Abstract
The phylogenetic structure of ecological communities can shed light on assembly processes, but the focus of phylogenetic structure research thus far has been on mature ecosystems. Here, I present the first investigation of phylogenetic community structure during succession. In a replicated chronosequence of 30 sites in northeastern Costa Rica, I found strong phylogenetic overdispersion at multiple scales: species present at local sites were a non-random assemblage, more distantly related than chance would predict. Phylogenetic overdispersion was evident when comparing the species present at each site with the regional species pool, the species pool found in each age category to the regional pool or the species present at each site to the pool of species found in sites of that age category. Comparing stem size classes within each age category, I found that during early succession, phylogenetic overdispersion is strongest in small stems. Overdispersion strengthens and spreads into larger size classes as succession proceeds, corroborating an existing model of forest succession. This study is the first evidence that succession leaves a distinct signature in the phylogenetic structure of communities.
Keywords: community assembly, phylogenetic structure, succession, chronosequence, tropical forest
1. Introduction
The phylogenetic structure of ecological communities has become a major focus of recent research (Webb 2000; Webb et al. 2002; Cavender-Bares et al. 2004, 2006; Kembel & Hubbell 2006). Newly available tools (Webb et al. 2008) make it possible to examine ecological datasets from an evolutionary point of view. Here, I present the first assessment of changes in community phylogenetic structure during succession, using vegetation data from a chronosequence of sites in a tropical rainforest (Letcher & Chazdon 2009). Natural forest regeneration in the tropics provides an ideal system to examine phylogenetic structure during succession because tropical forests are highly diverse in terms of both species composition and phylogenetic make-up. Structural and floristic changes during tropical forest succession have been well studied (Finegan 1996; Guariguata & Ostertag 2001; Chazdon 2003, 2008; Letcher & Chazdon 2009), but the dynamics of community phylogenetic structure during succession have not yet been investigated comprehensively in any system. Incorporating phylogenetic community structure into studies of succession will bridge a gap between ecological and evolutionary questions, and greatly increase our understanding of community assembly processes.
The phylogenetic structure of communities can be assessed by comparing the species composition of a community with that of a regional species pool, where the pool ideally constitutes all the potential colonists in the community under study (Webb 2000; Webb et al. 2002; Swenson et al. 2006, 2007). Examined in this manner, community phylogenetic structure can take three basic forms: random, clustered or overdispersed (Webb 2000). With random phylogenetic structure, organisms in a community are no more or less related to one another than to random draws from the regional species pool. With clustering, species in the community are more closely related than expected by chance, and with overdispersion they are less closely related.
Depending on the conservatism or lability of functional traits in the community under study, these three types of community phylogenetic structure suggest different mechanisms of community assembly (Webb 2000; Webb et al. 2002; Kembel & Hubbell 2006; Silvertown et al. 2006). Random phylogenetic structure is predicted by neutral theory (Hubbell 2001), and reflects a community assembled solely by stochastic processes (Webb et al. 2002). Phylogenetic clustering is a predicted consequence of abiotic filtering, where functional traits are conserved (Webb 2000), because shared evolutionary history predisposes organisms to share habitat tolerances and preferences (Harvey & Pagel 1991; Tofts & Silvertown 2000; Webb et al. 2002; Silvertown et al. 2006; Webb et al. 2006). Where functional traits are strongly divergent, phenotypic (rather than phylogenetic) clustering is indicative of abiotic filtering (Cavender-Bares et al. 2004). Phylogenetic overdispersion can result from several different processes. Theoretically, closely related species are more prone to competitive exclusion, so phylogenetic overdispersion within a community could be seen as evidence for biotic filtering (Harvey & Pagel 1991; Tofts & Silvertown 2000; Webb 2000; Webb et al. 2002; Kembel & Hubbell 2006; Silvertown et al. 2006). Facilitation interactions among distantly related species can also produce phylogenetic overdispersion (Valiente-Banuet & Verdú 2007), although evidence for this phenomenon comes from Mediterranean shrublands where established plants provide shady germination microsites for distantly related taxa. This mechanism is unlikely to act in tropical forest communities, where light is often a limiting factor (Wright 2002). Finally, where strong trait divergences occur between lineages, abiotic filtering may contribute to phylogenetic overdispersion as lineages radiate into different habitats. In a simplified scenario, species A, B and C each radiates into a pair of sister species, one in habitat alpha and one in habitat beta. The species within habitat alpha or beta will appear overdispersed because their closest relatives occur outside the habitat (Webb et al. 2002; Fine et al. 2006; Baraloto et al. 2007).
The detection of clustering or overdispersion is sensitive to the taxonomic and spatial scales examined (Cavender-Bares et al. 2004, 2006; Swenson et al. 2006, 2007). Local populations of species from a narrowly defined lineage (e.g. one genus at a site) tend to be phylogenetically overdispersed (Cavender-Bares et al. 2006), but when a broader phylogenetic sample is included (e.g. all angiosperms present at a site), plant neighbourhoods tend to show phylogenetic clustering (Cavender-Bares et al. 2006; Swenson et al. 2006). The choice of species pool also impacts the detection of phylogenetic structure. When the species pool is very large compared with the sample, clustering is detected, but when the species pool is drawn from the local area, random phylogenetic structure is observed (Swenson et al. 2006).
