Abstract
Despite decades of study, some aspects of Phocidae (Pinnipedia, Carnivora) phylogeny still remain unresolved. Using the largest novel dataset to date, including all extant phocids and comprising 15 nuclear and 13 mitochondrial genes, we illustrate the utility of including multiple individuals per species in resolving rapid radiations, and provide new insight into phocid phylogeny. In line with longstanding morphological views, Pusa is recovered as monophyletic for the first time with genetic data. The data are also used to explore the relationship between genetic distance and taxonomic rank. Intraspecific sampling also highlights the discrepancy between molecular and morphological rates of evolution within Phocidae.
Keywords: rapid radiation, taxonomy, species tree
1. Introduction
The phylogeny of the true seals (Phocidae, Pinnipedia, Carnivora) remains to be completely resolved, despite decades of study. Higher-level classifications (figure 1) are widely accepted based on both morphological and molecular data (King 1966; Burns & Fay 1970; Davis et al. 2004; Árnason et al. 2006; Higdon et al. 2007) but some species relationships remain contentious.
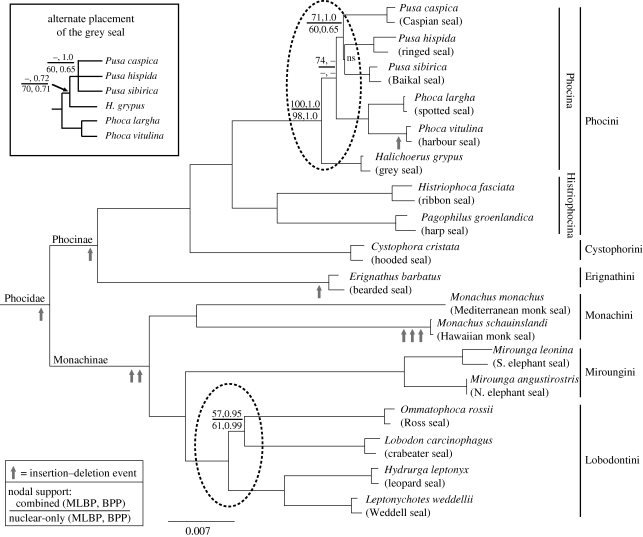
Figure 1.
Phylogeny of the Phocidae. Maximum likelihood topology combining nuclear and mtDNA. Branch lengths optimized in RAxML. BI of this dataset and nuclear-only analyses recovered the same topology, except within Phocina, where most analyses recovered a different placement for Halichoerus (upper left). All nodes were supported by BPP = 1.0 or MLBP = 99–100% in combined analyses except where noted, ns = not supported. Tribal structure (on right) sensu Burns & Fay (1970) and dashed circles represent contentious clades discussed in text.
Only two phocid tribes, Lobodontini and Phocini, contain multiple genera. Both clades have proved difficult to resolve, probably resultant from rapid radiations. Similar situations are common across Carnivora, including the origin of Pinnipedia (Flynn et al. 2005; Fulton & Strobeck 2006; Sato et al. 2006; Árnason et al. 2007). Even when large nuclear datasets are applied, some species relationships remain difficult to disentangle, such as for bears (Pagès et al. 2008), fur seals and sea lions (Yonezawa et al. 2009) and mustelids (Koepfli et al. 2008). The northern true seals (Phocinae) are comprised of pagophilic (ice-loving) species, while the ‘southern’ true seals (Monachinae) include pagophilic (Lobodontini), temperate (Miroungini) and warm water (Monachini) tribes. Thus, unraveling these species relationships is key to understanding the timing and patterns of invasion and diversification in new environments.
