Abstract
Inhibitory interneurons participate in local processing circuits, playing a central role in executive cognitive functions of the prefrontal cortex. Although humans differ from other primates in a number of cognitive domains, it is not currently known whether the interneuron system has changed in the course of primate evolution leading to our species. In this study, we examined the distribution of different interneuron subtypes in the prefrontal cortex of anthropoid primates as revealed by immunohistochemistry against the calcium-binding proteins calbindin, calretinin and parvalbumin. In addition, we tested whether genes involved in the specification, differentiation and migration of interneurons show evidence of positive selection in the evolution of humans. Our findings demonstrate that cellular distributions of interneuron subtypes in human prefrontal cortex are similar to other anthropoid primates and can be explained by general scaling rules. Furthermore, genes underlying interneuron development are highly conserved at the amino acid level in primate evolution. Taken together, these results suggest that the prefrontal cortex in humans retains a similar inhibitory circuitry to that in closely related primates, even though it performs functional operations that are unique to our species. Thus, it is likely that other significant modifications to the connectivity and molecular biology of the prefrontal cortex were overlaid on this conserved interneuron architecture in the course of human evolution.
Keywords: language, theory of mind, prefrontal cortex, chimpanzee, great ape
1. Introduction
Direct comparison of the human genome with that of other species has led to important insight into the molecular changes underlying human brain evolution (Dorus et al. 2004; Haygood et al. 2007; Uddin et al. 2008). Understanding how genetic evolution relates to distinctive anatomical specializations of the human brain, however, requires a more complete description of the human neural phenotype in comparison to our closest relatives, especially chimpanzees and other great apes. Comparative studies of mRNA transcript levels using high-throughput methodologies, such as microarrays, have provided information about human-specific differences in gene expression and networks of co-expression in the brain (Caceres et al. 2003; Khaitovich et al. 2004; Uddin et al. 2004; Oldham et al. 2006), shedding light on physiological pathways and molecular mechanisms that have been important in human brain evolution. Yet, the results of such microarray studies have been difficult to link to species-specific variation in neuronal connectivity. In part, this is due to complications associated with interpreting species differences in overall levels of mRNA within regions of the brain that are composed of heterogeneous population of cells, distributed across functionally distinct layers (Geschwind 2000). Because of these challenges, a parallel line of inquiry is needed to elucidate how homologous neocortical areas vary between humans and other species in terms of neuronal morphology, distribution of neuron types, expression of receptors and innervation by different neurotransmitter systems (Preuss 2007; Sherwood et al. 2008).
It has been proposed that human cognition is uniquely distinguished by traits such as an enhanced ability to represent the mental states of oneself and others in social cognition (Herrmann et al. 2007) and to employ a hierarchical recursive syntactic organization in language (Hauser et al. 2002). However, it is not known to what extent the cerebral cortex carries out these complex operations using machinery that is evolutionarily conserved versus specialized. Beyond the more than threefold enlargement in neocortical size during human evolution (Holloway et al. 2004), how has its architecture and connectivity changed to support human-specific functions? One possibility, raised initially by Ramón y Cajal (1923), is that human brain evolution was associated with an increased number and diversity of ‘short-axon’ interneurons. Inhibitory GABAergic interneurons are fundamental to the distribution of thalamic input in the neocortex and comprise the local circuitry that influences the activity of pyramidal cells (Hendry 1987). The diverse cortical interneuron population can be subdivided into subtypes on the basis of morphology, biochemical phenotype, post-synaptic target, developmental origin and expression of transcription factors (Zaitsev et al. 2009). The calcium-binding proteins, calbindin D-28k (CB), calretinin (CR) and parvalbumin (PV), are particularly useful markers for revealing the architecture of the cortical interneuron network because they are colocalized with GABA in 90–95% of interneurons and occur in morphologically and physiologically distinct subtypes (DeFelipe 1997). One important role of interneurons, particularly PV-immunoreactive (ir) fast-spiking subtypes, is to control the phase of rhythmic oscillations across ensembles of pyramidal neurons (Sohal et al. 2009). Such coordination of spike timing is critical to the temporal precision underlying processes ranging from perception to cognition (Bartos et al. 2007).
We used stereological methods to quantify the density of CB-, CR- and PV-ir interneurons within the cerebral cortex from a diversity of anthropoid primates, focusing special attention on prefrontal regions that are involved in cognitive abilities such as working memory (Brodmann's area 9), mental state attribution (area 32) and language (area 44) (Gallagher & Frith 2003; Friederici et al. 2006), relative to the primary motor cortex (area 4). We hypothesized that interneuron proportions might be selectively increased in prefrontal areas that underlie our species' cognitive specializations (areas 9, 32 and 44). In addition, we aligned vertebrate orthologues of 20 genes that are important for the regulation of neocortical interneuron development and analysed their protein-coding sequence for evidence of positive Darwinian selection along the human lineage. Through examination of the phenotype and related genes, we tested whether there is evidence of evolutionary modification of the prefrontal interneuron system of humans, or alternatively, whether this cellular network is conserved.
2. Material and methods
(a). Specimens, preparation and immunohistochemical staining
Formalin-fixed brain samples representing 23 anthropoid primate species, from 51 individuals, were used for the phenotype analyses (see the electronic supplementary material). A 1 : 10 series of 40 μm-thick sections was stained for Nissl substance with a solution of 0.5 per cent cresyl violet. Immunohistochemistry was performed on adjacent 1 : 20 series of sections with monoclonal antibodies against PV (dilution 1 : 10 000) and CB (dilution 1 : 8000), or with a polyclonal antibody against CR (dilution 1 : 10 000; Swant, Bellinzona, Switzerland) (see the electronic supplementary material).
(b). Quantification of neuron densities
All quantitative analyses were restricted to layers II and III because CB- and CR-ir interneurons predominate in the superficial layers of the primate neocortex (Hof et al. 1999). Our previous studies of species differences in quantitative cyto- and chemoarchitecture of the cerebral cortex in anthropoid primates provide detailed descriptions of the histological criteria used to identify cortical areas (Sherwood et al. 2006; Raghanti et al. 2008b). Analyses of allometric scaling and variance partitioning across the 23 species used neuron densities in layers II and III of dorsolateral prefrontal cortex (DLPFC), corresponding to area 9. We also analysed the percentage of the total neuron population in layers II and III that contains each calcium-binding protein in humans, chimpanzees and macaque monkeys (n = 6 each species) from a larger number of frontal areas, including primary motor cortex (area 4 in the region of the hand representation), DLPFC (area 9), anterior paracingulate cortex (area 32) and inferior prefrontal cortex (area 44) (see electronic supplementary material for more details).
