Abstract
Peroxiredoxin 5 is a distinct isoform of the peroxiredoxin gene family. The antioxidative and anti-apoptotic functions of peroxiredoxin 5 have been extensively demonstrated in cell culture experiments. In the present paper, we provide the first functional analysis of peroxiredoxin 5 in a multicellular organism, Drosophila melanogaster. Similar to its mammalian, yeast or human counterparts, dPrx5 (Drosophila peroxiredoxin 5) is expressed in several cellular compartments, including the cytosol, nucleus and the mitochondrion. Global overexpression of dPrx5 in flies increased resistance to oxidative stress and extended their life span by up to 30% under normal conditions. The dprx5−/− null flies were comparatively more susceptible to oxidative stress, had higher incidence of apoptosis, and a shortened life span. TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling) analysis revealed that the dprx5−/− null mutant had discernible tissue-specific apoptotic patterns, similar to those observed in control flies exposed to paraquat. In addition, apoptosis was particularly notable in oenocytes. During development the dPrx5 levels co-varied with ecdysone pulses, suggesting inter-relationship between ecdystreroids and dPrx5 expression. The importance of dPrx5 for development was further underscored by the embryonic lethal phenotype of progeny derived from the dprx5−/− null mutant. Results from the present study suggest that the antioxidant and anti-apoptotic activities of dPrx5 play a critical role in development and aging of the fly.
Keywords: aging, apoptosis, development, oxidative stress, peroxiredoxin
INTRODUCTION
The peroxiredoxin family of proteins is comprised of six subtypes, 1–6, distinguishable by their structure, subcellular location and the mode of action. Experimental variations in the expression levels of peroxiredoxins in cultured cells suggest that they act both as antioxidants, as well as redox regulators of signal transduction. Knock-out of peroxiredoxins 1, 2, 3 or 6 results in abnormalities, such as anaemia, increased incidence of malignancies, disruption of haematologic homoeostasis, stillbirth, enhanced susceptibility to pro-inflammatory responses, LPS (lipopolysaccharide)-induced lethal shock, accelerated cellular senescence and shortened life span [1–10]. Such effects are reportedly reversible upon overexpression of peroxiredoxins or catalase, implying a mechanistic role for H2O2 [1,2,11,12].
All of the six peroxiredoxin subtypes found in mammals have also been identified in Drosophila [13,14], including the most recently identified, dPrx5 (Drosophila peroxiredoxin 5) [15]. In a series of investigations in mammalian cultured cells, peroxiredoxin 5 has been shown to possess both peroxidase as well as peroxynitrite reductase activity [16,17]. Compared with the other peroxiredoxins, it has a relatively wider intracellular distribution: e.g. in mitochondria, peroxisomes, nuclei and the cytosol [18,19]. To date, peroxiredoxin 5 has been implicated in a broad range of functions, such as the protection of nuclear and mitochondrial DNA against damage by oxidants or anti-cancer drugs [20–22], inhibition of apoptosis induced by p53 or anti-cancer drugs [16,23], and the modulation of signalling pathways [18]. While the basis of the peroxiredoxin 5 functions has been generally ascribed to its antioxidant activity and the capacity to participate in redox regulation [24,25], an additional function, namely interaction with specific transcriptional complexes, which is not based on redox activity, has also been suggested [26].
In the present study, we provide an analysis of peroxiredoxin 5 function in a whole organism for the first time, using Drosophila strains that under- or over-express peroxiredoxin 5. We describe the effects of dPrx5 modulation on development, apoptosis, stress resistance, and life span of the flies.
EXPERIMENTAL
Fly strains and procedures
The y w reference strain has been maintained in this laboratory for over 14 years. The tubulin (Tub)-GAL4, daughterless (Da)-GAL4, Appl (β-amyloid precursor protein-like)-GAL4 and armadillo (Arm)-GAL4 driver lines were supplied by Dr Blanka Rogina (Department of Genetics and Developmental Biology, University of Connecticut Health Center, Farmington, CT, U.S.A.). PSwitch 106 driver was obtained from the Bloomington Stock Center. The dprx5 mutation is associated with a P-element transposon insertion located in the first exon of the coding region (stock number 15852, Bloomington Stock Center). Homozygous dprx5 flies (−/−) have no detectable dPrx 5 product, but are viable [15]. Nevertheless, it appears that some dPrx5 product is required in the early stages of embryogenesis, based on the observation that homozygous mutants can only be obtained if one or both of the parents are heterozygous. In other words some parental dPrx5 product can be transmitted to the (−/−) zygote via either the egg or the sperm and this is sufficient to support early developmental events.
To generate UAS-dPrx5 transgenic lines, the entire dPrx5 coding region derived from cDNA (CG7217) was inserted into the pUAST vector and the recombinant DNA was then sent for P-element transformation (BestGene). To exclude background effects on survivorship and other traits, UAS-dPrx5 transgenic and GAL4 driver lines and the dprx5−/− mutant were backcrossed to y w for 6–8 generations to obtain genetically homogeneous stocks.
For experimental studies, flies were collected within 1–2 days after hatching and reared on a standard sucrose-cornmeal medium at 25°C. To induce oxidative stress, flies were fed appropriate concentrations of either H2O2 or paraquat in 1% sucrose. Survivorship studies were performed as described previously [27]. Developmental stages were synchronized by collecting eggs at 2 h intervals or by selecting white prepupae.
Over- and under-expression of dPrx5 in Drosophila S2 cells and heterologous expression of dPrx5 in Escherichia coli
To overexpress peroxiredoxin 5 in Drosophila S2 cells, the entire coding region of the Drosophila dPrx5 gene, derived from CG7217 cDNA, was cloned into the pIB/V5-His vector (Invitrogen), upstream of the V5 and His6 epitopes [15]. Plasmid DNA was transfected into S2 cells and stable cell lines were obtained by selection with 100 µg/ml blasticidin, in accordance with the manufacturer’s protocol (Invitrogen). Underexpression of dPrx5 in S2 cells was achieved by RNA interference, as described previously in [15]. For expression in E. coli, the dPrx5 coding domain was cloned into the pProEX™ HT vector (Gibco BRL) in-frame with the upstream polyhistidine His6 sequence and the resulting construct was introduced into E. coli via standard transformation protocol.
Subcellular fractionation and immunoblotting
Subcellular fractionation was performed using gradient centrifugation as described in Radyuk et al. [13]. Alternatively, nuclear and mitochondrial fractions were isolated using the NEPER® Nuclear and Cytoplasmic Extraction Reagents and the Mitochondria Isolation Kit (Pierce). Proteins for immunoblot analysis were extracted with T-PER Tissue Protein Extraction Reagent (Pierce) containing protease inhibitors (Roche). Protein concentrations were determined by the Bio-Rad Protein Assay reagent (Bio-Rad) and 10 µg aliquots of the protein extracts were then resolved by SDS/10% PAGE, followed by transfer to PVDF membrane (Millipore). Immunoblots were performed as described previously in [13,15]. Anti-actin antibodies (MP Biomedicals) were used as a control for loading. Antibodies raised against aconitase (mitochondrial), GCLm (modulatory subunit of glutamate-cysteine ligase; cytosolic) and HDAC (histone deacetylase) (nuclear, BD Biosciences) were used to assess purity of the subcellular fractions.