Phocini relationships have not been solidified using morphology (Burns & Fay 1970; de Muizon 1982) and molecular studies have yet to recover all genera as reciprocally monophyletic (figure 2; Bininda-Emonds et al. 1999; Árnason et al. 2006; Fulton & Strobeck 2006; Palo & Väinölä 2006; Higdon et al. 2007; Dasmahapatra et al. 2009). Difficulty in resolving Phocina relationships is complicated not only by rapid radiation, but also by extremely large population sizes: the ringed seal exists in extremely high numbers (N ≈ 5–7 million) in a largely panmictic population (King 1983; Palo et al. 2001; Sasaki et al. 2003), suggesting large effective population size. Phocina species radiated approximately 1.4–2.6 Ma (Fulton & Strobeck in press), and with generation times of approximately 11–17 years, it is possible that sufficient effective generations have not passed for complete lineage sorting. Increasing both the number of loci and the number of individuals should help to disentangle these close relationships (Maddison & Knowles 2006; Carstens & Knowles 2007). Here, we employ multiple nuclear loci plus complete mitochondrial (mt) genomes for two individuals per species to generate the largest dataset yet applied to estimate the Phocidae species tree.
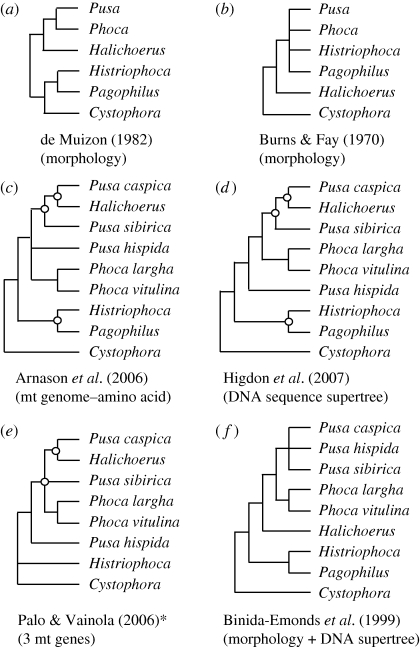
Figure 2.
Review of proposed phocine relationships. (a,b) Examples of morphological studies of the Phocini + Cystophorini; (c–f) recent molecular hypotheses including all Pusa species. Open circles represent nodes not strongly supported for the molecular studies c, d and e. Nomenclature used is that of this study (for consistency) and not necessarily that of the original studies c, e and f. *Adapted—the basal polytomy indicates discrepancy between analysis methods, not a lack of resolution indicated by the authors.
Using multiple individuals also provides the opportunity to assess problematic taxonomy from a molecular perspective. Within Phocini, Halichoerus (grey seal) is consistently recovered within Pusa (ringed, Baikal and Caspian seals), leading to the argument that it should be reassigned to Phoca (harbour and spotted seals, plus Pusa as a subgenus) on genetic grounds (Árnason et al. 1995, 2006). Conversely, it has been argued to be the only species warranting its own genus within all Phocini based on dentition and snout morphology (Burns & Fay 1970). Assessment of inter- and intraspecific variation levels provide insight into this taxonomic issue, as well as highlight intriguing differences between the rates of morphological and molecular evolution across Phocidae.
2. Material and methods
Forty-seven species were included in this study including seven non-pinniped carnivores. Two individuals from each phocid species were sequenced for each gene, except Phoca sibirica and Monachus monachus (one individual each), and Mirounga leonina (no individuals) for the nuclear dataset and M. monachus and Mirounga angustirostris (one individual each) for the mt dataset, due to sample availability. The same two individuals were not always represented, though the same individuals were used when possible. Fifteen nuclear genes from 14 unlinked regions were selected (see the electronic supplementary material, table S1). Extraction, amplification and sequencing conditions are listed in the electronic supplementary material.
