Abstract
Resistance emerging after pefloxacin therapy was investigated in an experimental Enterobacter cloacae infection. Mice were inoculated intraperitoneally (mean inoculum, 0.9 X 10(8) CFU) with one of four strains initially susceptible to quinolones and treated with a single 25-mg/kg dose of pefloxacin. This therapy produced a net decrease of bacterial counts in the peritoneal fluid, but with the of the isolates, posttherapy (PT1) strains emerged with decreased susceptibilities to quinolones (4- to 1,024-fold), to the structurally unrelated antibiotics (4- to 16-fold) chloramphenicol and trimethoprim, and sometimes to tetracycline and beta-lactam compounds. In a second set of experiments, new mice were similarly infected with PT1 strains and treated with up to five 25-mg/kg doses of pefloxacin. Compared with parent isolates, PT1 strains produced similar disease and peritoneal bacterial count in the control animals. In treated mice posttherapy (PT2) strains emerged that showed 8- to 64-fold increases in quinolone MICs compared with the PT1 strains inoculated. All PT1 and PT2 strains showed altered outer membrane protein patterns, principally marked by a decreased 37,000-molecular-weight band generally accompanied by an increased 42,000-molecular-weight band. Whole cells from all PT1 and PT2 strains, exposed to [14C]pefloxacin for 15 to 60 s, bound significantly less radioactivity than the corresponding parent strains. After partial purification, DNA gyrase extracted from the most resistant isolates (one PT1 and the PT2 strains) showed a 100- to 450-fold 50% inhibitory concentration increase for pefloxacin. Altogether, pefloxacin can select in vivo two types of resistant strain, one with only decreased permeability and another with decreased permeability combined with altered DNA gyrase.
Full text
PDF

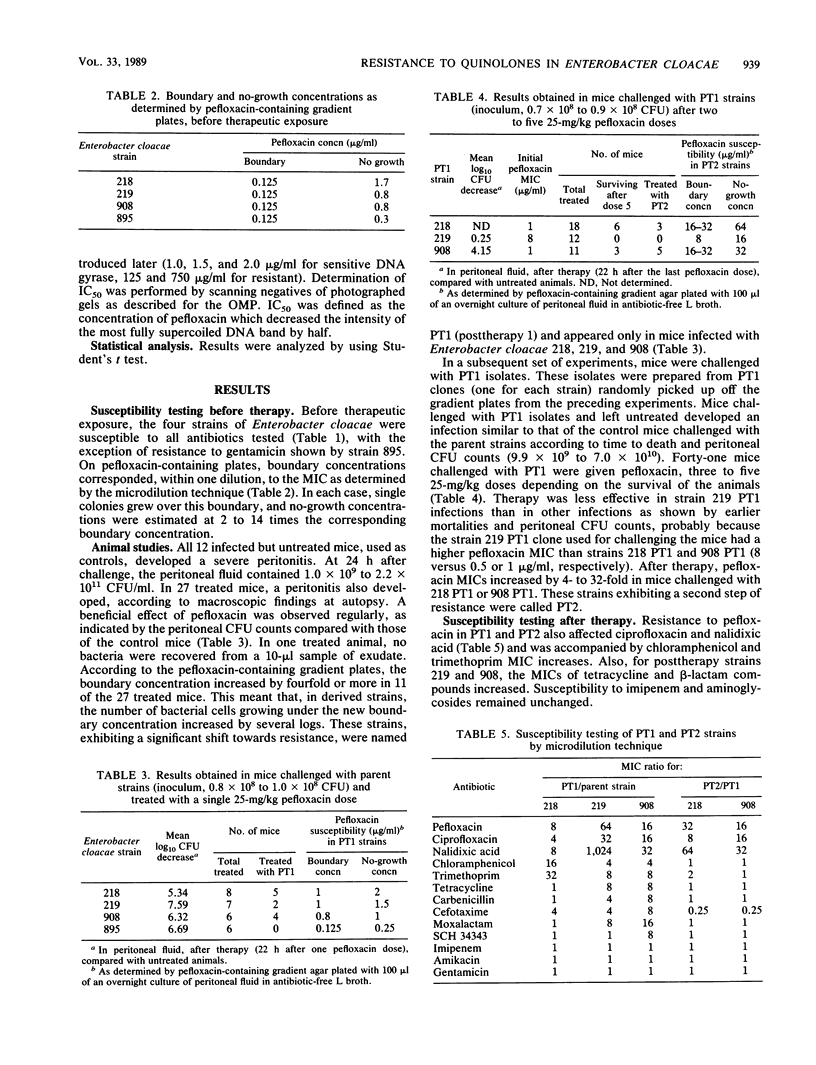
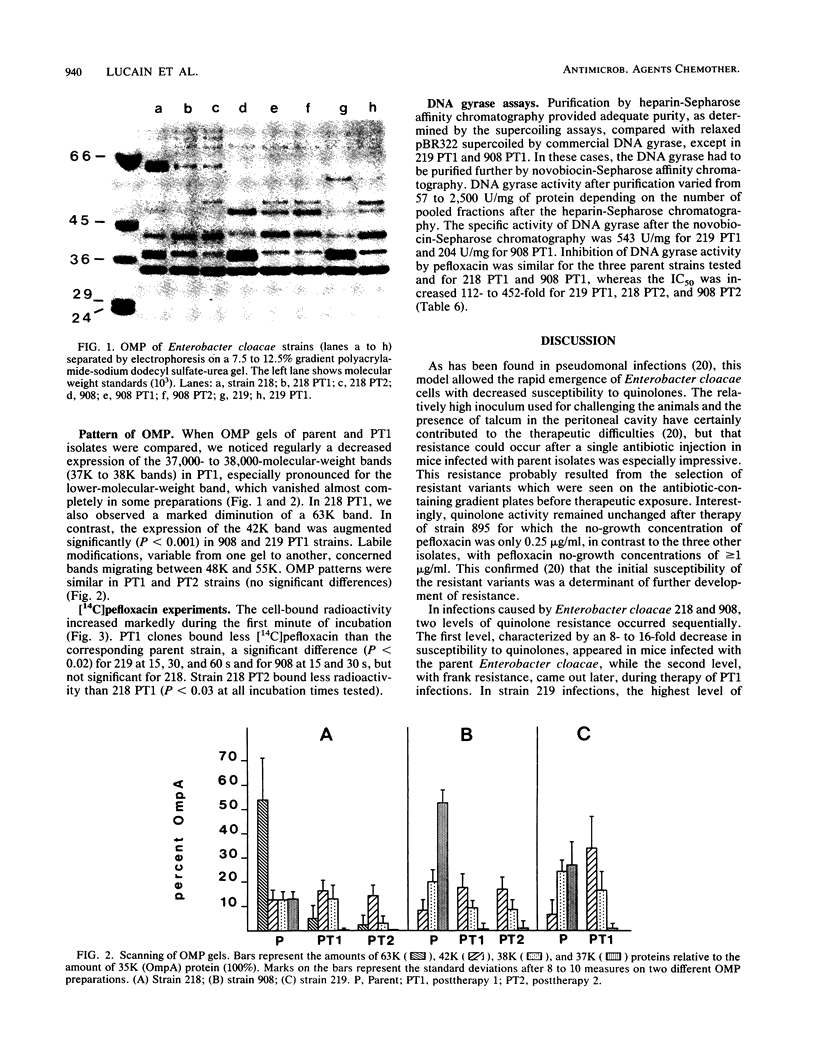
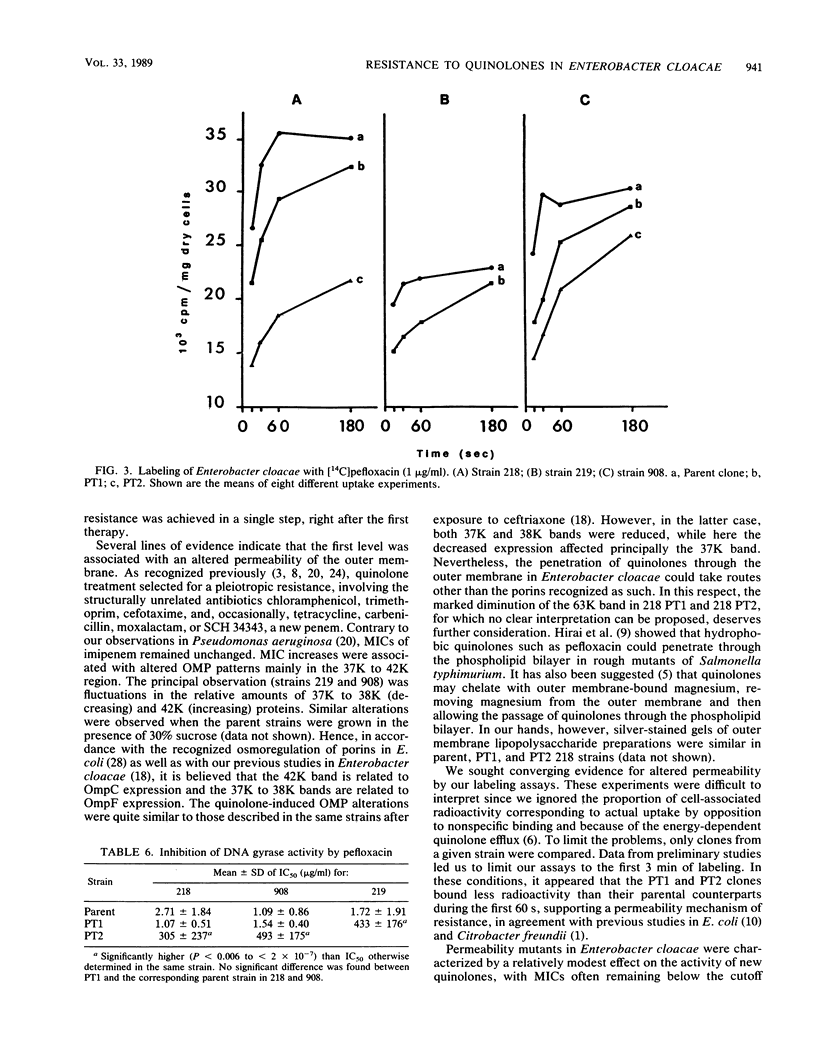


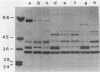