Flow cytometry, Comet assay and HPLC analysis of mitochondrial DNAs
For viability and apoptosis assays, overnight cell cultures with 1 × 106 cells/ml density were exposed to specified concentrations of H2O2 (Sigma), paraquat (Sigma) or SNAP (S-nitroso-N-acetylpenicillamine) (Molecular Probes) and incubated at room temperature (21–22°C) for different durations. Flow cytometric analysis and Comet assays were performed essentially as described in Radyuk et al. [28]. Mitochondrial DNA was extracted from cells using the mtDNA Extractor CT Kit (Wako) and dissolved in 100 µl of TE buffer, followed by preparation of DNA hydrolysates and HPLC analysis, as described in [28].
TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling) analysis
Tissue-specific assessment of apoptosis-induced DNA fragmentation was made in cryosections prepared from whole flies. Slides were fixed with 2% paraformaldehyde (Sigma) for 5 min at room temperature and incubated with DNA-labelling reagents using the In Situ Cell Death Detection Kit, TMR red (Roche), following the manufacturer’s recommendations. Images were acquired by fluorescence microscopy (Nikon), using MetaMorph software.
RESULTS
Subcellular localization of dPrx5
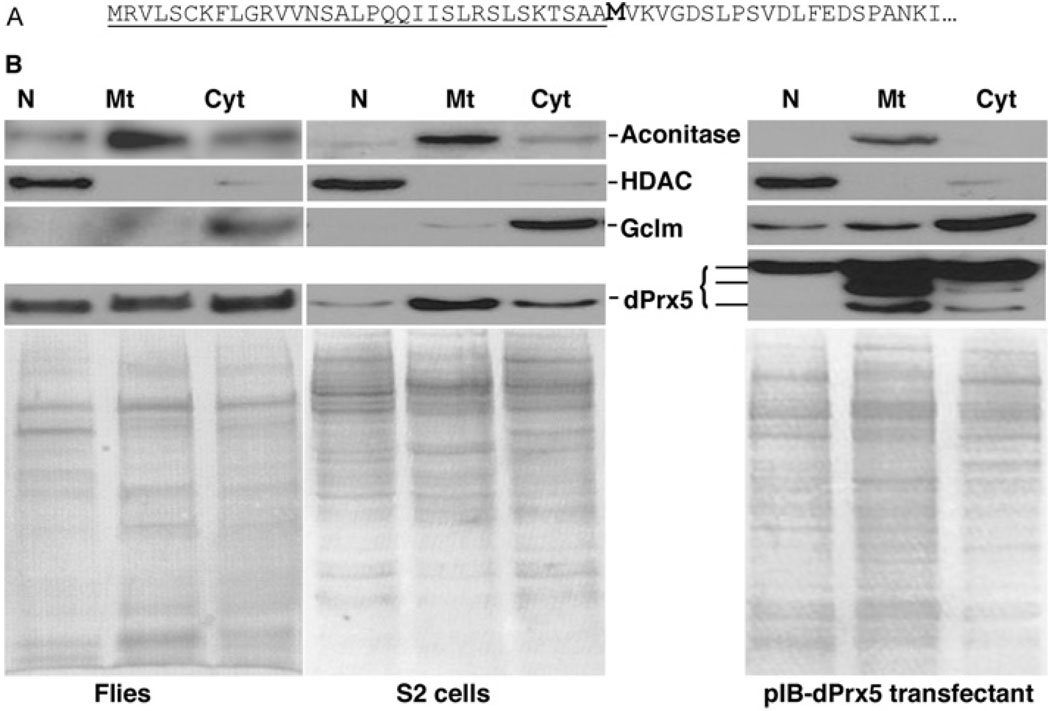
Homogenates of Drosophila S2 cells and adult flies were separated into mitochondrial, nuclear and cytosolic fractions, and subjected to immunoblot analysis in order to establish the subcellular location of dPrx5. A single band of approx. 17 kDa was detected in all three fractions (Figure 1B), which is similar to that observed in mammalian cells [18,19]. The molecular mass of the detected protein was ~3 kDa smaller than the molecular mass of 19.9 kDa, calculated from the deduced amino acid sequence. However, when dPrx5 was expressed in E. coli, only the longer full-length peptide was observed, suggesting post-translational processing in the eukaryotic cell milieu. Notably, the deduced amino acid sequence of dPrx5 contains a potential mitochondrial-targeting signal peptide with a cleavage site between positions 33 and 34 (Figure 1A). Cleavage of the signal peptide could thus result in a peptide of the observed length. Nevertheless, our assumption that this cleavage is mitochondria-specific requires an explanation of why a dPrx5 peptide of a similar size is also detectable in the cytosolic as well as the nuclear fraction. Foremost, these observations are not unique to Drosophila. In mammalian cells [18,22,24,29], there exists a second in-frame AUG codon in close proximity to the mitochondrial-specific cleavage site and initiation at this alternate AUG codon can result in a peptide that is indistinguishable in size from the mitochondrially-targeted form [24]. A similar situation might prevail in Drosophila, since it also has a second in-frame AUG codon at position 34 (Figure 1A).
Figure 1. Analysis of subcellular localization of Drosophila dPrx5 protein.
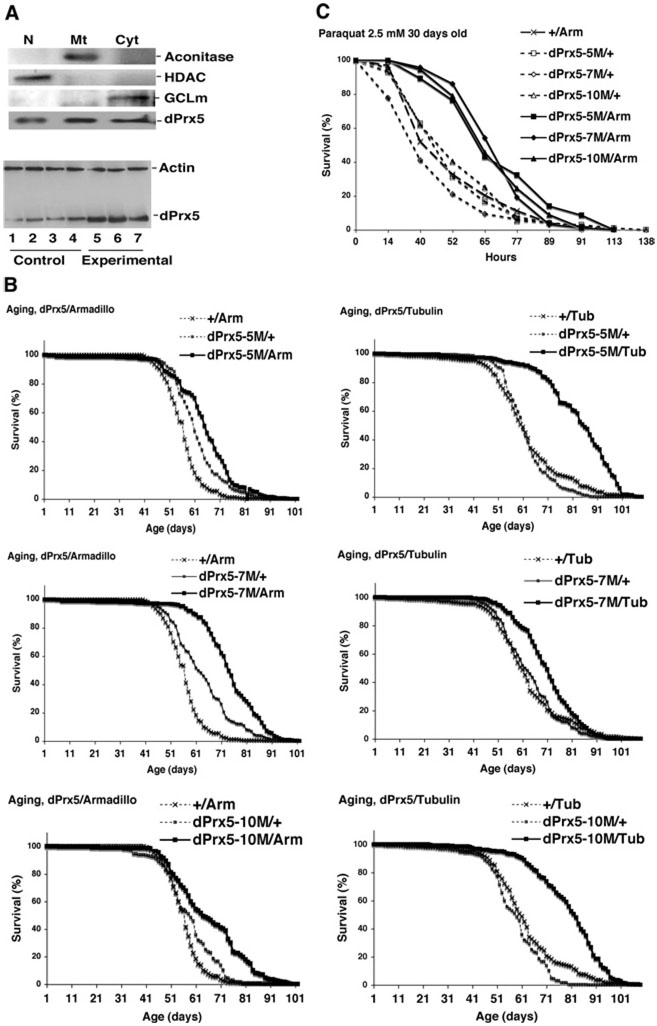
(A) The N-terminal part of the deduced amino acid sequence of dPrx5 protein. The predicted mitochondrial targeting sequence is underlined and the putative second start site is bold. (B) Immunoblot analysis of protein lysates extracted from nuclear (N), mitochondrial (Mt) and cytosolic (Cyt) fractions prepared from flies, S2 cells and S2 cells overexpressing dPrx5 tagged with V5/His6 epitopes (pIB-dPrx5 transfectant). Signals were obtained with anti-dPrx5 antibodies and with anti-aconitase (mitochondrial protein), anti-HDAC (nuclear protein) and anti-GCLm (cytosolic protein) antibodies to control for a specificity of subcellular fractions. Staining with Coomassie Blue (lower panels) was performed to control for a loading.
