Abstract
The dispersal of individuals among fragmented populations is generally thought to prevent genetic and demographic isolation, and ultimately reduce extinction risk. In this study, we show that a century of reduction in coastal old-growth forests, as well as a number of other environmental factors, has probably resulted in the genetic divergence of marbled murrelets (Brachyramphus marmoratus) in central California, despite the fact that 7 per cent of modern-sampled murrelets in this population were classified as migrants using genetic assignment tests. Genetic differentiation appears to persist because individuals dispersing from northern populations contributed relatively few young to the central California population, as indicated by the fact that migrants were much less likely to be members of parent–offspring pairs than residents (10.5% versus 45.4%). Moreover, a recent 1.4 per cent annual increase in the proportion of migrants in central California, without appreciable reproduction, may have masked an underlying decline in the resident population without resulting in demographic rescue. Our results emphasize the need to understand the behaviour of migrants and the extent to which they contribute offspring in order to determine whether dispersal results in gene flow and prevents declines in resident populations.
Keywords: dispersal, genetic variation, habitat fragmentation, marbled murrelet, old-growth forest, rescue effects
1. Introduction
Habitat loss and fragmentation are among the leading contemporary causes of extinction in vertebrates (Wilcove et al. 1998). Small populations in remnant habitat patches may have elevated risks of local extinction owing to deterministic factors, such as edge effects, predation and nest parasitism (Wilcove 1985; Chalfoun et al. 2002; Fahrig 2003), as well as increased susceptibility to demographic and environmental stochasticity (Lande 1993). Inbreeding can also increase the probability of extinction in fragmented populations (Keller & Waller 2002), and the loss of additive genetic variation can constrain adaptation to future environmental change (Lande & Shannon 1996).
Movements of individuals among fragmented populations is thought to reduce extinction risk by increasing both local abundance (Brown & Kodric-Brown 1977) and genetic diversity (Westemeier et al. 1998; Madsen et al. 1999). However, demographic frameworks for describing spatially structured populations generally assume that migrant and resident individuals are equivalent with respect to their contributions to subsequent generations (Pulliam 1988; Thomas & Kunin 1999). Specifically, rescue effects expected from interpopulation movements are based on the assumption that migrants recruit into the breeding population and contribute sufficient offspring to increase population growth (Pulliam 1988). Similarly, genetic rescue requires that migrants produce enough offspring so that gene flow balances the loss of genetic diversity in small populations (Allendorf & Luikart 2007).
Migrants, however, may not recruit into the breeding population or successfully produce offspring (Dearborn et al. 2003). Such individuals may simply represent temporary visitors (e.g. prospectors for breeding sites), seasonal migrants that return to their original breeding population the subsequent breeding season, or immigrants that attempt to recruit into the population but produce fewer young than residents (Nosil et al. 2005). Dispersal among populations without the production of young by migrants is not expected to result in genetic and demographic rescue, may mask the continued erosion of a resident population, and may delay the implementation of conservation measures that could prevent extinction.
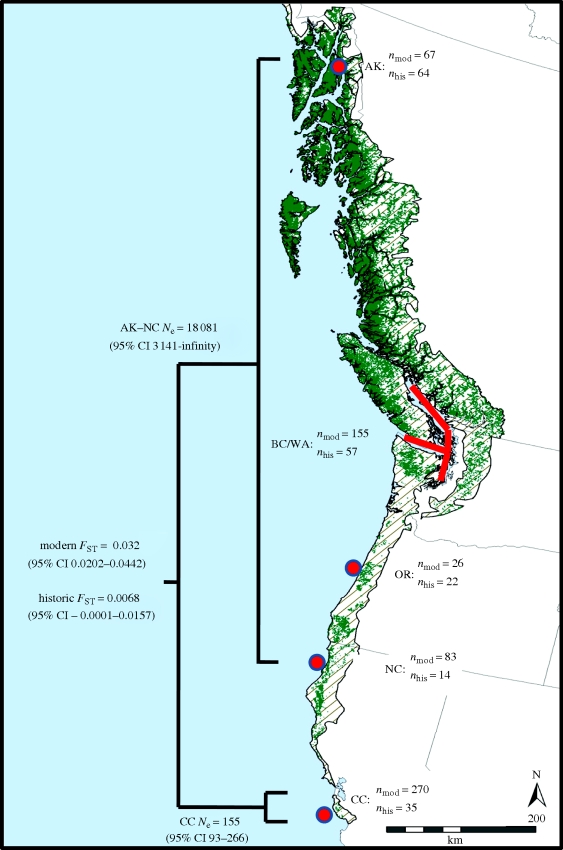
The marbled murrelet (Brachyramphus marmoratus) is a threatened seabird that nests in old-growth forests along the west coast of North America. This species has declined throughout much of its range owing to the loss of old-growth nesting habitat, nest predation by synanthropic species and changes in prey resources in the coastal waters where they forage (Ralph et al. 1995; Peery et al. 2004b; Becker & Beissinger 2006; Piatt et al. 2007). Extensive harvesting of old-growth forest since the late nineteenth century, particularly in the southern portion of the murrelet's range, has resulted in the geographical isolation of the central California population, which is now separated from the nearest substantial population to the north by several hundred kilometres (figure 1). Population size in central California estimated with at-sea surveys remained stable at approximately 600 individuals from 1999 to 2003, despite the fact that reproductive rates have declined by an order of magnitude over the past century (Beissinger & Peery 2007). Reproductive rates are now well below mortality rates, suggesting that the population should decline rapidly in the absence of immigration from northern populations (Peery et al. 2006), and the number of close relatives is lower than expected according to a closed population model (Peery et al. 2008a). Both results suggest that immigration from larger populations to the north helps sustain this population. Recently, Hall et al. (in press) used genetic assignment methods to show that migrants compose a significant component of the central California population. However, in spite of the murrelet's propensity to disperse among populations, the central California population is genetically distinct from northern populations (Friesen et al. 2005; Piatt et al. 2007).
