Abstract
Identifying the rules and mechanisms that determine the composition and diversity of naturally co-occurring species assemblages is a central topic in community ecology. Although micro-organisms represent the ‘unseen majority’ of species, individuals and biomass in many ecosystems and play pivotal roles in community development and function, the study of the factors influencing the assembly of microbial communities has lagged behind that of plant and animal communities. In this paper, we investigate experimentally the mechanisms accounting for the low species richness of yeast communities inhabiting the nectar of the bumble-bee-pollinated Helleborus foetidus (Ranunculaceae), and explore the relationships between community assembly rules and phylogenetic relatedness. By comparing yeast communities on the glossae of foraging bumble-bees (the potential species pool) with those eventually establishing in virgin nectar probed with bee glossae (the realized community), we address the questions: (i) does nectar filter yeast inocula, so that the communities eventually established there are not random subsamples of species on bumble-bee glossae? and (ii) do yeast communities establishing in H. foetidus nectar exhibit some phylogenetic bias relative to the species pool on bumble-bee glossae? Results show that nectar filtering leads to species-poor, phylogenetically clustered yeast communities that are a predictable subset of pollinator-borne inocula. Such strong habitat filtering is probably due to H. foetidus nectar representing a harsh environment for most yeasts, where only a few phylogenetically related nectar specialists physiologically endowed to tolerate a combination of high osmotic pressure and fungicidal compounds are able to develop.
Keywords: community assembly rules, environmental filtering, Helleborus foetidus, Metschnikowia, phylogenetic structure, nectar yeast communities
1. Introduction
Identifying the rules and mechanisms that determine the composition and diversity of co-occurring species assemblages is a focal topic in community ecology. Current knowledge on these aspects, however, is largely confined to animal and plant communities (Begon et al. 2006; Gurevitch et al. 2006; Emerson & Gillespie 2008; Vamosi et al. 2009), possibly because practical difficulties (e.g. species identification) have until recently hindered the progress of microbial community ecology (Fierer et al. 2007; Jackson et al. 2007). Micro-organisms, however, represent the ‘unseen majority’ of species, individuals and biomass in many ecosystems, and play pivotal roles in community development and function (Whitman et al. 1998). The structure of microbial communities has important functional implications in ecosystems (Fuhrman 2009), thus elucidating the rules and mechanisms involved in the assembly of microbial communities will be essential to a better understanding of aspects of ecosystem functioning in which micro-organisms are involved (Horner-Devine & Bohannan 2006; Horner-Devine et al. 2007; Maherali & Klironomos 2007; Bryant et al. 2008; Fuhrman 2009).
Ecological studies on microbial communities have mostly focused on hyperdiverse microbiota associated with soil, plant surfaces and aquatic environments (Lambais et al. 2006; Schloss & Handelsman 2006b; Fierer et al. 2007; Peay et al. 2008; Shaw et al. 2008). The formidable complexity of these systems, however, limits the possibilities of unravelling the mechanisms influencing microbial composition and diversity at biologically relevant spatial scales in the field. Such questions could be profitably addressed in ‘scaled-down’ natural microbial communities made up of relatively few, easily identifiable species confined to well-defined microhabitats amenable to experimental manipulation (Maherali & Klironomos 2007). Dense yeast communities often occur in the floral nectar of animal-pollinated plants, where they can behave as parasites of plant-pollinator mutualisms (Brysch-Herzberg 2004; Canto et al. 2008; Herrera et al. 2008, 2009). Yeast communities of floral nectar seem to be characterized by lower species richness (Vörös-Felkai 1957; Eisikowitch et al. 1990; Brysch-Herzberg 2004) than those occurring in other environments like soil, plant surfaces or invertebrate guts (Lachance et al. 2001; Suh et al. 2005; Connell et al. 2008). In addition, species concepts and systematics are reasonably well developed for yeasts (Barnett et al. 2000), further contributing to render nectar yeast communities a manageable study system, particularly suited to address detailed ecological questions that are hard to tackle in more complex microbiota.
In this paper, we adopt an experimental approach to investigate the mechanisms responsible for the low species richness of yeast communities inhabiting the floral nectar of Helleborus foetidus (Ranunculaceae), a bumble-bee-pollinated plant, and to explore the possible relationships between community assembly mechanisms and phylogenetic patterns. Yeasts colonize H. foetidus floral nectar following probing of the nectaries by foraging bumble-bees that carry inocula on their glossae (Brysch-Herzberg 2004; Canto et al. 2008). Consequently, the potential composition of yeast communities in the nectar can be determined by examining the composition of yeast inocula that are ‘travelling’ on the glossae of foraging bumble-bees (i.e. the potential species pool). We will compare the yeast communities on bumble-bees' glossae with those becoming established in nectar (i.e. the realized community) previously inoculated by probing with bee glossae. This allows us to answer whether yeast communities in experimentally inoculated nectar mirror those on bumble-bee glossae or, alternatively, nectar filter the immigrant inocula so that a different community eventually builds up there. Environmental filtering has been suggested to result in phylogenetically clustered communities, i.e. made up of taxa that are more closely related among themselves than expected by chance (Webb et al. 2002; Horner-Devine & Bohannan 2006; Vamosi et al. 2009). This hypothesis motivates the second question addressed in this paper: do yeast communities experimentally established in H. foetidus nectar exhibit some phylogenetic bias in relation to the species pool on bumble-bee glossae?
