Abstract
Proteasome dysfunction has been repeatedly reported in alcoholic liver disease. Ethanol metabolism end-products affect the structure of the proteasome, and, therefore, change the proteasome interaction with its regulatory complexes 19S and PA28, as well as its interacting proteins. Chronic ethanol feeding alters the ubiquitin-proteasome activity by altering the interaction between the 19S and the 20S proteasome interaction. The degradation of oxidized and damaged proteins is thus decreased and leads to accumulation of insoluble protein aggregates, such as Mallory-Denk bodies. Ethanol also affects the immunoproteasome formation. PA28a/b interactions with the 20S proteasome are decreased in the proteasome fraction isolated from the liver of rats fed ethanol chronically, thus affecting the cellular antigen presentation and defense against pathogenic agents. Recently, it has been shown that ethanol also affects the proteasome interacting proteins (PIPs). Interaction of the proteasome with Ecm29 and with deubiquitinating enzymes Rpn11, UCH37, and Usp14 has been found to decrease. However, the two UBL-ubiquitin-associated domain (UBA) PIPs p62 and valosin-containing protein are upregulated when the proteasome is inhibited. The increase of these UBL-UBA proteins, as well as the increase in Hsp70 and Hsp25 levels, compensated for the proteasome failure and helped in the unfolding/docking of misfolded proteins. Chronic alcohol feeding to rats causes a significant inhibition of the proteasome pathway and this inhibition results from a decreases of the interaction between the 20S proteasome and the regulatory complexes, PIPs, and the ubiquitin system components.
Keywords: Alcoholic liver diseases, Proteasome, Proteasome interacting proteins
INTRODUCTION
Proteasome dysfunction is well established in alcoholic liver injury. However, the mechanism by which ethanol feeding causes proteasome dysfunction is still unclear. Proteasome chymotrypsin-like activity decrease has been the major finding to explain the proteasome dysfunction and accumulation of misfolded and ubiquitinated proteins in the liver of chronic ethanol-fed animals. Previous studies reported by the author[1,2] have shown that the dysfunction of the proteasome system in alcoholic liver disease is caused by structural changes in the α type subunits of the proteasome. However, the dysfunction of the proteasome pathway in alcoholic liver disease is more complicated. There are multiple levels of proteasome activity and specificity regulation. The ubiquitin system and the proteasome interacting proteins (PIPs) are part of the regulation of proteasome activity and specificity. The effects of ethanol feeding on the ubiquitin system and the proteasome interacting protein are still unknown.
PROTEASOME SYSTEM
The proteasome system is a sophisticated, selective, and highly specific proteolytic pathway. It includes the 20S proteasome, also called the catalytic core particle (CP), the regulatory complexes, and interacting proteins. The catalytic core is formed by 28 subunits, arranged into four heteroheptameric rings made of seven subunits each (7α7β7β7α). The α-type subunits form the external rings, and the β subunits the internal rings.
CP BINDING TO THE 19S REGULATORY COMPLEX
The CP of the proteasome system binds to the regulatory complex 19S to form the 26S proteasome (Figure 1). The 26S proteasome is involved in the ubiquitin proteasome pathway (UPP), and is responsible for the ubiquitin-targeted protein degradation[3] (Figure 1).
Figure 1.
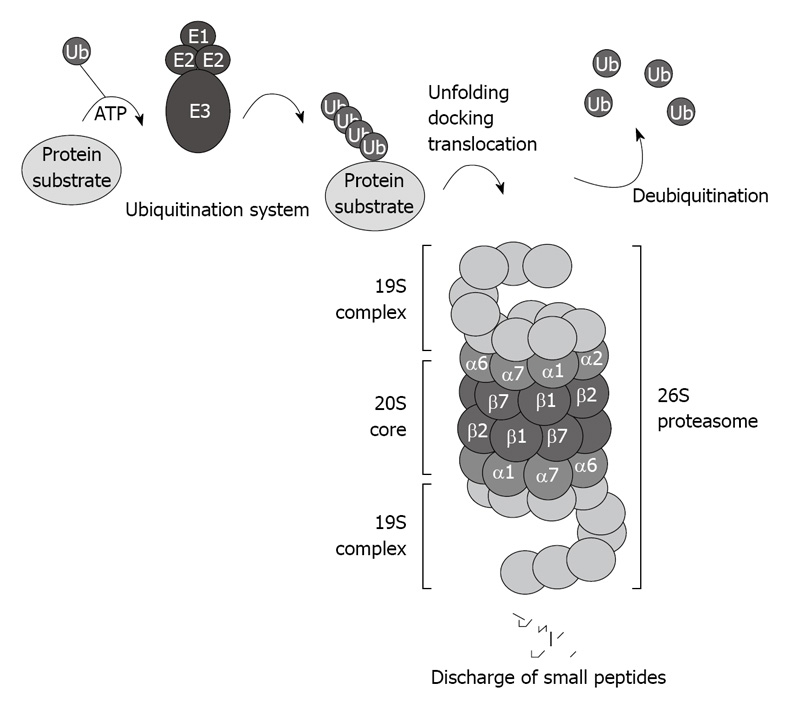
Schematic illustration of the ubiquitin proteasome pathway. The 26S proteasome consists of the 20S core capped with the 19S regulatory complexes that recognize ubiquitinated protein substrates designated for proteolysis.