Figure 1.
Historic (pre-European settlement; hatched areas) and current (green areas) distribution of potential murrelet nesting habitat in northwestern North America. The map of the distribution of murrelet habitat from Oregon to Alaska was developed by Inforain's rainforest mapping project (http://www.inforain.org/rainforestatlas/rainforestatlas/page4.html), and the distribution of murrelet habitat in California was developed by Fox (1996). Also depicted are sample sizes for genetic analyses (nmod = modern sample size, nhis = historic sample size), estimates of effective population size (Ne), and estimates of genetic population differentiation (FST) for marbled murrelets (B. marmoratus) sampled at five locations (solid red circles and red line) during both modern (1997–2007) and historic (1888–1940) sampling periods. Sampling locations included southeast Alaska (AK), British Columbia/Washington (BC/WA), Oregon (OR), northern California (NC) and central California (CC).
In this study, we determined how (i) habitat change and other factors have affected genetic population structure in marbled murrelets, (ii) structure has persisted despite measurable dispersal into central California, and (iii) dispersal affects both the viability and our perception of the viability of the central California population. To do so, we compared patterns of genetic variation in modern and historic murrelet samples and used genetic assignment and parentage methods to assess the reproductive contribution of migrants. We demonstrate that a century of habitat loss and other environmental factors have resulted in the genetic differentiation of marbled murrelets in central California despite regular dispersal from northern populations, probably because migrants contribute fewer offspring than residents.
2. Material and methods
(a). Sampling and laboratory methods
For modern analyses, we captured and sampled blood from 601 marbled murrelets at sea in five locations from southeast Alaska to central California in waters adjacent to known concentrations of old-growth nesting habitat from 1 April to 15 October 1997–2007 following methods described in Peery et al. (2006) (figure 1). For historic analyses, we sampled a small amount of tissue from the toepads of 192 murrelets that were collected at sea from central California to southeast Alaska and held in North American museum collections. Historical samples were collected from 1888 to 1940 in the same five regions and months of the year that modern sampling was conducted (figure 1). Murrelets were sampled at sea because capturing or collecting individuals inland near nests is not feasible given this species’ secretive and crepuscular behaviour at cryptic nests placed high in the canopy (Whitworth et al. 1997). However, multiple forms of evidence indicate that murrelets sampled at sea during the breeding season directly represent breeding populations and that loss of nesting habitat is expected to influence the genetic composition of individuals sampled at sea. First, murrelets captured and radio-marked at sea in central California (the population of primary interest) and northern California (the most probable source of migrants) generally nested or flew inland to prospect for nest sites in adjacent forests (Peery et al. 2004a,b; Hébért & Golightly 2008). Second, radio-marked murrelets foraged on average only 6 and 20 km from the mouth of flyways used to access inland nesting habitat in central and northern California, respectively (Hébért & Golightly 2008; Peery et al. 2009). In comparison, there is an approximately 300 km gap in the at-sea distribution of murrelets between central and northern California populations, with very few birds found in intervening waters (Ralph & Miller 1995; Becker et al. 1997; Raphael 2006; Falxa et al. 2008). While murrelets occasionally make long-distance movements of several hundreds of kilometres during the nesting season (Hébért & Golightly 2008; Peery et al. 2008b), it was the objective of this paper to determine if such movements influenced population structure and estimates of population viability. Finally, the number of murrelets occurring at sea and flying inland to nest in a given inland area is strongly and positively correlated with the acreage of old-growth nesting habitat in that area at multiple spatial scales (Burger 2001; Raphael et al. 2002; Raphael 2006). For these reasons, loss of nesting habitat is expected to reduce the number of murrelets occurring in adjacent waters and influence the genetic composition of individuals sampled at sea accordingly.
The 1 April–15 October sampling window was chosen so that results would be comparable to previous demographic and genetic analyses in the region (Peery et al. 2006, 2008a). Restricting sampling to this window also increases the likelihood that sampled individuals with foreign genotypes represented individuals attempting to recruit into the population that they were sampled from rather than post-breeding murrelets dispersing into the region to exploit foraging opportunities. The proportion of the population classified as migrants in central California does not increase until November, when murrelets from other populations begin arriving in the region en masse (Hall et al. 2009). All sampled murrelets were included in analyses regardless of breeding status because brood patches (the only recognizable external indicator of reproduction) are not visible in museum specimens and the probability of being of migrant origin does not differ between brood patch and non-brood patch murrelets (Hall et al. 2009). All age classes were included because, even though young birds have a greater dispersal propensity (Hall et al. 2009), they are the most probable age class to attempt to recruit into another population. The pattern of increasing genetic divergence in central California was evident regardless of the inclusion of juveniles (see below).
Modern samples were amplified and genotyped at up to 16 tetranucleotide microsatellite loci using primers developed by Rew et al. (2006; see the electronic supplementary material, table S2). Historic samples were amplified at a subset of nine of these loci. Tissue sampling and DNA extraction procedures; steps used to minimize contamination, errors in allele scoring and allelic dropout; as well as polymerase chain reaction conditions are described in appendix S1 in the electronic supplementary material.
(b). Temporal comparisons of within- and among-population genetic variation
To test for changes in among-population genetic variation over the past century, we used the program GDA v. 1.1 (Lewis & Zaykin 2001) to estimate FST for each pairwise comparison among the five populations in both the historic and modern sampling periods. To test for a loss in within-population genetic diversity, we compared the number of alleles present per locus (allelic richness) in modern versus historically sampled marbled murrelet populations for the nine loci sampled in both periods. The absolute number of alleles detected was not directly comparable between periods because more individuals were sampled in the modern period. Therefore, we randomly re-sampled a number of gene copies from the modern sample that was equal to the number of historically sampled gene copies with replacement and repeated this process 500 times. We then determined how many loci had more alleles in the historic sample than the upper 95% CL for the bootstrapped modern sample.