Total neuron density in layers II and III was quantified from adjacent series of Nissl-stained sections and was used as a reference variable for analyses of interneuron subtype densities. Calcium-binding protein-ir interneuron subtypes were counted separately in quantitative analyses. Because we were only interested in the distribution of interneurons, we did not count lightly stained CB-ir somata with distinct apical dendrites or pyramidal morphology. Densities of neurons within layers II and III were estimated using an unbiased stereological design employing the optical disector technique combined with fractionator sampling (Mouton 2002). The stereological analyses resulted in sampling an average of 143 counting frames per region in each individual for each cell type, with a total of 59 472 counting frames investigated and 24 557 neurons sampled (see the electronic supplementary material). It should be noted that our estimates of densities for all neuron types in humans were considerably higher than expected based on previous literature and in comparison to the other species in this study. It is likely that different tissue handling procedures at the Northwestern University Alzheimer's Disease Center Brain Bank contributed to this effect. Nonetheless, because of our concern about uncontrolled and unknown degrees of tissue shrinkage for the entire sample, all subsequent statistical analyses only considered interneuron subtype density relative to total neuron density within the same individual, i.e. calculated as percentages or as regressions of interneuron density against total neuron density. Thus, equal amounts of fixation and histological artefact are included in both independent and dependent variables. Consideration of the effect of post-mortem interval, fixation condition and age on quantitative results revealed no significant relationships (see the electronic supplementary material).
(c). Allometric scaling analyses
Logarithm (base 10)-transformed species means were used in allometric scaling analyses of interneuron density against total neuron density in DLPFC. To determine the exponent of scaling relationships, we used reduced major axis (RMA) line-fitting to bivariate data to allow for error in both independent and dependent variables. All RMA tests were calculated using (S)MATR software v. 2.0.
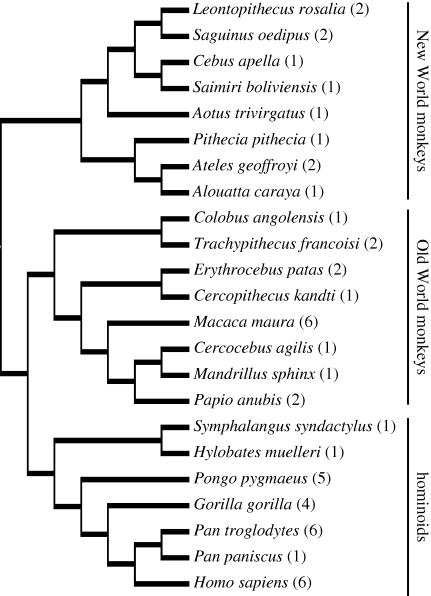
Phylogenetic independent contrasts were also calculated from the data to examine scaling relationships while controlling for the effects of phylogenetic relatedness in the dataset (Felsenstein 1985). Standardized independent contrasts were calculated using the PDAP:PDTREE module of Mesquite software v. 1.12 (Maddison & Maddison 2005) from log-transformed data based on a phylogeny of primates in Goodman et al. (2005) (figure 1). Branch lengths were transformed according to the method of Pagel (1992), which assigns all branch lengths to 1 with the constraint that tips are contemporaneous.
Figure 1.
Phylogenetic relationships among the primate species used in the analysis of phenotype. This phylogeny was used to calculate independent contrasts. Sample sizes for each species used in stereological analyses are shown in parentheses.
We also examined whether human interneuron densities represent significant deviations from allometric expectations based on other anthropoid primates. We calculated least-squares regression equations and 95 per cent prediction intervals for humans based on the non-human data using both contemporary ‘tip’ species data and independent contrasts according to the method of Garland & Ives (2000). After logarithmic detransformation of predictions, the percentage difference between observed and predicted values was calculated as the ratio of (observed−predicted)/observed.
(d). Quantifying the partitioned variation
We employed a variance partitioning method to dissect further the interaction between the phenotype and phylogeny (Desdevises et al. 2003). Westboy et al. (1995) proposed the theoretical partitioning of variance in a dataset into three components: a, b and c—where a is a part strictly owing to adaptation to the environment, b is a part owing to the common influence of environment and phylogeny and c is a part strictly owing to phylogeny. Desdevises et al. (2003) described a multiple regression method for partitioning this variation by expressing the phylogeny as a distance matrix. In the current study, the decomposition of the variation for interneuron subtypes across anthropoid species in DLPFC was undertaken in accordance with the procedural steps outlined by Desdevises et al. (2003). For further details see the electronic supplementary material.
(e). Analysis of coding sequence evolution of interneuron-important genes among mammals
Genes important to the transcriptional regulation of cortical interneuron specification and differentiation were identified from the literature (Wonders & Anderson 2006). Human RefSeq IDs corresponding to these genes were identified using NCBI's Gene database. Where more than one transcript variant was available for a particular gene, the longest variant was retained for further analysis. This process resulted in a total of 20 human RefSeq IDs that were used in subsequent investigations (see the electronic supplementary material).
Multiple sequence alignments of the coding regions for the 20 interneuron-important genes were obtained using OCPAT, an online codon-preserved alignment tool for evolutionary genomic analysis of protein-coding sequences (Liu et al. 2007). Briefly, the tool first identifies putative orthologues from up to 14 taxa (chimpanzee, macaque, rabbit, mouse, rat, dog, cow, tenrec, elephant, armadillo, opossum, platypus, chicken and frog) that correspond to the queried human RefSeq ID. Next, it creates an alignment from the identified putative orthologues that retains the codon structure of all included sequences. Aligned sequences were subsequently analysed for patterns of sequence evolution using the PAML 3.15 package (Yang 1997). The phylogenetic relationships used to infer these patterns were obtained from the literature (Hallstrom et al. 2007; Wildman et al. 2007). Sequences were analysed by comparing the likelihood score of the data in a model that assumes the same dN/dS (i.e. non-synonymous changes per non-synonymous site/synonymous changes per synonymous site) ratio among all branches (i.e. the one omega or M0 model) to the likelihood score of the data in a model that permits dN/dS ratios to vary freely among all branches in the phylogenetic tree (i.e. the free ratio model or M1 model). All models were run with three different starting omega values (0.5, 1 and 2) to ensure optimal likelihood scores. Models were compared using the likelihood ratio test, using the highest maximum likelihood value obtained among the three different starting omega values. Patterns of selection were further investigated for genes passing the M0 versus M1 test by mapping the PAML-inferred M1 dN/dS ratios onto the phylogenetic tree.