Cellular key proteins, such as cyclins, cyclin-dependent kinase inhibitors, IκB, hypoxia-inducible factor-1α, Nrf2, and p53, are substrates for the 26S proteasome, and their ubiquitin-dependent degradation is highly controlled[4-6].
The UPP is also responsible for the clearance of misfolded and oxidized proteins. Therefore , proteasome failure is the cause of numerous diseases associated with poor clearance of these often deleterious proteins.
There are two major steps involved in the ubiquitin-proteasome-dependent degradation pathway: (1) the enzymatic polyubiquitination of protein substrates; and (2) the docking and recognition by the 26S proteasome prior to degradation. A cascade of enzymes, including the ubiquitin-activating E1 enzymes, the ubiquitin-carrier protein E2 enzymes, and the ubiquitin-protein ligases E3, which conjugate the ubiquitin residues to the target protein substrate for degradation, are responsible for the ubiquitination of the protein substrate[7]. There are at least four different E1s, 24 different E2s, and at least 100 E3s ligases[8].
Several rounds of ubiquitination yield an ubiquitin chain that includes at least four ubiquitin residues, which designates the target protein for degradation by the 26S proteasome. Once the polyubiquitinated protein is delivered to the 26S proteasome, the polyubiquitin chain is freed, and the ubiquitin residues are recycled. Several deubiquitinating enzymes play an important regulatory role in regenerating the ubiquitin protein[9], and in certain cases, represent a rate-limiting step for proteasome-mediated protein degradation[10]. When change occurs and interferes with these rounds of ubiquitination, deleterious proteins accumulate and cause cellular dysfunction. For instance, mutation of the E3 ligases and misreading of ubiquitin have been reported to modify the ubiquitin system level and cause a significant inhibition of proteasome activity[11].
This great diversity in the ubiquitination system and in the interactions of the CP with its regulatory complexes reflects the complexity and the specificity of the UPP. Disruption of the UPP has been implicated in a wide range of human diseases. Therefore, the proteasome and ubiquitination components are highly attractive targets for pharmaceutical intervention.
CP BINDING TO THE PA28 REGULATORY COMPLEX
The 20S proteasome can also bind to the regulatory complex PA28 (also known as REG and 11S) to form the immunoproteasome (Figure 2). PA28 is a heteromeric complex of 28-kDa subunits. It binds to the cylinder end of the 20S, thus opening the gate channel to the catalytic chamber[12,13].
Figure 2.
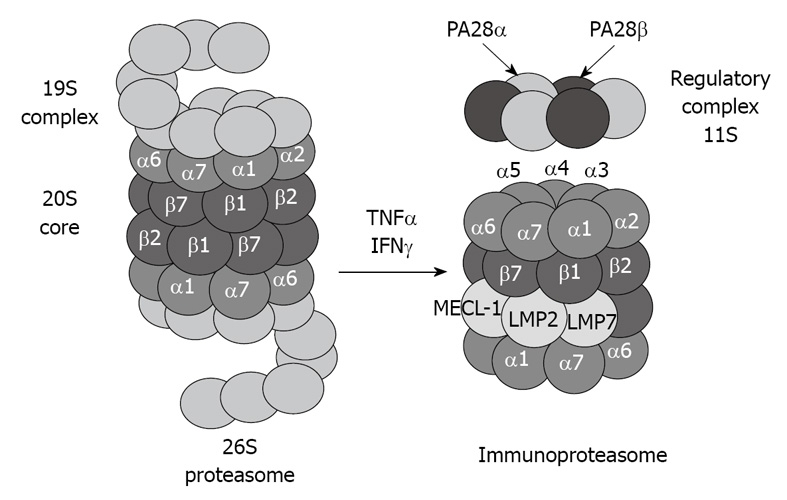
Tumor necrosis factor (TNF) α and interferon (IFN) γ induce formation of immunoproteasome subunits. LMP7, LMP2 and MECL-1 subunits replace the constitutive catalytic subunits β5, β1 and β2, respectively, which shift the catalytic properties of the proteasome to generate MHC-I-binding peptides.
The immunoproteasome forms under the influence of high levels of cytokines, such as interferon (IFN) γ[14]. The immunoproteasome formation also consists of a replacement of the catalytic β subunits of the 20S proteasome, i.e. chymotrypsin-like (β5), trypsin-like (β1), and peptidylglutamyl peptide-hydrolase, recently called caspase-like activity (β2), by the immunoproteasome subunits LMP7, LMP2, and MECL-1, respectively[15,16]. This replacement is required for the cleavage site specificity of the immunoproteasome for efficient antigen processing and presentation by major histocompatibility complex class I molecules[17,18]. However, despite the induction of the immunoproteasome subunits, if the α subunits are modified, for instance by chronic ethanol feeding[1,2], the binding between the catalytic core 20S and the regulatory complex PA28 is blocked, and antigen presentation is thus lowered, which alters the host defense.