We used coalescent simulations implemented in program Simcoal2 (Excoffier et al. 2000) to determine if observed changes in genetic population structure between central California and larger populations to the north (see below) were plausible given several possible models of demographic history for marbled murrelets. When simulating multilocus genotypes, we assumed that, 12 generations in the past, a small population representing central California split from a large historic and equilibrium population of Ne = 367 500. We simulated two different levels of migration between these populations following the bottleneck, one in which the two populations exchanged no migrants and one in which they exchanged five migrants per generation. A historic Ne of 367 500 was derived using Ne=θ/4μ assuming that (i) the mutation rate, μ, equalled 10−5 mutations per locus per generation and microsatellites evolved according to a stepwise mutation model, and (ii) genetic diversity, θ, equalled 14.7 (95% CI = 4.7–24.7) based on the magnitude of variation in microsatellite repeats among gene copies in the historic sample (θ = 2Vs, where Vs was the variance in repeat number; Di Rienzo et al. 1994). The estimator based on the variance in repeat number was used because it is robust to deviations from a stepwise mutation model (Kimmel & Chakraborty 1996). We explored a range of Ne's for the smaller population ranging from 50 to 500, by increments of 50, reflecting probable post-bottleneck Ne's in central California (see below). FST was then calculated for each simulated dataset and compared with FST estimated from the empirical data.
(c). Estimating effective population size
We estimated temporal-Ne based on the magnitude of change in allele frequencies between historic and modern sampling periods (Waples 1989). We estimated temporal-Ne for the four populations from northern California to southeast Alaska combined because no population structure was detected based on modern or historic samples in this region (see below). To estimate temporal-Ne, we used pseudo-likelihood estimation procedures implemented in program Mlne 2.3 (Wang 2001; Wang & Whitlock 2003). This approach provides an estimate of temporal-Ne assuming that a relatively small population and a population of infinite size were connected by migration; the smaller population was sampled on multiple occasions and the larger population was sampled on a single occasion. We applied this logic to the estimation of effective population size in central California, where samples from northern California to southeast Alaska were considered to represent the population of infinite size. The approach also provides an estimate of temporal-Ne for a single isolated population assuming Nm = 0 and temporally spaced samples are available. We applied this logic to northern populations because the most probable rate of gene flow was equal to zero based on coalescent simulations (see below). Temporal-Ne estimators assume nonoverlapping generations, an assumption that was violated for murrelets. The direction of the bias depends on the species’ life history and the sampling design, but biases are small when samples are separated by five or more generations (Waples & Yokota 2007), and approximately 12 generations separated the average sampling date in the modern and historic sampling periods (see the electronic supplementary material, table S3) assuming a generation time of eight years (generation time was estimated using equation 2.6 in Ebert (1999) and demographic parameters in Peery et al. (2006)). Temporal-Ne provides an estimate of the harmonic-mean Ne between sampling periods (Wang 2005), and is therefore expected to produce an estimate more closely reflecting the population size in the second sampling period for declining species.
(d). Identifying migrants and measuring reproductive contribution
We used genetic assignment methods to identify putative migrant and resident individuals with program Geneclass2 (Piry et al. 2004) following methods described in Hall et al. (2009). We then assessed the extent to which migrant individuals recruited into and successfully reproduced in the central California breeding population (i.e. gene flow occurs) by determining how many migrants had a parent or offspring present in the population. Parents do not appear to care for young after fledging (Peery et al. 2007), so it is unlikely that both a parent and offspring originating from another population would be sampled in the relatively small central California population. Pairs of individuals sharing at least one allele at all loci were considered to be probable parent–offspring dyads. In a previous study, we used Monte Carlo simulations to demonstrate that less than 10 per cent of putative parent–offspring dyads share an allele at all loci by chance and represent ‘false matches’ given the 16 microsatellite loci considered here (Peery et al. 2008a).
3. Results
Mean observed and expected heterozygosity were very similar between modern and historic populations (see the electronic supplementary material, table S3), suggesting that null alleles and allelic dropout were rare in historic samples. Moreover, observed heterozygosity did not deviate from expectations under Hardy–Weinberg equilibrium at any locus for any population in either the modern or the historic sampling period, based on exact tests conducted in the program Arlequin v. 3.01 (Excoffier et al. 2006) after applying Bonferonni's sequential corrections for multiple comparisons (Rice 1989).
We estimated that temporal-Ne between modern and historic sampling in central California was 155 (95% CI 93–266; figure 1). This effective population size was compatible with the mean estimate of census population size (572, 95% CL 413–731; Peery et al. 2006) from 1999 to 2003 in the region, given typical ratios of effective to census population size of 0.2–0.3 in wildlife populations (Allendorf & Luikart 2007). Temporal-Ne for the combined northern populations was much larger (Ne = 18 081, 95% CI 3141-infinity).
Genetic divergence among historic murrelet populations was weak or nonexistent (table 1), with the only statistically significant pairwise FST estimate occurring between southeast Alaska and central California (FST = 0.0101, 95% CI = 0.0038–0.0173). All other historic pairwise FST estimates were 0.0068 or less and not statistically significant. However, central California appears to have diverged genetically from northern populations over the past century, as pairwise FST estimates between these two groups in the modern sampling period ranged from 0.0277 to 0.0387 and all estimates were statistically greater than zero (table 1). All pairwise FST-estimates involving central California were significantly greater (based on 95% CIs) in the modern than the historic sampling period. Moreover, the pairwise FST estimate for central California versus northern populations combined increased from 0.0068 (95% CI −0.0001 to 0.0157) to 0.0320 (95% CI 0.0202–0.0442) between sampling periods (table 1, figure 1). The magnitude of the difference between historic and modern FST estimates for central California was essentially the same when juveniles were excluded (historic FST = 0.0116, 95% CI 0.0006–0.0270; modern FST = 0.0358, 95% CI 0.0234–0.0484), although 95% CIs overlapped somewhat because of reduced sampled sizes.
