Abstract
Recent research on plant responses to bacterial attack has identified extracellular and intracellular host receptors that recognize conserved pathogen-associated molecular patterns and more specialized virulence proteins, respectively. These findings have shed light on our understanding of the molecular mechanisms by which bacteria elicit host defences and how pathogens have evolved to evade or suppress these defences.
Plants are a rich source of nutrients and water for microbes, and they are infected by many bacterial pathogens from both the Proteobacteria and Actinobacteria phyla (Supplementary information 1 (table)). Because of their broad host range, serious economic consequences and experimental tractability, the most intensively studied bacteria are members of the Proteobacteria phylum (such as Agrobacterium, Erwinia, Pseudomonas, Ralstonia and Xanthomonas)1–4. These pathogens are spread by wind, rain, insects or cultivation practices. They enter plant tissues either by wounds or through natural openings such as lenticels, hydathodes or stomata5, and they occupy the inter cellular spaces (apoplast) of various plant tissues or the xylem.
Plant-pathogenic members of the Proteobacteria cause diverse disease symptoms, including specks, spots, blights, wilts, galls and cankers, and they can cause host-cell death in roots, leaves, flowers, fruits, stems and tubers (FIG. 1). These symptoms affect both yield and quality of agricultural crops and bacterial diseases can have serious economic, social and even political consequences6–8. Control of bacterial diseases is only partially effective and consists of copper-based sprays, antibiotics, biocontrol strategies, large-scale removal of infected plants and, most importantly, host genetic resistance5. Therefore, research on bacterial diseases of plants helps to elucidate fundamental aspects of microbial pathogenesis and associated host responses and also to develop more effective and sustainable disease-control methods.
Figure 1. Disease symptoms caused by some bacterial pathogens of plants and representative virulence mechanisms used by these pathogens.
Top panels (left to right): bacterial speck of tomato caused by Pseudomonas syringae pathovar (pv.) tomato; crown gall of grape caused by Agrobacterium tumefaciens; blackleg of potato caused by Erwinia carotovora subspecies atroseptica; and bacterial wilt of tomato caused by Ralstonia solanacearum. Bottom panels (left to right): P. syringae pv. tomato enters the leaf apoplastic space through stomata or wounds, and uses a type III secretion system to inject a large number of virulence (effector) proteins into the plant cell. Agrobacterium tumefaciens uses a type IV secretion system to inject a tumour-inducing transfer DNA (tDNA) into the plant cell cytoplasm. This tDNA is integrated into the plant genome and leads to the development of crown gall disease. Erwinia carotovora subspecies atroseptica uses a type II secretion system to deliver cell wall-degrading enzymes (for example, cellulases and pectinases) to the plant cell wall. Ralstonia solanacearum enters plant roots through wounds and multiplies in the xylem vessels in which it produces exopolysaccharides that are believed both to interfere with recognition and to inhibit water transport through the vascular system. Each of these four pathogens also uses other virulence mechanisms (Supplementary information 1 (table)). Ti, tumour inducing. Photo credits for top panels, left to right: G.B.M., T. Burr, A. Charkowski and P. Frey.
Plant-pathogenic bacteria use virulence strategies that are either specialized to plant tissues or are broadly conserved among pathogens of both plants and animals (FIG. 1). Bacterial virulence is manifested as increases in the rate of growth or final population size, as well as by enhanced disease symptoms, which promote the spread of the pathogen through the plant or the broader environment.
Unlike mammals, plants have a complex cell wall that bacteria must surmount to gain access to water and nutrients. Bacteria attack this barrier with extracellular virulence factors, such as cell wall degrading enzymes, and bypass it by the secretion of cell wall-permeable toxins5. However, perhaps the most effective virulence strategy, and one shared with animal bacterial pathogens, is to breach the wall by use of the type III secretion system (T3SS), an elaborate protein-delivery system that consists of more than 20 proteins9. The T3SS delivers into the plant cell a wide array of proteins, called effectors. The activities of effector proteins are just now beginning to be understood and probably hold significant clues about how the pathogen disrupts host signalling and commandeers host metabolism for its own benefit10.
A pivotal development in the past three years has been the elucidation of an essentially complete inventory of type III effectors that are present in several plant-pathogenic bacteria1,11,12. This work revealed that these bacteria express a far larger number of type III effectors with greater sequence diversity than bacterial pathogens of animals, and it has opened up exciting new possibilities for investigating how these bacterial pathogens manipulate host processes to promote virulence10. In this article, we introduce the range of virulence strategies that are used by plant-pathogenic bacteria. We then discuss our current understanding of the roles of pathogen-associated molecular patterns (PAMPs) and pathogen recognition receptors (PRRs) in the activation of plant basal defence responses. Last, we turn to a series of recent developments that are shedding light on the sophisticated molecular mechanisms that are used by bacterial pathogens to interfere with PRR-mediated basal defences and to manipulate other important plant processes to promote pathogenesis.
Overview of bacterial virulence factors
Secreted proteins
Bacterial pathogens contain a well stocked armoury of virulence factors that facilitate their growth and disease-causing capabilities in plant tissues. An important step of bacterial pathogenesis is the delivery of virulence proteins from the bacterium into the plant’s apoplast or cytoplasm. Indeed, in many early genetic screens for mutants of bacterial pathogenesis, mutations that disrupted the function of protein-secretion systems were identified rather than effector proteins and enzymes that are direct modulators of plant biology (reviewed in REF. 13).
Three distinct protein-secretion pathways have been extensively studied in plant pathogens. The type II secretion system (T2SS), or the Out system, is essential for microbes with a soft-rotting lifestyle, characteristic of bacteria in the genus Erwinia14,15 (FIG. 1). Using a two-step process, the T2SS exports enzymes that are involved in degrading the plant cell wall, including pectinases, endo-glucanases and cellulases. These and other exoenzymes are believed to be responsible for causing the rotting and macerating phenotypes that are associated with these pathogens.
Perhaps the most widely studied secretion system in plant pathogens is the T3SS. The T3SS is related to the bacterial flagellum, and forms a pilus that injects effectors into the plant cell. Inside the plant cell, these effectors modulate the plant’s physiology to benefit the pathogen11. Bacteria of different lifestyles, including biotrophic, soft-rotting bacterial pathogens, and even some symbiotic bacteria, rely on the T3SS to successfully interact with their hosts. The effectors delivered by the T3SS have a prominent role in promoting the virulence of pathogenic bacteria in plants and animals10,11,15,16. There is a great diversity of effectors both within and among bacterial species based on sequence-level comparisons12; over thirty effectors are likely to be delivered by Pseudomonas syringae pathovar (pv.) tomato (Pst)12,17. These effectors have diverse enzymatic activities, such as cysteine protease18–21, ubiquitin-like protease22,23, E3 ubiquitin ligase24,25 and protein phosphatase activity26,27, and studies of subcellular localization and host-mediated post-translational modifications have provided further clues regarding effector function28–30. However, most effectors have no sequence similarity to known proteins and their functions remain unknown.