CP BINDING TO THE PA200 REGULATORY COMPLEX
The 20S proteasome CP also associates with other activating complexes such as the HEAT-repeat protein PA200 (the homologue of yeast Blm10)[19], which also opens the gate and stimulates peptide entry. PA200 is present within hybrid complexes (19S-20S-PA200), and is an ATP-independent proteasome regulatory complex. It is known to be involved mainly in the nucleus and DNA repair[20].
In summary, the role of these different regulatory complexes, binding to the 20S proteasome CP, is to open the 20S proteasome gate at the α subunits, then to confer specificity to the proteolytic activity of the catalytic chamber formed by the β subunits. Numerous studies have focused on the effects of chronic ethanol feeding on the ubiquitin-proteasome pathway (26S proteasome activity) and the consequence of its dysfunction in liver cells[21,22]. These effects still need further understanding. Substantial studies need to be undertaken to elucidate the effects of chronic ethanol feeding on the immunoproteasome and the nuclear proteasome, with respect to host defense and the epigenetic mechanism regulation.
WHAT ARE THE PIPS?
As much as the proteasome is complex, it does not work alone. There is an orchestra of proteins involved in the regulation of the proteasome activity. An increasing number of PIPs have been reported, which indicates that the 20S proteasome CP is a dynamic structure that interacts with specific proteins for specific functions. Therefore, any interference with the 20S proteasome and with its interacting proteins would affect the proteasome specific functions.
It has been a decade since Verma et al[23] have reported and analyzed PIPs. These authors used a one-step affinity method to purify intact 26S proteasome and its interacting proteins from budding yeast cells, and reported the existence of PIPs. These newly discovered PIPs were classified in four groups that include proteasome subunits, chaperone, transcription, and ribosomal proteins. New methods to purify the proteasome were then developed. Scanlon et al[24] have used the GST UBL (ubiquitin-like domain as an affinity chromatography matrix), and purified the proteasome from human cell lines. These authors have classified the 26S PIPs into four groups of proteins that include de-ubiquitinases, ubiquitin ligases, ubiquitin domain-containing proteins and conjugating/ubiquitin, and proteins involved in DNA repair. Using the GST UBL matrix to purify the PIPs has helped find proteins dominantly related to the proteasome and ubiquitination systems; probably because of the affinity of UBL for the ubiquitin-proteasome pathway.
Among identified PIPs are a number of abundant cellular proteins, such as heat shock proteins (Hsps), elongation factors, and ribosomal proteins. The Hsps interaction with the proteasome is highly specific because they are induced to compensate for the proteasome failure, when the proteasome activity is decreased. In higher eukaryotes, it has been shown that Hsc70/Hsp70 members facilitate the delivery of aggregation-prone substrates for degradation by interacting with the proteasome through an adaptor protein[25,26]. Along with Hsp70, Hsp27 assists in the unfolding of the proteins designated for degradation by the proteasome[27]. In addition, Hsp90 family members have been suggested to play a role in the proteasome structural integrity and assembly through their interactions with the 26S proteasome[28].
Other proteins work upstream of the ubiquitin system to recognize, unfold, shuttle, dock, and deubiquitinate the protein substrate designated for degradation. The chaperone system Bip/PDI is associated with endoplasmic reticulum (ER)-associated degradation[29]. Other proteins that contain a ubiquitin-associated domain (UBA), such as Rad23 and p62, are involved in carrying and docking the protein substrates at the proteasome[30]. These interacting proteasome partners differ in their biological roles, and are chosen to interact with the proteasome according to their specific cellular function. Therefore, different chaperone members may play distinct roles in modulating protein degradation by the proteasome.
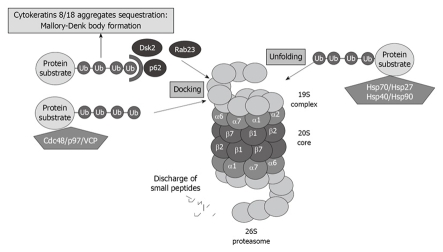
A great number of yet-to-be-identified proteins can mediate ubiquitin recognition at the proteasome. UBL-UBA domain-containing proteins associate with substrates designated for degradation, as well as with subunits of the proteasome, thus regulating the proper turnover of proteins. The best known UBL-UBA proteins of PIPs are Cdc48/p97/valosin-containing protein (VCP), which present misfolded ER-proteins to the proteasome. P62, also called sequestosome 1, is also involved in presenting ubiquitinated proteins to the proteasome (Figure 3).
Figure 3.
Proteasome interacting proteins (PIPs) involved in unfolding and docking of protein substrates designated for degradation by the proteasome. VCP: Valosin-containing protein.
VCP and P62 have been reported to be involved in cytokeratin 8/cytokeratin 18 aggregate sequestration and Mallory-Denk body (MDB) formation in alcoholic liver disease[31] (Figure 3). Both proteins are significantly upregulated when the proteasome activity is inhibited using PS-341, thus indicating that these proteins play a crucial role in proteasome activity[32].
The 26S stabilizing protein, Ecm29, has been found in the fraction of the purified proteasome, and is well established as a PIP. Ecm29 tethers the proteasome CP to the 19S regulatory particle, and confers stability of 19S-20S binding in yeast[33]. It may play a crucial role in the 26S proteasome dysfunction in alcoholic liver disease[34].