Table 1.
Pairwise FST estimates (95% CIs) for five marbled murrelet populations. (Historical (1888–1940) FST estimates are given below the diagonal, modern (1997–2007) FST estimates are given above the diagonal, and FST estimates for comparisons between historic and modern sampling periods are on the diagonal. Statistically significant FST estimates (p < 0.05) are in bold.)
| region | southeast Alaska | British Columbia—Washington | Oregon | northern California | central California |
|---|---|---|---|---|---|
| southeast Alaska | 0.0068 (−0.0021, 0.0038) | −0.0004 (−0.0014, 0.0075) | 0.0010 (−0.0049, 0.0086) | 0.0051 (0.0016, 0.0090) | 0.0317 (0.0183, 0.0462) |
| British Columbia—Washington | −0.0008 (−0.0030, 0.0014) | −0.0014 (−0.0031, 0.0006) | 0.0015 (−0.0025, 0.0064) | 0.0067 (0.0029, 0.0109) | 0.0387 (0.0224, 0.0558) |
| Oregon | 0.0037 (−0.0019, 0.0096) | 0.0038 (−0.0034, 0.0116) | −0.0072 (−0.0124, −0.0011) | −0.0003 (−0.0039, 0.0030) | 0.0278 (0.0179, 0.040) |
| northern California | −0.0045 (−0.0103, 0.0021) | 0.0002 (−0.0033, 0.0041) | −0.0018 (−0.0085, 0.0049) | −0.0052 (−0.0006, −0.0103) | 0.0277 (0.0189, 0.0363) |
| central California | 0.0101 (0.0038, 0.0173) | 0.0061 (−0.0049, 0.0190) | 0.0017 (−0.0067, 0.0106) | −0.0010 (−0.0103, 0.0082) | 0.0148 (0.0055, 0.0263) |
Estimates of FST can be sensitive to the level of within-population heterozygosity and comparisons of differentiation among groups using FST can be misleading if, for example, markers, species or populations with different levels of genetic variation are compared (Jost 2008). However, all five murrelet populations had very similar levels of heterozygosity in both the modern and historic samples, and differentiation estimated with Dest (Jost 2008) yielded similar inference as FST (see the electronic supplementary material, table S4).
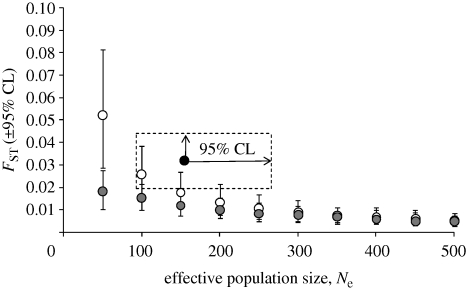
The observed increase in FST between central California and northern populations was consistent with expectations given our estimate of temporal-Ne in central California (Ne = 155, 95% CI 93–266; figure 2). For example, the 95% CI for the estimated FST overlapped the 95% CI for the FST of simulated populations with Nem = 0 when Ne ranged from 100 to 200 (figure 2). The 95% CI for the estimated FST also overlapped the 95% CI for the FST of simulated populations with Nem = 5, but only very slightly and only when Ne = 100 (i.e. very near the lower 95% CL of the estimated temporal-Ne). Thus, the observed increase in FST could have occurred even under the homogenizing influence of a low level of gene flow, but is unlikely to have occurred if Nem > 5.
Figure 2.
Observed and expected population structure (FST) at nine microsatellite loci between marbled murrelets (B. marmoratus) sampled in central California versus northern California–southeast Alaska, based on a range of possible effective population sizes (Ne) and number of migrants per generation (Nem) for central California (open circle, expected FST when Nem = 0 for a given Ne; filled grey circle, expected FST when Nem = 5 for a given Ne; filled black circle, observed FST and Ne in central California).
Concurrent with the increase in genetic divergence between central California and northern populations, alleles were lost at three of the nine microsatellite loci between historic and modern sampling in central California, and allelic richness declined by an average of 6.9 per cent across loci (see the electronic supplementary material, table S5). Alleles were only lost at one locus between historic and modern sampling for the northern populations, and allelic richness declined by an average of 4.5 per cent across loci (see the electronic supplementary material, table S5).
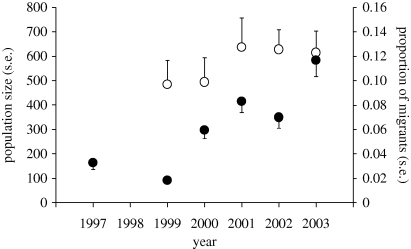
Despite the increase in genetic population structure and the loss of genetic diversity, the dispersal of individuals into central California continues to occur regularly. Seven per cent (19 of 270) of murrelets in the modern central California sample were classified as migrants based upon the likelihood of their multi-locus genotype. Moreover, based on linear regression analysis, the proportion of migrants in this population increased by 1.4 per cent per year from 1997 to 2003 (F1,4 = 3.39, p = 0.028, r2 = 0.74; figure 3). The increase in the proportion of migrants was not owing simply to a decline in the size of the resident population because the number of migrants captured generally increased over the six years that sampling was conducted (from one in 1997 to seven in 2003), despite relatively constant sampling effort across years.
Figure 3.
Estimates of population size (open circle, ±1 s.e.) and the proportion (filled black circle, ±1 s.e.) of migrant marbled murrelets (B. marmoratus) in central California in 1999–2003 and 1997–2003, respectively.