In addition to shedding light on the mechanisms associated with the assembly of microbial communities, our study will also contribute to a better understanding of the interactions between plants and nectar yeasts, an intriguing relationship whose ecological and evolutionary significance is still far from being well established (Eisikowitch et al. 1990; Herrera et al. 2008, 2009). Recent quantitative surveys have documented very high frequencies of occurrence and extraordinary population densities of yeasts in nectar of animal-pollinated plants from three continents (Brysch-Herzberg 2004; Herrera et al. 2009; de Vega et al. 2009). Nectar yeasts, particularly at high densities, induce metabolic degradation of nectar, which can be detrimental to plant reproduction through reduced pollinator service (Herrera et al. 2008). This might originate selective pressures on plants to defend their nectars from exploiters through, e.g. antimicrobial secondary compounds (Adler 2000; Irwin et al. 2004). This so-called ‘antimicrobial hypothesis’ would explain the seemingly paradoxical presence of toxic substances in floral nectar as a defensive adaptation against nectarivorous microbes (González-Teuber & Heil 2009). The hypothesis relies on the as-yet-untested assumption that nectars with toxic substances are actually defended from nectarivorous microbes. By showing that nectar can be a poor growing place from many incoming yeasts, our study provides evidence supporting the antimicrobial hypothesis.
2. Material and methods
(a). Field and laboratory methods
The diversity and composition of yeast communities naturally occurring in the floral nectar of H. foetidus, an early-blooming, bumble-bee-pollinated herb whose nectar harbours dense yeast populations (Herrera et al. 2008), was studied in 2008 at one locality of Sierra de Cazorla, southeastern Spain (‘Las Navillas’, 1220 m elevation). On 25 February and 10 March, single-nectary nectar samples were collected with sterile microcapillaries from flowers of 10 widely spaced H. foetidus plants exposed to natural pollinator visitation (two flowers per plant, two nectaries per flower, on each collection date). A 1 µl aliquot of each nectar sample (n = 80) was streaked onto a Yeast Mould (YM) + chloramphenicol agar plate (1.0% glucose, 0.5% peptone, 0.3% malt extract, 0.3% yeast extract, 2.0% agar, 0.01% chloramphenicol, pH = 6.0), and incubated at 25°C. For each cultured nectar sample, distinct yeast isolates were obtained from all the resulting colonies following standard methods and criteria described in Yarrow (1998). For each isolate, approximately 500 nucleotides of the D1/D2 domain of the 26S subunit ribosomal DNA, the gene most commonly used for yeast identification and phylogenetic studies, were two-way sequenced following methods in Kurtzman & Robnett (1998) and Lachance et al. (1999). DNA sequences were obtained for a total of 39 yeast isolates, which were analysed following the methods described in §2b. The rest of each nectar sample (mean ± s.d. = 1.6 ± 0.6 µl, n = 80) was examined microscopically and yeast cell density determined following methods described by Herrera et al. (2009).
During the study period, the only species flowering locally were H. foetidus and Rosmarinus officinalis (Lamiaceae), and bumble-bees visited flowers of both species when growing at close range. To determine as comprehensively as possible the composition of the yeast species pool potentially arriving at H. foetidus flowers, 45 bumble-bees (Bombus terrestris) were hand-netted between 23 February and 17 March 2008 while foraging at flowers of H. foetidus (n = 30) and R. officinalis (n = 15) within a radius of approximately 1.5 km around the Las Navillas site. Netting was not done on the same H. foetidus plants used for sampling natural nectar yeast communities to avoid interference with other studies. Because we were interested in the overall composition of the local bumble-bee-borne yeast community, all netted bees were treated in the analyses as a single sample irrespective of the plant where they had been captured. Nevertheless, to test for the robustness of results to differences in the origin of samples, some analyses were repeated separately for data from bumble-bees collected from H. foetidus and R. officinalis plants.
Immediately after capture, bees were placed individually in sterile containers and anaesthetized by placing them inside a refrigerator at 4°C until used in the experiments, generally within a few hours of collection. In these experiments, bees were used as sources of inocula to generate two types of artificial yeast communities, by inoculating either sterile yeast culture media (‘bee-only communities’ hereafter) or virgin natural nectar from H. foetidus flowers (‘bee + nectar communities’ hereafter) from 10 plants that had been previously excluded from pollinator visitation by bagging inflorescences at the bud stage. Since individual H. foetidus plants may differ in the susceptibility of nectar to yeast growth (C. M. Herrera 2009, unpublished data), two or three virgin flowers were collected from each plant, pooled in a container and a random sample of nectaries drawn for use in the inoculation experiments. The glossa of each individual bee was extended using fine forceps beyond the tip of the maxillary galeae and, depending on the treatment, either rubbed against the surface of a YM + chloramphenicol agar culture plate (‘bee-only’ treatment) or briefly introduced into a nectary of a virgin flower containing natural nectar to mimic natural nectar probing (‘bee + nectar’ treatment). The forceps used to handle bee tongues were cleaned between assays using 99 per cent ethanol. Bumble-bee individuals were assigned to experimental treatments in one of two ways. Each bee was used either to inoculate culture media (n = 15) or nectar (n = 15) alone, or to inoculate both culture media and nectar in succession (n = 15), the order being alternated in successive assays. Results obtained with the two methods were similar and have been combined for the analyses. Culture plates from the bee-only treatment were handled as described above for wild-collected nectar samples. Inoculated nectaries (bee + nectar treatment) were kept within a sealed container at room temperature for 48 h, and then the nectar in each nectary was streaked onto a YM + chloramphenicol agar plate, incubated and yeast isolates obtained from each nectar sample following the same protocols as for cultures from wild-collected nectar and the bee-only treatment. The D1/D2 domain of the 26S subunit ribosomal DNA was also two-way sequenced for all the isolates obtained from experimental treatments.