Expression of dPrx5 in Drosophila during development and aging
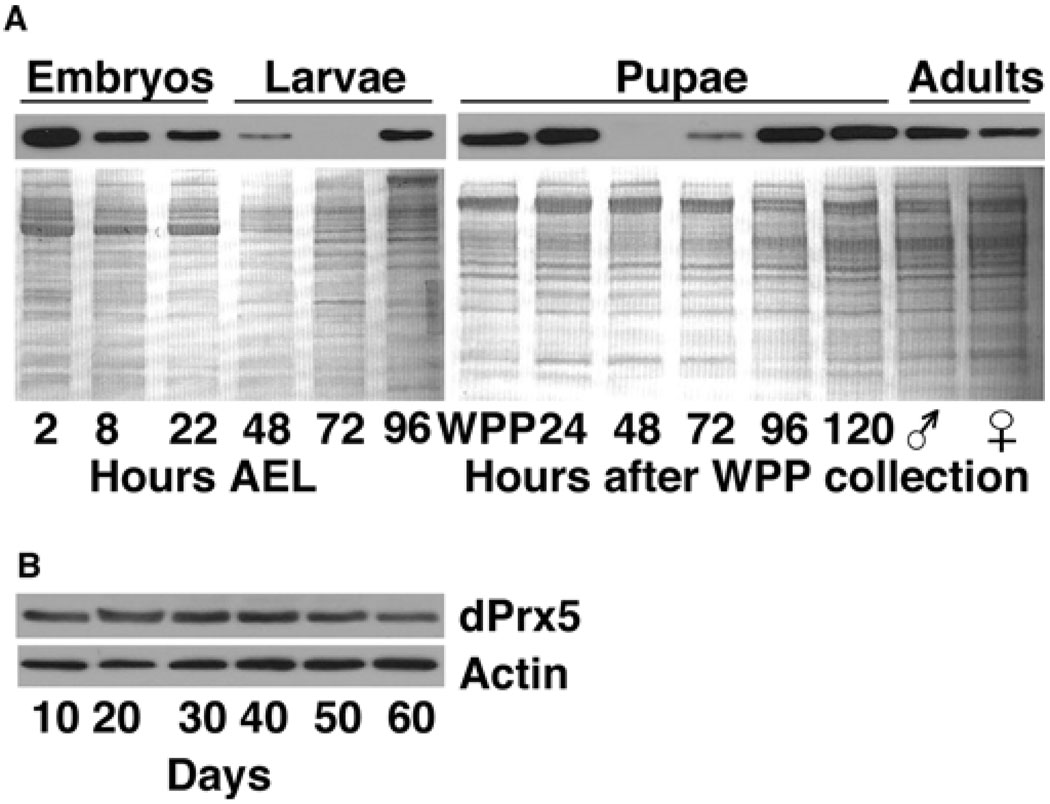
Expression patterns were examined at the whole animal level using immunoblot analysis. dPrx5 protein levels varied dramatically during development, with the highest amounts expressed in embryos (Figure 2A). The level of expression remained relatively low during larval development until the late third instar, followed by an increase in early pupariation with a subsequent dramatic drop-off, and then rising to a peak during late pupariation. These alterations in dPrx5 levels largely paralleled the pulses of ecdysone synthesis [30], indicating a possible interrelationship between dPrx5 expression and ecdysteroids. In adult flies, dPrx5 levels declined during aging, reaching their lowest level in the older flies (50–60 days, Figure 2B).
Figure 2. Immunoblot analysis of dPrx5 expression during development (A) and aging (B).
(A) Protein was extracted from embryos, larvae and pupae, which were allowed to develop for different periods of time after egg laying (AEL) or after white prepupae (WPP) collection. Male and female symbols indicate freshly hatched animals (0–3 h). (B) Adults were collected at different ages. Immunoblots were performed using anti-dPrx5 antibodies and re-probed with anti-actin antibodies to control for a loading. Control for a loading in samples isolated during development was performed with Coomassie Blue staining, as actin levels vary more significantly at these stages.
To understand the physiological role of dPrx5, expression levels were determined under conditions of oxidative stress. RT–PCR (reverse transcription–PCR) and immunoblot analyses indicated that dPrx5 expression is not affected by paraquat, H2O2, or elevated temperatures (33°C and 37°C, results not shown). These results are in agreement with computer analysis of the functional promoter of the dPrx5 gene [15], which found no evidence of ARE (antioxidant response element)- or HSF (heat-shock factor)-binding sites. However, the analysis revealed NF (nuclear factor)-κB-binding motifs (−410 to −400 and −340 to −330), normally found in immune or inflammation response genes, and a binding site (−170 to −150) for the BRC (Broad-Complex), a transcription factor that mediates ecdysone effects (Figure 3). There were also potential binding sites for the transcription factors Sp-1 (specificity protein 1), GATA-1 and NF-1 (Figure 3), which act synergistically and are essential for basal transcription of many genes [31,32].
Figure 3. Analysis of the 5′-flanking region of dPrx5 gene.
The transcription start site is indicated by an arrow. ATG translation start codons are indicated by bold capital letters. Predicted transcription factor binding sites are underlined. The prediction was performed using TFsearch and AliBaba2 softwares (www.gene-regulation.com). alp/alph, alpha (α); AP-2, activator protein-2; BRCZ, BRC zinc finger; C/EBP, CCAAT/enhancer-binding protein; COUP, chicken ovalbumin upstream promoter; ENKTF, enkephalin transcription factor; GCN4, general control non-derepressible 4; GLO, GLOBOSA; HNF, hepatocyte NF; HOXA, homeobox A4; PR, progesterone receptor; RAR-a1, retinoicacid receptor α1; SRF, serum-response factor; T3R-a1, tri-iodothyronine receptor α1; TBP, TATA-box-binding protein.