The type IV secretion system (T4SS) has a critical role in the pathogenesis of Agrobacterium tumefaciens and its capability to form galls on plants (FIG. 1). Related to the bacterial F-pilus, the T4SS mediates the trafficking of bacterial proteins and DNA into the plant cell31. The bacterial DNA is integrated into the host genome and produces plant hormones that induce the characteristic gall symptoms. It also promotes the biosynthesis of nutrient-rich opine compounds that can be catabolized by A. tumefaciens but not by most other organisms. Several bacterial proteins are transported through the T4SS to enable the efficient transfer and integration of bacterial DNA31–33. It is important to note that many pathogens rely on multiple mechanisms of protein secretion13. For example, many Erwinia species require both a T2SS and a T3SS to cause disease15, and several strains of Xanthomonas have T2SS, T3SS and T4SS34.
Small molecules as virulence factors
Small molecules, such as toxins, plant hormones, autoinducers and exopolysaccharides (EPS), are used by bacteria to promote disease. Bacterial toxins that have an important role in virulence and symptom development include: coronatine, syringomycin, syringopeptin, tabtoxin and phaseolotoxin35. Using diverse mechanisms of action, including mimicking plant hormones, forming pores in plant membranes or inhibiting host metabolic enzymes, these toxins can cause necrotic or chlorotic symptoms on affected plants. Many strains of Pseudomonas and Xanthomonas produce the plant hormone auxin36. Recently, it was discovered that plants gain enhanced disease resistance by downregulating auxin levels in response to pathogen challenge37. Therefore, it is possible that bacterial-derived auxin might function to counter this plant response to suppress plant defences. Bacteria also produce hormone-like molecules called autoinducers to detect the local population density of a particular bacterial strain or species38. This process, quorum sensing, is believed, among other things, to enable bacteria to regulate their gene expression such that they only induce the expression of virulence factors when they have reached high enough levels to effectively parasitize the plant. Many phytobacteria, including bacteria in the genera Ralstonia and Xanthomonas, secrete large amounts of EPS, which are high molecular-mass sugar molecules that can clog the xylem and cause characteristic wilting symptoms39 (FIG. 1). EPS enhances pathogen virulence, perhaps by protecting the bacteria from antimicrobial environments in the plant, such as antimicrobial factors that might be present in the xylem. In many cases, the above, diverse mechanisms work together to promote pathogenesis. For example, quorum sensing was recently shown to regulate the formation of EPS in the pathogen Pantoea stewartii subspecies stewartii40, indicating that temporal control of EPS expression is important for bacterial pathogenesis.
Bacterial elicitation of host basal defences
Plants, like mammals and invertebrates, have evolved PRRs, which function to recognize certain PAMPs41,42. PAMPs are important molecules for the microbial lifestyle, and they contain a conserved structural feature that is recognized by a PRR. Recognition of a PAMP activates several early, ‘frontline’ plant defences against bacterial pathogens (BOX 1)41,43–45. Recognition capacity for certain PAMPs such as flagellin is conserved among diverse plant taxa, whereas the perception of others such as cold-shock protein and elongation factor Tu (EF-Tu) is limited to only certain plant families46–48.
Box 1. Basal and R-gene-mediated defences in plants.
Plants lack mammalian-like adaptive immunity and therefore their various inducible defence responses are collectively referred to as ‘innate immunity’. Plant responses to pathogen attack can be differentiated into ‘basal’ and ‘resistant (R)-gene-mediated’ defences. These two defence responses can be distinguished experimentally by several assays, but they share some similar features and might even share some common molecular mechanisms62.
Basal defences occur early in the plant–pathogen interaction (<10 minutes after contact) in response to the perception by plant pattern recognition receptors (PRRs) of extracellular pathogen-associated molecular patterns (PAMPs). They are elicited experimentally by exposing whole leaves, suspension cells or protoplasts to bacteria or purified PAMPs such as flagellin, lipopolysaccharide or elongation factor Tu (EF-Tu)45–47. Assays for basal defences in suspension cell or protoplast systems include detection of increased extracellular pH (caused by a rapid efflux of K+), increases in Ca2+, ethylene, reactive oxygen and nitrogen species, activation of mitogen-activated protein kinases (MAPKs) and increased expression of certain genes (for example, FRK1 and WRKY29)48,51,52. Several of these assays are also applied to intact plants and can detect other phenotypes that are associated with basal defence, such as callose deposition at the cell periphery, exclusion of certain dyes from the vascular system and the inhibition of seedling growth50,67,110.
R-gene-mediated defences are typically detectable later in the plant–pathogen interaction (2–3 hours) after the delivery of type III effectors into the host cytoplasm78. They are elicited experimentally by either the inoculation of R-gene-expressing leaves with bacteria that are expressing a cognate effector gene or by the expression of an effector transgene in the plant cell through transient agroinfiltration or particle bombardment. Depending on the resistancegene–effector-gene combination, these responses, like basal defence, might include the generation of reactive oxygen and nitrogen species, changes in gene expression and activation of MAPKs 45,111,112. However, the most characteristic feature of R-gene-mediated defences, which is generally not associated with basal defence, is the development of localized programmed cell death (the hypersensitive response)77,113.
Plant perception of flagellin
The best characterized phytobacterial PAMP is flagellin, a structural component of the bacterial flagellum43,46. In a series of seminal studies, a 22-amino-acid epitope of flagellin, flg22, was found to be recognized by the Arabidopsis FLS2 leucine-rich repeat (LRR) receptor kinase46,49,50 (FIG. 2). Treatment of Arabidopsis leaves with flg22 activates multiple defence responses, including mitogen-activated protein kinase (MAPK, also abbreviated MPK) cascades51 (BOX 1), and decreases growth of subsequently inoculated Pst52,53. Plants that lack FLS2 are more susceptible to Pseudomonas infection when the pathogen is sprayed on plant leaves but not when it is infiltrated directly into the apoplast. This finding indicates that FLS2 can function at an early stage to interfere with bacterial entry into the apoplast53.
Figure 2. Model depicting the activation of PRR-mediated basal defences and their suppression by type III effectors.
Plants possess plasma-membrane-localized pattern recognition receptors (PRRs) that consist of an extracellular leucine-rich repeat (LRR) domain and a cytoplasmic serine/threonine kinase domain. In Arabidopsis, FLS2 and elongation factor Tu (EF-Tu) receptor (EFR) are PRRs that recognize the flg22 peptide of flagellin or the elf18 peptide of EF-Tu, respectively 47,49,54,59,119. Other bacterial elicitors such as lipopolysaccharide (LPS), harpins, and cold-shock protein have been identified11,45,48,120. PRR activation triggers signalling events that lead to the upregulation of over 300 plant genes51,52,56,57. A complete mitogen-activated protein kinase (MAPK) pathway and several WRKY transcription factors that function downstream of FLS2 and induce the expression of genes such as FRK1 and NHO1 have been identified51. Phenotypes that are associated with activated basal defences include cell wall fortifications and the production of reactive oxygen species (ROS) and nitrogen species (NO). Delivery of effector proteins through the type III secretion system (T3SS) into plant cells is one strategy that is used by bacteria to suppress PRR-mediated defences. As many as 16 effectors have been identified that suppress basal defences42,65–68,110. The model highlights the effector AvrPto that is required for suppressing the recognition of flg22 and other pathogen-associated molecular patterns (PAMPs)42,87. In Arabidopsis, AvrPto functions upstream of MAPK kinase kinase (MAPKKK), indicating that it targets components that function early on in the PRR pathway42. The residue I96, which resides within an extended Ω-loop, is required for the basal defence suppression of AvrPto in Arabidopsis. See text for more details.