(b). Data analyses
DNA sequences of yeast isolates were used to assess the composition and diversity of yeast communities found in the natural nectar samples of H. foetidus and in the artificial assemblages generated by the bee-only and bee + nectar experimental treatments. The nucleotide collection databases at GenBank were queried with the Basic Local Alignment Search Tool (BLAST; Altschul et al. 1997) to look for described yeast species with DNA sequences matching those of our isolates. All sequences queried (n = 136) yielded highly significant alignments with named yeast accessions in GenBank databases, most often at very high levels of sequence identity and coverage (99–100%). In a few cases (5.2% of queries, all from experimental groups), sequence identity was slightly lower (96–98%), which might denote the presence of undescribed taxa or species without accessions in the GenBank databases. To evaluate possible biases in richness estimates caused by the presence of undescribed species, we also considered operational taxonomic units (OTUs) defined on the basis of similarity of DNA sequences (Hughes et al. 2001; Fierer et al. 2007; Shaw et al. 2008). Determination of the number of distinct OTUs occurring in a set of DNA sequences, and assignment of sequences to OTUs, was done with the program DOTUR (Schloss & Handelsman 2005), using a DNA dissimilarity cut-off of 3 per cent. Although this cut-off is more conservative than the 1 per cent threshold suggested for species-level rDNA differentiation in yeasts (Kurtzman & Robnett 1998), it is the threshold commonly used to distinguish ‘molecular’ species of micro-organisms in recent environmental studies (Fierer et al. 2007; Peay et al. 2008).
Although both the number of bumble-bees assayed in the experiments and the number yielding some yeast colony were similar for the bee-only (n = 30 and 22 bumble-bees, respectively) and bee + nectar treatments (n = 30 and 20, respectively), the number of yeast isolates obtained differed between treatments, hence rarefaction methods were applied to compare diversities (Gotelli & Colwell 2001). Expected yeast species richness was estimated using individual-based rarefaction curves and the Chao2 non-parametric asymptotic estimator, treating each DNA sequence as a separate individual (Hughes et al. 2001). Expected richness and standard errors were computed with the program EstimateS (Colwell 2005).
The phylogenetic bias in the assembly of nectar yeast communities was examined by testing (i) whether isolates from the bee-only and bee + nectar treatments are unequally distributed over the common phylogenetic tree for all isolates combined, i.e. the two experimental groups harbour different lineages, and (ii) whether yeasts in the bee + nectar treatment represent a phylogenetically structured subset of those in the bee-only one, i.e. the nectar yeast community is phylogenetically clustered. The whole set of DNA sequences from the two experimental groups (n = 97) was aligned with ClustalW, and Gblocks was then used to eliminate poorly aligned positions and divergent regions (Castresana 2000). A phylogenetic tree was constructed for all isolates using MrBayes 3.1 (Ronquist & Huelsenbeck 2003), assuming a general time-reversible model and gamma-distributed rates. The hypothesis that the bee-only and bee + nectar communities harbour different lineages was tested with the parsimony test implemented in the program TreeClimber (Schloss & Handelsman 2006a). Given the phylogenetic tree constructed for all isolates, each tip was associated with one experimental group and the minimum number of transitions between groups was obtained using Fitch's (1971) algorithm. Statistical significance was tested by comparing that value with a null-model distribution obtained from randomly distributed experimental groups on the same phylogenetic tree. The hypothesis of phylogenetic clustering was tested with the ‘comstruct’ function of the Phylocom program (Webb et al. 2008). Net relatedness index (NRI), a standardized form of mean phylogenetic distance, was computed for yeast communities in the two experimental groups, and their significance assessed by comparison with randomly generated samples. Results obtained with the four null models available in the program were similar, and only those from the ‘phylogeny shuffle’ method will be presented.
3. Results
Nectar samples from H. foetidus flowers exposed to natural pollinator visitation contained yeasts quite frequently. Yeasts were present in 72.5 per cent of the 80 nectar samples from the Las Navillas site that were examined microscopically. In yeast-containing samples, cell density ranged between 34 and 127 622 yeast cells per mm3 (mean ± s.e. = 17 821 ± 2844 cells per mm3, n = 58). A total of 39 isolates were sequenced, all of which belonged to a single species, Metschnikowia reukaufii. Per cent identity scores between their DNA sequences and the best alignments from BLAST queries were very high, ranging between 98 and 100 per cent (mean ± s.e. = 99.5 ± 0.1%).