Functional impact of dPrx5 overexpression in S2 cells
Transfection of S2 cells with the pIB/His-V5 constitutive expression vector containing the entire dPrx5 coding region (see the Experimental section) resulted in 2- to 3-fold elevation in the total levels of dPrx5 (results not shown). Increased dPrx5 levels were observed in cytosol, nuclei and mitochondria (Figure 1B, right panels). Among the three visible dPrx5 cross-reacting bands, the lower band had the same mobility as an endogenous dPrx5, while the two upper bands seemed to correspond to the processed and unprocessed peptides derived from the recombinant dPrx5 gene. Relatively bigger sizes of the recombinant products than the endogenous one are presumably due to the presence of the V5/His6 epitopes in the former. A mitochondria-enriched fraction was found to contain considerable amounts of unprocessed dPrx5–His-V5. This may be due in part to the level of purity of mitochondrial preparations, which contained detectable levels of cytosolic proteins, as indicated by the cytosolic marker, GCLm (Figure 1B). In addition, it is also possible that small amounts of overexpressed dPrx5 are retained in the mitochondrial membrane. The absence of a discernable intermediate band in the nuclear and cytosolic fractions suggested that the second AUG codon in the dPrx5 recombinant coding region was not as effective as a secondary start codon; nevertheless the prominence of the intermediate mitochondrial band implied that processing of the mitochondrial targeting pre-sequence remained operative.
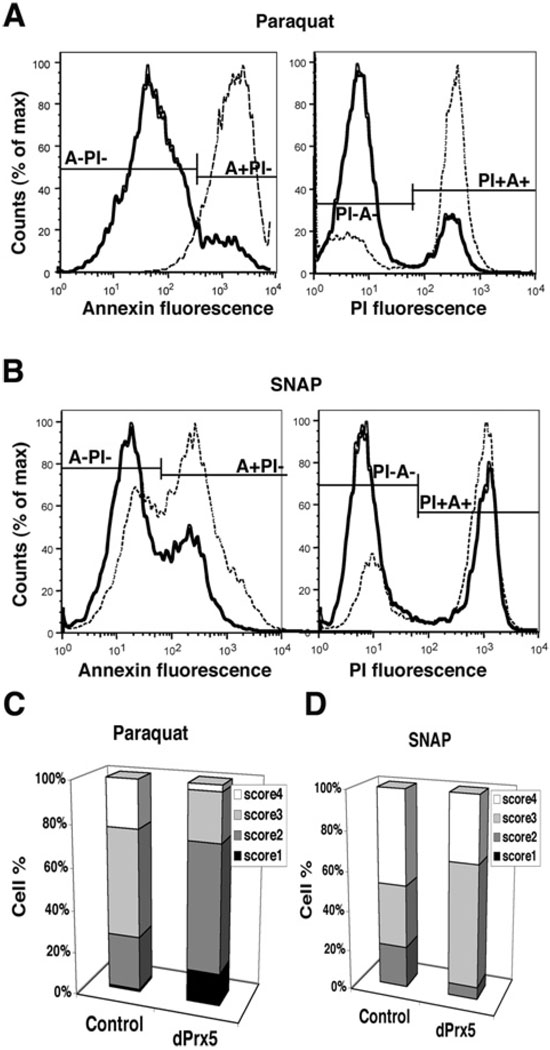
Overexpression of dPrx5 was found to protect S2 cells from death caused by free radical generators, such as paraquat or SNAP (Figure 4). Flow cytometry analysis revealed that the number of cells undergoing apoptosis (annexin-positive cell fraction) or found dead [PI (propium iodide)-positive] following exposure to paraquat or SNAP was significantly reduced among the dPrx5-overexpressing cells compared with the controls (Figure 4A and 4B). However, this protective effect was less evident in cells exposed to H2O2 (results not shown). Single-cell gel electrophoresis (Comet assay) also indicated that the negative effects of paraquat or SNAP on DNA integrity were significantly alleviated in dPrx5 overexpressing cells (Figure 4C and 4D). Among the paraquat-exposed cells, the numbers scoring 1 (intact nucleus) or 2 (showing a modest degree of DNA damage) was substantially higher in dPrx5 over-expressing cells than in the controls, whereas the number of the cells scoring 4 (cells showing total DNA degradation) was significantly diminished.
Figure 4. dPrx5 overexpression protects S2 cells from oxidants.
(A and B) Flow cytometry analysis of cell death after exposure to paraquat and SNAP. Control cells (transfected with empty vector) and cells overexpressing dPrx5 were exposed to 10 mM paraquat or 1 mM SNAP for 8 h followed by incubation with anti-Annexin-V-FITC antibodies and PI. Cells undergoing apoptosis (Annexin-V-FITC-positive, PI negative; A + PI−) are seen in the right peaks; non-apoptotic viable cells (Annexin-V-FITC- and PI-negative; A−PI−) are seen in the peaks on the left. PI-positive cells (right peaks) are dead cells or cells in the late stage of apoptosis (Annexin-V-FITC- and PI-positive; PI + A+). Continuous lines represent cells overexpressing dPrx5 and broken lines represent control cells transfected with empty vector. (C and D) Evaluation of DNA damage in control cells and cells expressing dPrx5 by single cell Comet assay. Cells were treated with 10 mM paraquat or 1 mM SNAP for 24 h. Frequency distribution of nuclei in four different classes of increasing total DNA damage is shown (see the Experimental section). The results are mean percentage values of three separate experiments. Differences in scores 1–4 for dPrx5 overexpressor compared with control exposed to paraquat (C) were statistically significant (P < 0.05).In samples treated with SNAP, statistically significant differences were determined for cells at the score 2 and 3 (D).
Overexpression of dPrx5 also conferred a protective effect against oxidative damage to mitochondrial DNA, as indicated by the 8-oxodG levels. When cultured under normal conditions, cells overexpressing dPrx5 exhibited a 44.5% decrease in the 8-oxodG levels relative to controls (results not shown). Ameliorative effects were also observed when cells were exposed to paraquat or H2O2 (9.1–54.5% decrease in 8-oxodG). Altogether, these results suggested a significant protective role of dPrx5 on cell survival, particularly under stressful conditions.