Two recent studies address how FLS2 recognizes flg22 and the molecular mechanisms downstream of this receptor in plant cells. In the first report, the FLS2 protein was shown by crosslinking and immunoprecipitation to directly bind to the flg22 peptide54. Specific features of this binding capacity were observed upon heterologous expression of the Arabidopsis FLS2 in tomato cells. A point mutation in one LRR of FLS2 abolishes binding to flg22. However, it remains to be established whether the LRR region is sufficient for flg22 binding or whether additional host proteins contribute to this process.
A second report investigated the tissue-specific expression and subcellular localization of a GFP-tagged FLS2 protein55. FLS2 localized in roots, stems and flowers. Consistent with a role in early pathogen detection, FLS2 was also present in leaf epidermal cells and stomatal guard cells — typical entry points for bacterial pathogens. Specific exposure of Arabidopsis cells to flg22 induces FLS2 receptor internalization and accumulation in intracellular vesicles, from where it subsequently disappears. A possible role for FLS2 autophosphorylation during endocytosis was supported by the fact that the mutation of a threonine located in the juxtamembrane region of the protein did not affect binding to flg22, but did significantly reduce FLS2 internalization. The mechanism of FLS2 internalization and degradation and the possible role for its endocytosis in FLS2 signalling remains unknown.
The signalling events and gene-expression changes that occur after flagellin recognition have been investigated in several studies51,52,56,57. MAPK cascades have an important role in transmitting PAMP recognition to the plant cell51,58. Arabidopsis mutants that are defective in salicylic acid, jasmonic acid or ethylene signalling retain flagellin-mediated resistance to bacterial infection, indicating either some redundancy among these pathways or that previously uncharacterized pathways mediate PRR signalling53. Three extensive studies of PAMP-induced transcriptional reprogramming52,56,57 found rapid induction of genes that encode transcription factors, proteins that are associated with protein degradation, hormone-related proteins, phosphatases and diverse protein kinases including a large number of LRR-receptor-like kinases (RLKs). Also notable, because PAMPs elicit host cell wall fortifications, was the identification of a large numbers of genes that are associated with cell wall reorganization, crosslinking and various secretory pathways. Many of these processes seem to be targets for downregulation by bacterial type III effectors (see discussion below).
Plant perception of elongation factor
Arabidopsis plants also respond to a second bacterial PAMP, EF-Tu47. EF-Tu is the most abundant protein in bacterial cells, and an N-acetylated 18-amino-acid region (elf18) from the N terminus of this protein from Escherichia coli was sufficient to elicit FLS2-like basal defences and to trigger Arabidopsis resistance to Pst when it was applied prior to infection59. Transcriptional profiling showed that many similar plant genes are induced after treatment of Arabidopsis with either flg22 or elf18. One of these genes, encoding an LRR-RLK that is similar to FLS2, was found to encode the EF-Tu receptor (EFR) 59. The expression of EFR in the wild tobacco species Nicotiana benthamiana, that is normally unresponsive to EF-Tu, resulted in recognition of elf18 and activation of typical basal defences. Remarkably, compared to wild-type plants, Arabidopsis leaves with a mutated Efr were significantly more susceptible to Agrobacterium transformation59. This discovery raises the exciting possibility that PRRs might be manipulated both to increase the efficiency of Agrobacterium-mediated genetic engineering of certain crop plants and to enhance plant resistance to bacterial pathogens.
Bacterial responses to host basal defences
Evasion of host basal defences
One plausible strategy to evade PRR-mediated detection is for bacteria to evolve unrecognizable PAMPs or to lose them entirely. However, PAMPs seem to be targeted by PRRs because they are highly conserved and indispensable bacterial features. Therefore, bacteria are limited to alterations that do not significantly diminish PAMP function. Despite these constraints, several studies have discovered variations in PAMPs, including post-translational modifications, between bacterial species that prevent detection by host PRRs46,60,61.
Significant variations in PAMPs can also occur at the subspecies level. Flagellin proteins isolated from twelve strains of the black rot pathogen Xanthomonas campestris pv. campestris (Xcc) vary dramatically in their capability to elicit FLS2-mediated responses in Arabidopsis62. A single amino-acid polymorphism in the flg22 region of flagellin is primarily responsible for the observed variability. Furthermore, the eliciting activity of each Xcc flagellin directly correlates with the capability of individual Xcc strains to grow on susceptible Arabidopsis plants. These data indicate that variations in flagellin allow Xcc to avoid PRR-mediated detection. Surprisingly, FLS2 is not required for the observed variability of growth among Xcc strains on Arabidopsis. However, an FLS2-dependent restriction in Xcc growth was observed if plants were pre-treated with recombinant elicitation-active Xcc flagellins. It seems that despite the presence of elicitation-active flagellin in some Xcc strains, Xcc has developed extra mechanisms to either mask flagellin recognition or suppress FLS2-dependent responses, or both. Despite this complication, this work raises the possibility that PAMP variability might make meaningful contributions to the specificity of plant–bacteria interactions at the subspecies level. It also highlights the difficulty in assigning function to PAMP recognition due to the multiple layers of defence evasion.
Suppression of host basal defences
Limitations on the extent of variation (or loss) of specific PAMPs that can be tolerated by bacteria might be a key driving force in the evolution of more mechanisms to suppress PAMP recognition. Initial observations that otherwise virulent bacteria elicit plant defences if they lack a functional T3SS indicated that effector proteins function in part to suppress plant defences63,64. However, the effector proteins that were responsible for the suppression activity were unknown.
An important breakthrough was the discovery that transgenic Arabidopsis plants that overexpress the Pst effector AvrPto were compromised in their capability to deposit callose at the cell wall when they were challenged with a non-pathogenic T3SS-defective Pst strain65. In addition, a similar group of genes was suppressed in the transgenic AvrPto plants as compared to wild-type plants treated with virulent Pst. Importantly, the overexpression of AvrPto in Arabidopsis allowed increased growth of the T3SS mutant.
In the past two years, the catalogue of effectors that suppress basal defences has expanded dramatically. Nine effectors, including AvrPto, were identified in a screen for Pst effectors that suppress the expression of the flagellin-induced NHO1 gene in Arabidopsis protoplasts66 (FIG. 2). Similar to AvrPto, the expression of some of these effectors in transgenic Arabidopsis resulted in increased growth of a non-pathogenic Pst mutant strain. Effectors that suppress basal defences in N. benthamiana have also been identified67,68.
It remains unknown why so many effectors with similar suppression activities are delivered into the plant cell, whether the suppression activities of effectors are the result of targeting the same host protein or whether interfering with distinct host targets can produce similar phenotypes. Moreover, most studies have used either transgenic plants or protoplast expression systems that overexpress individual effectors to evaluate basal defence suppression. For many effectors, it remains to be seen if the observed suppression activity is biologically relevant when delivered by the T3SS. Identifying the host targets of effectors will be important in determining the specific function of each effector and whether or not their functions are truly redundant.