A total of nine different yeast species were represented in the isolates obtained from the bee-only experimental treatment, belonging to the genera Metschnikowia (two species), Candida (two), Hanseniaspora (two), Kluyveromyces (one), Saccharomyces (one) and Cryptococcus (one) (table 1). Isolates from the bee + nectar treatment had lower observed species richness. Only four species were identified, representing a subset of those found in the bee-only treatment (table 1). The DOTUR-based analysis of DNA sequence data yielded similar results. A total of 10 OTUs were recognized in the bee-only isolates, while only six OTUs were identified in the bee + nectar isolates. Given the similarity between richness estimates based on named species and OTUs, we will consider hereafter only the results obtained using the first method.
Table 1.
Taxon richness and species composition of the two groups of experimentally assembled nectar yeast communities considered in this study. The bee-only and bee + nectar experimental groups refer, respectively, to assemblages obtained by rubbing the glossae of bumble-bees on culture media and from probing H. foetidus virgin nectar with the bees’ glossae, incubating for 48 h and then streaking the inoculated nectar on culture media.
| bee-only | bee + nectar | |
|---|---|---|
| yeast isolates sequenced | 37 | 60 |
| observed number of taxa: | ||
| named species, BLAST search | 9 | 4 |
| OTUs, 3% DNA dissimilarity cut-off | 10 | 6 |
| BLAST-identified, named species (number of sequences): | ||
| Metschnikowia reukaufii | 18 | 45 |
| M. gruessii | 3 | 11 |
| Candida bombi | 7 | 3 |
| Kluyveromyces dobzhanskii | 3 | 0 |
| Hanseniaspora sp.a | 2 | 1 |
| H. osmophila | 1 | 0 |
| Saccharomyces bayanus | 1 | 0 |
| Cryptococcus saitoi | 1 | 0 |
| Candida friedrichii | 1 | 0 |
aGenBank accession EF653942.1.
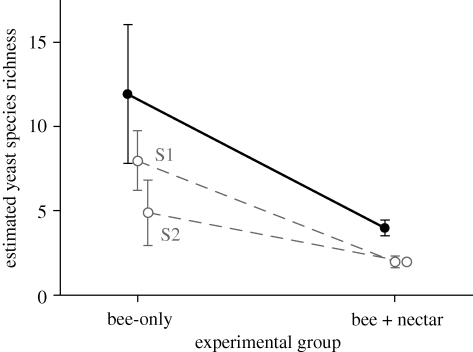
The Chao2 non-parametric estimator of species richness reached (bee + nectar) or closely approached (bee-only) a plateau for the total number of isolates examined for each experimental group (results not shown). This indicates that the number of isolates sampled for each group was sufficient to provide reliable estimates of the expected total species richness. Chao2 richness estimates (±s.e.) were higher for the bee-only (11.9 ± 4.1 species) than for the bee + nectar group (4 ± 0.4 species), and the sign of the difference was robust to splitting the sample into subsamples consisting of the bumble-bees captured at H. foetidus and R. officinalis flowers (figure 1). In addition to differences in observed and estimated yeast species richnesses, the two experimental groups differed in evenness, with the most abundant species (M. reukaufii) accounting for 48.6 and 75 per cent of isolates in the bee-only and bee + nectar groups, respectively (table 1).
Figure 1.
Estimated yeast species richness (Chao2 non-parametric estimator) for bee-only and bee + nectar experimental communities (black dots and continuous line; vertical segments denote ±1 s.e.). Bee-only and bee + nectar yeast assemblages refer, respectively, to those resulting from rubbing the glossae of bumble-bees on culture media, and from probing virgin H. foetidus nectar with the bees’ glossae, incubating for 48 h, and then streaking the inoculated nectar on culture media. Grey dots and dashed lines depict species richness estimates computed separately for data subsets corresponding to bumble-bees captured at flowers of H. foetidus (S1) and R. officinalis (S2).
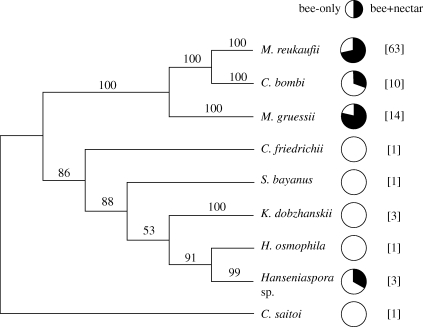
The phylogenetic tree constructed for the combined set of 97 isolates from the two experimental groups is shown in the electronic supplementary material, appendix. Yeast isolates associated with the bee-only and bee + nectar experimental groups harboured different lineages, as denoted by the highly significant result of the parsimony test (p = 0.0006). In addition, bee + nectar samples were clustered relative to random communities of equal richness assembled from the pool of all yeasts found in bee-only samples (i.e. the pool of all yeast species in all samples), as revealed by their NRI departing significantly from null model expectations (table 2). Results were nearly identical when separate analyses were conducted on data from bumble-bees collected from H. foetidus and R. officinalis (table 2). Taken together, these results show that yeasts communities experimentally assembled in the nectar comprised a phylogenetically biased, clustered subset of isolates occurring on bumble-bee glossae, as illustrated in the simplified, species-level phylogenetic tree shown in figure 2. With the only exception of a single isolate of Hanseniaspora sp., all isolates from the bee + nectar group clustered into a distinct ‘nectarivorous’ lineage with 100 per cent Bayesian credibility comprising M. reukaufii, M. gruessii and Candida bombi.
Table 2.