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alphen W. V., Lugtenberg B. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J Bacteriol. 1977 Aug;131(2):623–630. doi: 10.1128/jb.131.2.623-630.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama H., Fujimaki K., Sato K., Fujii T., Inoue M., Hirai K., Mitsuhashi S. Clinical isolate of Citrobacter freundii highly resistant to new quinolones. Antimicrob Agents Chemother. 1988 Jun;32(6):922–924. doi: 10.1128/aac.32.6.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama H., Sato K., Fujii T., Fujimaki K., Inoue M., Mitsuhashi S. Purification of Citrobacter freundii DNA gyrase and inhibition by quinolones. Antimicrob Agents Chemother. 1988 Jan;32(1):104–109. doi: 10.1128/aac.32.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama H., Sato K., Kato T., Hirai K., Mitsuhashi S. Norfloxacin resistance in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1987 Oct;31(10):1640–1641. doi: 10.1128/aac.31.10.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A. S., Hirano L., Yih J. Development of beta-lactam resistance and increased quinolone MICs during therapy of experimental Pseudomonas aeruginosa endocarditis. Antimicrob Agents Chemother. 1988 Feb;32(2):231–235. doi: 10.1128/aac.32.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. S., Georgopapadakou N. H. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother. 1988 Apr;32(4):438–442. doi: 10.1128/aac.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., Hooper D. C., Wolfson J. S., Souza K. S., McMurry L. M., Levy S. B. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988 Aug;32(8):1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann L., Williamson R., Moreau N., Kitzis M. D., Collatz E., Acar J. F., Goldstein F. W. Cross-resistance to nalidixic acid, trimethoprim, and chloramphenicol associated with alterations in outer membrane proteins of Klebsiella, Enterobacter, and Serratia. J Infect Dis. 1985 Mar;151(3):501–507. doi: 10.1093/infdis/151.3.501. [DOI] [PubMed] [Google Scholar]