In addition, the phylogenetic structure of a forest community may differ among stem size classes, a putative proxy for forest dynamics over time. Swenson et al. (2007) found that phylogenetic overdispersion increased with stem size in five Neotropical forest plots, a result that they interpret as a signature of biotic filtering as tree cohorts age. In contrast, Webb et al. (2006) found increased clustering from the sapling to adult size class in a forest in Borneo. They interpret this result as abiotic filtering, but it is important to note that this study was conducted in a Dipterocarp forest, where that single family dominates the canopy (Cannon & Leighton 2004 and references therein).
Most of the research on community phylogenetic structure has been conducted in mature, presumably undisturbed ecosystems (Webb et al. 2002, 2006; Cavender-Bares et al. 2004, 2006; Silvertown et al. 2006). Although several studies have included secondary forest (Kembel & Hubbell 2006; Swenson et al. 2006, 2007), they did not specifically investigate successional changes in phylogenetic structure. This study, combining a chronosequence approach and a multiple size class approach, provides the first investigation of the phylogenetic structure of a community undergoing succession.
2. Material and Methods
(a). Study site and data collection
The study area, in the Sarapiquí region of northeastern Costa Rica, is classified as tropical lowland wet forest (McDade et al. 1994). The landscape matrix consists of old-growth forest fragments surrounded by secondary forest and agricultural lands (McDade et al. 1994; Guariguata et al. 1997). I selected 30 study sites, seven in old-growth forest and 23 in secondary forests ranging from 10 to 44 years old on abandoned agricultural lands at 40–200 m elevation. I verified forest age and prior land use via satellite images, aerial photos and interviews with landowners and neighbours. The sites are mapped and described in detail by Letcher & Chazdon (2009).
In each site, I performed a 0.1 ha vegetation inventory using a modified Gentry transect (Phillips & Miller 2002): a series of five parallel 2 × 100 m strips, each separated by 10 m, and at least 10 m from the forest edge. In the survey area, I recorded the species identity and diameter at 1.3 m (dbh) of all woody stems 2.5 cm or more dbh for trees/shrubs and 0.5 cm or more dbh for lianas (Letcher & Chazdon 2009). Following the methods of Gentry (Phillips & Miller 2002), I also recorded giant herbs (Zingiberales) if their pseudostem diameter exceeded 2.5 cm dbh at 1.3 m height. For each species at each site, I collected a voucher specimen for verification at the Instituto Nacional de Biodiversidad (INB), Santo Domingo de Heredia, Costa Rica. All voucher specimens are on file at INB, with duplicates of fertile specimens filed in the Museo Nacional de Costa Rica.
(b). Analytical methods
Sites were divided into five successional age categories (table 1), such that each age category contained at least five sites. Originally, I had planned to have enough sites so that each age category would be 5 years, but it proved impossible to find enough appropriate sites. The areas sampled represent all of the sites in this region with large enough patches of even-aged forest where I could verify the land-use history and obtain permission to work (Letcher & Chazdon 2009).
Table 1.
Sites and species pools for analysis.
| forest 10–15 years | forest 16–20 years | forest 21–30 years | forest 31–44 years | old-growth forest | |
|---|---|---|---|---|---|
| number of sites | 6 | 6 | 6 | 5 | 7 |
| number of species in pool | 196 | 226 | 218 | 250 | 275 |
| average number of species per site ± s.e. | 52.8 ± 4.0 | 54.7 ± 4.0 | 64.6 ± 4.5 | 85.2 ± 4.8 | 90.7 ± 5.7 |
To construct an appropriate regional species pool, I attempted to identify all the potential colonists to these sites that would have been detected by this inventory method. I used a database of vascular plants in the Sarapiquí region (Proyecto Flora Digital de La Selva 2007) including 2016 species, and used herbarium records (INB, NY, MO, USA) to identify species that could have entered my dataset (trees and shrubs that reach a diameter of 2.5 cm or more dbh, woody vines that reach a diameter of 0.5 cm or more and giant herbs with pseudostems that reach 1.3 m and have a diameter of 2.5 cm or more). I excluded non-angiosperm taxa in the phylogenetic analyses, because they are rare in the region and would contribute disproportionately to phylogenetic structure owing to their low relatedness to the majority of species. The final list for the community species pool included 750 species (electronic supplementary material, appendix I).
I used Phylomatic (Webb & Donoghue 2005) to map the community species pool onto a maximally resolved supertree of angiosperms (R20080417; available online at www.phylodiversity.net). Interior nodes in the phylogeny were assigned ages from Wikström et al. (2001), and I implemented the BLADJ algorithm to reduce variance between branch lengths by evenly spacing nodes of unknown ages. I used this tree (henceforth ‘regional supertree’) to investigate the phylogenetic community structure at each site. I also constructed a tree for each successional age category (henceforth ‘age-category supertree’), using the pool of taxa found in sites in that category (table 1).