More recently, Kautto et al[35] have developed a rapid method of 26S proteasome isolation using chromatography, and have identified over 100 proteins in the proteasome purified fraction, including the 26S proteasome and 32 proteasome subunits. 14-3-3-like proteins have been identified to interact with the proteasome, and also have been classified as PIPs. In our laboratory, when chromatography and high salt concentration purification was used, only a few proteins were identified by mass spectrometry, which indicates that the salt disrupted the proteasome interactions with its associating proteins. The identified proteins included the 14-3-3 proteins, the kinases protein kinase A (PKA) and transglutaminase, and the phosphatases PP2A and PP1[36].
The role and function of the 14-3-3 proteins, in the regulation of proteasome function, remain to be elucidated. This protein has been identified by mass spectrometry in the 20S fraction purified chromatographically with a high salt gradient, which reflects the strength of 14-3-3 interaction with the 20S proteasome. 14-3-3, which is a major scaffolding protein and a phosphobinding protein in the cell, plays a major role in cellular mechanisms, such as signal transduction and regulation of transcription factors[37-39]. A pilot study has shown that chronic ethanol feeding increases the interaction of 14-3-3 with the 20S proteasome, probably to regulate the 20S proteasome ratio of phosphorylation/dephosphorylation to modulate the proteasome activity changes due to ethanol feeding[36].
The proteins kinases associated with the 20S proteasome, such as casein kinase II[40], transglutaminase (TG2), and PKA also co-isolated with the 20S proteasome through multiple chromatographic steps and high salt concentration, as well as the phosphatases PP2A and PP1[41,42]. These proteins regulate the 20S proteasome activity via phosphorylation/dephosphorylation and are believed to regulate also the 20S proteasome binding to its regulatory complexes. TG2 has been found in the fraction of highly purified 20S proteasome[36], and is known to be responsible for stabilizing macromolecular assemblies[43,44]. It is possible that TG2 is involved in the stabilization of the proteasome macromolecules via its kinase activity. These kinases and phosphatases are crucial for the function of the different types of proteasomes because they regulate the phosphorylation of the α type subunit of the proteasome, thus determining the binding of the 20S proteasome to its regulatory complexes (Figure 4).
Figure 4.
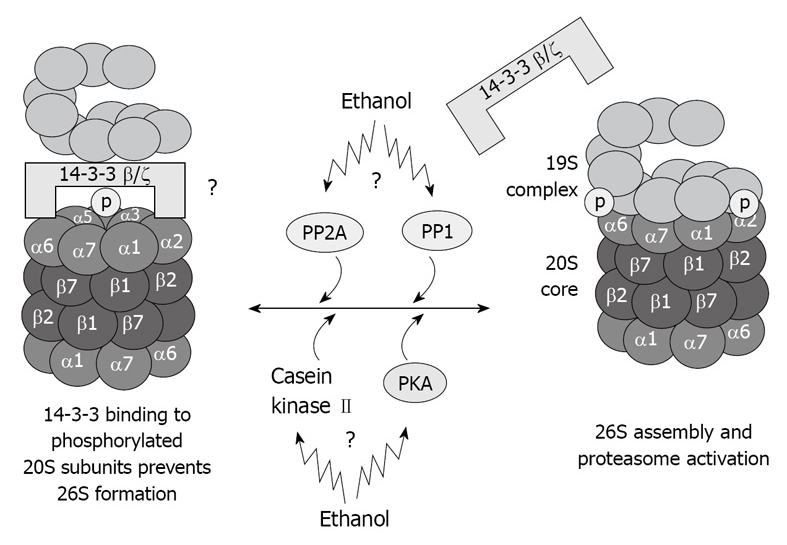
Illustration of hypothetical chronic ethanol feeding effects in 26S dysfunction. As a result of phosphorylation deregulation, 20S and 19S binding is altered, which causes 26S dysfunction. Total dephosphorylation of proteasome subunits by phosphatases is prevented by 14-3-3 protein sequestration of the 20S proteasome.
Similarly to PKA, TG2 and PP2A, the enzyme δ-aminolevulinate dehydratase (ALAD) has been identified by mass spectrometry in the highly purified 20S proteasome fraction[45]. ALAD, also called porphobilinogen synthase, is a cytosolic sulfhydryl-containing enzyme that catalyzes the condensation of two molecules of aminolevulinic acid (ALA). It has been reported that blood ALAD activity is significantly decreased when rats are chronically fed ethanol, which indicates that ethanol feeding causes an alteration in blood[46] as well as liver ALAD activity[47].
When this enzyme is inhibited, the ALA accumulates, which may impair heme biosynthesis and cause porphyria in the liver and pro-oxidant activity in the brain[48,49]. ALAD was among the first PIPs to be identified and its role in the proteasome pathway still needs to be clarified[50].
The recent and most productive method to investigate the PIPs is the QTAX-based tag-team technique, as used by Guerrero et al[51,52]. These authors have identified at least 471 proteins in the network of the proteasome from yeast. The first group of proteins that have been characterized with high affinity was the ubiquitin receptor proteins. The rest of the identified proteins were grouped in the 35 distinct gene ontology protein complexes that are involved in various biological processes, such as chromatin remodeling, metabolism, translation, DNA replication, endocytosis, and protein folding.