3. Results
(a). Scaling relationships of interneurons in DLPFC
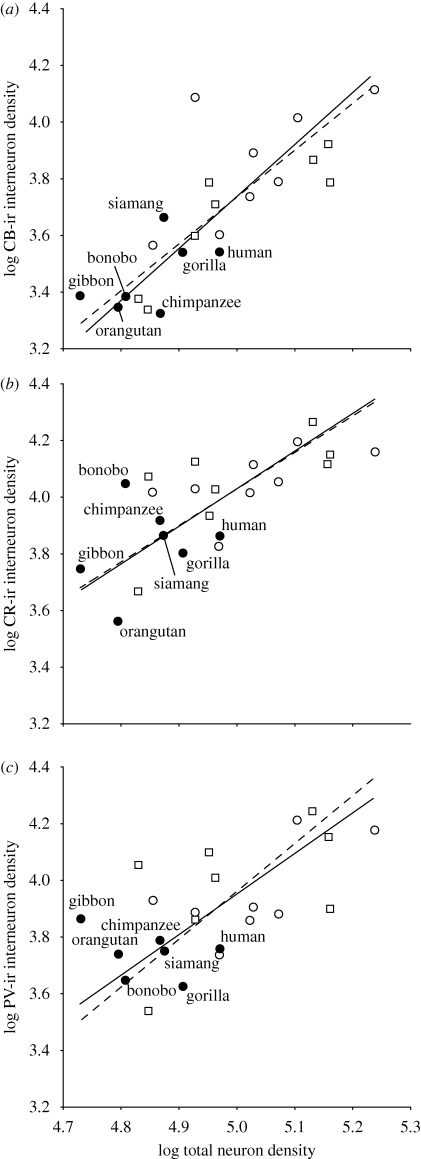
Figure 2 shows the allometric scaling of interneuron subtype densities against total neuron density from layers II and III of DLPFC (area 9) across 23 anthropoid primate species, including humans. The scaling exponents based on independent contrasts for each interneuron subtype were contained within the 95 per cent confidence intervals of those calculated from contemporary tip species data, indicating that there was not a strong effect of phylogenetic bias (table 1). We also tested for differences in the slope and elevation of the scaling function for each interneuron subtype among hominoids (n = 7 species), Old World monkeys (n = 8 species) and New World monkeys (n = 8 species). There were no significant differences among these phylogenetic groups as revealed by a likelihood ratio test for common slopes or analysis of variance (ANOVA) for elevation differences. Notably, all interneuron subtypes scaled against total neuron density with a positive allometric exponent as indicated by both tip species data and independent contrasts (table 1).
Figure 2.
Scatterplots showing the allometric scaling relationship between the density of (a) CB-ir interneurons, (b) CR-ir interneurons and (c) PV-ir interneurons against total neuron density in layers II and III of DLPFC. Solid lines indicate the RMA using tip species data. Dashed lines indicate the RMA using independent contrasts according to the method of Garland & Ives (2000). Open circle, New World monkeys; square, Old World monkeys and filled circle, hominoids.
Table 1.
The allometric scaling of interneuron subtype densities versus total neuron density in DLPFC of anthropoid primates. RMA, reduced major axis; CI, confidence intervals.
| contemporary tip species data |
independent contrasts |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| difference from H0 slope = 1 |
||||||||||
| interneuron subtype | r2 | p-value | RMA slope | lower 95% CI | upper 95% CI | F | p-value | r2 | p-value | RMA slope |
| CB | 0.66 | 0 | 1.84 | 1.41 | 2.38 | 25.86 | 0 | 0.74 | 0 | 1.69 |
| CR | 0.50 | 0 | 1.32 | 0.96 | 1.82 | 3.37 | 0.081 | 0.43 | 0 | 1.29 |
| PV | 0.39 | 0.001 | 1.44 | 1.01 | 2.03 | 4.70 | 0.042 | 0.40 | 0.001 | 1.69 |
The variance partitioning analyses provided additional support for the conclusion that phylogeny plays a relatively weak role in determining the density of interneuron types in the DLPFC (table 2). The exclusive phylogenetic component (c) explained only a small proportion of the variance in interneuron subtype densities (between 7% and 13%), whereas a greater proportion of the variance was explained by either total neuron density alone (a) (between 13% and 28%), or the interaction between neuron density and phylogeny (b) (19–38%). It is notable, however, that the unexplained component of variance (d) was usually greater than any of the defined predictors.
Table 2.
Results of variance partitioning analysis of interneurons in DLFPC.
| % explained by total neuron density |
||||
|---|---|---|---|---|
| % explained by phylogeny |
% unexplained | |||
| dependent variable | a | b | c | d |
| CB interneuron density | 28.3 | 37.9 | 9.5 | 24.3 |
| CR interneuron density | 13.3 | 36.5 | 7.2 | 43.0 |
| PV interneuron density | 20.2 | 18.9 | 13.4 | 47.5 |
| % explained by brain mass | ||||
| % CB interneurons | 1.2 | 13.4 | 14.4 | 71.0 |
| % CR interneurons | 0.7 | 8.9 | 15.3 | 75.1 |
| % PV interneurons | 1.3 | 5.7 | 18.9 | 74.1 |
We also tested whether brain size correlates with interneuron distributions among species using variance partitioning (table 2). When the percentage of interneuron subtypes in DLPFC was considered in relation to brain mass and phylogeny, a very large fraction of variance remained unexplained (71–75%). Congruent with this result, the simple bivariate relationship between species mean brain mass and the percentage of each interneuron subtype in the DLPFC also showed no significant correlations using tip species data and independent contrasts.
(b). Human allometric departures of interneuron proportions in DLPFC
Given the regular scaling of interneurons, we tested whether human interneuron densities in DLPFC deviate from expectations for an anthropoid primate of the same total neuron density (figure 2). Because there was no evidence of a significant phylogenetic effect on the scaling relationships, we calculated least-squares prediction equations based on the total non-human anthropoid sample. Human interneuron subtype densities were all contained within the 95 per cent prediction intervals of the non-human tip species data, but always fell below the expected values (see electronic supplementary material). Next, we used independent contrasts to generate predicted interneuron densities for a hypothetical species attached to the branch leading to humans in the phylogenetic tree. From this phylogenetically based prediction, the interneuron densities in humans also fell within the 95 per cent prediction intervals and were lower than expected (see electronic supplementary material).