I used Phylocom 4.0.1 (Webb et al. 2008) to investigate phylogenetic structure. The program implements techniques from Webb (2000) based on mean pairwise distance (MPD), the average branch length on the supertree between each pair of taxa in a sample. The net relatedness index (NRI) is a standardized metric of phylogenetic clustering (Webb 2000; Webb et al. 2002), defined as
| 2.1 |
where Ms is the MPD in a sample of taxa, Mr is the average MPD obtained from a set of randomized samples (I used 9999 permutations) and σr is the standard deviation from the randomized set. Species for the randomized samples are drawn from the terminal taxa in the supertree (i.e. the entire community species pool). The NRI indicates whether taxa in a sample are phylogenetically clustered (positive NRI) or overdispersed (negative NRI; Webb 2000).
Several different randomization models can be used to calculate the NRI (Webb et al. 2002), and in some datasets the models give divergent results (Kembel & Hubbell 2006; Hardy & Senterre 2007). To test the effects of model selection, I calculated the NRI with the four different randomization models available in Phylocom (Webb et al. 2008). I calculated the NRI (using the regional supertree) for all 30 sites with each of the four models and compared the outcome using linear regression in the R Statistical Package (R Development Core Team 2008). Correlation coefficients between each possible set of models were very high (adjusted R2 = 0.96–0.99). I chose a model 2 randomization, in which the number of taxa in the sample is kept constant and the taxa used in randomizations are a random draw from the species pool, for all analyses.
In many studies of phylogenetic clustering, the nearest taxon index (NTI) is also calculated (Webb 2000; Webb et al. 2002; Kembel & Hubbell 2006; Swenson et al. 2006, 2007). The NTI measures phylogenetic clustering at the tips of the branches, i.e. intrafamilial clustering (Webb 2000; Webb et al. 2002). It is calculated in a manner analogous to the NRI, but using mean nearest neighbour distance rather than MPD. The NTI is more difficult to interpret in trees with little intrafamilial resolution because branch lengths of many taxa are approximate. The Phylomatic algorithm maps unknown relationships as polytomies on the supertree, with congeners clustering together within a family (Webb et al. 2008). In my regional supertree, 56/94 families (59.6%) had intrafamilial resolution (i.e. more than one genus in the family). To test the effects of tree resolution on NTI, I used Mesquite v. 2.01 (Maddison & Maddison 2007) to construct a version of the regional supertree with all families collapsed into polytomies. I compared the NRI and NTI for each of the 30 sites in the maximally resolved Phylomatic tree versus the polytomized tree using linear regression in R (R Development Core Team 2008). The NRI values calculated using the Phylomatic tree and the polytomized tree were very highly correlated (adjusted R2 = 0.99, p < 0.00001). The NTI values showed a significant correlation, but it was much weaker (adjusted R2 = 0.34, p < 0.001). Details of this analysis are included in electronic supplementary material, appendix II, and NTI results are reported in electronic supplementary material, appendix III. Because a metric so sensitive to tree topology may not be appropriate for comparing among multiple trees, in subsequent analyses, I only used the NRI.
I calculated the phylogenetic community structure of each successional age category in three ways. To investigate the species pool for each age category, I pooled the species found in all sites belonging to each age category (e.g. all species found in old-growth forest) and calculated the NRI for this pool using the regional supertree. To investigate the site-specific phylogenetic structure, I calculated NRI values for each site versus the regional supertree, and averaged the values for sites in each age category. These analyses are designed to reveal whether the species pools of each site and each age category are drawn at random, with respect to phylogeny, from the regional species pool. I also calculated the NRI in each site using the appropriate age-category supertree, to reveal whether the community of a local site is drawn at random from the pool of species that inhabit sites of that age.
I also examined the phylogenetic structure of diameter classes within each age category. I divided all stems into three size classes (less than 5, 5–9.9, or 10 cm or more dbh) and calculated the NRI for each size class at each site using the regional supertree, to investigate changes within a site over time in the manner of Webb et al. (2006) and Swenson et al. (2007).
To identify clades that contributed significantly to the phylogenetic structure, I used the procedure nodesig in Phylocom (Webb et al. 2008) to compare the species pool of each age category with the regional supertree. Nodesig tests whether a particular node in the phylogeny has significantly more or less descendent taxa in a sample than a null model predicts. The null model is a random draw of n taxa from the supertree, where n is the number of taxa in the sample (Webb et al. 2008). I tested the sensitivity of nodesig to tree resolution by comparing the results for the maximally resolved versus polytomized trees. The nodesig results were identical except in the cases where infrafamilial divisions contributed significant structure (table 2), showing that the nodesig procedure is robust to changes in tree resolution. Results shown are from the maximally resolved tree.
Table 2.
Nodes in the phylogeny with significantly more/less daughter taxa in the given age category when compared with a random draw of n taxa from the supertree, where n is the number of taxa in each age category (Phylocom procedure nodesig; Webb et al. 2008). Where several consecutive nodes were significant, they are listed on the same line in the table. Clades without accepted names are listed in parentheses. Where the significant node occurs below the family level, I have provided a family name in the next column for reference. ‘Life form’ indicates prevalent life forms for this clade in this region (excluding herbs, epiphytes and parasites). Where multiple life forms are listed, the first is the most common. Sig. more/less, significantly more or less daughter taxa in the given age category.