Another method to purify the proteasome is to use multiple centrifugation with ATP and a final glycerol gradient zonal centrifugation. This procedure separates the proteasomes and preserves its binding to its regulatory complexes and interacting proteins[34]. Then, mass spectrometry analysis is used to identify the PIPs, as well as to quantify their levels, and thus their interaction with the proteasome. The PIPs identified with the other above-mentioned method have also been characterized by our approach, and in addition, the effects of chronic ethanol feeding have been analyzed[34].
Proteasome activity is regulated at multiple levels. Chronic ethanol feeding, which causes dysfunction of the proteasome pathway, may also occur at the level of PIPs, which could lead to the failure of the proteasome pathway in the liver of alcoholic patients.
EFFECT OF ETHANOL ON THE PIPs
The regulatory control of cellular protein levels by the UPP is essential because inhibition of proteasome activity leads to the loss of cellular regulation and development of many pathological disorders[53]. In experimental alcoholic liver disease, chronic ethanol feeding causes significant inhibition of the 26S proteasome in liver cells[54-57], which results in accumulation of oxidatively damaged and ubiquitinated proteins.
Aggregates of ubiquitinated proteins accumulate and form Mallory-Denk-like bodies[58], which are characteristic of alcoholic liver disease[59,60]. Ethanol-induced inhibition of proteasome activity is also associated with a decrease in misfolded protein degradation at the ER[61,62], which leads to an increased demand on resident chaperones, even though the chaperones and the docking proteins, such as p62[31,32,63], are helping to unfold and deliver proteins designated to be degraded by the proteasome. The immunoproteasome activity is also altered in the liver of alcoholic patients[64,65].
The cause of the proteasome system failure in alcoholic liver disease is not fully understood. The mechanism that causes the failure of the 26S proteasome to remove the oxidized and damaged protein resides in the incapacity of the 20S proteasome to bind the 19S regulatory complex[53]. The proteasome system is under a sophisticated regulation that prevents uncontrolled proteolysis in the cell. The binding between the regulatory complex 19S and the 20S proteasome occurs via a phosphorylation/dephosphorylation-dependent interaction between the α subunits at the 20S and the ATPase subunits at the 19S lid, which leads to the opening of the channel at the α subunit, which favors the translocation of protein substrate to the catalytic chamber. The α subunits of the 20S proteasome play a crucial role in proteolysis mediated by the ubiquitin-proteasome pathway. They form the gate to the catalytic chamber, which is composed of the β subunits. Köhler et al[66] have shown that the amino-terminal sequences of α subunits block this channel, thus keeping the opening of the proteasome in the closed state, i.e. when the 20S proteasome is not bound to an activator[67]. Therefore, any modifications of the phosphorylation level at the α subunits, i.e. modifications caused by ethanol treatment, lead to the closed state of the proteasome because of the dissociation of 20S and 19S. Post-translational modifications at the proteasome subunit have been reported as one mechanism that causes this proteasome inhibition in alcoholic liver disease[1]. Consequently, there is a decrease in 26S formation, thus leading to a decrease in ubiquitin protein degradation[1].
In inflammatory and infectious conditions, the induction of the pro-inflammatory cytokines, such as tumor necrosis factor α and IFNγ, causes the three catalytic subunits of the 20S proteasome CP to be replaced by their IFNγ-inducible counterparts, LMP2, LMP7, and MECL-1, which results in alternatively assembled immunoproteasomes[14,68]. The assembly of the immunoproteasome consists of the binding of the catalytic core that carries the inducible subunits with the regulatory complex PA28. The immunoproteasomes possess enhanced proteolytic activity, and are expressed constitutively by professional antigen-presenting cells, including B cells[69]. It is well known that the host defense is reduced in alcoholic liver disease[64]. This is possibly due to the failure of the immunoproteasome assembly. Recently, it has been shown that chronic ethanol feeding causes a decrease in the binding between the 20S catalytic core and the regulatory complex PA28; binding that is required for the immunoproteasome function, despite the induction of the immunoproteasome subunits[34]. Results from the latest study have indicated that, similar to the mechanism that causes 26S proteasome dysfunction, the immunoproteasome is also altered in alcoholic liver disease. Post-translational modifications of α-type subunits are the key mechanism that regulates the binding of the 20S proteasome catalytic core to its regulatory complexes 19S or PA28. Phosphorylation/dephosphorylation of these subunits is known to regulate their interaction with the 19S ATPases subunits, and thus, 26S formation[70]. Chronic ethanol feeding alters α-type subunit phosphorylation[1]. It is possible that the immunoproteasome assembly is regulated via the interaction between the 20S and the α and β subunits of the PA28 regulatory complex.
Studies to date have focused on the changes that can occur either to the ubiquitin system, or to the proteasomes subunits themselves. However, there is increasing evidence that this pathway is also regulated by other proteins that are just as important as the ubiquitin-proteasome pathway elements[71], and the regulatory complexes 19S, PA28, or PA200.
