygen species production in mitochondria but also with a significant reduction in glutathione concentration in both neurons and astrocytes. The level of MCB fluorescence in CY3-I neurons was 59 ± 4% of control (ES-I), and 62 ± 5% for astrocytes (P < 0.001 for both; n = 4 experiments; Fig. 5). Thus, the severe complex I mutation in CY3-I cells induces oxidative stress.
Figure 5.
Glutathione in both neurons and astrocytes. MCB was used to assess astrocyte and neuronal glutathione concentration. Mean intensities of MCB-glutathione adduct fluorescence (arb.U) at a steady state are presented. In CY3-I glutathione concentration was decreased compared with other cell lines in both astrocytes and neurons.
Cell viability
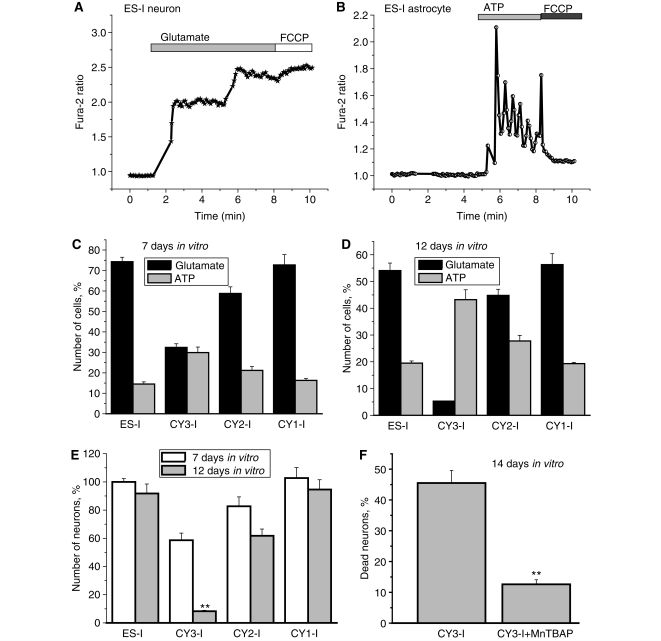
The number of functional neurons and astrocytes in a co-culture can be estimated by counting cells that show [Ca2+]c signals in response to physiological stimuli, KCl or ATP, which give characteristic and distinct responses in either neurons or astrocytes (see above). The application of glutamate (50 μM) induced robust [Ca2+]c responses in neurons (Fig. 6A), while 100 μM ATP gave characteristic responses in astrocytes while neurons generally did not respond to ATP (Fig. 6B). At 7 days in vitro, the cultures showed relative proportions of neurons (74.6 ± 6.1%) to astrocytes (15.9 ± 0.9%) (ES-I, n = 297) and 71.3 ± 4.5% to 17.8 ± 1.1% (CY1-I, n = 320 cells). The co-cultures of CY2-I and CY3-I cells contained more astrocytes than control ES-I cells (24.7 ± 2.1% of astrocytes in CY2-I cells, n = 265; and 32.7 ± 2.9% in CY3-I with only 34.9 ± 2.1% neurons; n = 232 cells; Fig. 6C). With increasing time in culture, there was a progressive increase in cell death of neurons, resulting in a change in the proportion between cell types (Fig. 6D). At 7 days in vitro the CY2-I culture contained fewer neurons (82.6 ± 6.9% of control) and even less in 12 days in vitro culture (64.6% of control in 7 days in vitro). The most dramatic loss of neurons was observed in CY3-I culture, with 59.6 ± 4% of control cultures at 7 days in vitro, but only 6.2 ± 0.4% at 12 days in vitro.
Figure 6.
Cell death in neurons and astrocytes. The number of functional neurons and astrocytes in a co-culture was estimated by counting cells that show [Ca2+]c signals in response to physiological stimuli, which give characteristic and distinct responses in either neurons (glutamate, 50 μM) (A) or astrocytes (ATP, 100 μM) (B). With increasing time in culture, there was a progressive increase in cell death, seen predominantly in neurons, resulting in a change in the proportion between cell types (C–E). The antioxidant MnTBAP significantly protected CY3-I cells against the progressive increase in cell death; the number of dead neurons in this experiment was estimated as percentage of propidium iodide-positive neurons in the culture (F). *P < 0.05.