Alignment details, indel information and model selection methodology are listed in the electronic supplementary material. Base composition homogeneity across taxa was assessed in PAUP* v.4.0b10 (Swofford 2003) for each dataset as a whole and partitioned by codon position. The nuclear data showed no bias among taxa (p = 0.9999), while mtDNA data did (p = 0), but this bias was not observed after the mt 3rd codon position bases were removed (p = 0.9999). The mt 3rd codon position bases were thus excluded. Locus congruence was tested using the ILD test (Farris et al. 1995) as the partition homogeneity test (100 replicates) in PAUP*. No locus was significantly incongruent at p = 0.01, nor were the mt and nuclear partitions from each other (p = 0.84).
Maximum likelihood (ML) tree searching and bootstrapping (MLBP) was performed using RAxML v.7.0.0 (Stamatakis 2006a,b). Bayesian inference (BI) was performed in MrBayes v.3.1.2 (Huelsenbeck & Ronquist 2001; Ronquist & Huelsenbeck 2003). Details are included in the electronic supplementary material.
The effect of intraspecific sampling was examined by randomly excluding different sets of individuals (including all possible combinations in Phocina) so each species was only represented by a single taxon. Maximum parsimony bootstrap (MPBP) searches were performed in PAUP*. Inter- and intraspecific variation was measured by the distance (logdet for mtDNA, GTR for nuclear) between each taxon pair in PAUP*.
3. Results
(a). Sequencing and phylogenetic results
GenBank accession numbers GU167671–GU167877 and GU174591–GU174608 were assigned to new sequences (see the electronic supplementary material, table S3). Amplification could not be achieved in six cases (see the electronic supplementary material). Only relationships within Phocina and Lobodontini were not clearly resolved (figure 1). Nuclear and mt datasets analysed separately or together recovered Pusa as monophyletic. Pusa and Halichoerus were recovered as sister by nuclear-only analysis (MLBP = 70, Bayesian Posterior Probability (BPP) = 1.0) and BI of the combined dataset (BPP = 0.72), while Pusa and Phoca were sister in mt analysis (MLBP = 82) and ML inference of combined data (MLBP = 74). All analyses recovered two clades within Lobodontini (figure 1).
(b). Use of multiple individuals per species
Topological changes using different datasets including only one individual/species were only observed within Phocini. MPBP analyses including two individuals/species in the nuclear dataset did not support any clades more than 50 per cent within Pusa + Halichoerus, except species monophyly. The first individual of Halichoerus grypus was always sister to Pusa caspica either weakly (MPBP = 51) or moderately (MPBP = 77), depending on the included Pusa individuals. Using only the second H. grypus individual yielded either a polytomy of all Pusa spp. + H. grypus or recovered P. caspica + Phoca hispida (MPBP = 71–76). In the complete mtDNA dataset, MPBP analysis using two individuals/species recovered a monophyletic Pusa (MPBP = 59) with sister H. grypus. When one individual per species was used, regardless of the combination of individuals, the two Phoca species were drawn into the Pusa + Halichoerus clade with moderate support (MPBP = 61–76).
Only Phoca largha was not consistently recovered as monophyletic owing to occasional inclusion of Phoca vitulina. Monophyly of both species was supported by AFLP analysis (Dasmahapatra et al. 2009), but they occasionally interbreed (Shaughnessy & Fay 1977), which may have contributed to occasional nuclear gene paraphyly here.
(c). Inter- and intraspecific variation
Pair-wise nuclear distances between taxa were approximately 15–20 times lower in all comparisons than for mtDNA, when LogDet distance was used for both datasets. Average genetic distance between relevant taxa provided comparisons within species (between individuals), within genus (between species, with individuals averaged/species), between genera of the same subtribe (between species, with individuals averaged/species), between subtribes (genus comparisons, with species averaged/genus), and between tribes (genera averaged within subtribe). Genetic distances were not corrected for physical distance within the species range, population size or structure or any other factors that may affect intraspecific genetic distance.