It is now well established that PIPs are significantly involved in the regulation of proteasome activity. However, the effect of chronic alcohol feeding on these PIPs remains to be investigated. A change in the proteasome interaction with its interacting proteins or modulators, concomitant with the proteasome inhibition due to ethanol feeding, could result in significant inclusion body formation, a decrease in the anti-inflammatory and immune responses, and apoptotic conditions that lead to liver cell injury. Therefore, it is important to determine the effects of ethanol on the proteasome activity, especially the effects of alcohol feeding on the PIPs. The UBA-UBL docking proteins p62 and VCP have been shown to be induced by chronic ethanol feeding, which reflects the failure of the proteasome system to clear the accumulated damaged proteins[31,32]. Similar to the UBA-UBLA proteins, Hsps are also affected by alcohol intake[72], and are generally upregulated when the proteasome is inhibited, mainly to compensate for proteasome failure[31,32].
Recently, the effect of chronic ethanol feeding has been investigated on PIPs, and it has been shown that chronic ethanol feeding affects PIPs[34]. Most importantly, the level of Ecm29, a PIP known to stabilize 20S proteasome interaction with the 19S regulatory complexes[73], is decreased after ethanol feeding[34]. Proteasomes that lack Ecm29 are prone to dissociate from their regulatory complex 19S[74] and most likely from the regulatory complexes PA28a/b and PA200. However, the mechanism of the ethanol effect remains unknown. It is possible that the oxidative stress caused by alcohol-induced detoxifying CYP2E1 is one of the mechanisms that modifies the proteasome subunits[57] and impedes binding between the CP and Ecm29[74].
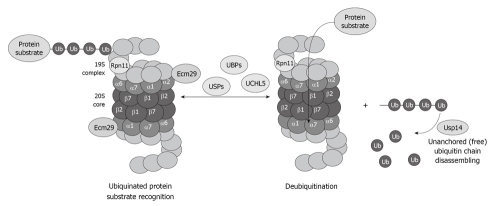
The deubiquitination system is also affected by chronic ethanol feeding. Enigmatically, the three deubiquitinases Rpn11, Usp14 and UCHL5 (Figure 5), are decreased in the 26S proteasome fraction that is purified from ethanol-fed animals[34]. Why the deubiquitination requires at least three enzymes and why alcohol feeding causes a decrease in the three enzymes is not known. However, a decrease in the proteasome activity and the deubiquitination process leads to serious cellular dysfunction that is reflected by accumulation of the ubiquitinated proteins that aggregate and form MDBs.
Figure 5.
Diagram showing the important role of the deubiquitinases in the process of proteasome function and removal of ubiquitinated protein substrates, as well as recycling of free ubiquitin.
CONCLUSION
Chronic ethanol feeding modifies the structure of the proteasome subunits, but also alters proteasome interaction with its proteins partners, thus contributing to serious dysfunction in liver cells. However, we are still at the very beginning of understanding the effects of chronic ethanol feeding on the proteasome pathway. The focus has been to determine the post-translational modifications of the 20S proteasome α type subunits caused by ethanol feeding, because modification of these subunits regulates the 26S proteasome and immunoproteasome formation. As Ecm29 is a key protein involved in 26S proteasome formation, and because it plays a crucial role in stabilizing these proteasomal macromolecules, ethanol-induced Ecm29 downregulation merits further analysis. The determination of the effect of chronic ethanol feeding on the ubiquitination/deubiquitination system to maximize clearance of the altered and ubiquitinated proteins and prevent MDB formation associated with alcoholic liver disease should also be the focus of future research.
Acknowledgments
The author thanks Emmanuel Gorce for typing and editing the manuscript, and drawing the illustrations.
Footnotes
Supported by NIH/NIAAA 8116 and by Alcohol Center Grant on Liver and Pancreas P50-011999, Morphology Core
Peer reviewers: Filip Braet, Associate Professor, Australian Key Centre for Microscopy and Microanalysis, Madsen Building (F09), The University of Sydney, Sydney NSW 2006, Australia; Hartmut Jaeschke, Professor, Liver Research Institute, University of Arizona, College of Medicine, 1501 N Campbell Ave, Room 6309, Tucson, AZ 85724, United States; Mark J Czaja, MD, Liver Research Center, Albert Einstein College of Medicine, 1300 Morris Park Ave, Bronx, NY 10461, United States
S- Editor Tian L L- Editor Kerr C E- Editor Zheng XM
References
- 1.Bardag-Gorce F, Venkatesh R, Li J, French BA, French SW. Hyperphosphorylation of rat liver proteasome subunits: the effects of ethanol and okadaic acid are compared. Life Sci. 2004;75:585–597. doi: 10.1016/j.lfs.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Bardag-Gorce F, Li J, French BA, French SW. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp Mol Pathol. 2005;78:109–115. doi: 10.1016/j.yexmp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Liu CH, Goldberg AL, Qiu XB. New insights into the role of the ubiquitin-proteasome pathway in the regulation of apoptosis. Chang Gung Med J. 2007;30:469–479. [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays. 2008;30:1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 8. Available from: http://www.genome.jp/kegg/