| forest age category | sig. more/ less | node | family | clade | life forms |
|---|---|---|---|---|---|
| 10–15 years | more | Smilax | Smilacaceae | monocot | lianas |
| Zingiberales | monocot | giant herbs | |||
| Olacaceae | core Eudicot | small trees, lianas | |||
| Dilleniaceae | core Eudicot | lianas | |||
| Mendoncia | Acanthaceae | asterid I | lianas | ||
| Paullinia | Sapindaceae | rosid II | lianas | ||
| (Luehea, Apeiba, Byttneria) | Malvaceae | rosid II | soft-wooded trees, lianas | ||
| less | Ericales, Sapotaceae | basal Asterid | trees | ||
| (Ixoroideae, Cinchonoideae) | Rubiaceae | asterid I | trees and shrubs | ||
| (Oxalidales, Malpghiales, Celastrales) | rosid I | trees, lianas, shrubs | |||
| 15–20 years | more | Smilax | Smilacaceae | monocot | lianas |
| Siparunaceae | magnoliid | shrubs, trees | |||
| (Dilleniales, Caryophyllales) | core Eudicot | lianas, small trees | |||
| Sapindales, Sapindaceae, Paullinia | rosid II | lianas | |||
| less | Ficus | Moraceae | rosid I | trees | |
| (Myrtaceae, Vochysiaceae) | basal Rosid | trees | |||
| Ericales | basal Asterid | trees | |||
| Rubiaceae, (Ixoroideae, Cinchonoideae) | asterid I | trees, shrubs | |||
| 21–30 years | more | Smilax | Smilacaceae | monocot | lianas |
| Lamiales | asterid I | shrubs, trees, lianas | |||
| Vismia | Hypericaceae | rosid I | trees | ||
| Paullinia | Sapindaceae | rosid II | lianas | ||
| (Anacardiaceae, Burseraceae), Protium | Burseraceae | rosid II | trees | ||
| less | (Sapotaceae, Lecythidaceae) | basal Asterid | trees | ||
| Eugenia | Myrtaceae | basal Rosid | trees | ||
| more than 30 years | more | Smilax Magnoliids, (Laurales, | Smilacaceae Annonaceae | monocot magnoliid | lianas trees |
| Magnoliales), Guatteria | |||||
| less | (Myrtaceae, Melastomataceae, Vochysiaceae) | basal Rosid | trees, shrubs | ||
| Myrtaceae, Eugenia | basal Rosid | trees | |||
| old growth | more | Arecaceae, Geonoma | monocot | understorey palms | |
| Myristicaceae | magnoliid | trees | |||
| (Coussarea, Faramea) | Rubiaceae | asterid I | trees, shrubs | ||
| less | (Ixoroideae,Cinchonoideae) | Rubiaceae | asterid I | trees, shrubs | |
| (Fabales, Rosales), Moraceae, Ficus | rosid I | trees | |||
| Solanaceae | asterid I | shrubs, trees |
I used indicator value (IV) analysis (Dufrêne & Legendre 1997), implemented in PC-ORD 4 (McCune & Mefford 1999), to investigate the distribution of species, genera and families along the chronosequence, using false discovery rate procedures (Benjamini & Hochberg 1995) to control the familywise error rate. I compared the species characteristic of each successional age category with the species driving the phylogenetic structure detected for each age category.
3. Results
Omitting the non-angiosperms, I recorded data for 8856 individuals of 474 species from 94 families (APG classification; Stevens 2008). Nine individuals were completely unidentified, and five could only be identified to genus level. I also collected 98 specimens of consistent morphospecies corresponding to as-yet undescribed species in eight genera (N. Zamora 1997, personal communication; Letcher & Chazdon 2009). Overall, 8749 individuals (98.8%) were positively identified and matched to described species. Including the morphospecies, this figure rises to 99.9 per cent.
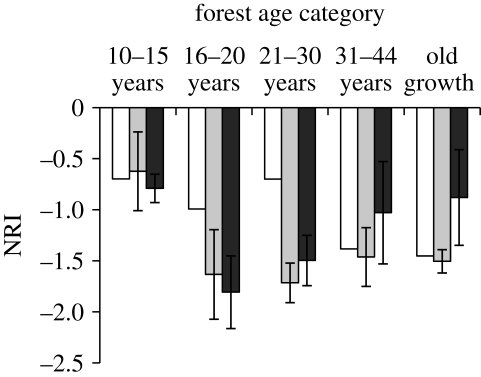
Comparing sites to the regional supertree, I found phylogenetic overdispersion (NRI values below zero) at all sites and for all age categories (figure 1). For the species pools, overdispersion was most pronounced in the oldest sites (30–44 years and old growth). For the site averages, the youngest sites (less than 15 years) had relatively little phylogenetic structure, but all the older sites showed strong overdispersion. Thus, for intermediate-aged sites (16–30 years), site-level overdispersion exceeded the overdispersion observed in the species pool for the age category. Comparing each site with its respective age-category supertree, I also found phylogenetic overdispersion (figure 1), with the strongest negative values in sites of 16–30 years.
Figure 1.
The NRI for species found in successional forests at three different scales: (a) the NRI of the species pool in each age category compared with the regional supertree (white bars); (b) the average NRI of sites within an age category compared with the regional supertree (pale grey bars); (c) the average NRI for sites within an age category compared with a supertree constructed from species present in that age category (dark bars). Error bars represent ±1 s.e.