A significantly lower proportion of murrelets classified as migrants in central California were involved in parent–offspring pairs (2 of 19 or 10.5%) than residents (114 of 251 or 45.4%, 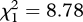 , p < 0.003) in the modern sample, indicating that migrants contributed fewer offspring to this population than residents. Two migrants possessed a parent or offspring in the population, but approximately two residents were expected to be mistakenly classified as migrants and about 10 per cent of putative parent–offspring dyads were probably type I errors (Peery et al. 2008a; Hall et al. 2009). Thus, it is possible that no migrant actually possessed a parent or offspring in the sample.
, p < 0.003) in the modern sample, indicating that migrants contributed fewer offspring to this population than residents. Two migrants possessed a parent or offspring in the population, but approximately two residents were expected to be mistakenly classified as migrants and about 10 per cent of putative parent–offspring dyads were probably type I errors (Peery et al. 2008a; Hall et al. 2009). Thus, it is possible that no migrant actually possessed a parent or offspring in the sample.
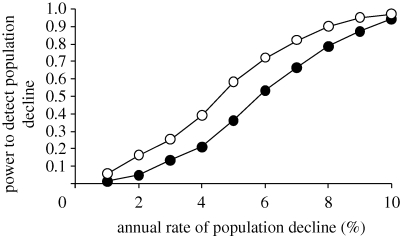
Murrelet population size averaged 572 individuals from 1999 to 2003 based on at sea surveys, and experienced a non-significant increase during this period using simple linear regression (F1,3 = 2.51, p = 0.087, r2 = 0.68; figure 3; Peery et al. 2006). To determine if increasing dispersal compromised our ability to detect a population decline from 1999 to 2003, we conducted Monte Carlo simulation-based power analyses with and without a 1.4 per cent annual increase in the proportion of migrants in the population (see appendix S2 in the electronic supplementary material). Indeed, the increase in the proportion of migrants that we observed from 1999 to 2003 appreciably reduced statistical power to detect 3–8% annual declines. The reduction in power to detect population declines was greatest for 5 per cent annual declines when dispersal reduced power by 71 per cent (figure 4).
Figure 4.
Statistical power to detect 1–10% annual declines in the resident central California marbled murrelet (B. marmoratus) population, with and without a 1.4% annual increase in the proportion of migrants in the population, using estimates of abundance from at-sea surveys collected from 1999 to 2003 (filled black circle, 1.4% annual increase in migration; open circle, no migration).
4. Discussion
Historically, murrelets from southeast Alaska to central California constituted a single genetically undifferentiated population, but during the twentieth century central California murrelets diverged from northern populations and are now moderately genetically differentiated (FST = 0.032). Coalescent simulations suggested that the current level of genetic differentiation between central California and northern populations was owing to (i) the enhanced effects of genetic drift given the small effective size of the central California population during the past century, and (ii) low levels of gene flow. Indeed, both a reduction in Ne and limited gene flow were needed to generate the observed change in FST (figure 2). A decline in Ne in central California is supported by the loss of alleles at three of nine loci between historic and modern sampling, compared with a loss of alleles at only one locus in northern populations, which despite recent declines still numbers in the hundreds of thousands (Piatt et al. 2007). Ecologically, a reduction in Ne in central California is supported by an order of magnitude decline in reproductive rates between historic and modern sampling (Beissinger & Peery 2007; Hall et al. 2009). In addition, approximately 90 per cent of nesting habitat was removed, nest predator populations increased dramatically, prey availability declined and significant mortalities owing to oil spills and gillnetting occurred (Ralph et al. 1995; Peery et al. 2004b; Becker & Beissinger 2006).
Limited gene flow between northern populations and central California could have occurred owing to the removal of large amounts of old-growth forest in the approximately 300 km long region between central and northern California populations (figure 1). This region represents the most probable source of migrants based on proximity, but now only contains about 135 murrelets based on at-sea surveys (Falxa et al. 2008). The existing gap in nesting habitat in this region could also reduce gene flow from populations farther to the north if, historically, migration occurred according to an isolation-by-distance model (Friesen et al. 2005).
The increase in genetic divergence occurred despite regular dispersal of individuals into central California. Specifically, the increase in divergence suggested that five or lesser effective migrants entered the population per generation (i.e. approx. 0.63 migrants per year). By contrast, the proportion of migrants detected in central California with assignment tests in April–October (0.07) suggested that 41 migrants were present during this period, assuming a population size of 582 (Peery et al. 2006). However, the former approach measures the number of effective migrants (i.e. gene flow or the number of migrants that reproduce at the same rate as residents), whereas the latter approach measures the number of migrant individuals present, regardless of their reproductive contribution. The discrepancy between the two estimates very probably occurred because migrants contributed proportionally fewer young than residents, as indicated by the fact that migrants were less likely to be members of a parent–offspring pair than residents (10.5% versus 45.4%). In theory, the effective migration rate could have been higher than indicated based on the small number of migrants in central California (two) that were detected with a parent or offspring also present in the population if murrelets in central California experienced inbreeding depression, and matings between migrants and residents resulted in offspring with comparatively high fitness (Ingvarsson & Whitlock 2000). However, there is little evidence that inbreeding occurred in central California as mean observed heterozygosity in central California was high (0.753, n = 270) and nearly identical to expected heterogygosity (0.763).
Does dispersal into central California result in insignificant levels of gene flow because migrants represent (i) temporary visitors that do not attempt to breed, or (ii) immigrants into the breeding population that have comparatively low reproductive success? Either scenario would explain the observation that migrants were involved in proportionately fewer parent–offspring pairs than residents. Some migrants probably do breed in central California, as the only migrant for which breeding information was available (from radio-telemetry) attempted to nest in the Santa Cruz Mountains (Peery et al. 2004b). If significant numbers of migrants do attempt to breed in central California, their reproductive success could be lower than residents owing to outbreeding depression (Nosil et al. 2005), competition with residents for breeding sites (Fretwell & Lucas 1970) or a lack of adequate knowledge of local predators and foraging resources (Stamps 2001). Although we restricted our sampling period to reduce the number of post-breeding murrelets dispersing from other populations, some migrants could have represented temporary visitors from other populations that exploited central California waters for foraging. Indeed, approximately 9–13% of nonbreeders and post-breeders in central California made temporary long-distance movements up to several hundred kilometres along the coast during the period in which genetic sampling was conducted (April–October; Peery et al. 2008b). Without long-term observations of the behaviour and breeding histories of migrant individuals, the relative proportion of breeding versus nonbreeding migrants in central California is uncertain.