(c). Interneuron variation across the frontal cortex of humans, chimpanzees and macaque monkeys
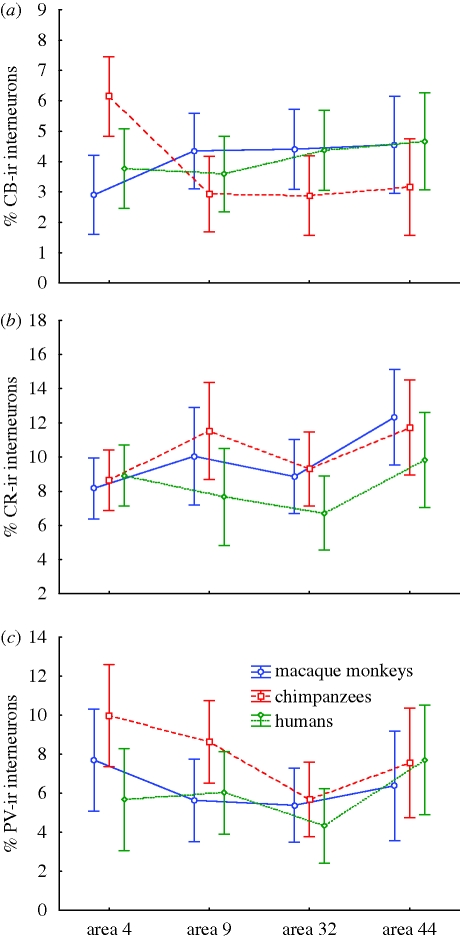
Our next analyses investigated whether humans, chimpanzees and macaque monkeys vary from each other in the proportions of interneuron subtypes across different frontal cortical areas. Individual data on the percentage of interneuron subtypes in areas 4, 9, 32 and 44 of these species (see electronic supplementary material) were analysed using repeated-measures ANOVA. None of the ANOVAs revealed a significant main effect of species (figure 3), demonstrating an overall similarity in the proportions of interneuron subtypes among these taxa. However, the percentage of CB-ir interneurons displayed an interaction effect between species and cortical area. Results of Bonferroni post hoc comparisons indicated that chimpanzees diverged from the other two species in the regional distribution of CB-ir interneurons, with the most remarkable difference being a greater percentage of CB-ir interneuron in primary motor cortex (area 4).
Figure 3.
Inter-regional variation in the proportion of (a) CB-ir (area * species: F6,45 = 4.22, p = 0.002; chimpanzee: area 4 > area 9, area 32), (b) CR-ir (area: F3,45 = 4.88, p = 0.005; area 44 > area 4, area 32) and (c) PV-ir (area: F3,45 = 4.88, p = 0.005; area 4, area 44 > area 32) interneurons among humans, chimpanzees and macaque monkeys. Means and 95% confidence intervals are shown. The significant effects of the repeated-measures ANOVA models and Bonferroni post hoc results are displayed in the legend for each interneuron subtype.
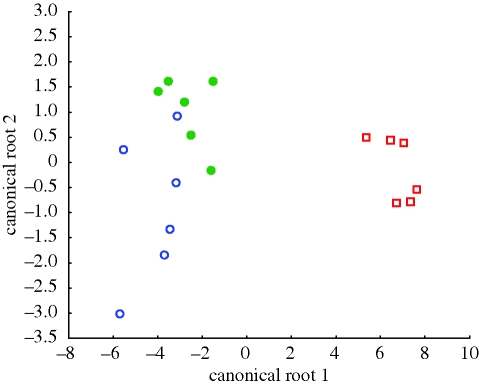
To determine further which variables distinguish among species most clearly, we employed a forward stepwise discriminant function analysis. The final discriminant function was significant (Wilks' Λ = 0.020, F14,18 = 7.90, p < 0.00001) and classified 94.4 per cent of the cases to the correct species. Among the seven variables that were retained in the final discriminant function (see electronic supplementary material), three represented the distribution of CB-ir interneurons, three represented the distribution of PV-ir interneurons and one represented the distribution of CR-ir interneurons. The canonical variates plot shows that chimpanzees were the most distinct of the three species in their frontal cortex interneuron phenotype (figure 4). The largest absolute correlations with canonical root 1, which separates chimpanzees from the other two species, were the percentage of CB-ir interneurons in area 4 (r = 0.19) and the percentage of PV-ir interneurons in area 9 (r = 0.11).
Figure 4.
Canonical variates plot of the full interneuron subtype dataset from areas 4, 9, 32 and 44 in humans, chimpanzees and macaques. Note that chimpanzees show the most distinct separation from the other species. Open circle, macaque monkeys; square, chimpanzees and closed circle, humans.
(d). Evolution of genes important to the transcriptional regulation of cortical interneuron specification and differentiation
For the 20 analysed genes, alignments were obtained that included a minimum of eight species, with the majority (n = 11) of alignments including all 14 possible species. Likelihood ratio tests indicated that for all but four genes (i.e. DLX5, EMX2, LHX8 and NPAS1), the M1 model had a better fit to the data than did the M0 model (table 3) indicating that there was variation in the rate of evolution of these genes across the lineages represented in our phylogenetic tree; for the four genes that did not pass the M0 versus M1 test, inferred dN/dS values ranged from a low of 0.013 to a high of 0.354 indicating a conserved rate of amino acid changing substitutions (i.e. dN/dS < 1) which did not vary significantly across the sampled phylogenetic lineages.
Table 3.
M0 versus M1 likelihood ratio tests of interneuron-important genes. The likelihood of a model in which dN/dS is equivalent on all branches (M0) was compared with the likelihood of a model in which dN/dS was allowed to vary on all branches of a phylogenetic tree (M1) using a likelihood ratio test. If the likelihood of M1 was significantly greater (p ≤ 0.05) than the likelihood of M0, the gene can be interpreted to have varying rates of amino acid changing substitutions during the descent of mammals.
| gene symbol | RefSeq | lnl1 M0 | lnl2 M1 | 2(diff) = chi square | degrees of freedom | probability |
|---|---|---|---|---|---|---|
| ARX | NM_139058 | −5580.7 | −5566.1 | 29.2 | 15 | 0.0151 |
| ASCL1 | NM_004316 | −1835.8 | −1805.5 | 60.6 | 21 | 0 |
| DLX1 v1 | NM_178120 | −3410.0 | −3383.7 | 52.7 | 27 | 0.0022 |
| DLX2 | NM_004405 | −5330.3 | −5278.1 | 104.4 | 23 | 0 |
| DLX5 | NM_005221 | −4403.8 | −4397.1 | 13.4 | 25 | 0.9717 |
| DLX6 | NM_005222 | −2037.0 | −2016.8 | 40.3 | 27 | 0.0476 |
| EMX2 v1 | NM_004098 | −2366.1 | −2355.6 | 20.9 | 27 | 0.7903 |
| ETV1 v1 | NM_004956 | −6952.5 | −6898.9 | 107.2 | 27 | 0 |
| GLI3 | NM_000168 | −31 240.6 | −31 202.5 | 76.0 | 27 | 0 |
| GSX2 | NM_133267 | −5211.0 | −5189.8 | 42.5 | 21 | 0.0036 |
| LHX6 v1 | NM_014368 | −2502.1 | −2471.3 | 61.6 | 27 | 0.0001 |
| LHX8 | NM_001001933 | −4484.9 | −4475.2 | 19.4 | 27 | 0.8548 |
| NKX2-1 v2 | NM_003317 | −4670.4 | −4646.8 | 47.1 | 19 | 0.0003 |
| NPAS1 | NM_002517 | −5799.0 | −5792.7 | 12.6 | 13 | 0.4784 |
| NPAS3 v3 | NM_173159 | −12 587.4 | −12 508.3 | 158.2 | 27 | 0 |
| NR2E1 | NM_003269 | −4416.7 | −4383.5 | 66.5 | 27 | 0 |
| PAX6 v1 | NM_000280 | −4790.2 | −4681.1 | 218.1 | 27 | 0 |
| SHH | NM_000193 | −8600.3 | −8541.4 | 117.9 | 21 | 0 |
| SIX3 | NM_005413 | −3466.4 | −3438.7 | 55.4 | 27 | 0 |
| VAX1 v2 | NM_199131 | −2168.5 | −2150.5 | 35.9 | 23 | 0.0423 |
Among the remaining 16 genes, there was an overwhelming pattern of conservation (i.e. dN/dS < 1) observed among the majority of sampled mammalian lineages, including primates. In most instances where dN/dS was greater than 1, the actual number of PAML-estimated non-synonymous and/or synonymous changes was less than 1. For two genes that showed exceptions to this general rule among primates (ETV1 and SHH), results are tempered by the inclusion of bioinformatically predicted ‘XM’ or ‘XR’ gene sequences, which do not have experimentally verified transcripts as do ‘NM’- or Ensembl-based sequences. Thus, among genes for which higher quality non-human sequences were available, the overall pattern of conservation among primates remained consistent.