Phenotypes of the dprx5−/− mutant
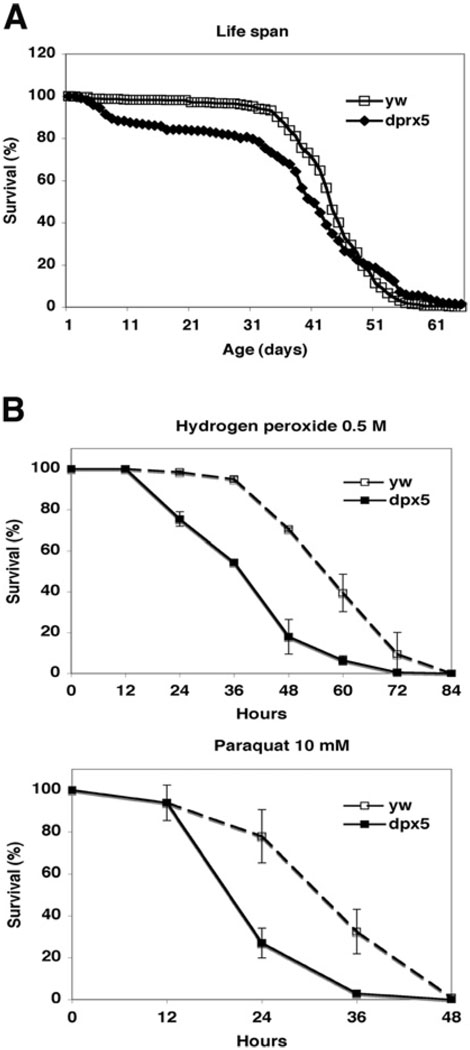
We have identified previously a P-element mutant, which is homozygous viable and has no detectable expression of peroxiredoxin 5 protein [15]. When crossed among themselves, dprx5−/− flies produced progeny that were largely arrested during early embryogenesis, at stage 8. This phenotype could be rescued, provided that either of the two parents contributed the wild-type dprx5/+ (−/+) allele, indicating that dPrx5 is required for embryonic development. There was a modest reduction in the mean life span of the mutant flies, due largely to an accelerated death rate during 5–10 days post-eclosion, which was observed in all three independent experiments, suggesting that dPrx5 is required at stages when tissues still proliferate. Subsequently, when flies became fully developed, the mortality rate stabilized and the surviving flies reached a maximum life span similar to that of the controls (Figure 5A).
Figure 5. Survival of dprx5−/− mutant under normal (A) and oxidative stress (B) conditions.
(A) Pooled data of three independent experiments are shown for each line. In all experiments, 200–250 flies for each fly strain were used. The median age was 43 ± 16 and 40 ± 12 days and mean life span was 65.3 and 57.4 days for y w and dprx5 strains correspondingly. There were no significant differences in maximum age (10% survival) between y w strain and dprx5−/− mutant, which in both cases was 53–54 days. (B) Approx. 100–150 flies of each group (dprx5 mutant and y w control) were fed 1% sucrose solution containing 10 mM paraquat or 0.5 M H2O2. Results are means ± S.D. of two independent experiments.
Although the lack of dPrx5 expression had only a moderate effect on fly survival under normal conditions, it dramatically affected viability under conditions of oxidative stress. The mean survival time of the dprx5−/− mutants was 36 ± 4 h compared with 60 ± 8 h of controls when treated with H2O2, and 22 ± 1 h compared with 31 ± 4 h in control following exposure to paraquat (Figure 5B). The phenotype was fully rescued by expressing dPrx5 from the transgene (results not shown), thus confirming that the observed effects on fly viability were indeed due to the loss of peroxiredoxin 5 expression.
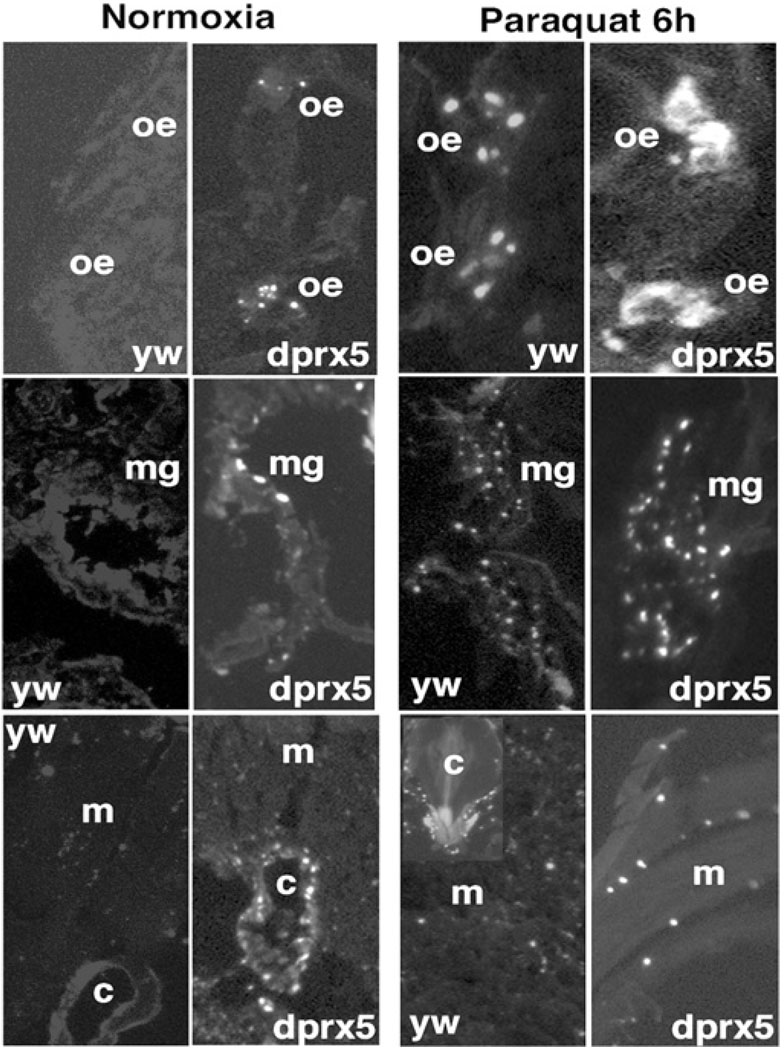
The cell culture studies described above revealed a possible role for dPrx5 in protecting cells from apoptosis. To test the hypothesis that apoptosis could contribute to the enhanced mortality observed among young flies and those exposed to oxidants, the incidence of apoptosis was compared between mutant dprx5−/− flies and the controls in the presence or absence of paraquat. Since newly eclosed adult flies exhibited significant fluctuations in apoptotic activity [33], flies were allowed to mature until at least 5 days of age. While the control exhibited virtually no discernable apoptosis, indicated by in situ TUNEL, the mutant showed apoptotic activity in oenocytes, tissues of the digestive system (mostly in the midgut), and cardia (Figure 6, left panels). Apoptotic incidence in these tissues was higher in the presence of paraquat in both mutant and control flies, although the dprx5−/− mutant had a relatively higher proportion of labelled cells and also exhibited more intense labelling than the control (Figure 6, right panels). Consistent with previous observations [33], exposure to paraquat also induced wide-spread apoptosis in the flight muscles and the digestive tract. Again, relatively greater numbers of labelled cells were present in the dprx5−/− mutant (Figure 6, right lower panel). Thus it seems that the apoptotic response due to the absence of dPrx5 resembles that elicited by paraquat in both intensity and tissue specificity, with the strongest effects occurring in the oenocytes. Altogether, these data indicated that one function of dPrx5 is to protect cells from oxidant-induced apoptosis.