- Hirai K., Aoyama H., Irikura T., Iyobe S., Mitsuhashi S. Differences in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob Agents Chemother. 1986 Mar;29(3):535–538. doi: 10.1128/aac.29.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K., Aoyama H., Suzue S., Irikura T., Iyobe S., Mitsuhashi S. Isolation and characterization of norfloxacin-resistant mutants of Escherichia coli K-12. Antimicrob Agents Chemother. 1986 Aug;30(2):248–253. doi: 10.1128/aac.30.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper D. C., Wolfson J. S., Ng E. Y., Swartz M. N. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987 Apr 27;82(4A):12–20. [PubMed] [Google Scholar]
- Hooper D. C., Wolfson J. S., Souza K. S., Tung C., McHugh G. L., Swartz M. N. Genetic and biochemical characterization of norfloxacin resistance in Escherichia coli. Antimicrob Agents Chemother. 1986 Apr;29(4):639–644. doi: 10.1128/aac.29.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Ohue T., Yamagishi J., Nakamura S., Shimizu M. Mode of incomplete cross-resistance among pipemidic, piromidic, and nalidixic acids. Antimicrob Agents Chemother. 1978 Aug;14(2):240–245. doi: 10.1128/aac.14.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Sato K., Fujii T., Hirai K., Inoue M., Iyobe S., Mitsuhashi S. Some properties of subunits of DNA gyrase from Pseudomonas aeruginosa PAO1 and its nalidixic acid-resistant mutant. J Bacteriol. 1987 May;169(5):2322–2325. doi: 10.1128/jb.169.5.2322-2325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaatz G. W., Barriere S. L., Schaberg D. R., Fekety R. The emergence of resistance to ciprofloxacin during treatment of experimental Staphylococcus aureus endocarditis. J Antimicrob Chemother. 1987 Nov;20(5):753–758. doi: 10.1093/jac/20.5.753. [DOI] [PubMed] [Google Scholar]
- Kocher O., Skalli O., Cerutti D., Gabbiani F., Gabbiani G. Cytoskeletal features of rat aortic cells during development. An electron microscopic, immunohistochemical, and biochemical study. Circ Res. 1985 Jun;56(6):829–838. doi: 10.1161/01.res.56.6.829. [DOI] [PubMed] [Google Scholar]
- Marchou B., Bellido F., Charnas R., Lucain C., Pechère J. C. Contribution of beta-lactamase hydrolysis and outer membrane permeability to ceftriaxone resistance in Enterobacter cloacae. Antimicrob Agents Chemother. 1987 Oct;31(10):1589–1595. doi: 10.1128/aac.31.10.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchou B., Michea-Hamzehpour M., Lucain C., Pechère J. C. Development of beta-lactam-resistant Enterobacter cloacae in mice. J Infect Dis. 1987 Aug;156(2):369–373. doi: 10.1093/infdis/156.2.369. [DOI] [PubMed] [Google Scholar]