Regardless of the extent to which migrants represented seasonal dispersers or immigrants that attempted to recruit into the breeding population, the increasing trend in the proportion of migrants may have compromised our ability to detect an underlying decline in the resident central California population with at-sea surveys between 1999 and 2003. In the absence of immigration, the population was projected to decline rapidly (9.5% annually), because very low reproductive rates did not compensate for mortality rates (Peery et al. 2006). However, there was considerable sampling uncertainty around the estimate of the rate of decline (95% CL 0–20%), and we suspect that the actual rate of decline was less than 8 per cent because we had 80 per cent or more power to detect 8 per cent or more declines. Assuming the population decline indeed was less than 8 per cent per year, the 1.4 per cent annual increase in the proportion of migrants reduced our ability to detect population declines appreciably (figure 4). For example, a 5 per cent annual population decline was 71 per cent more likely to be detected when no increase in migrants occurred than when the proportion of migrants increased by 1.4 per cent per year. Thus, the confounding effects of dispersal on population counts may have contributed to a false impression that central California was a stable (albeit sink) population (Peery et al. 2006, 2008a).
Previous demographic and genetic research yielded contradictory results, at least superficially, with respect to the level of demographic independence among marbled murrelet populations. Specifically, Friesen et al. (2005) suggested that central California should be designated as a separate management unit because population structure was detected using neutral genetic markers. By contrast, Peery et al. (2006, 2008a) suggested that central California may be a sink population sustained by immigration based on demographic and kinship analyses. The present study clearly reconciles these differences by demonstrating that significant dispersal does occur into central California, but also that dispersing individuals, whether they be seasonal migrants or immigrants that attempted to breed, are involved in few parent–offspring pairs and result in too little reproduction to rescue the population (0.63 or lesser breeding migrants per year). Moreover, the population is not viable because mortality rates greatly exceed birth rates, and dispersal will only mask the underlying decline in the resident population. For these reasons, we agree with Friesen et al. (2005) that central California should be treated as a separate management unit.
Changes in genetic population structure detected in this study are supported by a growing body of research indicating that the spatial distribution of genetic variation in species occurring in fragmented landscapes is due, in part, to the disruption of gene flow across historically contiguous landscapes (Keyghobadi 2007). Significant changes in structure can occur within short time frames, even when effective population size remains reasonably large (e.g. within 12 generations for an effective population size in the hundreds of individuals for marbled murrelets). Moreover, dispersal ability does not necessarily translate into genetic resilience in the face of habitat fragmentation, even for vagile species that can and do move among populations (Hoehn et al. 2007; Lindsay et al. 2008).
It is well accepted in conservation biology that dispersal among fragmented populations can result in the genetic and demographic rescue of endangered populations. However, our study indicates that the movement of individuals does not necessarily prevent the loss of genetic diversity and maintain population viability if dispersers do not recruit into the breeding population and produce offspring. Without information on the contribution of migrants to local population dynamics and genetic diversity, dispersal can confound monitoring efforts by masking population declines. Consequently, assessing the reproductive contribution of both residents and migrants in endangered populations may be required to detect local population declines in sufficient time to implement appropriate conservation measures. Identifying migrants and their offspring is now comparatively straightforward using genetic methods and in many cases requires only a modest increase in effort. This can be true even in elusive species such as the marbled murrelet for which we were able to identify migrants and their offspring using a combination of assignment methods and kinship approaches.
Acknowledgements
The work presented herein was authorized by IACUC protocol no. R233 from the University of California, Berkeley and US Fish and Wildlife Service permit no. TE R233 issued to S.B.
Support was provided by the Humboldt Redwood Company. Samples were provided by the Burke Museum, California Academy of Sciences, Museum of Vertebrate Zoology, American Natural History Museum, San Diego Natural History Museum, Donald R. Dickey Collection, Museum of Comparative Zoology and Humboldt State University. Beth Galleher provided GIS support. Robin Waples, Vicki Friesen and two anonymous reviewers provided comments that improved earlier drafts of this paper. Lydia Smith, Jim Harvey and Jon Geller provided laboratory space and technical assistance.