NRI (a standardized measurement of mean pairwise phylogenetic distance) for yeast isolates in the two groups of communities assembled experimentally. Subsets 1 and 2 refer to data from bumble-bees captured at flowers of H. foetidus and R. officinalis, respectively. p-Values were calculated by comparing the observed mean pairwise phylogenetic distance with a null distribution for 105 randomly generated phylogenies, obtained by shuffling yeast isolates across the tips of the phylogeny shown in the electronic supplementary material, appendix (phylogeny shuffle method in the Phylocom program). The p-values shown correspond to one-tailed tests, and stand for the probability of obtaining by chance alone a mean phylogenetic distance as extreme as the observed one. NRI significantly greater than 0 denote phylogenetic clustering.
| bee-only |
bee + nectar |
|||
|---|---|---|---|---|
| dataset | NRI | p-value | NRI | p-value |
| whole sample | −4.26 | <0.00001 | +5.17 | <0.00001 |
| subset 1 | −2.17 | 0.013 | +6.54 | <0.00001 |
| subset 2 | −2.18 | 0.013 | +0.23 | 0.41 |
Figure 2.
Phylogenetic tree summarizing the evolutionary relationships between the nine yeast species identified in this study, obtained by the Bayesian inference analysis of DNA sequence data from individual isolates. The simplified tree and Bayesian clade credibility (posterior probability) values presented here were generated by culling taxa from the larger phylogram depicting the relationships between the 97 isolates from the two experimental groups combined, shown in the electronic supplementary material, appendix. Clade credibility values, expressed as percentages, are shown next to the branches (are missing from branches represented by single isolates). Pie charts denote the proportion of isolates of each species in the bee-only (open sectors) and bee + nectar (filled sectors) experimental treatments. Total numbers of isolates per species are shown in brackets (see also table 1).
4. Discussion
Our inventories of yeast species in natural nectar samples of H. foetidus corroborate the results of previous studies showing that, at least in temperate habitats, culturable yeast communities associated with floral nectar are characterized by low species richness (Vörös-Felkai 1957; Eisikowitch et al. 1990; Brysch-Herzberg 2004). During the two-week sampling period, yeast communities in H. foetidus nectar from the Las Navillas site consisted of a single species, M. reukaufii. The experimental yeast assemblages obtained by inoculating nectar with yeasts on bumble-bee glossae were slightly more diverse, containing three additional species (M. gruessii, C. bombi and Hanseniaspora sp.). Since bumble-bees used in the experiments were collected over a broader area than natural nectar samples, the difference in yeast diversity between natural H. foetidus nectar and experimental bee + nectar samples probably reflect variation over the sampling area in the composition of yeast communities on bumble-bee glossae. Another question raised by our results concerns the origin of inocula of yeast species that were unable to grow in H. foetidus: if they are not found in H. foetidus nectar, then where did bumble-bee glossae get contaminated with inocula of these species? Their most likely origins are honey pots at the bumble-bees’ nests, pollen and flower surfaces contacted during foraging bouts, and floral nectar of other plants (Lachance et al. 2001; Brysch-Herzberg 2004).
Both observed and estimated yeast species richness was lower in nectar (bee + nectar treatment) than on the bumble-bee glossae used to inoculate it (bee-only treatment), which demonstrates that H. foetidus nectar is filtering the multi-species set of inocula brought in by foraging bumble-bees. Circumstantial evidence suggests that such habitat filtering is most likely due to H. foetidus nectar representing a strongly inhibitory environment that constrains the growth of most yeast species, and that only a few nectar specialists possessing certain physiological abilities can successfully develop there. Helleborus foetidus nectar contains protoanemonin (R. Pérez, I. M. García & C. M. Herrera 2009, unpublished data), an unsaturated lactone that inhibits the growth of some yeasts even at extremely low concentrations (Mares 1987; Kyung et al. 2007; A. Canto 2009, unpublished data). In addition, sugar concentration of virgin H. foetidus nectar is often very high (mean ± s.e. = 40.5 ± 1.2% w/w, range = 28–58%, n = 50; C. M. Herrera 2009, unpublished data), as usual among bee-pollinated plants (Nicolson et al. 2007). The osmotic stress associated with these high sugar concentrations is probably a limiting environmental factor, since tolerance to high sugar concentrations is a specialized physiological trait (Tokuoka 1993) possessed by only a small fraction of yeast species (around 13%, according to data in Barnett et al. 2000). Consistent with our interpretation that osmotic stress is an ecological factor contributing to nectar filtering is the observation that the three main species unaffected by filtering (M. reukaufii, M. gruessii, C. bombi) can grow on culture media containing 50 per cent glucose, while at least three of the species filtered by nectar cannot (e.g. Saccharomyces bayanus, Cryptococcus saitoi, Hanseniaspora osmophila) (Barnett et al. 2000). In a comparative screening of 252 strains of micro-organisms, M. reukaufii exhibited the highest arabitol production rate when subjected to intense osmotic stress (Nozaki et al. 2003). Since arabitol is one of several osmolytes playing an essential role in osmoregulation by osmotolerant yeasts (Grant 2004), that finding supports the suggested association between the ability of a yeast species to tolerate high osmotic pressure and its capacity to go through the ecological filter set by nectar.