- Michéa-Hamzehpour M., Auckenthaler R., Regamey P., Pechère J. C. Resistance occurring after fluoroquinolone therapy of experimental Pseudomonas aeruginosa peritonitis. Antimicrob Agents Chemother. 1987 Nov;31(11):1803–1808. doi: 10.1128/aac.31.11.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter R., Cozzarelli N. R. Escherichia coli DNA gyrase. Methods Enzymol. 1983;100:171–180. doi: 10.1016/0076-6879(83)00053-1. [DOI] [PubMed] [Google Scholar]
- Robillard N. J., Scarpa A. L. Genetic and physiological characterization of ciprofloxacin resistance in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1988 Apr;32(4):535–539. doi: 10.1128/aac.32.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. C., Sanders W. E., Jr, Goering R. V., Werner V. Selection of multiple antibiotic resistance by quinolones, beta-lactams, and aminoglycosides with special reference to cross-resistance between unrelated drug classes. Antimicrob Agents Chemother. 1984 Dec;26(6):797–801. doi: 10.1128/aac.26.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Then R. L., Angehrn P. Trapping of nonhydrolyzable cephalosporins by cephalosporinases in Enterobacter cloacae and Pseudomonas aeruginosa as a possible resistance mechanism. Antimicrob Agents Chemother. 1982 May;21(5):711–717. doi: 10.1128/aac.21.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J. S., Hooper D. C., Ng E. Y., Souza K. S., McHugh G. L., Swartz M. N. Antagonism of wild-type and resistant Escherichia coli and its DNA gyrase by the tricyclic 4-quinolone analogs ofloxacin and S-25930 stereoisomers. Antimicrob Agents Chemother. 1987 Nov;31(11):1861–1863. doi: 10.1128/aac.31.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi J., Yoshida H., Yamayoshi M., Nakamura S. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol Gen Genet. 1986 Sep;204(3):367–373. doi: 10.1007/BF00331012. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Kojima T., Yamagishi J., Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet. 1988 Jan;211(1):1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]