Figure 6. TUNEL analysis of cell death in y w control and dprx5 mutant.
Representative images show DNA fragmentation in cryosections made from 10-day-old flies reared under normal conditions (left panels) or fed 10 mM paraquat for 6 h (right panels). Shown are selected images of the abdomen and thoraces regions, where differences in DNA fragmentation between dprx5−/− mutant and control were detected. oe, oenocytes; mg, midgut; c, cardia and m, muscles.
Effects of dPrx5 overexpression in whole flies
To determine the effects of dPrx5 on longevity and resistance to oxidative stress, dPrx5 was overexpressed under the control of the UAS promoter, using a set of tissue-specific and global GAL4 drivers. In fly lines with global GAL4 drivers (Tub-GAL4 or Arm-GAL4), dPrx5 expression levels were elevated in the cytosolic, nuclear and mitochondrial compartments (Figure 7A). Moreover, all three UAS-dPrx5 responder lines exhibited significant increases in life span (Figure 7B). In Arm-GAL4/UAS-dPrx5 flies, increases in median age ranged from 13% to 24%, whereas in Tub-GAL4/UAS-dPrx5 flies, the increases were 29% to 30%. The low-level global overexpression of dPrx5, driven by Arm-GAL4, also conferred relatively higher resistance to paraquat, which was particularly evident around 30 days of age (Figure 7C). Similar, but smaller effects, were observed with the Tub-GAL4 driver or upon exposure to H2O2 or hyperoxia (results not shown). Broad overexpression of dPrx5 in neuronal tissues (Appl-GAL4 driver) or overexpression in fat bodies, using the PSwitch106 driver, had no effect on fly survival under normal or oxidative stress conditions (results not shown).
Figure 7. The effects of dPrx5 overexpression on aging and resistance to oxidative stress.
(A) Immunoblot analysis of dPrx5 overexpression. Subcellular fractions (upper panel) were prepared as described in Figure 1. Lower panel, samples were prepared from whole flies. Lanes 1, 2, 3 and 4, driver (Tub-GAL4/+) and three different transgene (UAS-dPrx5/+) controls; lanes 5, 6, and 7, three different experimental (UAS-dPrx5/Tub-GAL4) lines. (B) Life spans of flies overexpressing dPrx5 globally at low levels (with Arm-GAL4 driver) and at high levels (with Tub-GAL4 driver). Pooled data of two independent experiments are shown for each line. The median ages were extended in dPrx5/Arm experimental flies by 24.7 ± 3.7% compared with driver and by 13.4 ± 3.9% compared with transgenic controls. With Tub-GAL4 driver, median age was extended by 29.2 ± 7.7% compared with driver and by 30.0 ± 4.6% compared with transgenic controls. (C) Resistance of 30-day-old flies to 2.5 mM paraquat. Approx. 100–150 flies of each group were fed 1% sucrose solution containing 5 mM paraquat. Similar results were obtained in two independent experiments. The median survival time in experimental flies was increased by 52.6% to 60.0% compared with driver control and by 41.5% to 52.3% compared with transgene controls.
DISCUSSION
One notable feature of peroxiredoxin 5 in the mammalian cells is its occurrence in multiple subcellular compartments [18,19,34,35]. In Drosophila, dPrx5 is also quite widely distributed, being detectable in cytosol, nucleus and the mitochondrion (Figure 1 and Figure 7), but unlike mammals, it is undetectable in peroxisomes. Consistent with this pattern, computer analysis of the deduced amino acid sequence revealed the existence of a putative N-terminal mitochondrial targeting domain, but the signature peroxisomal targeting tag (SQL), normally found at the C-terminus, was absent. On the other hand, there was no indication of the existence of the classical nuclear localization signal, thus the mechanism by which dPrx5 enters the nucleus remains unclear at present. It is possible that dPrx5 passively diffuses into the nucleus, as <50 kDa proteins normally do [36].
The present study on flies under- and over-expressing dPrx5, the first to provide a functional analysis of peroxiredoxin 5 at the organismal level, indicate that dPrx5 has a broad range of effects on development, longevity and resistance to oxidative stress. Similar to knock-outs for other peroxiredoxins [2,6,8,10], the Drosophila dprx5−/− null is also homozygous viable. Nevertheless, while disruption of peroxiredoxins 1, 2, 3 or 6 in mice had no obvious effects on fertility, peroxiredoxin 5-null flies are sterile. The development of the progeny is arrested during the early stages of embryogenesis. Within the peroxiredoxin gene family, peroxiredoxin 5 appears to be unique for its role in development.
Significant variations in the levels of dPrx5 were observed during development, with peak expressions occurring during embryogenesis, puparium formation and in late pupae (Figure 2). This pattern is similar to that observed for other ecdysone-responsive genes [37]. It is also worth noting that CG7215, which was found to be an ecdysone-responsive gene [37], and dPrx5, which together form a dicistron, are co-transcribed during the early stages of development [15,37]. It is thus plausible that during development dPrx5 is a target of ecdysone signalling and is involved in mediating hormonal effects. Additional support for the possible existence of a relationship between dPrx5 and ecdysteroids is provided by the finding that oenocytes are highly sensitive to apoptosis in the absence of dPrx5 (Figure 6). Oenocytes are cells, associated with fat metabolism, pheromone production and ecdysteroid synthesis [38–41]. They also express CuZnSOD (Cu/Zn superoxide dismutase) and catalase, in a pattern suggestive of ecdysone control, at relatively high levels [42,43]. The magnitude of oenocytic death in dprx5−/− mutant flies was comparable with that occurring under acute oxidative stress. Altogether, such characteristics suggest that dPrx5 may act within the context of hormonal pathway(s) to regulate ROS (reactive oxygen species) production and apoptosis.
The survivorship studies on overexpressors and mutants indicate that dPrx5 has a significant impact on longevity in the presence or absence of stressors. Global overexpression of dPrx5 at relatively low (Arm-GAL4) or moderately high (Tub-GAL4) levels was found to result in the prolongation of life span by up to 13% and 29%, respectively, in a long-lived background. In addition, such flies were significantly more resistant to paraquat. On the other hand, neuronal overexpression of dPrx5 had no beneficial effect on life span. Thus, dPrx5 does not appear to be a limiting or a “weak link” in neuronal tissue. The beneficial effects of dPrx5 overexpression were also evident in S2 cells. In response to acute stress, dPrx5 overexpressing cells exhibited enhanced viability, protection from apoptosis and attenuation of oxidative damage to both nuclear and mitochondrial DNAs (Figure 4D and results not shown). In contrast, the absence of dPrx5 had a demonstrably deleterious effect on survival. Under normal unstressful conditions, the life span of the dprx5−/− mutant was ~15% shorter than the y w control. Mortality was particularly notable during the first 10–15 days of adulthood (Figure 5A), however, the survival curve of the remaining flies soon converged with that of the control flies, and there was no difference in maximum life span (defined here as age at 90% mortality). Although the absence of dPrx5 had only a modest negative effect on survival under normal conditions, mutant flies were extremely sensitive to paraquat or H2O2 (Figure 5B), suggesting an important role for dPrx5 in protection against endogenous and exogenous oxidative stress.