4. Discussion
As with other molecular studies (Davis et al. 2004; Fyler et al. 2005; Árnason et al. 2006; Fulton & Strobeck 2006; Higdon et al. 2007), we confirm the generally accepted subfamily classification structure of Phocidae (figure 1). But unlike other molecular studies, we provide the first molecular evidence for the Phocina that is congruent with morphology, supporting Pusa as monophyletic. There are two Phocini subtribes (Chapskii 1955): Histriophocina (Histriophoca, Pagophilus) and Phocina (Phoca, Pusa, Halichoerus). Pusa was historically considered a subgenus of Phoca, but was elevated by Scheffer (1958) to generic status. Both Pusa (King 1966; de Muizon 1982) and Phoca (Pusa; Chapskii 1955; McLaren 1960; Burns & Fay 1970) have been applied since, and all the species are morphologically similar. Previous molecular work placed the grey seal (Halichoerus) within the Phoca–Pusa group, often rendering Pusa paraphyletic (figure 2). The use of multiple individuals per species proved key for unraveling the rapid Phocini divergences. Here, Pusa is supported as monophyletic (figure 1), in agreement with morphology. Placement of Halichoerus within Pusa can be recovered when particular combinations including only one individual per species are used, indicating that it is probably an artefact of something like compositional or rate heterogeneity (Ho & Jermiin 2004). Because of the power of intraspecific sampling in identifying this variation in phylogenetic reconstruction based on sampling, it would probably benefit all studies of recent rapid radiations to include multiple individuals per species (see also Syring et al. 2007; Brumfield et al. 2008; Willyard et al. 2009), to highlight areas of uncertainty and increase resolution.
Intraspecific sampling, particularly in combination with new gene tree/species tree analytical advances, can be a powerful tool in resolving recent rapid radiations (Carstens & Knowles 2007). These methods are primarily designed to estimate the species tree by reconciling conflict between genes when deep coalescences occur (Degnan & Rosenberg 2009). However, the data here were selected to represent a range of evolutionary rates to address phylogeny at all levels from species to subfamily. Inclusion of slowly evolving markers in this case does not lend well to analyses that first examine genes separately, as each nuclear gene alone provides very little resolution (see the electronic supplementary material). In a Bayesian framework (i.e. Liu 2008), analysing the many nuclear genes here with individually low information content leads to incredibly wide posterior distributions and difficulty in achieving convergence of all parameters. In a likelihood framework (i.e. Kubatko et al. 2009), choosing the single ML tree to represent each gene's history ignores thousands of nearly equally probable topologies and is not in the intended spirit of the method, which is to resolve incongruence, not to extract weak signals from individual genes. Instead, a concatenation approach, as presented here, seems synergistic, illustrating what appears to be an emergent signal (Gatesy & Baker 2005), highlighting the importance of sampling for this clade and providing an important step towards resolving Phocina relationships. However, application of new species tree estimation methods to a more focused dataset for Phocina including many more individuals per species will probably now be another key to improved resolution for Phocina.
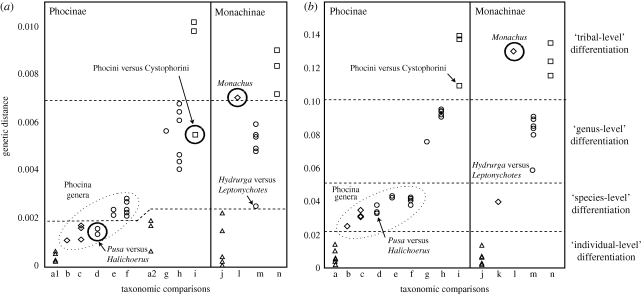
Intraspecific sampling also provided insight into taxonomic distinctions and a comparison of morphological versus molecular differentiation. All Phocina species are close genetically, particularly at nuclear loci, where their interspecific variation level is more similar to the intraspecific variation level of other phocids (figure 3a). Halichoerus is no more distant from any of the Pusa genera than Pusa species are to one another (figure 3; see also Davis et al. 2004). Although Phoca is more genetically distinct, the distance is still much smaller than between any other genera (except Hydrurga and Leptonychotes). From a purely genetic perspective, it is appropriate to roll all three genera, Pusa, Halichoerus and Phoca, into one (Phoca, Linnaeus 1758) as suggested previously (Árnason et al. 1995, 2006). But Halichoerus is morphologically distinct (Burns & Fay 1970; Nowak 1999), thus it seems premature to revoke its generic status, though its exact phylogenetic placement is not conclusive. The low genetic and high morphological divergence in Halichoerus is contrasted by the case of the monk seals (Monachus), which exhibit high morphological similarity, but extreme genetic divergence (figure 3). Monk seals are unquestionably monophyletic, but appear almost morphologically stagnant, resembling their extinct relatives (Hendey 1972; Repenning & Ray 1977; Wyss 1988). Whether the sister group of Halichoerus is Pusa (generally preferred genetically) or Phoca + Pusa (more preferred morphologically) depends on the analysis performed, but it is clear that including multiple individuals and multiple loci is critical to solidifying the phylogeny of this tribe, bringing molecules and morphology in line and opening an avenue of investigation into shifts in the rate of morphological evolution across Phocidae.