4. Discussion
The present data demonstrate that cellular distributions of interneurons in the human prefrontal cortex are similar to those in other anthropoid primates and can be explained by general scaling rules. Furthermore, genes related to the transcriptional regulation of neocortical GABAergic neuron development show evolutionary conservation among primates.
(a). The distribution of inhibitory interneurons across species
Given the crucial role played by local inhibitory circuits, some investigators have suggested the existence of a canonical network of neocortical interneurons that is relatively invariant across species (Silberberg et al. 2002; Douglas & Martin 2004). In the present study, scaling analyses and partitioning of variance did not reveal a strong effect of phylogeny (i.e. genetic relatedness) on the relationship between interneuron subtype densities with respect to total neuron density in DLPFC of anthropoid primates. Interestingly, the positive allometric exponent of interneuron scaling in DLPFC resembled that of visual areas V1 and V2 from a previous study (Sherwood et al. 2007), suggesting that common scaling rules govern interspecific variation in interneurons spanning disparate regions of the neocortex. Despite such local network scaling patterns, however, species differences in the proportion of DLPFC interneurons did not correlate with brain mass. Taken together, these results indicate that the distribution of interneuron subtypes in the neocortex of anthropoid primates may be constrained within an optimal range of functionality regardless of overall brain size. Indeed, the integrity of cortical inhibitory interneurons appears to be required for normal cognitive processing in humans, as evident by dysfunctions of this system in patients with schizophrenia and bipolar disorder (Benes & Berretta 2001; Gonzalez-Burgos & Lewis 2008).
Although our narrow phylogenetic analysis of anthropoid primates showed remarkable consistency among species, previous comparisons among representatives of higher order taxonomic groups have demonstrated substantial variation in proportions of neocortical interneurons (Glezer et al. 1993; DeFelipe et al. 2002). In particular, primates have a larger percentage of interneurons than rodents and other mammals (e.g. afrotherians and xenarthrans) (Hendry et al. 1987; Gabbott et al. 1997; Gonchar & Burkhalter 1997; Sherwood et al. 2009). Additionally, most interneurons in rodents originate from the ganglionic eminence, whereas in primates there are also substantial numbers of cortical GABAergic interneurons that migrate from the lateral ventricular neuroepithelium (Letinic et al. 2002). Thus, evolutionary changes in the developmental programme underlying neocortical interneuron proliferation seem to have occurred during the divergence of primates, but the complement of interneuron subtypes has remained relatively stable in anthropoids since that time.
Our evolutionary analyses of genes important to the transcriptional regulation of cortical interneuron specification and differentiation, furthermore, showed a strong pattern of conservation in the primate lineage, and in other mammals. This finding is consistent with the known functional importance of these genes in the development of cortical interneurons in mammals. Among humans, mutations of such genes, including SHH, SIX3, ARX, DLX2 and DLX5, have been linked to multiple diseases including holoprosencephaly, autism, schizophrenia and anxiety disorders (Wonders & Anderson 2006), further reinforcing the importance of sequence conservation in these loci. Moreover, it has been shown that several of the studied gene products coregulate one another (Sussel et al. 1999; Gulacsi & Anderson 2006). Nevertheless, the existence of phenotypic differences among mammals in the distribution of interneurons suggests that mechanisms other than sequence variation in protein-encoding loci underlie the observed variation and differences in developmental origin. Further studies should investigate the possibility that interneuron diversity arises from changes in promoter regions or from cell-specific and/or species-specific epigenetic regulation (Farcas et al. 2009).
(b). The regional variation of interneurons is similar among humans, chimpanzees and macaques
Although calcium-binding protein immunohistochemistry has been used in the parcellation of cortical areas (Nimchinsky et al. 1997; Öngür et al. 2003; Bourne et al. 2007; Ding et al. 2009), there are relatively few quantitative studies that examine the regional variation of interneuron subtype distributions in primates (Dombrowski et al. 2001). In the current study, we obtained data from three different catarrhine primate species, encompassing 25 Myr of evolution since the last common ancestor of macaque monkeys and humans. These data demonstrated that the percentage of CR- and PV-ir interneurons across areas of the frontal cortex does not differ among species, revealing certain fundamental characteristics of neurobiological organization that are shared within this branch of primate evolution. Remarkably, prefrontal regions that are involved in cognitive functions that are unique in humans, such as ‘theory of mind’ and language, did not show equally distinctive interneuron distributions. Such regional characteristics of GABAergic neuron populations might be related to the specific processing requirements of each cortical area. In particular, intercolumnar inhibition mediated by GABAergic interneurons has been implicated in shaping the temporal pattern of activation across neuronal ensembles (Constantinidis et al. 2002), and specializations of cortical inhibitory circuitry may contribute to differences in processing among sensory modalities (Pallas 2001). Similarly, our findings suggest that certain computational processes of areas in the frontal cortex of catarrhine primates are preferentially supported by specific inhibitory architecture.
(c). Humans do not have specialized cellular distributions of interneurons in the prefrontal cortex
We did not find evidence that the distribution of interneurons in the human prefrontal cortex is evolutionarily specialized. Human interneuron densities were contained within the 95 per cent prediction intervals for DLPFC based on scaling to total neuron density in non-human anthropoids. Humans also did not differ significantly from chimpanzees or macaques in the regional distribution of interneurons within the frontal cortex. Multivariate discriminant function analysis, moreover, demonstrated that the species with the most distinct frontal cortex interneuron phenotype was chimpanzees, not humans. In sum, we cannot conclude that alterations of interneuron distributions in the prefrontal cortex have made an important contribution to the evolution of human species-specific cognition.