- 9.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 11.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Köycü S, Ramdjielal RD, Salehi A, Martens GJ, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 12.Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Förster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Kloetzel PM. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol. 2004;5:661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- 15.Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 16.Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem. 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- 17.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel PM. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 18.Groettrup M, Standera S, Stohwasser R, Kloetzel PM. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 20.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osna NA, Donohue TM Jr. Implication of altered proteasome function in alcoholic liver injury. World J Gastroenterol. 2007;13:4931–4937. doi: 10.3748/wjg.v13.i37.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.French SW. Mechanisms of alcoholic liver injury. Can J Gastroenterol. 2000;14:327–332. doi: 10.1155/2000/801735. [DOI] [PubMed] [Google Scholar]
- 23.Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlon TC, Gottlieb B, Durcan TM, Fon EA, Beitel LK, Trifiro MA. Isolation of human proteasomes and putative proteasome-interacting proteins using a novel affinity chromatography method. Exp Cell Res. 2009;315:176–189. doi: 10.1016/j.yexcr.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Lüders J, Demand J, Höhfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- 26.Wiederkehr T, Bukau B, Buchberger A. Protein turnover: a CHIP programmed for proteolysis. Curr Biol. 2002;12:R26–R28. doi: 10.1016/s0960-9822(01)00644-3. [DOI] [PubMed] [Google Scholar]
- 27.Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 28.Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 31.Nan L, Wu Y, Bardag-Gorce F, Li J, French BA, Fu AN, Francis T, Vu J, French SW. p62 is involved in the mechanism of Mallory body formation. Exp Mol Pathol. 2004;77:168–175. doi: 10.1016/j.yexmp.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Bardag-Gorce F, Francis T, Nan L, Li J, He Lue Y, French BA, French SW. Modifications in P62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci. 2005;77:2594–2602. doi: 10.1016/j.lfs.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 34.Bousquet-Dubouch MP, Nguen S, Bouyssié D, Burlet-Schiltz O, French SW, Monsarrat B, Bardag-Gorce F. Chronic ethanol feeding affects proteasome-interacting proteins. Proteomics. 2009;9:3609–3622. doi: 10.1002/pmic.200800959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kautto L, Grinyer J, Birch D, Kapur A, Baker M, Traini M, Bergquist P, Nevalainen H. Rapid purification method for the 26S proteasome from the filamentous fungus Trichoderma reesei. Protein Expr Purif. 2009;67:156–163. doi: 10.1016/j.pep.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Bardag-Gorce F, Basillian S, French BA, Whitelegge J, French SW. The proteasome and its interacting proteins. Mol Biol Cell. 2006;10 Suppl:A89. [Google Scholar]
- 37.Brunelli L, Cieslik KA, Alcorn JL, Vatta M, Baldini A. Peroxisome proliferator-activated receptor-delta upregulates 14-3-3 epsilon in human endothelial cells via CCAAT/enhancer binding protein-beta. Circ Res. 2007;100:e59–e71. doi: 10.1161/01.RES.0000260805.99076.22. [DOI] [PubMed] [Google Scholar]
- 38.Schoonheim PJ, Veiga H, Pereira Dda C, Friso G, van Wijk KJ, de Boer AH. A comprehensive analysis of the 14-3-3 interactome in barley leaves using a complementary proteomics and two-hybrid approach. Plant Physiol. 2007;143:670–683. doi: 10.1104/pp.106.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park DJ, Freitas TA, Wallick CJ, Guyette CV, Warn-Cramer BJ. Molecular dynamics and in vitro analysis of Connexin43: A new 14-3-3 mode-1 interacting protein. Protein Sci. 2006;15:2344–2355. doi: 10.1110/ps.062172506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J. 2004;378:177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- 42.Pereira ME, Wilk S. Phosphorylation of the multicatalytic proteinase complex from bovine pituitaries by a copurifying cAMP-dependent protein kinase. Arch Biochem Biophys. 1990;283:68–74. doi: 10.1016/0003-9861(90)90613-4. [DOI] [PubMed] [Google Scholar]
- 43.LaCelle PT, Lambert A, Ekambaram MC, Robinson NA, Eckert RL. In vitro cross-linking of recombinant human involucrin. Skin Pharmacol Appl Skin Physiol. 1998;11:214–226. doi: 10.1159/000029830. [DOI] [PubMed] [Google Scholar]
- 44.Mehta K, Fok JY, Mangala LS. Tissue transglutaminase: from biological glue to cell survival cues. Front Biosci. 2006;11:173–185. doi: 10.2741/1789. [DOI] [PubMed] [Google Scholar]
- 45.Bardag-Gorce F, Banerjee A, Li J, French BA, French SW. Delta aminolevulinic acid dehydratase regulates proteasome activity. Mol Biol Cell. 2005;16 Suppl:A130. [Google Scholar]