Host targets of bacterial effectors
Several studies have provided insights into the host proteins and signalling pathways that are targeted by type III effectors to suppress PAMP signalling. Two type III effectors, AvrRpt2 and AvrRpm1, have been shown to inhibit flg22-induced defences in Arabidopsis and promote the growth of T3SS-deficient bacteria69. In previous studies, both AvrRpt2 and AvrRpm1, along with a third effector AvrB, were each shown to interact with RIN4, a negative regulator of resistance (R) protein-mediated defences in Arabidopsis70–72 (as discussed below). The overexpression or absence of RIN4 in Arabidopsis led, respectively, to the inhibition or enhancement of flg22-stimulated callose deposition and growth of T3SS-deficient bacteria. Therefore, RIN4 might be a negative regulator of FLS2 signalling that is targeted by AvrRpt2 and AvrRpm1 to suppress basal defences. Indeed, RIN4 was shown to be cleaved by AvrRpt2, which has cysteine protease activity19,20. Also, AvrRpm1 and the effector AvrB induce the phosphorylation of RIN4 (REF. 71). The mechanism by which the degradation or phosphorylation of this negative regulator affects the inhibition of this signalling pathway remains unclear. Both AvrRpt2 and AvrRpm1 still promote Pseudomonas virulence in plants that lack RIN4, showing that both effectors must have other host targets73–75.
A recent screen for effectors that suppress flg22-induced basal defences provided further insights into the host pathways that are suppressed by effectors. The effectors AvrPto and AvrPtoB (also known as HopAB2) were identified as potent suppressors of FRK1 expression, an Arabidopsis gene that is induced by flg22 treatment42. Both AvrPto and AvrPtoB suppress the activation of the MAPKs MPK3 and MPK6 downstream of several distinct elicitors; however, the effectors were unable to block MPK3 and MPK6 activation due to overexpression of the MAPK kinase kinase MEKK1. These data indicate that both effectors block PAMP signalling upstream of FLS2-dependent MAPK signalling. Ultrastructural analyses and suppression of basal-resistance-associated gene expression studies also showed that AvrPtoB suppresses basal defences76. AvrPtoB only enhanced pathogen growth in specific Arabidopsis ecotypes that lack FLS2, indicating that the capability of AvrPtoB to overcome basal defences might quantitatively depend on the strength of recognition by PRRs. Surprisingly, several effectors that were previously identified as suppressors of FLS2-dependent NHO1 expression66, including AvrRpm1, HopAO1, HopE1, and HopK1, were unable to suppress the expression of FRK1 (REF. 42). These different results might imply that the induction of FRK1 and NHO1 occur through distinct pathways downstream of FLS2, and that each pathway is targeted by only a subset of effectors.
Resistance proteins counteract effectors
Plants have evolved a defence strategy based on disease R genes, which functions, in part, to counteract the suppression of PRR-mediated defences by type III effectors. R genes have been studied for decades because they are easily manipulated by breeders to provide resistance in normally susceptible plant cultivars. In the gene-for-gene model of disease resistance, R genes are only effective if a specific avirulence (avr) gene is present in the pathogen. R-protein-mediated defences include the hypersensitive response (HR), a rapidly induced, localized programmed cell death (PCD; BOX 2) response that is believed to limit pathogen spread77. Over forty R genes have been cloned to date from diverse plant species and their study is an active and broad area of research78. Here we discuss some recent developments that relate to defence against bacterial pathogens.
Box 2. Cell death during plant–bacterium interactions.
Cell death is associated with both immunity and disease susceptibility in plant–pathogen interactions. Hypersensitive response (HR) and disease-associated cell death can both be controlled by common cell death regulators21,113, indicating that the cell death processes might be mechanistically linked. In a resistant plant, HR-based programmed cell death (PCD) is initiated by plant resistance (R)-protein-mediated recognition of avirulence proteins. The cell death is rapid, typically microscopic and localized near the site of recognition, and it kills both the plant cell and the attacking pathogen in the process of limiting pathogen spread77. By contrast, disease-associated cell death is visible, macroscopic cell death that generates many of the characteristic symptoms of bacterial diseases (for example, specks, spots and blights) and it is associated with substantial, 100–10,000-fold multiplication of the pathogen. Therefore, the timing of the cell death is believed to be a key determinant of its role in disease outcome. Although PCD is closely associated with hypersensitive response (HR)-based resistance, it is still not clear if PCD is mechanistically responsible for a successful resistance response. It is possible that HR-associated cell death is a consequence of a strongly activated defence response and that the observed cell death is simply a byproduct of this strongly activated response. Nevertheless, HR-based PCD remains an excellent marker for assessing if a plant can mount a successful resistance response, and it is often an accurate indicator that a defence response is mediated by an R protein.
The guard hypothesis and RIN4
The main class of R proteins are intracellular and have a nucleotide-binding site, leucine-rich repeats (NBS-LRR), with either a coiled-coil domain or a Toll-interleukin-1-like domain at the N terminus. Because most bacterial avr genes encode cytoplasmic type III effectors, it has been postulated that R proteins function as intracellular receptors that directly interact with type III effectors after they are delivered into the host cell. Despite considerable effort by many labs, a direct interaction between an R protein and a type III effector has been identified in only two cases79–81.
This lack of evidence gave rise to the ‘guard’ hypothesis, whereby R proteins are postulated to indirectly detect the presence of Avr proteins (in this case type III effectors) by monitoring effector-mediated changes in host targets, rather than the effectors themselves82. The most direct evidence to support this hypothesis centres around the RIN4 protein in Arabidopsis. The NBS-LRR R proteins RPM1 and RPS2 both interact with RIN4 in uninfected plants70–72. As discussed above, during infection, RIN4 is targeted by the Pseudomonas effectors AvrRpt2, AvrRpm1 or AvrB. Potentially, as a result of AvrRpm1 or AvrB virulence activity, RIN4 becomes phosphorylated71. This phosphorylation event in turn activates RPM1-dependent resistance. On the other hand, RPS2-dependent resistance is activated if RIN4 is degraded by the proteolytic activity of AvrRpt2 (REFS. 19,20,70,72,83). Importantly, RIN4-null plants seem to constitutively activate the RPS2 pathway, indicating that the loss of RIN4 is indeed sufficient to trigger a defence response. These observations together indicate that both RPM1 and RPS2 guard RIN4 and monitor the modifications of RIN4 that occur (either phosphorylation or degradation) as a result of effector activity.
The guard hypothesis and Pto
From the studies described earlier, it is clear that the type III effector AvrPto is an important suppressor of basal defences in Arabidopsis. However, AvrPto not only suppresses basal defences in Arabidopsis, it also contributes to the virulence of P. syringae in tomato84. The tomato R protein, Pto, recognizes AvrPto and the sequence-dissimilar protein AvrPtoB, and provides effective resistance against Pst infection85. In contrast to the typical NBS-LRR R protein, Pto is a cytoplasmic serine/threonine kinase that is believed to interact directly with AvrPto in the plant cell. Resistance to AvrPto also requires a second gene, Prf, that is genetically linked to the Pto locus and encodes an NBS-LRR protein86.
The structure of AvrPto was recently resolved by NMR, and showed that the central region of the 18-kDa effector adopts an α-helical bundle fold, whereas ~30 amino acids at the N terminus and C terminus are unstructured and flexible87. Residues that are required for the interaction with Pto, for example I96, reside within an extended Ω-loop between two α-helices of AvrPto. I96 is required for the basal defence suppression of AvrPto in Arabidopsis, and the overexpression of Pto in Arabidopsis can partially block AvrPto suppression activity42. An attractive hypothesis from these data is that a Pto-like kinase in Arabidopsis is the host protein that is targeted by AvrPto to suppress basal defences. Further experiments are needed to determine if Pto is a host target of AvrPto in susceptible tomato plants.