Figure 3.
Genetic versus ‘taxonomic’ distance. For (a) nuclear genes with GTR distance and (b) mitochondrial genes with logdet distance, a series of pairwise distances were calculated from between individuals (a,a1,a2,j) up to between tribes (i,n). Distances were averaged for nested taxonomic comparisons. The approximate breaks in genetic distance between different taxonomic levels are indicated by dashed lines and outliers are noted. The range of genetic distance within and between species for nuclear loci was overlapping and a natural break was not observed (a, Phocinae spp.; a1, Pusa spp.; a2, other Phocinae spp.; b, Phoca, c, Pusa; d, Pusa versus Halichoerus; e, Phoca versus Halichoerus; f, Pusa versus Phoca; g, Histriophocina; h, Phocini; i, Phocinae; j, Monachinae spp.; k, Mirounga, l, Monachus, m, Lobodontini; triangles, within species; diamonds, within genus; circles, within tribe; squares, within subfamily).
In contrast, DNA evidence for the Antarctic Lobodontini supports a movement away from traditional morphological groups. Morphological analyses using primarily cranial and dental characters (Hendey 1972; de Muizon 1982) placed the four Antarctic species into two clades: leopard + crabeater and Weddell + Ross. Molecules strongly disagree with morphology, placing the leopard and Weddell seals together (figure 1), but vary in placing the crabeater (Fyler et al. 2005; Higdon et al. 2007) or Ross (Davis et al. 2004; Árnason et al. 2006; Fulton & Strobeck 2006) seal as most basal. AFLP analysis (Dasmahapatra et al. 2009) supported the same two sister groupings as recovered here. Leopard and Weddell seals are very close genetically (figure 3), and share some superficial morphological similarity, such as their spotted coats. The Weddell seal has also been called the ‘sea leopard’ (Scheffer 1958) and the ‘false sea leopard’ (Hince 2000). Leopard seals are highly predatory but also rely heavily on krill (Rogers 2002). The crabeater seal relies almost solely on krill (Bengtson 2002) and both have specialized dentition for this food source, thus, a re-evaluation of morphology with a decreased focus on dental characters would be useful. The placement of the Ross and crabeater seals is less clear. Analysing nuclear loci alone or in combination with mtDNA provided a novel hypothesis for sequence analyses: that these species are sister (figure 1). A morphology + DNA supertree also recovered this topology (Bininda-Emonds et al. 1999), thus, this grouping may be more congruent with morphology than thought. Whatever the final resolution, the Lobodontini lineages diverged rapidly from one another, presumably near the time of their entry into the Antarctic (Fulton & Strobeck in press). Increasing nuclear markers and morphological review will be required to solidify their relationships and investigate the processes of how species invade such a divergent environment from their ancestral habitat, bringing us closer to understanding the links between environment, specialization and speciation.