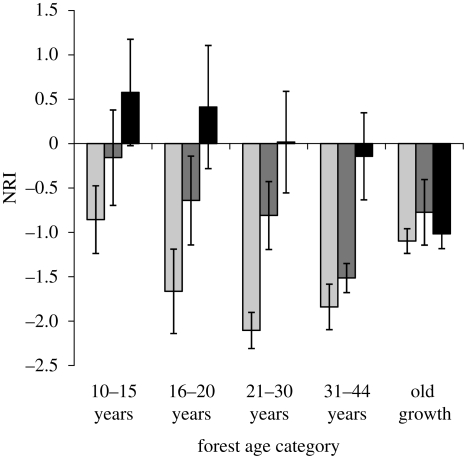
Phylogenetic overdispersion was highest in small stems and declined with increasing stem size in all age categories except old-growth forests, where overdispersion was roughly equal across all diameter classes (figure 2). Stems 10 cm or more dbh showed slight clustering (NRI values greater than 0) in younger sites (10–20 years), NRI values close to zero in intermediate-aged sites (21–44 years) and overdispersion in old-growth forest.
Figure 2.
The average NRI for stems within a diameter class in each age category, based on comparisons with the regional supertree. Error bars show ±1 s.e. Light grey bars, 0.5–5 cm dbh; dark grey bars, 5–9.9 cm dbh; black bars, 10 cm or more dbh.
In the IV analysis, 23 of 94 families (24.5%), 56 of 283 genera (19.8%) and 67 of 474 species (14.1%) had significant IVs after FDR error correction (electronic supplementary material, appendix IV). Characteristic species and genera were found for each age category, and characteristic families were found for each category except forests 21–30 years (electronic supplementary material, appendix IV). The indicator taxa for young forests were mainly lianas, shrubs or soft-wooded trees. Indicator taxa for older forests were generally palms (Arecaceae) and large-seeded, animal-dispersed trees (e.g. Annonaceae, Burseraceae, Elaeocarpaceae, Lecythidaceae, Sabiaceae and Sapotaceae; Chazdon et al. 2003).
The nodesig analysis (Webb et al. 2008) revealed taxa contributing to the phylogenetic structure for each age category (table 2). Clades with significantly more taxa in young forests were generally rich in lianas, shrubs and soft-wooded trees, though nodesig reveals a set of important groups different from that revealed by the IV analysis. Giant herbs were important in the youngest age category. Clades with significantly fewer taxa in young forests were mostly trees. The absence of Ericales, particularly Sapotaceae and Lecythidaceae, characterized forests less than 30 years. In old forests, clades with more taxa were generally trees and lianas, and clades with significantly fewer taxa were mainly trees and shrubs (table 2). Taxa with significant nodesig values came from all the major clades of the angiosperm phylogeny (table 2).
Taxa contributing significantly to the phylogenetic structure showed some overlap with the indicator taxa (table 2 and electronic supplementary material, appendix IV): Dilleniaceae in the youngest sites, Laurales and the genus Smilax in forests 30–44 years, Arecaceae and the Coussarea–Faramea clade (Rubiaceae) in old-growth forests contributed to the phylogenetic structure and showed high IVs.
4. Discussion
The strong phylogenetic overdispersion evident at almost every scale suggests that succession generates non-random patterns of phylogenetic structure. The taxa present at a site are a non-random subset of the taxa that could potentially occupy the site, and as succession proceeds, the taxa present become more distantly related than chance would predict. This pattern is unlikely to be an artefact of the methods I employed: according to the phylogenetic scale dependence identified by Cavender-Bares et al. (2006) and Swenson et al. (2006, 2007), the scales used in this study (a broad sample of the angiosperm phylogeny, narrow geographical range) more often reveal clustering than overdispersion. The power of NRI-based methods to detect non-random phylogenetic structure tends to decline with the size of the phylogeny (Swenson 2009), and the phylogeny in this study is twice as large as any of the examples provided by Swenson (2009; 750 taxa versus a maximum of 320). In addition, I omitted non-angiosperm taxa, which would have contributed to unusually high MPD values in the samples where they were observed and increased the overall likelihood of detecting overdispersion. Even with these parameters, I detected significant phylogenetic overdispersion instead of clustering. Taxa responsible for the overdispersion came from all regions of the angiosperm phylogeny (table 2), showing that no single group is driving the patterns observed.
Ultimately, the interpretation of phylogenetic signal depends on the traits of the organisms under study (Webb 2000; Webb et al. 2002; Cavender-Bares et al. 2004, 2006; Fine et al. 2006; Losos 2008). There are two broad possibilities to explain phylogenetic overdispersion during succession: (i) if traits are conserved, phylogenetic overdispersion indicates strong biotic filtering during succession. (ii) If traits diverge strongly among closely related taxa, abiotic filtering could produce the overdispersion as sister taxa sort into different habitats along an age gradient. It is likely that both these mechanisms act during succession, and future compilations of trait data will be useful in teasing apart the importance of these two processes for different groups (Losos 2008). Trait conservatism has been demonstrated for a number of functional, defence-related and reproductive traits in Neotropical forest trees (Chazdon et al. 2003; Chave et al. 2006; Fine et al. 2006; Swenson et al. 2007). Alternately, studies of species pairs have shown that strong divergences in functional and defence traits can occur within a genus (Fine et al. 2006; Baraloto et al. 2007). Certain tropical genera have undergone rapid radiations, with species diverging into many habitats; e.g. Inga (Fabaceae; Richardson et al. 2001), Piper (Piperaceae; Jaramillo & Manos 2001), Ocotea (Lauraceae; Chanderbali et al. 2001) and Miconia (Melastomataceae; Renner et al. 2001).