Although it has been argued by some investigators that executive cognitive functions mediated by the prefrontal cortex have been modified in human evolution (Coolidge & Wynn 2005; Aboitiz et al. 2006), there is a paucity of data addressing whether there has been corresponding neuroanatomical reorganization. It is possible that the prefrontal cortex (or subdivisions of it) has become disproportionately enlarged in humans (Semendeferi et al. 2001; Schoenemann et al. 2005; Rilling 2006; Schenker et al. 2009); however, many of the reported differences are actually within the expected range for allometric scaling at human brain size (Holloway 2002; Sherwood et al. 2005). Aside from the possible differential enlargement of areas within the prefrontal cortex, there is currently only minimal evidence of histological or connectional reorganization that would serve as a neurobiological basis for the unique cognitive abilities of humans. Indeed, several recent studies have demonstrated notable commonalities in the neocortical architecture of humans relative to other primates. For example, humans and chimpanzees are similar in having a greater density of axons containing serotonin, dopamine and acetylcholine innervating the prefrontal cortex as compared with macaque monkeys (Raghanti et al. 2008a,b,c). Furthermore, the total number of neurons in the human neocortex accords with allometric scaling predictions from other primate brains (Azevedo et al. 2009).
Such evidence of continuity between humans and our close relatives, however, is complemented by other data indicating that corticocortical connectivity and descending subcortical projections have changed considerably in recent human evolution (Kuypers 1958; Rilling 2008). With the expansion of the forebrain in humans, novel long-range neuronal projection patterns have emerged that link previously unconnected processing modules, such as areas in the inferior frontal cortex and the middle temporal gyrus which are important in language (Rilling et al. 2008). Interestingly, several molecules involved in cell adhesion and axon guidance show evidence of selection in their amino acid sequence and surrounding non-coding regions in humans as compared with other primates (Prabhakar et al. 2006; Uddin et al. 2008). In this light, the modern human neocortex appears to combine both conserved and specialized architectural features into an evolutionary mosaic that underlies our species' uniqueness.
Acknowledgements
We thank Amy R. Garrison for assistance with histology. Brain materials used in this study were loaned by the Great Ape Ageing Project (NIH grant AG14308), the Foundation for Comparative and Conservation Biology, the Cleveland Metroparks Zoo, the New England Primate Research Center, Dr Antoine Mudakikwa from Office Rwandais du Tourisme et des Parcs Nationaux, Dr Mike Cranfield from the Mountain Gorilla Veterinary Project, the Northwestern University Alzheimer's Disease Center Brain Bank (NADC grant P30 AG13854) and Dr Todd M. Preuss. This work was supported by the National Science Foundation (BCS-0515484, BCS-0549117, BCS-0827531, BCS-0550209, BCS-0827546, DGE-0801634), the National Institutes of Health (NS42867), the Wenner-Gren Foundation for Anthropological Research and the James S. McDonnell Foundation (22002078).
References
- Aboitiz F., García R. R., Bosman C., Brunetti E.2006Cortical memory mechanisms and language origins. Brain Lang. 98, 40–56 (doi:10.1016/j.bandl.2006.01.006) [DOI] [PubMed] [Google Scholar]
- Azevedo F. A., Carvalho L. R., Grinberg L. T., Farfel J. M., Ferretti R. E., Leite R. E., Jacob Filho W., Lent R., Herculano-Houzel S.2009Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541 (doi:10.1002/cne.21974) [DOI] [PubMed] [Google Scholar]
- Bartos M., Vida I., Jonas P.2007Synaptic mechanisms of synchronized gamma oscillations in inhibitory interneuron networks. Nat. Rev. Neurosci. 8, 45–56 (doi:10.1038/nrn2044) [DOI] [PubMed] [Google Scholar]
- Benes F. M., Berretta S.2001GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology 25, 1–27 (doi:10.1016/S0893-133X(01)00225-1) [DOI] [PubMed] [Google Scholar]
- Bourne J. A., Warner C. E., Upton D. J., Rosa M. G.2007Chemoarchitecture of the middle temporal visual area in the marmoset monkey (Callithrix jacchus): laminar distribution of calcium-binding proteins (calbindin, parvalbumin) and nonphosphorylated neurofilament. J. Comp. Neurol. 500, 832–849 (doi:10.1002/cne.21190) [DOI] [PubMed] [Google Scholar]
- Caceres M., Lachuer J., Zapala M. A., Redmond J. C., Kudo L., Geschwind D. H., Lockhart D. J., Preuss T. M., Barlow C.2003Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl Acad. Sci. USA 100, 13 030–13 035 (doi:10.1073/pnas.2135499100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinidis C., Williams G. V., Goldman-Rakic P. S.2002A role for inhibition in shaping the temporal flow of information in prefrontal cortex. Nat. Neurosci. 5, 175–180 (doi:10.1038/nn799) [DOI] [PubMed] [Google Scholar]
- Coolidge F. L., Wynn T.2005Working memory, its executive functions, and the emergence of modern thinking. Camb. Archaeol. J. 15, 5–26 (doi:10.1017/S0959774305000016) [Google Scholar]
- DeFelipe J.1997Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28k, parvalbumin and calretinin in the neocortex. J. Chem. Neuroanat. 14, 1–19 (doi:10.1016/S0891-0618(97)10013-8) [DOI] [PubMed] [Google Scholar]
- DeFelipe J., Alonso-Nanclares L., Arellano J. I.2002Microstructure of the neocortex: comparative aspects. J. Neurocytol. 31, 299–316 (doi:10.1023/A:1024130211265) [DOI] [PubMed] [Google Scholar]
- Desdevises Y., Legendre P., Azouzi L., Morand S.2003Quantifying phylogenetically structured environmental variation. Evolution 57, 2647–2652 [DOI] [PubMed] [Google Scholar]
- Ding S. L., Van Hoesen G. W., Cassell M. D., Poremba A.2009Parcellation of human temporal polar cortex: a combined analysis of multiple cytoarchitectonic, chemoarchitectonic, and pathological markers. J. Comp. Neurol. 514, 595–623 (doi:10.1002/cne.22053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski S. M., Hilgetag C. C., Barbas H.2001Quantitative architecture distinguishes prefrontal cortical systems in the rhesus monkey. Cereb. Cortex 11, 975–988 (doi:10.1093/cercor/11.10.975) [DOI] [PubMed] [Google Scholar]