Acknowledgements
Thanks to B. Shapiro, W. Murphy, I. Delisle, D. Coltman, S. Moore, G. Wilson and two reviewers for helpful comments and to the many people/organizations who provided samples. This work was funded by an NSERC research grant to CS and fellowship to TLF.
References
- Árnason Ú., Bodin K., Gullberg A., Ledje C., Mouchaty S.1995A molecular view of pinniped relationships with particular emphasis on the true seals. J. Mol. Evol. 40, 78–85 (doi:10.1007/BF00166598) [DOI] [PubMed] [Google Scholar]
- Árnason Ú., Gullberg A., Janke A., Kullberg M., Lehman N., Petrov E. A., Väinölä R.2006Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol. Phylogenet. Evol. 41, 345–354 (doi:10.1016/j.ympev.2006.05.022) [DOI] [PubMed] [Google Scholar]
- Árnason Ú., Gullberg A., Janke A., Kullberg M.2007Mitogenomic analyses of caniform relationships. Mol. Phylogenet. Evol. 45, 863–874 (doi:10.1016/j.ympev.2007.06.019) [DOI] [PubMed] [Google Scholar]
- Bengtson J. L.2002Crabeater seal: Lobodon carcinophagus. In Encyclopedia of marine mammals (eds Perrin W. F., Wursig B., Thewissen J. G. M.), pp. 302–304 San Diego, CA: Academic Press [Google Scholar]
- Bininda-Emonds O. R. P., Gittleman J. L., Purvis A.1999Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia). Biol. Rev. 74, 143–175 (doi:10.1017/S0006323199005307) [DOI] [PubMed] [Google Scholar]
- Brumfield R. T., Liu L., Lum D. E., Edwards S. V.2008Comparison of species tree methods for reconstructing the phylogeny of bearded manakins (Aves: Pipridae, Manacus) from multilocus sequence data. Syst. Biol. 57, 719–731 (doi:10.1080/10635150802422290) [DOI] [PubMed] [Google Scholar]
- Burns J. J., Fay F. H.1970Comparative morphology of the skull of the ribbon seal, Histriophoca fasciata, with remarks on systematics of Phocidae. J. Zool. Lond. 161, 363–394 [Google Scholar]
- Carstens B. C., Knowles L. L.2007Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: an example from Melanoplus grasshoppers. Syst. Biol. 56, 400–411 (doi:10.1080/10635150701405560) [DOI] [PubMed] [Google Scholar]
- Chapskii K. K.1955An attempt at revision of the systematics and diagnostics of the subfamily Phocinae. Trud. Zool. Inst. Akad. Nauk SSSR 17, 160–199 [Google Scholar]
- Dasmahapatra K. K., Hoffman J. I., Amos W.2009Pinniped phylogenetic relationships inferred using AFLP markers. Heredity 103, 168–177 (doi:10.1038/hdy.2009.25) [DOI] [PubMed] [Google Scholar]
- Davis C. S., Delisle I., Stirling I., Siniff D. B., Strobeck C.2004A phylogeny of the extant Phocidae inferred from complete mitochondrial DNA coding regions. Mol. Phylogenet. Evol. 33, 363–377 (doi:10.1016/j.ympev.2004.06.006) [DOI] [PubMed] [Google Scholar]
- de Muizon C.1982Phocid phylogeny and dispersal. Ann. S. Afr. Mus. 89, 175–213 [Google Scholar]
- Degnan J. H., Rosenberg N. A.2009Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340 (doi:10.1016/j.tree.2009.01.009) [DOI] [PubMed] [Google Scholar]
- Farris J. S., Kallersjo M., Kluge A. G., Bult C.1995Constructing a significance test for incongruence. Syst. Biol. 44, 570–572 [Google Scholar]
- Flynn J. J., Finarelli J. A., Zehr S., Hsu J., Nedbal M. A.2005Molecular phylogeny of the Carnivora (Mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst. Biol. 54, 317–337 [DOI] [PubMed] [Google Scholar]