Bacterial suppression of HR-based PCD
R-protein-mediated recognition of basal defence suppressing effectors presents a strong selective challenge to the invading pathogen because loss of the recognized effector might also cause a significant decrease in its fitness. As a counter strategy to R proteins, it is believed that the pathogen evolved an alternative set of effectors that functions to suppress HR-based immunity 88,89 (FIG. 3). Indeed, at least nine effectors have been described that enable a pathogen to suppress or evade HR-based PCD21,26,27,90–94.
Figure 3. Suppression of R-protein-mediated defences by type III effectors.
Plants have evolved resistance (R) proteins to detect the presence of type III effectors and to signal hypersensitive response (HR)-based defences in response to effectors. Several effectors have been shown to suppress HR-based defences in plants21,26,27,90,91. Here we present a speculative model of how AvrPtoB suppresses HR-based defences. In tomato, the AvrPtoB N terminus is recognized by the R protein Rsb in a Prf-dependent manner to signal the HR. The AvrPtoB C terminus encodes E3 ubiquitin (Ub) ligase activity24,25, which includes a conserved E2 Ub-conjugating enzyme binding site. AvrPtoB might function as a scaffold to bind both to a tomato E2 Ub-conjugating enzyme and a positive regulator of HR-based programmed cell death (PCD). The E2 Ub-conjugating enzyme might then ubiquitylate the substrate to target it for degradation or alter the substrate’s localization, therefore interfering with HR-based signalling.
The effector protein VirPphA (also known as HopAB1) was the first example of an effector that modulated HR-based resistance92. It was observed that a normally virulent strain of P. syringae pv. phaseolicola (Pph) elicited the HR on beans when it was cured of a large plasmid. VirPphA was identified as the plasmid-borne gene that enabled Pph to evade HR-based resistance. Studies with the related Pst DC3000 effector protein AvrPtoB revealed that effectors might suppress HR-based immunity by suppressing PCD90,95 (BOX 2).
AvrPtoB overexpression in N. benthamiana leaves suppresses PCD that is otherwise elicited by diverse PCD inducers, indicating that this protein does not simply target R-protein-mediated signalling pathways. The anti-PCD activity was associated with the C terminus of AvrPtoB, and mutations generated in AvrPtoB that abrogated anti-PCD activity also caused a normally virulent strain of DC3000 to elicit resistance on previously susceptible tomato plants. This finding showed that the activity of a tomato R gene, termed Rsb, was normally hidden by the translocation of AvrPtoB into the plant cell, and indicates that other R-gene activities might be hidden by HR-suppressing effectors.
The enzymatic activities of several HR suppressing effectors indicate that these proteins are modifying or degrading targets in signalling pathways that are associated with the HR. In the case of the Pst effector HopAO1, its tyrosine phosphatase activity implicates MAPKs as likely host targets because tyrosine phosphorylation in plants is almost exclusively associated with MAPK signalling58.
Recently, we identified that AvrPtoB has E3 ubiquitin ligase activity in vitro and that this activity is required for suppression of HR-based PCD and plant immunity24,25. A crystal structure of AvrPtoB showed that the C terminus shares remarkable homology with the RING and U-box family of eukaryotic E3 ubiquitin ligases. The residues that are required for U-box E3 ubiquitin ligase function are conserved in AvrPtoB and are required for AvrPtoB E3 ubiquitin ligase activity and its capability to suppress plant immunity, which indicated that AvrPtoB might function as an E3 ubiquitin ligase in vivo. In a simple model, AvrPtoB can ubiquitylate and thereby induce the degradation of a host component that is required for HR-based resistance (FIG. 3). The discovery that AvrPtoB targets the host ubiquitin system highlights the importance of the ubiquitin pathway in disease resistance and cell death. Indeed, several recent papers show that plant U-box E3 ubiquitin ligases regulate plant cell death and immunity 96–98.
AvrPto and AvrPtoB share the virulence function of suppressing basal defences. This discovery is consistent with observations that single DC3000 deletion mutants of AvrPto or AvrPtoB exhibit similar levels of virulence99. AvrPtoB E3 ubiquitin ligase activity is probably not required for the suppression of basal defences, because mutations in the residues that are required for AvrPtoB E3 ubiquitin ligase activity and anti-PCD function do not abrogate the capability of AvrPtoB to suppress basal defences42,76. Structural data also support an E3-independent mechanism for basal defence suppression, because the AvrPto structure shows no homology to E3 ubiquitin ligases87. It is possible that an unidentified activity that suppresses basal defences is present in the AvrPtoB N terminus. Some homologues of AvrPtoB only have similarity to the N terminus of AvrPtoB100,101, indicating that the N terminus of AvrPtoB has a virulence activity that is independent of the C-terminal anti-PCD activity102.
Bacterial manipulation of hormone pathways
Plant hormones can quickly and potently affect plant physiology; therefore it is not surprising that pathogens manipulate plant hormone signalling to promote disease. The Pst toxin coronatine functions as a methyl-jasmonate homologue to alter jasmonic acid (JA)-dependent plant responses. Microarray experiments show that coronatine dramatically reprogrammes host gene expression, causing altered expression of hundreds of genes56,103, including the upregulation of genes that are involved in the synthesis of endogenous JA. Coronatine-dependent reprogramming of plant gene expression has been shown to induce systemic susceptibility to bacterial pathogens104, demonstrating that effector-mediated hormone regulation can broadly function as a virulence mechanism. Type III effectors have also been shown to modulate JA signalling to inhibit plant defence105,106.
AvrPto and AvrPtoB have also been shown to enhance the expression of the ethylene-forming enzyme ACC oxidase gene in susceptible tomato plants107. Ethylene is required for disease-associated cell death in plants, so it is possible that AvrPto and AvrPtoB induce the expression of ethylene to cause late-onset cell death that might enable better access to nutrients or improve dissemination in the environment.
Conclusions
Emerging models of plant immunity
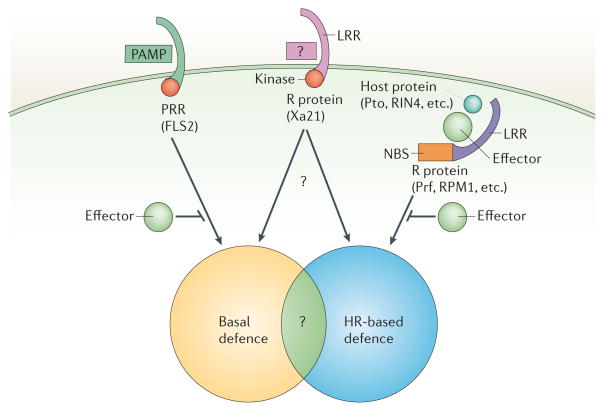
Recent advances have highlighted the role of PRR-mediated activation of basal defences as an important barrier to pathogen infection. PRRs share structural motifs with some R proteins, indicating that PRR- and R-protein-mediated defences might share common mechanisms (FIG. 4). For example, the PRR protein FLS2 and the R protein Xa21 are both receptor-like kinases, with an extracellular LRR domain and an intracellular kinase domain49,108. As proposed by Bent and colleagues62, the distinctions that are made between basal and R-protein-mediated defences might need to be revisited. It is proposed that the PRR–PAMP interaction shares many properties in common with Avr-R protein interactions. For example, similar to Avr-R protein interactions, FLS2 can only recognize specific variants of flagellin, and FLS2 is not conserved in all ecotypes of Arabidopsis. If Xa21 functions as a PRR, then in some cases, PRRs can signal strong enough defences to lead to what seems to be R-protein-mediated resistance. Also, the PRR and R-protein-mediated defence pathways might share some common signalling components or physical mechanisms of defence. For example, HR-like PCD has been reported in response to PAMPS61,109, and shared MAPK cascades are associated with both PRR and R-gene-mediated signalling51,58.