Since high sugar concentration and presence of secondary compounds are common features of floral nectars (Adler 2000; Nicolson et al. 2007; González-Teuber & Heil 2009), the low species diversity prevailing in nectar yeast communities so far studied could reflect a generalized environmental filtering similar to that documented here for H. foetidus. Very low nitrogen content, another characteristic feature of floral nectars (Nicolson et al. 2007), may be yet another factor limiting the suitability of floral nectars as habitats for yeasts other than highly specialized nectarivores. In this respect, it is interesting to note that the two species contributing most isolates in our study (M. reukaufii and M. gruessii) were also the most abundant ones in nectar samples from 143 insect-pollinated species from Central Europe, where they accounted for 73 per cent of all isolates (Brysch-Herzberg 2004). In a survey of the nectar yeasts of 22 species of southern Spanish plants, M. reukaufii and M. gruessii accounted altogether for 87 per cent of all isolates (M. I. Pozo 2009, unpublished data). Given the close phylogenetic relatedness of the two species (Hong et al. 2003), and the frequent association between phylogeny and physiology exhibited by yeasts (Middelhoven & Kurtzman 2003), the dominance of these two Metschnikowia species in western European nectar yeast communities is probably associated with their possessing some suite of physiological traits allowing them to overcome nectar filtering. A combination of osmotolerance, tolerance or resistance to secondary compounds and efficient nitrogen use possibly allows these specialists to exploit floral nectar.
Many studies have shown that patterns of co-occurrence of taxa can deviate from random expectations with regard to phylogenetic relatedness (Emerson & Gillespie 2008; Vamosi et al. 2009), but microbial communities have been infrequently examined in a phylogenetic context and the few studies available provide contrasting results. Anderson et al. (2004), working on yeast communities in decaying cactus tissues, found associations between yeast species abundance and phylogenetic relatedness to be variable and contingent on features of the environment. In contrast, bacterial communities are often phylogenetically clustered, a pattern generally interpreted as denoting habitat filtering when closely related taxa share some phylogenetically conserved trait(s) that allows them to tolerate the abiotic conditions of a given habitat (Webb et al. 2002; Emerson & Gillespie 2008; Vamosi et al. 2009). The interpretation of phylogenetic community structure in natural systems, however, is subject to many confounding factors, and experimental manipulative investigations can be essential to unravel the underlying mechanisms involved (Maherali & Klironomos 2007; Vamosi et al. 2009). By directly assessing, rather than inferring, the species composition of the regional species pool (bumble-bee glossae) and experimentally ensuring that all species in the pool were equally likely to colonize the focal habitat (nectar), our study provides compelling support for the connection between habitat filtering and phylogenetic clustering, and also allows the identification of the likely ecological mechanism underlying clustering. As discussed above, the phylogenetically clustered subset of species present on bumble-bee glossae that were able to grow in H. foetidus nectar apparently share some capabilities to tolerate the harsh nectar environment, as assumed by the hypothesis linking habitat filtering and phylogenetic clustering (Vamosi et al. 2009).
Certain floral traits involved in mutualistic interactions with pollinators can be partly explained as the outcome of selection to reduce the impact of exploiters on plant fitness (Irwin et al. 2004). In this context, the so-called antimicrobial hypothesis interprets the toxic substances often occurring in floral nectars as defences against nectarivorous microbes (Adler 2000; González-Teuber & Heil 2009). One key assumption underlying the antimicrobial hypothesis, that the deleterious effects of microbes on nectar can be sufficiently severe as to select for antimicrobial compounds, was recently supported by studies on southern Spanish and South African plants showing that yeasts induce substantial degradation of floral nectar (Herrera et al. 2008; de Vega et al. 2009). The present investigation adds support for another central assumption of the antimicrobial hypothesis, that nectars with secondary compounds are protected from nectarivorous micro-organisms. Toxic substances in H. foetidus nectar, however, do not confer protection against specialized microbial consumers (see also Manson et al. 2007), just as allelochemicals in other plant parts do not defend them from specialized herbivores (Bowers & Puttick 1988). Poisons in H. foetidus nectar possibly contribute to filter out unspecialized yeasts in the same way as plant allelochemicals filter out unspecialized herbivores. Under this hypothesis, the specialized nectarivores M. reukaufii, M. gruessii and C. bombi are predicted to tolerate protoanemonin and perhaps other inhibitory substances commonly found in nectar, just like specialist herbivores are immune to the allelochemicals of their host plants. This parallelism, if substantiated by other studies, would suggest an appealing similarity of the defensive mechanisms evolved by plants against microbial and non-microbial antagonists.
Acknowledgements
We are grateful to Pedro A. Tíscar and the Centro de Capacitación y Experimentación Forestal de Vadillo-Castril, Cazorla for essential laboratory space and facilities; José L. Garrido for advice on phylogenetic methods; André Lachance for useful advice; Clara de Vega for discussion and comments on the manuscript; and Lawrence Harder and two anonymous reviewers for suggestions on the manuscript. Permission to work in the Sierra de Cazorla was granted by the Consejería de Medio Ambiente, Junta de Andalucía. Funding for this work was provided by grants P06-RNM-01627 (Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía), CGL2006-01355 and EXPLORA CGL2007-28866-E/BOS (Ministerio de Educación y Ciencia, Gobierno de España) and FPI Predoctoral Fellowship BES-2007-17142 from Ministerio de Educación y Ciencia to M.I.P.