- Fulton T. L., Strobeck C.2006Molecular phylogeny of the Arctoidea (Carnivora): effect of missing data on supertree and supermatrix analyses of multiple gene data sets. Mol. Phylogenet. Evol. 41, 165–181 (doi:10.1016/j.ympev.2006.05.025) [DOI] [PubMed] [Google Scholar]
- Fulton T. L., Strobeck C.In press Multiple fossil calibrations, nuclear loci and mitochondrial genomes provide new insight into biogeography and divergence timing for true seals (Phocidae, Pinnipedia). J. Biogeogr [Google Scholar]
- Fyler C. A., Reeder T. W., Berta A., Antonelis G., Aguilar A., Androukaki E.2005Historical biogeography and phylogeny of monachine seals (Pinnipedia: Phocidae) based on mitochondrial and nuclear DNA data. J. Biogeogr. 32, 1267–1279 (doi:10.1111/j.1365-2699.2005.01281.x) [Google Scholar]
- Gatesy J., Baker R. H.2005Hidden likelihood support in genomic data: can forty-five wrongs make a right? Syst. Biol. 54, 483–492 (doi:10.1080/10635150590945368) [DOI] [PubMed] [Google Scholar]
- Hendey Q. B.1972The evolution and dispersal of the Monachinae (Mammalia: Pinnipedia). Ann. S. Afr. Mus. 59, 99–113 [Google Scholar]
- Higdon J. W., Bininda-Emonds O. R. P., Beck R. M. D., Ferguson S. H.2007Phylogeny and divergence of the pinnipeds (Carnivora: Mammalia) assessed using a mutigene dataset. BMC Evol. Biol. 7, 216 (doi:10.1186/1471-2148-7-216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hince B.2000The Antarctic dictionary: a complete guide to Antarctic English. Collingwood, Australia: CSIRO publishing [Google Scholar]
- Ho S. Y. W., Jermiin L. S.2004Tracing the decay of the historical signal in biological sequence data. Syst. Biol. 53, 623–637 (doi:10.1080/10635150490503035) [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F.2001MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- King J. E.1966Relationships of the Hooded and Elephant seals (genera Cystophora and Mirounga). J. Zool. 148, 385–398 (doi:10.1111/j.1469-7998.1966.tb02958.x) [Google Scholar]
- King J. E.1983Seals of the world, 2nd edn London and Oxford, UK: British Museum (Natural History) and Oxford University Press [Google Scholar]
- Koepfli K.-P., Deere K. A., Slater G. J., Begg C., Begg K., Grassman L., Lucherini M., Veron G., Wayne R. K.2008Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biol. 6 (doi:10.1186/1741-7007-6-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko L. S., Carstens B. C., Knowles L. L.2009STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics 25, 971–973 (doi:10.1093/bioinformatics/btp079) [DOI] [PubMed] [Google Scholar]
- Liu L.2008BEST: Bayesian estimation of species trees under the coalescent model. Bioinformatics 24, 2542–2543 (doi:10.1093/bioinformatics/btn484) [DOI] [PubMed] [Google Scholar]
- Maddison W. P., Knowles L. L.2006Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55, 21–30 (doi:10.1080/10635150500354928) [DOI] [PubMed] [Google Scholar]
- McLaren I. A.1960On the origin of the Caspian and Baikal seals and the paleoclimatological implication. Am. J. Sci. 258, 47–65 [Google Scholar]
- Nowak R. M.1999Walker's mammals of the world, 6th edn Baltimore, MD: The John Hopkins University Press [Google Scholar]