Figure 4. Factors that influence the outcome of plant–bacteria interactions.
The outcome of plant–pathogen interactions might be dependent on the complement of four important factors: pathogen-associated molecular patterns (PAMPS); pattern recognition receptors (PRRs); resistance (R) proteins; and effectors. If the host can recognize the PAMPS or effectors of the pathogen, then non-host or hypersensitive response (HR)-based resistance might be elicited. If the pathogen can vary PAMPS to avoid detection or has the correct complement of effectors to suppress PRR- or R-protein-mediated resistance then disease might be observed. LRR, leucine-rich repeats; NBS, nucleotide-binding site.
There is a broad spectrum of outcomes that have been defined for plant–pathogen interactions, ranging from non-host and R-protein-mediated resistance responses, to weakly or fully susceptible disease responses. At least for bacterial pathogens, it is likely that both PRR- and R-protein-mediated recognition restricts host range through a combination of basal defences (early responses) and HR-associated defences (later responses). The outcome of each interaction might be dependent on the complement of PAMPS and effectors in the pathogen, and the PRRs and R proteins in the host (FIG. 4).
Redundant functions of effectors
Redundant virulence functions of type III effectors have often been observed in studies of plant-pathogen effectors. It seems that the function of many effectors is to suppress plant defences, with two branches of defence targeted for suppression: PRR-mediated basal defences; and R-protein-mediated HR-based PCD. Effector proteins with apparently different biochemical functions target the same pathways, indicating that distinct components of these processes are modulated. For example, suppressors of basal defence might function directly at the PRR, at the level of signal transduction, or perhaps at a cell biological level by altering the mechanism of cell wall alteration. Identifying the host targets of type III effectors might offer clues about how effectors with different activities modulate common pathways.
Future Perspectives
To better understand the complex interactions between plants and bacterial pathogens, the field must continue to unravel the relative contributions of PRR-mediated and R-protein-mediated resistance to promoting plant immunity, and the role of PAMP variation and effector virulence activity in avoiding or suppressing plant defences. Eventually, it might be possible to predict the outcome of a given plant–bacterium interaction by simply knowing the complete complement of PAMPS, effectors, PRRs and R proteins that are in the system. It is possible that the observed differences between PRR-mediated and R-protein-mediated resistance might be due to the strength or timing of defence response elicitation or the relative recalcitrance to suppression by type III effectors. In practical terms, it might be useful to redefine plant defences as early, extracellular defences (PRRs) and later, intracellular defences (R proteins). As the distinctions between PRR-mediated and R-protein-mediated defences blur, it will be interesting to test to what degree the responses are shared between these plant defences.
Type III effectors represent excellent tools, using specific biochemical and cell biological assays, to dissect important processes that are associated with basal and HR-based defences. Presently, the biochemical activities of only a handful of type III effectors have been described, and the host targets of effectors remain mostly undiscovered (BOX 3). Given the present research landscape, it is clearly a very exciting time to study the bacterial pathogenesis of plants, and many new break-throughs in the mechanisms of disease are expected in the coming years.
Box 3. Molecular mimicry by type III effectors.
Type III effectors function within eukaryotic cells to promote virulence. Therefore, it is not surprising that many effectors from both plant and animal pathogens mimic eukaryotic enzymes88. Known enzymatic activities of effectors include: phosphatase, cysteine protease, ubiquitin-like protease and E3 ubiquitin ligase activities10. A subset of type III effectors, the AvrBs3 family, contain both functional nuclear-localization signals and acidic activation domains114, and they might function by mimicking eukaryotic transcription factors. Although the specific host targets of these effectors are unknown, AvrBs3 family members have recently been shown to suppress plant defences115. The recent cloning of the novel R gene Xa27 from rice indicates that plants might employ unique strategies to counter AvrBs3-like effectors116.
Type III effectors also mimic the substrates of host enzymes, and as a result become post-translationally modified. Plant-dependent effector modifications that have been identified include acylation28–30, phosphorylation117 and proteolytic cleavage19,75,118. These modifications contribute to the virulence function of effectors and therefore probably function as initial ‘activation’ steps that are necessary for subsequent interaction with host targets. Identifying the post-translational modifications of several different effectors has provided important insights into effector function. For example, clues about AvrPto function, an effector with no known enzymatic activity, have come from observations that AvrPto undergoes multiple distinct post-translational modifications by plant enzymes. Like several other known effectors, the N terminus of AvrPto contains a myristoylation motif that targets the effector to the plant plasma membrane, and it is strictly required for both AvrPto virulence and Pto-mediated recognition in tomato, and basal defence suppression in Arabidopsis29,42,87. In addition to acylation, a recent study shows that AvrPto is phosphorylated by a Pto-independent kinase activity117, and amino-acid substitutions that decrease AvrPto phosphorylation also decrease AvrPto virulence activity.
Supplementary Material
Acknowledgments
We thank Tracy Rosebrock and Ann Taylor for critical reading of the manuscript. We are grateful to Tom Burr, Amy Charkowski, and Paul Frey for providing photographs for Figure 1. Research in our laboratory is supported by the National Science Foundation, National Institutes of Health, United States Department of Agriculture-National Research Initiative, Binational Agriculture Research Fund, Binational Science Foundation and the Triad Foundation.