Considering the very high level of reactive oxygen species generation and low level of endogenous antioxidant in CY3-I neurons we investigated the effect of antioxidants on neuronal survival. Incubation of the CY3-I neurons with 200 μM manganese (III) tetrakis (4-benzoic acid)porphyrin (MnTBAP; a superoxide dismutase mimic and hydrogen peroxide scavenger) significantly reduced the number of dead neurons (measured as a % of propidium iodide positive neurons at 14 days in vitro). Thus, the proportion of dead neurons in untreated CY3-I culture was 45.5 ± 4.1%; n = 593 neurons; in cell cultures with MnTBAP pretreatment this fell to only 12.6 ± 1.1%, n = 651 neurons; (P < 0.01, Fig. 6F). Therefore, overproduction of reactive oxygen species in cells with severe mutation in the complex I is an important cause of specific neuronal death.
Discussion
We believe our approach to understanding the mechanisms involved in neurodegeneration due to mtDNA mutations has given us valuable insight into the neuronal consequences of these mutations. Whilst the majority of studies were performed on neurons and glia, limited studies were also performed on undifferentiated cells including exploration of one of the fundamental properties of mitochondria, mechanisms involved in the maintenance of membrane potential. For the severe complex I mutation there was a significant difference between undifferentiated cells and differentiated cells, glia and neurons. This highlights the need to study relevant models of disease whenever possible if we are to understand the mechanism of neurodegeneration.
Mitochondrial membrane potential
Maintenance of a membrane potential is an essential property of mitochondria. Cells lacking in mtDNA (rho0) maintain a modest membrane potential despite being unable to perform any oxidative metabolism. Discharge of the mitochondrial membrane potential has a number of consequences for the cell, including apoptosis. We have found that a severe mutation in complex I changes the mechanism of maintenance of mitochondrial membrane potential in neurons and astrocytes. In control cells and cells with complex IV mutation, Δψm is maintained by the proton pumping activity of the respiratory chain. The severe complex I mutation in neurons and glia significantly impaired oxidative phosphorylation. Hydrolysis of ATP by the F1Fo-ATPase (complex V) occurs under these conditions, pumping protons across the inner membrane and maintaining Δψm. Remarkably, non-differentiated embryonic stem cells with same mutation maintain mitochondrial membrane potential by respiratory chain activity—thus, the properties that determine the ability to maintain Δψm via hydrolysis of ATP neurons and glia seems to switch at the time of differentiation. Considering the low glycolytic activity of neurons compared with other cell types (Snyder and Wilson, 1983) enhanced consumption of ATP in response to pathological situations such as seizures, common in patients with mitochondrial disease, might be potentially harmful for neurons. This is of clinical relevance since epilepsy is a very prominent feature of mtDNA disease, and there is evidence that the stroke-like episodes seen in patients with the mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) syndrome are often associated with seizures (Betts et al., 2006).
Calcium handling
An interesting observation is the normal calcium handling by the cell lines under the experimental conditions used. Calcium ions are central to neuronal function, transducing electrical activity into molecular signals. In addition, altered cytosolic Ca2+ regulation underlies entry into various cell-death pathways. Consequently, factors that shape the Ca2+ transients are likely to assume great importance in normal neuronal function. A key role in this regard is played by mitochondria. The Δψm provides a powerful driving force to accumulate Ca2+ from the cytosol (Vasington and Murphy, 1962; Carafoli et al., 1964; Crompton and Heid, 1978). Agents which uncouple Δψm prevent mitochondria from taking up Ca2+ and thus alter the time course of cytosolic Ca2+ transients in neurons (Werth and Thayer, 1994) and astrocytes (Boitier et al., 1999). The preservation of Δψm in all cell lines under the conditions used is the likely explanation of the normal calcium handling in these cell lines.
Mitochondrial reactive oxygen species
It has been widely reported that inhibition of complex I and enhanced Δψm are classical inducers of reactive oxygen species generation in mitochondria (Abramov et al., 2007). The severe complex I deficient cells demonstrated unique combination of impaired complex I and high Δψm—combination of both factors dramatically increased production of reactive oxygen species into both the mitochondrial matrix and the cytosol. Excessive generation of free radicals induced glutathione depletion in both neurons and astrocytes, impairing neuronal viability as glutathione depletion leaves the neurons vulnerable to damage by oxidative stress. We believe this is the mechanism by which neurons die more than the astrocytes although features of mutations in complex I appeared in both cell types. Astrocytes contain far higher levels of glutathione than neurons (Dringen and Hirrlinger, 2003) and supply amino acid precursors for neuronal glutathione synthesis. Neurons depleted of glutathione die, probably through an inability to withstand their intrinsic pro-oxidants (Vesce et al., 2005). In contrast, astrocytes can sustain adequate ATP synthesis by glycolytic metabolism, seem not to be as significantly damaged by loss of mitochondrial function, and are also far more resistant to oxidative stress than are the neurons.