If we assume that trait conservatism is widespread (Prinzing et al. 2001) and that phylogenetic overdispersion indicates biotic filtering, the data presented here corroborate a model of forest succession developed by Chazdon (2008) based on Oliver & Larson's (1990) temperate forest model. The model describes three phases of tropical forest succession: stand initiation, stem exclusion and understorey reinitiation (Chazdon 2008). Strong biotic filtering mechanisms act in all but the first stage of succession to produce local species pools that are less closely related than would be expected by chance.
The stand initiation phase of succession is driven by stochastic factors, dispersal limitation and the harsh microclimatic conditions of open fields (Finegan 1996; Chazdon 2008). Early regeneration is dominated by light-wooded, fast-growing species (Finegan 1996; Chazdon 2003). The relatively few species of pioneer trees, coming from fairly closely related families (Urticaceae, Malvaceae), may have contributed to the positive NRI in trees 10 cm or more dbh in young forests (figure 2). The overall lack of phylogenetic structure in forests less than 15 years (figure 1) reflects the lack of strong biotic interactions during stand initiation.
During the stem exclusion phase, the forest gains floristic and structural complexity. Biotic interactions become important in the community assembly process as species richness increases (Finegan 1996; Guariguata & Ostertag 2001; Letcher & Chazdon 2009). Forests in this phase are highly dynamic, with high recruitment and high mortality (Chazdon 2008). Smaller stem size classes are more diverse than the canopy, as species characteristic of mature forest begin to recruit (Guariguata et al. 1997; Chazdon 2008). In this dataset, forests 16–30 years had the fewest characteristic taxa (electronic supplementary material, appendix IV). When compared with the species pool for their respective age categories, sites in this age range had the lowest NRI, even though the NRI of the species pools for these two age categories was not particularly low (figure 1). This pattern of phylogenetic overdispersion could be attributed to strong biotic filtering at the local scale.
In the understorey reinitiation phase, shade-intolerant pioneer trees die back, leading to gap formation, and the forest becomes more heterogeneous with respect to light availability (Finegan 1996). In species richness, species composition and structure, the forest converges with old growth (Chazdon 2008). In this dataset, the species richness and composition of forests greater than 30 years was not significantly different from that of old growth (Letcher & Chazdon 2009). Palms and large-seeded, animal-dispersed trees rise in importance (table 2; Chazdon 2008). Both sites and species pools showed strong phylogenetic overdispersion (figure 1). Forests 31–44 years still showed lower phylogenetic overdispersion in large stem size classes, though old-growth forests did not (figure 2). This pattern may be due to remnant cohorts of pioneer trees in older secondary forests (Finegan 1996; Chazdon 2008). In the understorey reinitiation phase, the phylogenetic overdispersion evident in both sites and species pools may indicate that strong biotic interactions have shaped both local and regional communities.
Thus, the interpretation that phylogenetic overdispersion indicates biotic filtering corroborates an established model of forest succession. What about the alternative explanation for phylogenetic overdispersion, in which traits are divergent and closely related taxa occupy different successional stages? This could also shape community structure. The disturbances that initiate succession have occurred throughout the evolutionary history of tropical forests, and plants could have diversified based on their differential utilization of habitats at various successional stages (van Steenis 1958). Evidence for phylogenetic overdispersion via abiotic filtering during succession would come from sister taxa that occupy different successional stages. In this dataset, few taxa had members that were indicative of distinct successional stages (electronic supplementary material, appendix IV). A notable exception is Miconia (Melastomataceae), which had species with high IVs in three different age categories. In the field, I also observed five other Miconia species that were characteristic of particular forest ages, though the numbers I recorded were too low for their specialization to be apparent. Likewise, Protium (Burseraceae) had species with high IVs in two age categories (electronic supplementary material, appendix IV), though in this case the categories were consecutive (forests 31–44 years and old growth). Fewer species of Protium than Miconia are present in the regional flora (five versus 28; Proyecto Flora Digital de La Selva 2007). Examining relatedness and successional stage specialization in species-rich tropical genera (e.g. Miconia, Ocotea, Inga and Piper) could provide more insight into this mechanism.
Theoretically, different filtering mechanisms govern community assembly at different points in succession (Connell & Slatyer 1977; Finegan 1996; Chazdon 2008), but this has proved difficult to test empirically. Here, I demonstrate that succession produces a distinctive signature in the phylogenetic structure of communities. Isolating the mechanisms behind this pattern will require trait data for a large number of species. Although efforts are underway, no dataset currently available combines trait data, phylogenies and successional dynamics for any plant community. Combining community phylogenetic structure and trait data to examine assembly processes will be a fruitful avenue for further study.