- 46.Jurczuk M, Moniuszko-Jakoniuk J, Brzóska MM. Involvement of some low-molecular thiols in the peroxidative mechanisms of lead and ethanol action on rat liver and kidney. Toxicology. 2006;219:11–21. doi: 10.1016/j.tox.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Solomon LR, Crouch JY. delta-Aminolevulinic acid dehydratase in rat liver: studies on the effects of ethanol, acetaldehyde, and B6 vitamers. J Lab Clin Med. 1990;116:228–236. [PubMed] [Google Scholar]
- 48.Demasi M, Penatti CA, DeLucia R, Bechara EJ. The prooxidant effect of 5-aminolevulinic acid in the brain tissue of rats: implications in neuropsychiatric manifestations in porphyrias. Free Radic Biol Med. 1996;20:291–299. doi: 10.1016/0891-5849(95)02035-7. [DOI] [PubMed] [Google Scholar]
- 49.Emanuelli T, Prauchner CA, Dacanal J, Zeni A, Reis EC, de Mello CF, de Souza DO. Intrastriatal administration of 5-aminolevulinic acid induces convulsions and body asymmetry through glutamatergic mechanisms. Brain Res. 2000;868:88–94. doi: 10.1016/s0006-8993(00)02327-1. [DOI] [PubMed] [Google Scholar]
- 50.Guo GG, Gu M, Etlinger JD. 240-kDa proteasome inhibitor (CF-2) is identical to delta-aminolevulinic acid dehydratase. J Biol Chem. 1994;269:12399–12402. [PubMed] [Google Scholar]
- 51.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci USA. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 54.Bardag-Gorce F, Yuan QX, Li J, French BA, Fang C, Ingelman-Sundberg M, French SW. The effect of ethanol-induced cytochrome p4502E1 on the inhibition of proteasome activity by alcohol. Biochem Biophys Res Commun. 2000;279:23–29. doi: 10.1006/bbrc.2000.3889. [DOI] [PubMed] [Google Scholar]
- 55.Fataccioli V, Andraud E, Gentil M, French SW, Rouach H. Effects of chronic ethanol administration on rat liver proteasome activities: relationship with oxidative stress. Hepatology. 1999;29:14–20. doi: 10.1002/hep.510290106. [DOI] [PubMed] [Google Scholar]
- 56.Donohue TM Jr, Zetterman RK, Zhang-Gouillon ZQ, French SW. Peptidase activities of the multicatalytic protease in rat liver after voluntary and intragastric ethanol administration. Hepatology. 1998;28:486–491. doi: 10.1002/hep.510280228. [DOI] [PubMed] [Google Scholar]
- 57.Bardag-Gorce F, French BA, Nan L, Song H, Nguyen SK, Yong H, Dede J, French SW. CYP2E1 induced by ethanol causes oxidative stress, proteasome inhibition and cytokeratin aggresome (Mallory body-like) formation. Exp Mol Pathol. 2006;81:191–201. doi: 10.1016/j.yexmp.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 59.Bardag-Gorce F, Riley NE, Nan L, Montgomery RO, Li J, French BA, Lue YH, French SW. The proteasome inhibitor, PS-341, causes cytokeratin aggresome formation. Exp Mol Pathol. 2004;76:9–16. doi: 10.1016/j.yexmp.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 60.French SW, Mayer RJ, Bardag-Gorce F, Ingelman-Sundberg M, Rouach H, Neve And E, Higashitsuji H. The ubiquitin-proteasome 26s pathway in liver cell protein turnover: effect of ethanol and drugs. Alcohol Clin Exp Res. 2001;25:225S–229S. doi: 10.1097/00000374-200105051-00036. [DOI] [PubMed] [Google Scholar]
- 61.Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 62.Kaplowitz N, Than TA, Shinohara M, Ji C. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27:367–377. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 63.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 64.Osna NA, White RL, Krutik VM, Wang T, Weinman SA, Donohue TM Jr. Proteasome activation by hepatitis C core protein is reversed by ethanol-induced oxidative stress. Gastroenterology. 2008;134:2144–2152. doi: 10.1053/j.gastro.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Osna NA. Hepatitis C virus and ethanol alter antigen presentation in liver cells. World J Gastroenterol. 2009;15:1201–1208. doi: 10.3748/wjg.15.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Köhler A, Bajorek M, Groll M, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. The substrate translocation channel of the proteasome. Biochimie. 2001;83:325–332. doi: 10.1016/s0300-9084(01)01242-1. [DOI] [PubMed] [Google Scholar]
- 67.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 68.Belich MP, Glynne RJ, Senger G, Sheer D, Trowsdale J. Proteasome components with reciprocal expression to that of the MHC-encoded LMP proteins. Curr Biol. 1994;4:769–776. doi: 10.1016/s0960-9822(00)00174-3. [DOI] [PubMed] [Google Scholar]
- 69.Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski UH, Kloetzel PM. Interferon gamma stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40:314–319. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 71.Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 72.Kaviarasan S, Ramamurthy N, Gunasekaran P, Varalakshmi E, Anuradha CV. Induction of alcohol-metabolizing enzymes and heat shock protein expression by ethanol and modulation by fenugreek seed polyphenols in Chang liver cells. Toxicol Mech Methods. 2009;19:116–122. doi: 10.1080/15376510802305039. [DOI] [PubMed] [Google Scholar]
- 73.Gorbea C, Goellner GM, Teter K, Holmes RK, Rechsteiner M. Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J Biol Chem. 2004;279:54849–54861. doi: 10.1074/jbc.M410444200. [DOI] [PubMed] [Google Scholar]
- 74.Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]