References
- Allendorf F. W., Luikart G.2007Conservation and the genetics of populations Malden, MA: Blackwell Publishing [Google Scholar]
- Becker B. H., Beissinger S. R.2006Centennial decline in the trophic level of an endangered seabird after fisheries decline. Conserv. Biol. 20, 470–479 (doi:10.1111/j.1523-1739.2006.00379.x) [DOI] [PubMed] [Google Scholar]
- Becker B. H., Beissinger S. R., Carter H. R.1997At-sea density monitoring of marbled murrelets in central California: methodological considerations. Condor 99, 743–755 (doi:10.2307/1370485) [Google Scholar]
- Beissinger S. R., Peery M. Z.2007Reconstructing the historic demography of an endangered seabird. Ecology 88, 296–305 (doi:10.1890/06-0869) [DOI] [PubMed] [Google Scholar]
- Brown J. H., Kodric-Brown A.1977Turnover rates in insular biogeography: effect of immigration and extinction. Ecology 58, 445–449 (doi:10.2307/1935620) [Google Scholar]
- Burger A. E.2001Using radar to estimate populations and assess habitat associations of marbled murrelets. J. Wildl. Mngmt 65, 696–715 [Google Scholar]
- Chalfoun A. D., Thompson F. R., III, Ratnaswamy M. J.2002Nest predators and fragmentation: a review and meta-analysis. Conserv. Biol. 16, 306–318 (doi:10.1046/j.1523-1739.2002.00308.x) [Google Scholar]
- Dearborn D. C., Anders A. D. A. S. E., Adams R. M. M., Mueller U. G.2003Inter-island movements and population differentiation in a pelagic seabird. Mol. Ecol. 12, 2835–2843 (doi:10.1046/j.1365-294X.2003.01931.x) [DOI] [PubMed] [Google Scholar]
- Di Rienzo A., Peterson A. C., Garza J. C., Valdes A.-M., Slatkin M., Freimer N. B.1994Mutational processes of simple sequence repeat loci in human populations. Proc. Natl Acad. Sci. USA 91, 3166–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert T. A.1999Plant and animal populations: methods in demography San Diego, CA: Academic Press [Google Scholar]
- Excoffier L., Novembre J., Schneider S.2000Simcoal: a general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J. Hered. 91, 506–509 (doi:10.1093/jhered/91.6.506) [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S.2006Arlequinv. 3.01 user manual: an integrated software package for population genetics data analysis Bernem, Switzerland: University of Berne; [PMC free article] [PubMed] [Google Scholar]
- Fahrig L.2003Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 34, 487–515 (doi:10.1146/annurev.ecolsys.34.011802.132419) [Google Scholar]
- Falxa G., et al. 2008Marbled murrelet effectiveness monitoring. Northwest forest plan: 2004–2007 summary report. US Fish and Wildlife Service, Arcata, CA, USA
- Fox L.1996Current status and distribution of coast redwood. In Coast redwood forest ecology and management (ed. Leblanc J.), pp. 18–19 Berkeley, CA: University of California [Google Scholar]
- Fretwell S. D., Lucas H. L.1970On the territorial behaviour and other factors influencing habitat distribution in birds. I. Acta Biotheoretica 19, 16–36 (doi:10.1007/BF01601953) [Google Scholar]
- Friesen V. L., Birt T. P., Piatt J. F., Golightly R. T., Newman S. H., Hebert P. N., Congdon B. C., Gissing G.2005Population genetic structure and conservation of marbled murrelets (Brachyramphus marmoratus). Conserv. Genet. 6, 607–614 (doi:10.1007/s10592-005-9012-x) [Google Scholar]
- Hall L. A., et al. In press Characterizing dispersal patterns in a threatened seabird with limited genetic structure. Mol. Ecol [DOI] [PubMed] [Google Scholar]
- Hébért P. N., Golightly R. T.2008At-sea distribution and movements of nesting and non-nesting marbled murrelets Brachyramphus marmoratus in northern California. Mar. Ornithol. 36, 99–105 [Google Scholar]
- Hoehn M., Sarre S. D., Henle K.2007The tales of two geckos: does dispersal prevent extinction in recently fragmented populations? Mol. Ecol. 16, 3299–3312 (doi:10.1111/j.1365-294X.2007.03352.x) [DOI] [PubMed] [Google Scholar]
- Ingvarsson P. K., Whitlock M. C.2000Heterosis increases the effective migration rate. Proc. R. Soc. Lond. B 267, 1321–1326 (doi:10.1098/rspb.2000.1145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L.2008G(ST) and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026 (doi:10.1111/j.1365-294X.2008.03887.x) [DOI] [PubMed] [Google Scholar]
- Keller L. F., Waller D. M.2002Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (doi:10.1016/S0169-5347(02)02489-8) [Google Scholar]
- Keyghobadi N.2007The genetic implications of habitat fragmentation. Can. J. Zool. 85, 1049–1063 (doi:10.1139/Z07-095) [Google Scholar]
- Kimmel M., Chakraborty R.1996Measures of variation at DNA repeat loci under a general stepwise mutation model. Theor. Popul. Biol. 50, 345–367 (doi:10.1006/tpbi.1996.0035) [DOI] [PubMed] [Google Scholar]
- Lande R.1993Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 142, 911–927 (doi:10.1086/285580) [DOI] [PubMed] [Google Scholar]
- Lande R., Shannon S.1996The role of genetic variation in adaptation and population persistence in a changing environment. Evolution 50, 434–437 (doi:10.2307/2410812) [DOI] [PubMed] [Google Scholar]
- Lewis P. O., Zaykin D.2001Genetic data analysis: computer program for the analysis of allelic data, version 1.0 Stores, CT: University of Connecticut [Google Scholar]
- Lindsay D. L., Barr K. R., Lance R. F., Tweddale S. A., Hayden T. J., Leberg P. L.2008Habitat fragmentation and genetic diversity of an endangered, migratory songbird, the golden-cheeked warbler (Dendroica chrysoparia). Mol. Ecol. 17, 2122–2133 (doi:10.1111/j.1365-294X.2008.03673.x) [DOI] [PubMed] [Google Scholar]
- Madsen T., Shine R., Olsson M., Wittzell H.1999Restoration of an inbred adder population. Nature 402, 34–35 (doi:10.1038/46941) [Google Scholar]
- Nosil P., Vines T. H., Funk D. J.2005Perspective: reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59, 705–719 [PubMed] [Google Scholar]