References
- Adler L. S.2000The ecological significance of toxic nectar. Oikos 91, 409–420 (doi:10.1034/j.1600-0706.2000.910301.x) [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J.1997Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T. M., Lachance M. A., Starmer W. T.2004The relationship of phylogeny to community structure: the cactus yeast community. Am. Nat. 164, 709–721 (doi:10.1086/425372) [DOI] [PubMed] [Google Scholar]
- Barnett J. A., Payne R. W., Yarrow D.2000Yeasts: characteristics and identification, 3rd edn Cambridge, UK: Cambridge University Press [Google Scholar]
- Begon M., Townsend C. R., Harper J. L.2006Ecology. From individuals to ecosystems, 4th edn Oxford, UK: Blackwell Publishing [Google Scholar]
- Bowers M. D., Puttick G. M.1988Response of generalist and specialist insects to qualitative allelochemical variation. J. Chem. Ecol. 14, 319–334 [DOI] [PubMed] [Google Scholar]
- Bryant J. A., Lamanna C., Morlon H., Kerkhoff A. J., Enquist B. J., Green J. L.2008Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl Acad. Sci. USA 105, 11 505–11 511 (doi:10.1073/pnas.0801920105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brysch-Herzberg M.2004Ecology of yeasts in plant–bumblebee mutualism in Central Europe. FEMS Microbiol. Ecol. 50, 87–100 [DOI] [PubMed] [Google Scholar]
- Canto A., Herrera C. M., Medrano M., Pérez R., García I. M.2008Pollinator foraging modifies nectar sugar composition in Helleborus foetidus L. (Ranunculaceae): an experimental test. Am. J. Bot. 95, 315–320 (doi:10.3732/ajb.95.3.315) [DOI] [PubMed] [Google Scholar]
- Castresana J.2000Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- Colwell R. K. EstimateS: statistical estimation of species richness and shared species from samples, version 7.5. 2005. See User's Guide and application at http://purl.oclc.org/estimates . [Google Scholar]
- Connell L., Redman R., Craig S., Scorzetti G., Iszard M., Rodriguez R.2008Diversity of soil yeasts isolated from South Victoria Land, Antarctica. Microb. Ecol. 56, 448–459 (doi:10.1007/s00248-008-9363-1) [DOI] [PubMed] [Google Scholar]
- de Vega C., Herrera C. M., Johnson S. D.2009Yeasts in floral nectar of some South African plants: quantification and associations with pollinator type. S. Afr. J. Bot. (doi:10.1016/j.sajb.2009.07.016) [Google Scholar]
- Eisikowitch D., Kevan P. G., Lachance M. A.1990The nectar-inhabiting yeasts and their effect on pollen germination in common milkweed, Asclepias syriaca L. Isr. J. Bot. 39, 217–225 [Google Scholar]
- Emerson B. C., Gillespie R. G.2008Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 23, 619–630 (doi:10.1016/j.tree.2008.07.005) [DOI] [PubMed] [Google Scholar]
- Fierer N., et al. 2007Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73, 7059–7066 (doi:10.1128/AEM.00358-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M.1971Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (doi:10.2307/2412116) [Google Scholar]
- Fuhrman J. A.2009Microbial community structure and its functional implications. Nature 459, 193–199 (doi:10.1038/nature08058) [DOI] [PubMed] [Google Scholar]
- González-Teuber M., Heil M.2009Nectar chemistry is tailored for both attraction of mutualists and protection from exploiters. Plant Signal. Behav. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotelli N. J., Colwell R. K.2001Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391 (doi:10.1046/j.1461-0248.2001.00230.x) [Google Scholar]
- Grant W. D.2004Life at low water activity. Phil. Trans. R. Soc. Lond. B 359, 1249–1266 (doi:10.1098/rstb.2004.1502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitch J., Scheiner S. M., Fox G. A.2006The ecology of plants, 2nd edn Sunderland, MA: Sinauer [Google Scholar]
- Herrera C. M., García I. M., Pérez R.2008Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89, 2369–2376 (doi:10.1890/08-0241.1) [DOI] [PubMed] [Google Scholar]
- Herrera C. M., de Vega C., Canto A., Pozo M. I.2009Yeasts in floral nectar: a quantitative survey. Ann. Bot. 103, 1415–1423 (doi:10.1093/aob/mcp026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. G., Bae K. S., Herzberg M., Titze A., Lachance M. A.2003Candida kunwiensis sp. nov., a yeast associated with flowers and bumblebees. Int. J. Syst. Evol. Microbiol. 53, 367–372 (doi:10.1099/ijs.0.02200-0) [DOI] [PubMed] [Google Scholar]
- Horner-Devine M. C., Bohannan B. J. M.2006Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87, S100–S108 [DOI] [PubMed] [Google Scholar]
- Horner-Devine M. C., et al. 2007A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88, 1345–1353 (doi:10.1890/06-0286) [DOI] [PubMed] [Google Scholar]
- Hughes J. B., Hellmann J. J., Ricketts T. H., Bohannan B. J. M.2001Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67, 4399–4406 (doi:10.1128/AEM.67.10.4399-4406.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin R. E., Adler L. S., Brody A. K.2004The dual role of floral traits: pollinator attraction and plant defense. Ecology 85, 1503–1511 (doi:10.1890/03-0390) [Google Scholar]