- Pagès M., Calvignac S., Klein C., Paris M., Hughes S., Hänni C.2008Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol. Phylogenet. Evol. 47, 73–83 (doi:10.1016/j.ympev.2007.10.019) [DOI] [PubMed] [Google Scholar]
- Palo J. U., Väinölä R.2006The enigma of the landlocked Baikal and Caspian seals addressed through phylogeny of phocine mitochondrial sequences. Biol. J. Linn. Soc. 88, 61–72 (doi:10.1111/j.1095-8312.2006.00607.x) [Google Scholar]
- Palo J. U., Makinen H. S., Helle E., Stenman O., Väinölä R.2001Microsatellite variation in ringed seals (Phoca hispida): genetic structure and history of the Baltic Sea population. Heredity 86, 609–617 (doi:10.1046/j.1365-2540.2001.00859.x) [DOI] [PubMed] [Google Scholar]
- Repenning C. A., Ray C. E.1977The origin of the Hawaiian monk seal. Proc. Biol. Soc. Wash. 89, 667–688 [Google Scholar]
- Rogers T. L.2002Leopard seal: Hydrurga leptonyx. In Encyclopedia of marine mammals (eds Perrin W. F., Wursig B., Thewissen J. G. M.), pp. 692–693 San Diego, CA: Academic Press [Google Scholar]
- Ronquist F., Huelsenbeck J. P.2003MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- Sasaki H., Numachi K., Grachev M. A.2003The origin and genetic relationships of the Baikal seal, Phoca sibirica, by restriction analysis of mitochondrial DNA. Zool. Sci. 20, 1417–1422 (doi:10.2108/zsj.20.1417) [DOI] [PubMed] [Google Scholar]
- Sato J. J., Wolsan M., Suzuki H., Hosoda T., Yamaguchi Y., Hiyama K., Kobayashi M., Minami S.2006Evidence from nuclear DNA sequences sheds light on the phylogenetic relationships of pinnipedia: single origin with affinity to musteloidea. Zool. Sci. 23, 125–146 (doi:10.2108/zsj.23.125) [DOI] [PubMed] [Google Scholar]
- Scheffer V. B.1958Seals, sea lions, and walruses: a review of the Pinnipedia Stanford, CA: Stanford University Press [Google Scholar]
- Shaughnessy P. D., Fay F. H.1977A review of the taxonomy and nomenclature of North Pacific Harbour seals. J. Zool. Lond. 182, 385–419 (doi:10.1111/j.1469-7998.1977.tb03917.x) [Google Scholar]
- Stamatakis A.2006aPhylogenetic models of rate heterogeneity: a high performance computing perspective. In 20th IEEE/ACM Int. Parallel and Distributed Processing Symp. (IPDPS2006), Rhodes, Greece, 25–29 April 2006, p. 278 (doi:10.1109/IPDPS.2006.1639535) [Google Scholar]
- Stamatakis A.2006bRAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- Syring J., Farrell K., Businsky R., Cronn R., Liston A.2007Widespread genealogical nonmonophyly in species of Pinus subgenus Strobus. Syst. Biol. 56, 163–181 (doi:10.1080/10635150701258787) [DOI] [PubMed] [Google Scholar]
- Swofford D. L.2003PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4 Sunderland, MA: Sinauer Associates [Google Scholar]
- Willyard A., Cronn R., Liston A.2009Reticulate evolution and incomplete lineage sorting among the ponderosa pines. Mol. Phylogenet. Evol. 52, 498–511 (doi:10.1016/j.ympev.2009.02.011) [DOI] [PubMed] [Google Scholar]
- Wyss A. R.1988On ‘retrogression’ in the evolution of the Phocinae and phylogenetic affinities of the monk seals. Am. Mus. Novit. 2924, 1–38 [Google Scholar]
- Yonezawa T., Kohno N., Hasegawa M.2009The monophyletic origin of sea lions and fur seals (Carnivora; Otariidae) in the Southern Hemisphere. Gene 441, 89–99 (doi:10.1016/j.gene.2009.01.022) [DOI] [PubMed] [Google Scholar]