Acknowledgements
I thank C. Webb, O. Hardy, J. Chave, P. Fine, C. Baraloto, B. Enquist, N. Swenson, N. Norden and five anonymous reviewers for comments; R. L. Chazdon for guidance; M. Luna, A. Thrall, B. Lewis and J. Foley for field assistance; the Organization for Tropical Studies and the Instituto Nacional de Biodiversidad for logistical support; the University of Connecticut, the Ronald Bamford Endowment, the UCONN Center for Conservation and Biodiversity, the Organization for Tropical Studies, an NSF Graduate Fellowship, and NSF DEB 0424767 to R. L. Chazdon for financial support. Data from this study are archived at http://www.salvias.net.
References
- Baraloto C., Morneau F., Bonal D., Blanc L., Ferry B.2007Seasonal water stress tolerance and habitat associations within four Neotropical tree genera. Ecology 88, 478–489 (doi:10.1890/0012-9658(2007)88[478:SWSTAH]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg R.1995Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (doi:10.2307/2346101) [Google Scholar]
- Cannon C. H., Leighton M.2004Tree species distributions across five habitats in a Bornean rain forest. J. Veg. Sci. 15, 257–266 (doi:10.1111/j.1654-1103.2004.tb02260.x) [Google Scholar]
- Cavender-Bares J., Ackerly D. D., Baum D. A., Bazzaz F. A.2004Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843 (doi:10.1086/386375) [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J., Keen A., Miles B.2006Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology 87, S109–S122 (doi:10.1890/0012-9658(2006)87[109:PSOFPC]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Chanderbali A. S., van der Werff H., Renner S. S.2001Phylogeny and historical biogeography of Lauraceae: evidence from the chloroplast and nuclear genomes. Ann. Mo. Bot. Gard. 88, 104–134 (doi:10.2307/2666133) [Google Scholar]
- Chave J., Müller-Landau H. C., Baker T. R., Easdale T. A., ter Steege H., Webb C. O.2006Regional and phylogenetic variation of woody density across 2456 Neotropical tree species. Ecol. Appl. 16, 2356–2367 (doi:10.1890/1051-0761(2006)016[2356:RAPVOW]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Chazdon R. L.2003Tropical forest recovery: legacies of human impact and natural disturbances. Perspec. Plant Ecol. 6, 51–71 (doi:10.1078/1433-8319-00042) [Google Scholar]
- Chazdon R. L.2008Chance and determinism in tropical forest succession. In Tropical forest community ecology (eds Carson W. P., Schnitzer S. A.), pp. 384–408 Oxford, UK: Blackwell Scientific [Google Scholar]
- Chazdon R. L., Careaga S., Webb C., Vargas O.2003Community and phylogenetic structure of reproductive traits of woody species in wet tropical forests. Ecol. Monogr. 73, 331–348 (doi:10.1890/02-4037) [Google Scholar]
- Connell J. H., Slatyer R. O.1977Mechanisms of succession in natural communities and their role in community stability and organization. Am. Nat. 111, 1119–1144 (doi:10.1086/283241) [Google Scholar]
- Dufrêne M., Legendre P.1997Species assemblages and indicator species: the need for a flexible asymmetric approach. Ecol. Monogr. 67, 345–366 (doi:10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2) [Google Scholar]
- Fine P. V. A., Miller Z. J., Mesones I., Irazuzta S., Appel H. M., Stevens M. H. H., Sääksjärvi I., Schultz J. C., Coley P. D.2006The growth-defense trade-off and habitat specialization by plants in Amazonian forests. Ecology 87, S150–S162 (doi:10.1890/0012-9658(2006)87[150:TGTAHS]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Finegan B.1996Pattern and process in neotropical secondary forests: the first 100 years of succession. Trends Ecol. Evol. 11, 119–124 (doi:10.1016/0169-5347(96)81090-1) [DOI] [PubMed] [Google Scholar]
- Guariguata M. R., Ostertag R.2001Neotropical secondary forest succession: changes in structural and functional characteristics. Forest Ecol. Manag. 148, 185–206 (doi:10.1016/S0378-1127(00)00535-1) [Google Scholar]
- Guariguata M. R., Chazdon R. L., Denslow J. S., Dupuy J. M., Anderson L.1997Structure and floristics of secondary and old-growth forest stands in lowland Costa Rica. Plant Ecol. 132, 107–120 (doi:10.1023/A:1009726421352) [Google Scholar]
- Hardy O. J., Senterre B.2007Characterizing the phylogenetic structure of communities by an additive partitioning of phylogenetic diversity. J. Ecol. 95, 493–506 (doi:10.1111/j.1365-2745.2007.01222.x) [Google Scholar]
- Harvey P. H., Pagel M. D.1991The comparative method in evolutionary biology Oxford, UK: Oxford University Press [Google Scholar]
- Hubbell S. P.2001The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- Jaramillo M. A., Manos P. S.2001Phylogeny and patterns of floral diversity in the genus Piper (Piperaceae). Am. J. Bot. 88, 706–716 (doi:10.2307/2657072) [PubMed] [Google Scholar]
- Kembel S. W., Hubbell S. P.2006The phylogenetic structure of a neotropical forest tree community. Ecology 87, S86–S99 (doi:10.1890/0012-9658(2006)87[86:TPSOAN]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Letcher S. G., Chazdon R. L.2009Rapid recovery of biomass, species richness, and species composition in a secondary forest chronosequence in northeastern Costa Rica. Biotropica 41, 608–617 (doi:10.1111/j.1744-7429.2009.00517.x) [Google Scholar]