- Dorus S., Vallender E. J., Evans P. D., Anderson J. R., Gilbert S. L., Mahowald M., Wyckoff G. J., Malcom C. M., Lahn B. T.2004Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 119, 1027–1040 (doi:10.1016/j.cell.2004.11.040) [DOI] [PubMed] [Google Scholar]
- Douglas R. J., Martin K. A.2004Neuronal circuits of the neocortex. Annu. Rev. Neurosci. 27, 419–451 (doi:10.1146/annurev.neuro.27.070203.144152) [DOI] [PubMed] [Google Scholar]
- Farcas R., et al. 2009Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol. Biol. Evol. 26, 1379–1389 (doi:10.1093/molbev/msp046) [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1985Phylogenies and the comparative method. Am. Nat. 125, 1–15 (doi:10.1086/284325) [Google Scholar]
- Friederici A. D., Bahlmann J., Heim S., Schubotz R. I., Anwander A.2006The brain differentiates human and non-human grammars: functional localization and structural connectivity. Proc. Natl Acad. Sci. USA 103, 2458–2463 (doi:10.1073/pnas.0509389103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbott P. L., Jays P. R., Bacon S. J.1997Calretinin neurons in human medial prefrontal cortex (areas 24a,b,c, 32', and 25). J. Comp. Neurol. 381, 389–410 (doi:10.1002/(SICI)1096-9861(19970519)381:4<389::AID-CNE1>3.0.CO;2-Z) [DOI] [PubMed] [Google Scholar]
- Gallagher H. L., Frith C. D.2003Functional imaging of ‘theory of mind’. Trends Cogn. Sci. 7, 77–83 (doi:10.1016/S1364-6613(02)00025-6) [DOI] [PubMed] [Google Scholar]
- Garland T., Ives A. R.2000Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am. Nat. 155, 346–364 (doi:10.1086/303327) [DOI] [PubMed] [Google Scholar]
- Geschwind D. H.2000Mice, microarrays, and the genetic diversity of the brain. Proc. Natl Acad. Sci. USA 97, 10 676–10 678 (doi:10.1073/pnas.97.20.10676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glezer I. I., Hof P. R., Leranth C., Morgane P. J.1993Calcium-binding protein-containing neuronal populations in mammalian visual cortex: a comparative study in whales, insectivores, bats, rodents, and primates. Cereb. Cortex 3, 249–272 (doi:10.1093/cercor/3.3.249) [DOI] [PubMed] [Google Scholar]
- Gonchar Y., Burkhalter A.1997Three distinct families of GABAergic neurons in rat visual cortex. Cereb. Cortex 7, 347–358 (doi:10.1093/cercor/7.4.347) [DOI] [PubMed] [Google Scholar]
- Gonzalez-Burgos G., Lewis D. A.2008GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophr. Bull. 34, 944–961 (doi:10.1093/schbul/sbn070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M., Grossman L. I., Wildman D. E.2005Moving primate genomics beyond the chimpanzee genome. Trends Genet. 21, 511–517 (doi:10.1016/j.tig.2005.06.012) [DOI] [PubMed] [Google Scholar]
- Gulacsi A., Anderson S. A.2006Shh maintains Nkx2.1 in the MGE by a Gli3-independent mechanism. Cereb. Cortex 16(Suppl. 1), i89–i95 (doi:10.1093/cercor/bhk018) [DOI] [PubMed] [Google Scholar]
- Hallstrom B. M., Kullberg M., Nilsson M. A., Janke A.2007Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol. Biol. Evol. 24, 2059–2068 (doi:10.1093/molbev/msm136) [DOI] [PubMed] [Google Scholar]
- Hauser M. D., Chomsky N., Fitch W. T.2002The faculty of language: what is it, who has it, and how did it evolve? Science 298, 1569–1579 (doi:10.1126/science.298.5598.1569) [DOI] [PubMed] [Google Scholar]
- Haygood R., Fedrigo O., Hanson B., Yokoyama K. D., Wray G. A.2007Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat. Genet. 39, 1140–1144 (doi:10.1038/ng2104) [DOI] [PubMed] [Google Scholar]
- Hendry S. H. C.1987Recent advances in understanding the intrinsic circuitry of the cerebral cortex. In Higher brain functions: recent explorations of the brain's emergent properties (ed. Wise S. P.), pp. 241–283 New York, NY: Wiley [Google Scholar]
- Hendry S. H. C., Schwark H. D., Jones E. G., Yan J.1987Number and proportions of GABA-immunoreactive neurons in different areas of monkey cerebral cortex. J. Neurosci. 7, 1503–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann E., Call J., Hernandez-Lloreda M. V., Hare B., Tomasello M.2007Humans have evolved specialized skills of social cognition: the cultural intelligence hypothesis. Science 317, 1360–1366 (doi:10.1126/science.1146282) [DOI] [PubMed] [Google Scholar]
- Hof P. R., Glezer I. I., Conde F., Flagg R. A., Rubin M. B., Nimchinsky E. A., Vogt Weisenhorn D. M.1999Cellular distribution of the calcium-binding proteins parvalbumin, calbindin, and calretinin in the neocortex of mammals: phylogenetic and developmental patterns. J. Chem. Neuroanat. 16, 77–116 (doi:10.1016/S0891-0618(98)00065-9) [DOI] [PubMed] [Google Scholar]
- Holloway R. L.2002Brief communication: how much larger is the relative volume of area 10 of the prefrontal cortex in humans? Am. J. Phys. Anthropol. 118, 399–401 (doi:10.1002/ajpa.10090) [DOI] [PubMed] [Google Scholar]
- Holloway R. L., Broadfield D. C., Yuan M. S.2004The human fossil record, volume 3, brain endocasts—the paleoneurological evidence. New York, NY: Wiley [Google Scholar]
- Khaitovich P., et al. 2004Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14, 1462–1473 (doi:10.1101/gr.2538704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers H. G. J. M.1958Some projections from the peri-central cortex to the pons and lower brainstem in monkey and chimpanzee. J. Comp. Neurol. 110, 221–251 (doi:10.1002/cne.901100205) [DOI] [PubMed] [Google Scholar]
- Letinic K., Zoncu R., Rakic P.2002Origin of GABAergic neurons in the human neocortex. Nature 417, 645–649 (doi:10.1038/nature00779) [DOI] [PubMed] [Google Scholar]
- Liu G., Uddin M., Islam M., Goodman M., Grossman L. I., Romero R., Wildman D. E.2007OCPAT: an online codon-preserved alignment tool for evolutionary genomic analysis of protein coding sequences. Source Code Biol. Med. 2, 5 (doi:10.1186/1751-0473-2-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R.2005Mesquite: a modular system for evolutionary analysis. Version 1.12. See http://mesquiteproject.org [Google Scholar]
- Mouton P. R.2002Principles and practices of unbiased stereology: an introduction for bioscientists Baltimore, MD: The Johns Hopkins University Press [Google Scholar]