- Jackson R. B., Fierer N., Schimel J. P.2007New directions in microbial ecology. Ecology 88, 1343–1344 (doi:10.1890/06-1882) [Google Scholar]
- Kurtzman C. P., Robnett C. J.1998Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73, 331–371 (doi:10.1023/A:1001761008817) [DOI] [PubMed] [Google Scholar]
- Kyung K. H., Woo Y. H., Kim D. S., Park H. J., Kim Y. S.2007Antimicrobial activity of an edible wild plant, apiifolia Virgin's Bower (Clematis apiifolia DC). Food Sci. Biotechnol. 16, 1051–1054 [Google Scholar]
- Lachance M. A., Bowles J. M., Starmer W. T., Barker J. S. F.1999Kodamaea kakaduensis and Candida tolerans, two new ascomycetous yeast species from Australian Hibiscus flowers. Can. J. Microbiol. 45, 172–177 (doi:10.1139/cjm-45-2-172) [DOI] [PubMed] [Google Scholar]
- Lachance M. A., Starmer W. T., Rosa C. A., Bowles J. M., Barker J. S. F., Janzen D. H.2001Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 1, 1–8 [DOI] [PubMed] [Google Scholar]
- Lambais M. R., Crowley D. E., Cury J. C., Bull R. C., Rodrigues R. R.2006Bacterial diversity in tree canopies of the Atlantic forest. Science 312, 1917 (doi:10.1126/science.1124696) [DOI] [PubMed] [Google Scholar]
- Maherali H., Klironomos J. N.2007Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748 (doi:10.1126/science.1143082) [DOI] [PubMed] [Google Scholar]
- Manson J. S., Lachance M. A., Thomson J. D.2007Candida gelsemii sp. nov., a yeast of the Metschnikowiaceae clade isolated from nectar of the poisonous Carolina jessamine. Antonie Van Leeuwenhoek 92, 37–42 (doi:10.1007/s10482-006-9132-4) [DOI] [PubMed] [Google Scholar]
- Mares D.1987Antimicrobial activity of protoanemonin, a lactone from ranunculaceous plants. Mycopathology 98, 133–140 (doi:10.1007/BF00437648) [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J., Kurtzman C. P.2003Relation between phylogeny and physiology in some ascomycetous yeasts. Antonie Van Leeuwenhoek 83, 69–74 (doi:10.1023/A:1022916630030) [DOI] [PubMed] [Google Scholar]
- Nicolson S. W., Nepi M., Pacini E. (eds) 2007Nectaries and nectar Dordrecht, The Netherlands: Springer-Verlag [Google Scholar]
- Nozaki H., Suzuki S., Tsuyoshi N., Yokozeki K.2003Production of d-arabitol by Metschnikowia reukaufii AJ14787. Biosci. Biotechnol. Biochem. 67, 1923–1929 (doi:10.1271/bbb.67.1923) [DOI] [PubMed] [Google Scholar]
- Peay K. G., Kennedy P. G., Bruns T. D.2008Fungal community ecology: a hybrid beast with a molecular master. BioScience 58, 799–810 (doi:10.1641/B580907) [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Handelsman J.2005Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506 (doi:10.1128/AEM.71.3.1501-1506.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Handelsman J.2006aIntroducing TreeClimber, a test to compare microbial community structures. Appl. Environ. Microbiol. 72, 2379–2384 (doi:10.1128/AEM.72.4.2379-2384.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Handelsman J.2006bToward a census of bacteria in soil. PLoS Comput. Biol. 2, e92 (doi:10.1371/journal.pcbi.0020092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. K., Halpern A. L., Beeson K., Tran B., Venter J. C., Martiny J. B. H.2008It's all relative: ranking the diversity of aquatic bacterial communities. Environ. Microbiol. 10, 2200–2210 (doi:10.1111/j.1462-2920.2008.01626.x) [DOI] [PubMed] [Google Scholar]
- Suh S. O., McHugh J. V., Pollock D. D., Blackwell M.2005The beetle gut: a hyperdiverse source of novel yeasts. Mycol. Res. 109, 261–265 (doi:10.1017/S0953756205002388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuoka K.1993Sugar and salt-tolerant yeasts. J. Appl. Bacteriol. 74, 101–110 [Google Scholar]
- Vamosi S. M., Heard S. B., Vamosi J. C., Webb C. O.2009Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18, 572–592 (doi:10.1111/j.1365-294X.2008.04001.x) [DOI] [PubMed] [Google Scholar]
- Vörös-Felkai G.1957Données sur les levures de fleurs répandues en Hongrie. Acta Bot. Acad. Sci. Hung. 3, 391–399 [Google Scholar]
- Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J.2002Phylogenies and community ecology. Ann. Rev. Ecol. Syst. 33, 475–505 (doi:10.1146/annurev.ecolsys.33.010802.150448) [Google Scholar]
- Webb C. O., Ackerly D. D., Kembel S. W.2008Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–2100 (doi:10.1093/bioinformatics/btn358) [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J.1998Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (doi:10.1073/pnas.95.12.6578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrow D.1998Methods for the isolation, maintenance and identification of yeasts. In The yeasts. A taxonomic study (eds Kurtzman C. P., Fell J. W.), pp. 77–100 Amsterdam, The Netherlands: Elsevier [Google Scholar]