- Losos J.2008Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 (doi:10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R.2007Mesquite: a modular system for evolutionary analysis, version 2.01. See http://mesquiteproject.org [Google Scholar]
- McCune B., Mefford M. J.1999PC-ORD: multivariate analysis of ecological data, version 4 Gleneden Beach, OR: MJM Software Design [Google Scholar]
- McDade L. A., Bawa K. S., Hespenheide H. A., Hartshorn G. S.(eds)1994La Selva: ecology and natural history of a Neotropical rain forest. Chicago, IL: University of Chicago Press [Google Scholar]
- Oliver C. D., Larson B. C.1990Forest stand dynamics. New York, NY: McGraw-Hill [Google Scholar]
- Phillips O., Miller J. S.2002Global patterns of plant diversity: Alwyn H. Gentry's forest transect data set. St. Louis, MO: Missouri Botanical Garden Press [Google Scholar]
- Prinzing A., Durka W., Klotz S., Brandl R.2001The niche of higher plants: evidence for phylogenetic conservatism. Proc. R. Soc. Lond. B 268, 2383–2389 (doi:10.1098/rspb.2001.1801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proyecto Flora Digital de La Selva 2007Lista de plantas vasculares, Estación Biológica La Selva Sarapiquí, Costa Rica: OET; See www.ots.ac.cr/local/florula [Google Scholar]
- R Development Core Team 2008R: a language and environment for statistical computing, version 2.7.2 Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- Renner S. S., Clausing G., Meyer K.2001Historical biogeography of Melastomataceae: the roles of tertiary migration and long-distance dispersal. Am. J. Bot. 88, 1290–1300 (doi:10.2307/3558340) [PubMed] [Google Scholar]
- Richardson J. E., Pennington R. T., Pennington T. D., Hollingsworth P. M.2001Recent and rapid diversification of a species-rich genus of neotropical trees. Science 293, 2242–2245 (doi:10.1126/science.1061421) [DOI] [PubMed] [Google Scholar]
- Silvertown J., Dodd M., Gowing D., Lawson C., McConway K.2006Phylogeny and the hierarchical organization of plant diversity. Ecology 87, S39–S49 (doi:10.1890/0012-9658(2006)87[39:PATHOO]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Stevens P. F.2008Angiosperm phylogeny website, version 8. See http://www.mobot.org/MOBOT/research/Apweb/ [Google Scholar]
- Swenson N. G.2009Phylogenetic resolution and quantifying the phylogenetic diversity and dispersion of communities. PLoS ONE 4, e4390 (doi:10.1371/journal.pone.0004390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson N. G., Enquist B. J., Pither J., Thompson J., Zimmerman J. K.2006The problem and promise of scale dependence in community phylogenetics. Ecology 87, 2418–2424 (doi:10.1890/0012-9658(2006)87[2418:TPAPOS]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Swenson N. G., Enquist B. J., Thompson J., Zimmerman J. K.2007The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology 88, 1770–1780 (doi:10.1890/06-1499.1) [DOI] [PubMed] [Google Scholar]
- Tofts R., Silvertown J.2000A phylogenetic approach to community assembly from a local species pool. Proc. R. Soc. Lond. B 267, 363–369 (doi:10.1098/rspb.2000.1010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Banuet A., Verdú M.2007Facilitation can increase the phylogenetic diversity of plant communities. Ecol. Lett. 10, 1029–1036 (doi:10.1111/j.1461-0248.2007.01100.x) [DOI] [PubMed] [Google Scholar]
- van Steenis C. G. G. J.1958Rejuvenation as a factor for judging the status of vegetation types. In Study of tropical vegetation: proceedings of the Kandy symposium, pp. 212–218 Paris, France: UNESCO [Google Scholar]
- Webb C. O.2000Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 156, 145–155 (doi:10.1086/303378) [DOI] [PubMed] [Google Scholar]
- Webb C. O., Donoghue M. J.2005Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Notes 5, 181–183 (doi:10.1111/j.1471-8286.2004.00829.x) [Google Scholar]
- Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J.2002Phylogenies and community ecology. Ann. Rev. Ecol. Syst. 33, 475–505 (doi:10.1146/annurev.ecolsys.33.010802.150448) [Google Scholar]
- Webb C. O., Gilbert G. S., Donoghue M. J.2006Phylodiversity-dependent seedling mortality, size structure, and disease in a Bornean rain forest. Ecology 87, S123–S131 (doi:10.1890/0012-9658(2006)87[123:PSMSSA]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Webb C. O., Ackerly D. D., Kembel S. W.2008Phylocom: software for the analysis of community phylogenetic structure and character evolution, version 4.01. See http://www.phylodiversity.net/phylocom [DOI] [PubMed] [Google Scholar]
- Wikström N., Savolainen V., Chase M. W.2001Evolution of the angiosperms: calibrating the family tree. Proc. R. Soc. Lond. B 268, 2211–2220 (doi:10.1098/rspb.2001.1782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. J.2002Plant diversity in tropical forests: a review of mechanisms of species coexistence. Oecologia 130, 1–14 [DOI] [PubMed] [Google Scholar]