The basis for the observed effects on survival may in part reside in the antioxidant/anti-apoptotic function(s) of dPrx5 in non-neuronal tissues. Zheng et al. [33] have shown that acute oxidative stress induced by paraquat increases the incidence of apoptosis in the muscles and digestive tract, and that this pattern was quite similar to that observed in the aged flies. We also detected DNA fragmentation inmuscles and the digestive tract under comparable conditions, but in addition identified a strong DNA fragmentation signal in oenocytes (Figure 6). These tissue-specific patterns were remarkably similar to those observed in the dprx5−/− mutants fed the normal food. Furthermore, tissue-specific apoptosis was significantly enhanced in the paraquat-administered dprx5−/− mutants, with the most dramatic changes occurring in the oenocytes. Altogether, the present results suggest that the absence of dPrx5 expression potentiates tissue-specific apoptosis induced by oxidants, the thereby supporting the hypothesis that the effects of dPrx5 on longevity occur via its antioxidant action, which also reduces apoptosis in critical tissues.
To conclude, it is presently unclear which particular subcellular isoform of dPrx serves the most critical role in the aging process and the protection of flies from oxidative stress and apoptosis. In a series of studies in mammalian cultured cells, peroxiredoxin 5 was found to bestow protection against reactive oxygen species, when overexpressed in the cytosol, the nucleus or the mitochondrion [20,21,23–25]. Overall, the results of the present study support the concept that peroxiredoxin 5 plays a role in survival under normal and stressful conditions.
Acknowledgments
FUNDING
This work was supported by a grant from the National Institute on Aging/National Institutes of Health [grant number RO1 AG20715].
Abbreviations used
- Arm
armadillo
- Appl
β-amyloid precursor protein-like
- BRC
Broad-Complex
- Da
daughterless
- dPrx5
Drosophila peroxiredoxin 5
- GCLm
modulatory subunit of glutamate-cysteine ligase
- HDAC
histone deacetylase
- NF
nuclear factor
- PI
propium iodide
- SNAP
S-nitroso-N-acetylpenicillamine
- Sp-1
specificity protein 1
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling
REFERENCES
- 1.Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, Lerner CP, Paigen B. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J. Biol. Chem. 2003;278:25179–25190. doi: 10.1074/jbc.M302706200. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Peroxiredoxin 6 gene-targeted mice show increased lung injury with paraquat-induced oxidative stress. Antioxid. Redox Signaling. 2006;8:229–237. doi: 10.1089/ars.2006.8.229. [DOI] [PubMed] [Google Scholar]
- 3.Nagy N, Malik G, Fisher AB, Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am. J. Physiol. 2006;291:H2636–H2640. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Shoji W, Oshima H, Obinata M, Fukumoto M, Kanno N. Crucial role of peroxiredoxin III in placental antioxidant defense of mice. FEBS Lett. 2008;582:2431–2434. doi: 10.1016/j.febslet.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 5.Egler RA, Fernandes E, Rothermund K, Sereika S, de Souza-Pinto N, Jaruga P, Dizdaroglu M, Prochownik EV. Regulation of reactive oxygen species, DNA damage, and c-Myc function by peroxiredoxin 1. Oncogene. 2005;24:8038–8050. doi: 10.1038/sj.onc.1208821. [DOI] [PubMed] [Google Scholar]
- 6.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 7.Yang HY, Jeong DK, Kim SH, Chung KJ, Cho EJ, Jin CH, Yang U, Lee SR, Lee DS, Lee TH. Gene expression profiling related to the enhanced erythropoiesis in mouse bone marrow cells. J. Cell. Biochem. 2008;104:295–303. doi: 10.1002/jcb.21620. [DOI] [PubMed] [Google Scholar]
- 8.Lee TH, Kim SU, Yu SL, Kim SH, Park DS, Moon HB, Dho SH, Kwon KS, Kwon HJ, Han YH, et al. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood. 2003;101:5033–5038. doi: 10.1182/blood-2002-08-2548. [DOI] [PubMed] [Google Scholar]
- 9.Moon EY, Noh YW, Han YH, Kim SU, Kim JM, Yu DY, Lim JS. T lymphocytes and dendritic cells are activated by the deletion of peroxiredoxin II (Prx II) gene. Immunol. Lett. 2006;102:184–190. doi: 10.1016/j.imlet.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Han YH, Kim HS, Kim JM, Kim SK, Yu DY, Moon EY. Inhibitory role of peroxiredoxin II (Prx II) on cellular senescence. FEBS Lett. 2005;579:4897–4902. doi: 10.1016/j.febslet.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 11.Yang CS, Lee DS, Song CH, An SJ, Li S, Kim JM, Kim CS, Yoo DG, Jeon BH, Yang HY, et al. Roles of peroxiredoxin II in the regulation of proinflammatory responses to LPS and protection against endotoxin-induced lethal shock. J. Exp. Med. 2007;204:583–594. doi: 10.1084/jem.20061849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumin A, Huber C, Rulicke T, Wolf E, Werner S. Peroxiredoxin 6 is a potent cytoprotective enzyme in the epidermis. Am. J. Pathol. 2006;169:1194–1205. doi: 10.2353/ajpath.2006.060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radyuk SN, Klichko VI, Spinola B, Sohal RS, Orr WC. The peroxiredoxin gene family in Drosophila melanogaster. Free Radical Biol. Med. 2001;31:1090–1100. doi: 10.1016/s0891-5849(01)00692-x. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez J, Agudo M, Van Damme J, Vandekerckhove J, Santaren JF. Polypeptides differentially expressed in imaginal discs define the peroxiredoxin family of genes in Drosophila. Eur. J. Biochem./FEBS. 2000;267:487–497. doi: 10.1046/j.1432-1327.2000.01022.x. [DOI] [PubMed] [Google Scholar]
- 15.Michalak K, Orr WC, Radyuk SN. Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem. Biophys. Res. Commun. 2008;368:273–278. doi: 10.1016/j.bbrc.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Kok KH, Chun AC, Wong CM, Wu HW, Lin MC, Fung PC, Kung H, Jin DY. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochem. Biophys. Res. Commun. 2000;268:921–927. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]
- 17.Dubuisson M, Vander Stricht D, Clippe A, Etienne F, Nauser T, Kissner R, Koppenol WH, Rees JF, Knoops B. Human peroxiredoxin 5 is a peroxynitrite reductase. FEBS Lett. 2004;571:161–165. doi: 10.1016/j.febslet.2004.06.080. [DOI] [PubMed] [Google Scholar]