- Stomate
A natural opening on leaves and stems. Stomata can open and close to ensure efficient exchange of gases and moisture in the apoplast
- Apoplast
The intercellular space in the plant tissue, including the cell wall, that is outside the plasma membrane, through which nutrients and water can freely diffuse
- Xylem
A network of cells in the vascular system of a plant that moves water and minerals
- Virulence
Increases in the rate of growth and final population size, or enhanced disease symptoms, that promote the spread of the pathogen through the plant or in nature
- Type III secretion system
A bacterial membrane-spanning protein complex, extended by a pilus. This complex functions like a syringe to inject bacterial proteins into the host cell cytoplasm
- Effector
A bacterial protein that is translocated by the type III secretion system into the plant cell cytoplasm
- Pathogen-associated molecular patterns
(PAMPs). Bacterial molecules that have an important role in the microbial lifestyle, and that contain a conserved feature that is recognized by a pathogen recognition receptor (PRR)
- Pathogen recognition receptor
(PRR). A host receptor, such as FLS2 or EFR, that can detect the presence of pathogens by recognizing conserved pathogen molecules (such as PAMPs)
- Basal defence
Plant defence that occurs early in the host–pathogen interaction in response to the perception by plant pattern recognition receptors (PRRs) of extracellular pathogen-associated molecular patterns (PAMPs)
- Biotrophy
A period of colonization during which a microorganism relies on living host tissue to grow
- Hormone
A signal molecule that is produced at specific locations and at low concentrations. Hormones can be transported throughout the plant and regulate biological processes
- Arabidopsis
A plant of the mustard family that is used as a model organism in plant molecular biology
- Callose
A polysaccharide that is a common plant cell wall constituent and that is deposited near infection sites in structures known as papillae. Callose deposition is associated with basal defences and is believed to limit pathogen virulence
- Resistance (R) protein
A plant protein that recognizes, either directly or indirectly, a specific pathogen avirulence protein (often a type III effector) to activate plant immunity
- Gene-for-gene model of disease resistance
A model for plant immunity in which plant resistance genes are only effective if a specific avirulence gene is expressed by the pathogen
- Avirulence (Avr) protein
A pathogen protein that elicits plant immunity in plants that express a specific resistance protein. Avr proteins are often type III effector proteins
- Hypersensitive response
(HR). A defence that is often associated with resistance (R)-protein-mediated immunity. During the HR, the plant initiates programmed cell death in cells that surround the pathogen to inhibit pathogen spreading
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
The following terms in this article are linked online to:
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
NHO1
UniProtKB: http://ca.expasy.org/sprot
AvrB | AvrRpt2 | EF-Tu | HopAB2| HopE1 | HopK1 | MPK3 | MPK6 | RIN4
FURTHER INFORMATION
Gregory B. Martin’s homepage: http://bti.cornell.edu/page.php?id=313
See online article: S1 (table)
References
- 1.Genin S, Boucher C. Lessons learned from the genome analysis of Ralstonia solanacearum. Annu Rev Phytopathol. 2004;42:107–134. doi: 10.1146/annurev.phyto.42.011204.104301. [DOI] [PubMed] [Google Scholar]
- 2.Nino-Liu DO, Ronald PR, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. doi: 10.1111/j.1364-3703.2006.00344.x. (in the press) [DOI] [PubMed] [Google Scholar]
- 3.Preston GM. Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol Plant Pathol. 2000;1:263–275. doi: 10.1046/j.1364-3703.2000.00036.x. [DOI] [PubMed] [Google Scholar]
- 4.Van Sluys MA, et al. Comparative genomic analysis of plant-associated bacteria. Annu Rev Phytopathol. 2002;40:169–189. doi: 10.1146/annurev.phyto.40.030402.090559. [DOI] [PubMed] [Google Scholar]
- 5.Agrios GN. Plant Pathology. Academic Press; San Diego: 1997. [Google Scholar]
- 6.Anonymous . Florida citrus canker eradication program. University of Florida, Institute of Food and Agricultural Sciences; 2006. [online] < http://edis.ifas.ufl.edu/FE532>. [Google Scholar]
- 7.Brown K. Florida fights to stop citrus canker. Science. 2001;292:2275–2276. doi: 10.1126/science.292.5525.2275. [DOI] [PubMed] [Google Scholar]
- 8.Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- 9.Buttner D, Bonas U. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr Opin Microbiol. 2006;9:193–200. doi: 10.1016/j.mib.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- 11.Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 12.Chang JH, et al. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci USA. 2005;102:2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preston GM, Studholme DJ, Caldelari I. Profiling the secretomes of plant pathogenic Proteobacteria. FEMS Microbiol Rev. 2005;29:331–360. doi: 10.1016/j.femsre.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Jha G, Rajeshwari R, Sonti RV. Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol Plant Microbe Interact. 2005;18:891–898. doi: 10.1094/MPMI-18-0891. [DOI] [PubMed] [Google Scholar]
- 15.Toth IK, Birch PR. Rotting softly and stealthily. Curr Opin Plant Biol. 2005;8:424–429. doi: 10.1016/j.pbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005;8:361–368. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Petnicki-Ocwieja T, et al. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000 . Proc Natl Acad Sci USA. 2002;99:7652–7657. doi: 10.1073/pnas.112183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 19.Coaker G, Falick A, Staskawicz B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005;308:548–550. doi: 10.1126/science.1108633. This study, along with reference 20, shows that the type III effector AvrRpt2 cleaves the plant defence regulator RIN4. [DOI] [PubMed] [Google Scholar]
- 20.Kim HS, et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Solanilla E, Bronstein PA, Schneider AR, Collmer A. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol Microbiol. 2004;54:353–365. doi: 10.1111/j.1365-2958.2004.04285.x. [DOI] [PubMed] [Google Scholar]
- 22.Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 23.Roden J, Eardley L, Hotson A, Cao Y, Mudgett MB. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol Plant Microbe Interact. 2004;17:633–643. doi: 10.1094/MPMI.2004.17.6.633. [DOI] [PubMed] [Google Scholar]
- 24.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci USA. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. References 24 and 25 show that AvrPtoB has E3 ubiquitin ligase activity that is required to suppress plant defences. [DOI] [PubMed] [Google Scholar]
- 26.Bretz JR, et al. A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol Microbiol. 2003;49:389–400. doi: 10.1046/j.1365-2958.2003.03616.x. [DOI] [PubMed] [Google Scholar]
- 27.Espinosa A, Guo M, Tam VC, Fu ZQ, Alfano JR. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol Microbiol. 2003;49:377–387. doi: 10.1046/j.1365-2958.2003.03588.x. [DOI] [PubMed] [Google Scholar]
- 28.Nimchuk Z, et al. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 29.Shan L, Thara VK, Martin GB, Zhou JM, Tang X. The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell. 2000;12:2323–2338. doi: 10.1105/tpc.12.12.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert-Seilaniantz A, Shan L, Zhou JM, Tang X. The Pseudomonas syringae pv. tomato DC3000 Type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol Plant Microbe Interact. 2006;19:130–138. doi: 10.1094/MPMI-19-0130. [DOI] [PubMed] [Google Scholar]
- 31.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 33.Gelvin SB. Agrobacterium and plant genes involved in t-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 34.da Silva AC, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 35.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glickmann E, et al. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact. 1998;11:156–162. doi: 10.1094/MPMI.1998.11.2.156. [DOI] [PubMed] [Google Scholar]
- 37.Navarro L, et al. A plant MiRNA contributes to antibacterial resistance by repressing auxin signalling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 38.Von Bodman SB, Bauer WD, Coplin DL. Quorum sensing in plant-pathogenic bacteria. Annu Rev Phytopathol. 2003;41:455–482. doi: 10.1146/annurev.phyto.41.052002.095652. [DOI] [PubMed] [Google Scholar]
- 39.Leigh JA, Coplin DL. Exopolysaccharides in plant–bacterial interactions. Annu Rev Microbiol. 1992;46:307–346. doi: 10.1146/annurev.mi.46.100192.001515. [DOI] [PubMed] [Google Scholar]
- 40.Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc Natl Acad Sci USA. 2006;103:5983–5988. doi: 10.1073/pnas.0509860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 42.He P, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. In Arabidopsis protoplasts, AvrPto and AvrPtoB suppress basal defence signalling upstream of MAPK activation. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Gomez L, Boller T. Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 2002;7:251–256. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- 44.Zipfel C, Felix G. Plants and animals: a different taste for microbes? Curr Opin Plant Biol. 2005;8:353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Zeidler D, et al. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA. 2004;101:15811–15816. doi: 10.1073/pnas.0404536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 47.Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felix G, Boller T. Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J Biol Chem. 2003;278:6201–6208. doi: 10.1074/jbc.M209880200. [DOI] [PubMed] [Google Scholar]