In conclusion, we have explored the mechanism of neuronal involvement with two different mtDNA mutations. These mutations were different both in terms of the site of the genetic defect and the severity of the biochemical defect—which probably reflects the different biochemical phenotype of these cells. In the cells with severe complex I deficiency there is evidence of marked cell loss which is entirely compatible with the neuropathological changes seen in patients with mtDNA disease. Interestingly this cell loss was decreased in the presence of antioxidants suggesting a role for reactive oxygen species in the cell death. This might have important implications for the management of patients with mtDNA disease where current treatment options are very limited.
Funding
Wellcome Trust, UK NIHR Biomedical Research Centre for Ageing and Age-related disease award to the Newcastle upon Tyne Hospitals NHS Foundation Trust, Newcastle University Centre for Brain Ageing and Vitality supported by BBSRC, EPSRC, ESRC and MRC; Ruth L. Kirschstein NRSA (NINDS, NIH, USA) (to T.K.S.); National Health and Medical Research Council of AUSTRALIA (NHMRC) CJ Martin Postdoctoral Training Fellowship (to D.M.K.). Work in the Duchen lab is supported by grants from the Wellcome Trust and MRC.
Acknowledgements
Sadly during the writing of this article Denise Kirby died after a short illness and the authors would like to dedicate this article to this outstanding scientist and great colleague.
Glossary
Abbreviations
- FCCP
carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
- MCB
monochlorobimane
- mtDNA
mitochondrial DNA
- Δψm
mitochondrial membrane potential
References
- Abramov AY, Duchen MR. Mechanisms underlying the loss of mitochondrial membrane potential in glutamate excitotoxicity. Biochim Biophys Acta. 2008;1777:953–64. doi: 10.1016/j.bbabio.2008.04.017. [DOI] [PubMed] [Google Scholar]
- Abramov AY, Scorziello A, Duchen MR. Three distinct mechanisms generate oxygen free radicals in neurons and contribute to cell death during anoxia and reoxygenation. J Neurosci. 2007;27:1129–38. doi: 10.1523/JNEUROSCI.4468-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Bender A, Schwarzkopf RM, McMillan A, Krishnan KJ, Rieder G, Neumann M, et al. Dopaminergic midbrain neurons are the prime target for mitochondrial DNA deletions. J Neurol. 2008;255:1231–5. doi: 10.1007/s00415-008-0892-9. [DOI] [PubMed] [Google Scholar]
- Betts J, Jaros E, Perry RH, Schaefer AM, Taylor RW, Abdel-All Z, et al. Molecular neuropathology of MELAS: level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathol Appl Neurobiol. 2006;32:359–73. doi: 10.1111/j.1365-2990.2006.00731.x. [DOI] [PubMed] [Google Scholar]
- Betts J, Lightowlers RN, Turnbull DM. Neuropathological aspects of mitochondrial DNA disease. Neurochem Res. 2004;29:505–11. doi: 10.1023/b:nere.0000014821.07269.8d. [DOI] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Pembrey M, Duncan A, Heron J, Ring SM, Hall A, et al. Prevalence of mitochondrial 1555A–>G mutation in European children. N Engl J Med. 2009;360:640–2. doi: 10.1056/NEJMc0806396. [DOI] [PubMed] [Google Scholar]
- Boitier E, Rea R, Duchen MR. Mitochondria exert a negative feedback on the propagation of intracellular Ca2+ waves in rat cortical astrocytes. J Cell Biol. 1999;145:795–808. doi: 10.1083/jcb.145.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E, Rossi CS, Lehninger AL. Cation and anion balance during active accumulation of Ca++ and Mg++ by isolated mitochondria. J Biol Chem. 1964;239:3055–61. [PubMed] [Google Scholar]
- Crompton M, Heid I. The cycling of calcium, sodium, and protons across the inner membrane of cardiac mitochondria. Eur J Biochem. 1978;91:599–608. doi: 10.1111/j.1432-1033.1978.tb12713.x. [DOI] [PubMed] [Google Scholar]
- Dringen R, Hirrlinger J. Glutathione pathways in the brain. Biol Chem. 2003;384:505–16. doi: 10.1515/BC.2003.059. [DOI] [PubMed] [Google Scholar]