- 18.Seo MS, Kang SW, Kim K, Baines IC, Lee TH, Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J. Biol. Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 19.Kropotov A, Sedova V, Ivanov V, Sazeeva N, Tomilin A, Krutilina R, Oei SL, Griesenbeck J, Buchlow G, Tomilin N. A novel human DNA-binding protein with sequence similarity to a subfamily of redox proteins which is able to repress RNA-polymerase-III-driven transcription of the Alu-family retroposons in vitro. Eur. J. Biochem./FEBS. 1999;260:336–346. doi: 10.1046/j.1432-1327.1999.00162.x. [DOI] [PubMed] [Google Scholar]
- 20.Banmeyer I, Marchand C, Clippe A, Knoops B. Human mitochondrial peroxiredoxin 5 protects from mitochondrial DNA damages induced by hydrogen peroxide. FEBS Lett. 2005;579:2327–2333. doi: 10.1016/j.febslet.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 21.Banmeyer I, Marchand C, Verhaeghe C, Vucic B, Rees JF, Knoops B. Overexpression of human peroxiredoxin 5 in subcellular compartments of Chinese hamster ovary cells: effects on cytotoxicity and DNA damage caused by peroxides. Free Radical Biol. Med. 2004;36:65–77. doi: 10.1016/j.freeradbiomed.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Kropotov A, Serikov V, Suh J, Smirnova A, Bashkirov V, Zhivotovsky B, Tomilin N. Constitutive expression of the human peroxiredoxin V gene contributes to protection of the genome from oxidative DNA lesions and to suppression of transcription of noncoding DNA. FEBS J. 2006;273:2607–2617. doi: 10.1111/j.1742-4658.2006.05265.x. [DOI] [PubMed] [Google Scholar]
- 23.Kropotov A, Gogvadze V, Shupliakov O, Tomilin N, Serikov VB, Tomilin NV, Zhivotovsky B. Peroxiredoxin V is essential for protection against apoptosis in human lung carcinoma cells. Exp. Cell Res. 2006;312:2806–2815. doi: 10.1016/j.yexcr.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Kropotov A, Usmanova N, Serikov V, Zhivotovsky B, Tomilin N. Mitochondrial targeting of human peroxiredoxin V protein and regulation of PRDX5 gene expression by nuclear transcription factors controlling biogenesis of mitochondria. FEBS J. 2007;274:5804–5814. doi: 10.1111/j.1742-4658.2007.06103.x. [DOI] [PubMed] [Google Scholar]
- 25.Tien Nguyen-nhu N, Knoops B. Mitochondrial and cytosolic expression of human peroxiredoxin 5 in Saccharomyces cerevisiae protect yeast cells from oxidative stress induced by paraquat. FEBS Lett. 2003;544:148–152. doi: 10.1016/s0014-5793(03)00493-9. [DOI] [PubMed] [Google Scholar]
- 26.Kropotov AV, Grudinkin PS, Pleskach NM, Gavrilov BA, Tomilin NV, Zhivotovsky B. Downregulation of peroxiredoxin V stimulates formation of etoposide-induced double-strand DNA breaks. FEBS Lett. 2004;572:75–79. doi: 10.1016/j.febslet.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J. Biol.Chem. 2005;280:37331–37338. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- 28.Radyuk SN, Michalak K, Rebrin I, Sohal RS, Orr WC. Effects of ectopic expression of Drosophila DNA glycosylases dOgg1 and RpS3 in mitochondria. Free Radical Biol. Med. 2006;41:757–764. doi: 10.1016/j.freeradbiomed.2006.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TH, Kim SJ, Kang SW, Lee KK, Rhee SG, Yu DY. Molecular cloning and characterization of the mouse peroxiredoxin V gene. Biochem.Biophys.Res. Commun. 2000;270:356–362. doi: 10.1006/bbrc.2000.2430. [DOI] [PubMed] [Google Scholar]
- 30.Richards G. The radioimmune assay of ecdysteroid titres in Drosophila melanogaster. Mol. Cell. Endocrinol. 1981;21:181–197. doi: 10.1016/0303-7207(81)90013-7. [DOI] [PubMed] [Google Scholar]
- 31.Rustighi A, Mantovani F, Fusco A, Giancotti V, Manfioletti G. Sp1 and CTF/NF-1 transcription factors are involved in the basal expression of the Hmgi-c proximal promoter. Biochem. Biophys. Res. Commun. 1999;265:439–447. doi: 10.1006/bbrc.1999.1680. [DOI] [PubMed] [Google Scholar]
- 32.Fischer KD, Haese A, Nowock J. Cooperation of GATA-1 and Sp1 can result in synergistic transcriptional activation or interference. J. Biol. Chem. 1993;268:23915–23923. [PubMed] [Google Scholar]
- 33.Zheng J, Edelman SW, Tharmarajah G, Walker DW, Pletcher SD, Seroude L. Differential patterns of apoptosis in response to aging in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita H, Avraham S, Jiang S, London R, Van Veldhoven PP, Subramani S, Rogers RA, Avraham H. Characterization of human and murine PMP20 peroxisomal proteins that exhibit antioxidant activity in vitro. J. Biol. Chem. 1999;274:29897–29904. doi: 10.1074/jbc.274.42.29897. [DOI] [PubMed] [Google Scholar]
- 35.Knoops B, Clippe A, Bogard C, Arsalane K, Wattiez R, Hermans C, Duconseille E, Falmagne P, Bernard A. Cloning and characterization of AOEB166, a novel mammalian antioxidant enzyme of the peroxiredoxin family. J. Biol. Chem. 1999;274:30451–30458. doi: 10.1074/jbc.274.43.30451. [DOI] [PubMed] [Google Scholar]
- 36.Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 37.Beckstead RB, Lam G, Thummel CS. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005;6:R99. doi: 10.1186/gb-2005-6-12-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romer F, Emmerich H, Nowock J. Biosynthesis of ecdysones in isolated prothoracic glands and oenocytes of Tenebrio molitor in vitro. J. Insect Physiol. 1974;20:1975–1987. doi: 10.1016/0022-1910(74)90105-x. [DOI] [PubMed] [Google Scholar]
- 39.Ferveur JF, Savarit F, O’Kane CJ, Sureau G, Greenspan RJ, Jallon JM. Genetic feminization of pheromones and its behavioral consequences in Drosophila males. Science. 1997;276:1555–1558. doi: 10.1126/science.276.5318.1555. [DOI] [PubMed] [Google Scholar]
- 40.Romer F, Gnatzy W. Arachnid oenocytes: ecdysone synthesis in the legs of harvestmen (Opilionidae) Cell Tissue Res. 1981;216:449–453. doi: 10.1007/BF00233631. [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 42.Klichko VI, Radyuk SN, Orr WC. Profiling catalase gene expression in Drosophila melanogaster during development and aging. Arch. Insect Biochem. Physiol. 2004;56:34–50. doi: 10.1002/arch.10142. [DOI] [PubMed] [Google Scholar]
- 43.Radyuk SN, Klichko VI, Orr WC. Profiling Cu,Zn-superoxide dismutase expression in Drosophila melanogaster–a critical regulatory role for intron/exon sequence within the coding domain. Gene. 2004;328:37–48. doi: 10.1016/j.gene.2003.12.016. [DOI] [PubMed] [Google Scholar]