- 49.Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 50.Gomez-Gomez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 51.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 52.Navarro L, et al. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 54.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. This paper provides biochemical evidence that the PRR FLS2 binds to flagellin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- 57.Truman W, de Zabala MT, Grant M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. 2006;46:14–33. doi: 10.1111/j.1365-313X.2006.02672.x. References 56 and 57 report microarray gene expression profiling of Arabidopsis that identified genes and pathways that are involved in basal defences. [DOI] [PubMed] [Google Scholar]
- 58.Pedley KF, Martin GB. Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol. 2005;8:541–547. doi: 10.1016/j.pbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. This paper, along with reference 47, reports that the PAMP EF-Tu is recognized by the Arabidopsis PRR EFR. [DOI] [PubMed] [Google Scholar]
- 60.Pfund C, et al. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant Microbe Interact. 2004;17:696–706. doi: 10.1094/MPMI.2004.17.6.696. [DOI] [PubMed] [Google Scholar]
- 61.Taguchi F, et al. Post-translational modification of flagellin determines the specificity of HR induction. Plant Cell Physiol. 2003;44:342–349. doi: 10.1093/pcp/pcg042. [DOI] [PubMed] [Google Scholar]
- 62.Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell. 2006;18:764–779. doi: 10.1105/tpc.105.037648. Variation of flagellin in Xanthomonas probably allows the pathogen to avoid PRR-mediated detection in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakobek JL, Smith JA, Lindgren PB. Suppression of bean defense responses by Pseudomonas syringae. Plant Cell. 1993;5:57–63. doi: 10.1105/tpc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown I, Mansfield JW, Bonas U. hrp genes in Xanthomonas campestris pv vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol Plant Microbe Interact. 1995;8:825–836. [Google Scholar]
- 65.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, et al. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA. 2005;102:12990–12995. doi: 10.1073/pnas.0502425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh HS, Collmer A. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J. 2005;44:348–359. doi: 10.1111/j.1365-313X.2005.02529.x. [DOI] [PubMed] [Google Scholar]
- 68.Metz M, et al. The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 2005;41:801–814. doi: 10.1111/j.1365-313X.2005.02338.x. [DOI] [PubMed] [Google Scholar]
- 69.Kim MG, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 70.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 71.Mackey D, Holt BF, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 72.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 73.Lim MT, Kunkel BN. The pseudomonas syringae avrRpt2 gene contributes to virulence on tomato. Mol Plant Microbe Interact. 2005;18:626–633. doi: 10.1094/MPMI-18-0626. [DOI] [PubMed] [Google Scholar]
- 74.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chisholm ST, et al. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci USA. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Torres M, et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006 June 22; doi: 10. 1111/j.1365–313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 77.Lam E. Controlled cell death, plant survival and development. Nature Rev Mol Cell Biol. 2004;5:305–315. doi: 10.1038/nrm1358. [DOI] [PubMed] [Google Scholar]
- 78.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 79.Tang X, et al. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 80.Scofield SR, et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 81.Deslandes L, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 83.Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- 84.Shan L, He P, Zhou JM, Tang X. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol Plant Microbe Interact. 2000;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- 85.Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol. 2003;41:215–243. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- 86.Salmeron JM, et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 87.Wulf J, Pascuzzi PE, Fahmy A, Martin GB, Nicholson LK. The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure. 2004;12:1257–1268. doi: 10.1016/j.str.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 88.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 89.Espinosa A, Alfano JR. Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell Microbiol. 2004;6:1027–1040. doi: 10.1111/j.1462-5822.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- 90.Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibiton of host programmed cell death. EMBO J. 2003;22:60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jamir Y, et al. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 2004;37:554–565. doi: 10.1046/j.1365-313x.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- 92.Jackson RW, et al. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsiamis G, et al. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 2000;19:3204–3214. doi: 10.1093/emboj/19.13.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Makino S, Sugio A, White F, Bogdanove AJ. Inhibition of resistance gene-mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol Plant Microbe Interact. 2006;19:240–249. doi: 10.1094/MPMI-19-0240. [DOI] [PubMed] [Google Scholar]
- 95.Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 96.Zeng LR, et al. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. This paper reports the cloning of a rice U-box E3 ubiquitin ligase that regulates plant cell death and defence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gonzalez-Lamothe R, et al. The u-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell. 2006;18:1067–1083. doi: 10.1105/tpc.106.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang CW, et al. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell. 2006;18:1084–1098. doi: 10.1105/tpc.105.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin NC, Martin GB. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe Interact. 2005;18:43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- 100.Guttman DS, et al. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science. 2002;295:1722–1726. doi: 10.1126/science.295.5560.1722. [DOI] [PubMed] [Google Scholar]
- 101.Lin NC, Abramovitch RB, Kim YJ, Martin GB. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl Environ Microbiol. 2006;72:702–712. doi: 10.1128/AEM.72.1.702-712.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abramovitch RB, Martin GB. AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol Lett. 2005;245:1–8. doi: 10.1016/j.femsle.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 103.Uppalapati SR, et al. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005;42:201–217. doi: 10.1111/j.1365-313X.2005.02366.x. [DOI] [PubMed] [Google Scholar]
- 104.Cui J, et al. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci USA. 2005;102:1791–1796. doi: 10.1073/pnas.0409450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao Y, et al. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003;36:485–499. doi: 10.1046/j.1365-313x.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 106.He P, et al. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 2004;37:589–602. doi: 10.1111/j.1365-313x.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- 107.Cohn JR, Martin GB. Pseudomonas syringae pv tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 2005;44:139–154. doi: 10.1111/j.1365-313X.2005.02516.x. [DOI] [PubMed] [Google Scholar]
- 108.Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 109.Shimizu R, et al. The DeltafliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol Genet Genomics. 2003;269:21–30. doi: 10.1007/s00438-003-0817-3. [DOI] [PubMed] [Google Scholar]
- 110.DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mysore KS, et al. Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002;32:299–315. doi: 10.1046/j.1365-313x.2002.01424.x. [DOI] [PubMed] [Google Scholar]
- 112.Pedley KF, Martin GB. Identification of MAPKs and their possible MAPKK activators involved in the Pto-mediated defense response of tomato. J Biol Chem. 2004;279:49229–49235. doi: 10.1074/jbc.M410323200. [DOI] [PubMed] [Google Scholar]
- 113.del Pozo O, Pedley KF, Martin GB. MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004;23:3072–3082. doi: 10.1038/sj.emboj.7600283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schornack S, Meyer A, Romer P, Jordan T, Lahaye T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol. 2006;163:256–272. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 115.Fujikawa T, Ishihara H, Leach JE, Tsuyumu S. Suppression of defense response in plants by the avrBs3/pthA gene family of Xanthomonas spp. Mol Plant Microbe Interact. 2006;19:342–349. doi: 10.1094/MPMI-19-0342. [DOI] [PubMed] [Google Scholar]
- 116.Gu K, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 117.Anderson JC, Pascuzzi PE, Xiao F, Sessa G, Martin GB. Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell. 2006;18:502–514. doi: 10.1105/tpc.105.036590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mudgett MB, Staskawicz BJ. Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol. 1999;32:927–941. doi: 10.1046/j.1365-2958.1999.01403.x. [DOI] [PubMed] [Google Scholar]
- 119.Meindl T, Boller T, Felix G. The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell. 2000;12:1783–1794. doi: 10.1105/tpc.12.9.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dow M, Newman MA, von Roepenack E. The Induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu Rev Phytopathol. 2000;38:241–261. doi: 10.1146/annurev.phyto.38.1.241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.