- Duchen MR. Mitochondria and calcium: from cell signalling to cell death. J Physiol. 2000;529 Pt 1:57–68. doi: 10.1111/j.1469-7793.2000.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR, Surin A, Jacobson J. Imaging mitochondrial function in intact cells. Methods Enzymol. 2003;361:353–89. doi: 10.1016/s0076-6879(03)61019-0. [DOI] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–60. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–62. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends Neurosci. 2008;31:251–6. doi: 10.1016/j.tins.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, et al. PINK1-associated Parkinson's; disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell. 2009;33:627–38. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Nakada K, Ogura A, Isobe K, Goto Y, Nonaka I, et al. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet. 2000;26:176–81. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- Keelan J, Allen NJ, Antcliffe D, Pal S, Duchen MR. Quantitative imaging of glutathione in hippocampal neurons and glia in culture using monochlorobimane. J Neurosci Res. 2001;66:873–84. doi: 10.1002/jnr.10085. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Rennie KJ, Smulders-Srinivasan TK, Acin-Perez R, Whittington M, Enriquez JA, et al. Transmitochondrial embryonic stem cells containing pathogenic mtDNA mutations are compromised in neuronal differentiation. Cell Prolif. 2009;42:413–24. doi: 10.1111/j.1365-2184.2009.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–20. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- McFarland R, Taylor RW, Turnbull DM. Mitochondrial disease–its impact, etiology, and pathology. Curr Top Dev Biol. 2007;77:113–55. doi: 10.1016/S0070-2153(06)77005-3. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Liolitsa D, Akinshina N, Campanella M, Sisodiya S, Hargreaves I, et al. Mitochondrial ND5 gene variation associated with encephalomyopathy and mitochondrial ATP consumption. J Biol Chem. 2007;282:36845–52. doi: 10.1074/jbc.M704158200. [DOI] [PubMed] [Google Scholar]
- Oldfors A, Fyhr IM, Holme E, Larsson NG, Tulinius M. Neuropathology in Kearns-Sayre syndrome. Acta Neuropathol (Berl) 1990;80:541–6. doi: 10.1007/BF00294616. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–9. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Snyder CD, Wilson JE. Relative levels of hexokinase in isolated neuronal, astrocytic, and oligodendroglial fractions from rat brain. J Neurochem. 1983;40:1178–81. doi: 10.1111/j.1471-4159.1983.tb08111.x. [DOI] [PubMed] [Google Scholar]
- Sparaco M, Simonati A, Cavallaro T, Bartolomei L, Grauso M, Piscioli F, et al. MELAS: clinical phenotype and morphological brain abnormalities. Acta Neuropathol (Berl) 2003;106:202–12. doi: 10.1007/s00401-003-0716-z. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 2009;10:137–43. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandebona H, Mitchell P, Manwaring N, Griffiths K, Gopinath B, Wang JJ, et al. Prevalence of mitochondrial 1555A–>G mutation in adults of European descent. N Engl J Med. 2009;360:642–4. doi: 10.1056/NEJMc0806397. [DOI] [PubMed] [Google Scholar]
- Vasington FD, Murphy JV. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237:2670–7. [PubMed] [Google Scholar]
- Vesce S, Jekabsons MB, Johnson-Cadwell LI, Nicholls DG. Acute glutathione depletion restricts mitochondrial ATP export in cerebellar granule neurons. J Biol Chem. 2005;280:38720–8. doi: 10.1074/jbc.M506575200. [DOI] [PubMed] [Google Scholar]
- Werth JL, Thayer SA. Mitochondria buffer physiological calcium loads in cultured rat dorsal root ganglion neurons. J Neurosci. 1994;14:348–56. doi: 10.1523/JNEUROSCI.14-01-00348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Cavelier L, Collins-Schramm HE, Seldin MF, McGrogan M, Savontaus ML, et al. Differentiation-specific effects of LHON mutations introduced into neuronal NT2 cells. Hum Mol Genet. 2002;11:431–8. doi: 10.1093/hmg/11.4.431. [DOI] [PubMed] [Google Scholar]