- Peery M. Z., Beissinger S. R., Newman S. H., Becker B. H., Burkett E., Williams T. D.2004aIndividual and temporal variation in inland flight behavior of marbled murrelets: implications for population monitoring. Condor 106, 344–353 (doi:10.1650/7398) [Google Scholar]
- Peery M. Z., Beissinger S. R., Newman S. H., Burkett E., Williams T. D.2004bApplying the declining population paradigm: diagnosing causes of poor reproduction in the marbled murrelet. Conserv. Biol. 18, 1088–1098 (doi:10.1111/j.1523-1739.2004.00134.x) [Google Scholar]
- Peery M. Z., Becker B. H., Beissinger S. R.2006Combining demographic and count-based approaches to identify source–sink dynamics of a threatened seabird. Ecol. Appl. 16, 1516–1528 (doi:10.1890/1051-0761(2006)016[1516:CDACAT]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Peery M. Z., Becker B. H., Beissinger S. R.2007Age ratios as estimators of productivity: testing assumptions on a threatened seabird, the marbled murrelet (Brachyramphus marmoratus). Auk 124, 224–240 (doi:10.1642/0004-8038(2007)124[224:ARAEOP]2.0.CO;2) [Google Scholar]
- Peery M. Z., Beissinger S. R., House R. F., Bérubé M., Hall L. A., Sellas A., Palsbøll P. J.2008aCharacterizing source–sink dynamics with genetic parentage assignments. Ecology 89, 2746–2759 (doi:10.1890/07-2026.1) [DOI] [PubMed] [Google Scholar]
- Peery M. Z., Henkel L. A., Newman S. H., Becker B. H., Harvey J. T., Thompson C., Beissinger S. R.2008bEffects of rapid flight-feather molt on post-breeding dispersal in a pursuit-diving seabird. Auk 125, 113–123 (doi:10.1525/auk.2008.125.1.113) [Google Scholar]
- Peery M. Z., Newman S. H., Storlazzi C. S., Beissinger S. R.2009Meeting reproductive demands in a dynamic upwelling system: foraging strategies of a pursuit-diving seabird, the marbled murrelet. Condor 111, 120–134 (doi:10.1525/cond.2009.080094) [Google Scholar]
- Piatt J. F., et al. 2007US geological survey open-file report 2006–1387 status review of the marbled murrelet (Brachyramphus marmoratus) in Alaska and British Columbia Reston, VA: US Geological Survey [Google Scholar]
- Piry S., Alapetite A., Cornuet J. M., Paetkau D., Baudouin L., Estoup A.2004Geneclass2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539 (doi:10.1093/jhered/esh074) [DOI] [PubMed] [Google Scholar]
- Pulliam H. R.1988Sources, sinks, and population regulation. Am. Nat. 132, 652–661 [Google Scholar]
- Ralph C. J., Miller S. L.1995Offshore population estimates of marbled murrelets in California. In Ecology and conservation of the marbled murrelet (eds Ralph C. J., Hunt G. L., Jr, Raphael M. G., Piatt J. F.), pp. 353–360 USDA Forest Service General Technical Report no. PSW-GTR-152, Albany, CA, USA [Google Scholar]
- Ralph C. J., Hunt G. L., Jr, Raphael M. G., Piatt J. F. 1995. Ecology and Conservation of the Marbled Murrelet. USDA Forest Service General Technical Report no. PSW-GTR-152, Albany, CA, USA.
- Raphael M. G.2006Conservation of the marbled murrelet under the Northwest Forest Plan. Conserv. Biol. 20, 297–305 (doi:10.1111/j.1523-1739.2006.00382.x) [DOI] [PubMed] [Google Scholar]
- Raphael M. G., Mack D. E., Cooper B. A.2002Landscape-scale relationships between abundance of marbled murrelets and distribution of nesting habitat. Condor 104, 331–342 (doi:10.1650/0010-5422(2002)104[0331:LSRBAO]2.0.CO;2) [Google Scholar]
- Rew M. B., Peery M. Z., Beissinger S. R., Berube M., Lozier J. D., Rubidge E. M., Palsboll P. J.2006Cloning and characterization of 29 tetranucleotide and two dinucleotide polymorphic microsatellite loci from the endangered marbled murrelet (Brachyramphus marmoratus). Mol. Ecol. Notes 6, 241–244 (doi:10.1111/j.1471-8286.2006.01206.x) [Google Scholar]
- Rice W. R.1989Analyzing tables of statistical tests. Evolution 43, 223–225 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- Stamps J. A.2001Habitat selection by dispersers: integrating proximate and ultimate approaches. In Dispersal (eds Clobert J., Danchin E., Dhondt A. A., Nichols J. D.), pp. 230–242 Oxford, UK: Oxford University Press [Google Scholar]
- Thomas C. D., Kunin W. E.1999The spatial structure of populations. J. Anim. Ecol. 68, 647–657 (doi:10.1046/j.1365-2656.1999.00330.x) [Google Scholar]
- Wang J.2001A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet. Res. 78, 243–257 (doi:10.1017/S0016672301005286) [DOI] [PubMed] [Google Scholar]
- Wang J.2005Estimation of effective population sizes from data on genetic markers. Phil. Trans. R. Soc. B 360, 1395–1409 (doi:10.1098/rstb.2005.1682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Whitlock M. C.2003Estimating effective population size and migration rates from genetic samples over space and time. Genetics 163, 429–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. S.1989A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. S., Yokota M.2007Temporal estimates of effective population size in species with overlapping generations. Genetics 175, 219–233 (doi:10.1534/genetics.106.065300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westemeier R. L., Brawn J. D., Simpson S. A., Esker T. L., Jansen R. W., Walk J. W., Kershner E. L., Bouzat J. L., Paige K. N.1998Tracking the long-term decline and recovery of an isolated population. Science 282, 1695–1698 (doi:10.1126/science.282.5394.1695) [DOI] [PubMed] [Google Scholar]
- Whitworth D. L., Takekawa J. Y., Carter H. R., McIver W. R.1997Night-lighting as an at-sea capture technique for Xantus’ Murrelets in the Southern California Bight. Colonial Waterbirds 20, 525–531 (doi:10.2307/1521603) [Google Scholar]
- Wilcove D. S.1985Nest predation in forest tracts and the decline of migratory songbirds. Ecology 66, 1211–1214 (doi:10.2307/1939174) [Google Scholar]
- Wilcove D. S., Rothstein D., Dubow J., Phillips A., Losos E.1998Quantifying threats to imperiled species in the United States. Bioscience 48, 607–615 (doi:10.2307/1313420) [Google Scholar]