- Nimchinsky E. A., Vogt B. A., Morrison J. H., Hof P. R.1997Neurofilament protein and calcium-binding proteins in the human cingulate cortex. J. Comp. Neurol. 384, 597–620 (doi:10.1002/(SICI)1096-9861(19970811)384:4<597::AID-CNE8>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- Oldham M. C., Horvath S., Geschwind D. H.2006Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc. Natl Acad. Sci. USA 103, 17 973–17 978 (doi:10.1073/pnas.0605938103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öngür D., Ferry A. T., Price J. L.2003Architectonic subdivision of the human orbital and medial prefrontal cortex. J. Comp. Neurol. 460, 425–449 (doi:10.1002/cne.10609) [DOI] [PubMed] [Google Scholar]
- Pagel M. D.1992A method for the analysis of comparative data. J. Theor. Biol. 156, 431–442 (doi:10.1016/S0022-5193(05)80637-X) [Google Scholar]
- Pallas S. L.2001Intrinsic and extrinsic factors that shape neocortical specification. Trends Neurosci. 24, 417–423 (doi:10.1016/S0166-2236(00)01853-1) [DOI] [PubMed] [Google Scholar]
- Prabhakar S., Noonan J. P., Pääbo S., Rubin E. M.2006Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786 (doi:10.1126/science.1130738) [DOI] [PubMed] [Google Scholar]
- Preuss T. M.2007Primate brain evolution in phylogenetic context. In The evolution of primate nervous systems. Evolution of nervous systems, vol. 4 (eds Kaas J. H., Preuss T. M.), pp. 1–34 Oxford, UK: Academic Press [Google Scholar]
- Raghanti M. A., Stimpson C. D., Marcinkiewicz J. L., Erwin J. M., Hof P. R., Sherwood C. C.2008aCholinergic innervation of the frontal cortex: differences among humans, chimpanzees, and macaque monkeys. J. Comp. Neurol. 506, 409–424 (doi:10.1002/cne.21546) [DOI] [PubMed] [Google Scholar]
- Raghanti M. A., Stimpson C. D., Marcinkiewicz J. L., Erwin J. M., Hof P. R., Sherwood C. C.2008bCortical dopaminergic innervation among humans, chimpanzees, and macaque monkeys: a comparative study. Neuroscience 155, 203–220 (doi:10.1016/j.neuroscience.2008.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti M. A., Stimpson C. D., Marcinkiewicz J. L., Erwin J. M., Hof P. R., Sherwood C. C.2008cDifferences in cortical serotonergic innervation among humans, chimpanzees, and macaque monkeys: a comparative study. Cereb. Cortex 18, 584–597 (doi:10.1093/cercor/bhm089) [DOI] [PubMed] [Google Scholar]
- Ramón y Cajal S.1923Recuerdos de mi vida: Historia de mi labor científica. Madrid, Spain: Alianza Editorial [Google Scholar]
- Rilling J. K.2006Human and nonhuman primate brains: are they allometrically scaled versions of the same design? Evol. Anthropol. 15, 65–77 (doi:10.1002/evan.20095) [Google Scholar]
- Rilling J. K.2008Neuroscientific approaches and applications within anthropology. Am. J. Phys. Anthropol. 137(Suppl. 47), 2–32 (doi:10.1002/ajpa.20947) [DOI] [PubMed] [Google Scholar]
- Rilling J. K., Glasser M. F., Preuss T. M., Ma X., Zhao T., Hu X., Behrens T. E. J.2008The evolution of the arcuate fasciculus revealed with comparative DTI. Nat. Neurosci. 11, 426–428 (doi:10.1038/nn2072) [DOI] [PubMed] [Google Scholar]
- Schenker N. M., Hopkins W. D., Spocter M. A., Garrison A. R., Stimpson C. D., Erwin J. M., Hof P. R., Sherwood C. C.2009Broca's area homologue in chimpanzees (Pan troglodytes): probabilistic mapping, asymmetry, and comparison to humans. Cereb. Cortex (doi:10.1093/cercor/bhp138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemann P. T., Sheehan M. J., Glotzer L. D.2005Prefrontal white matter volume is disproportionately larger in humans than in other primates. Nat. Neurosci. 8, 242–252 (doi:10.1038/nn1394) [DOI] [PubMed] [Google Scholar]
- Semendeferi K., Armstrong E., Schleicher A., Zilles K., Van Hoesen G. W.2001Prefrontal cortex in humans and apes: a comparative study of area 10. Am. J. Phys. Anthropol. 114, 224–241 (doi:10.1002/1096-8644(200103)114:3<224::AID-AJPA1022>3.0.CO;2-I) [DOI] [PubMed] [Google Scholar]
- Sherwood C. C., Holloway R. L., Semendeferi K., Hof P. R.2005Is prefrontal white matter enlargement a human evolutionary specialization? Nat. Neurosci. 8, 537–538 (doi:10.1038/nn0505-537) [DOI] [PubMed] [Google Scholar]
- Sherwood C. C., et al. 2006Evolution of increased glia-neuron ratios in the human frontal cortex. Proc. Natl Acad. Sci. USA 103, 13 606–11 611 (doi:10.1073/pnas.0605843103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood C. C., Raghanti M. A., Stimpson C. D., Bonar C. J., de Sousa A. A., Preuss T. M., Hof P. R.2007Scaling of inhibitory interneurons in areas V1 and V2 of anthropoid primates as revealed by calcium-binding protein immunohistochemistry. Brain Behav. Evol. 69, 176–195 (doi:10.1159/000096986) [DOI] [PubMed] [Google Scholar]
- Sherwood C. C., Subiaul F., Zawidzki T. W.2008A natural history of the human mind: tracing evolutionary changes in brain and cognition. J. Anat. 212, 426–454 (doi:10.1111/j.1469-7580.2008.00868.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood C. C., Stimpson C. D., Butti C., Bonar C. J., Newton A. L., Allman J. M., Hof P. R.2009Neocortical neuron types in Xenarthra and Afrotheria: implications for brain evolution in mammals. Brain Struct. Funct. 213, 301–328 (doi:10.1007/s00429-008-0198-9) [DOI] [PubMed] [Google Scholar]
- Silberberg G., Gupta A., Markram H.2002Stereotypy in neocortical microcircuits. Trends Neurosci. 25, 227–230 (doi:10.1016/S0166-2236(02)02151-3) [DOI] [PubMed] [Google Scholar]
- Sohal V. S., Zhang F., Yizhar O., Deisseroth K.2009Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698–702 (doi:10.1038/nature07991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L., Marin O., Kimura S., Rubenstein J. L.1999Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370 [DOI] [PubMed] [Google Scholar]
- Uddin M., Wildman D. E., Liu G., Xu W., Johnson R. M., Hof P. R., Kapatos G., Grossman L. I., Goodman M.2004Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl Acad. Sci. USA 101, 2957–2962 (doi:10.1073/pnas.0308725100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M., et al. 2008Distinct genomic signatures of adaptation in pre- and postnatal environments during human evolution. Proc. Natl Acad. Sci. USA 105, 3215–3220 (doi:10.1073/pnas.0712400105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westboy M., Leishman M. R., Lord J. M.1995On misinterpreting the ‘phylogenetic correction’. J. Ecol. 83, 531–534 (doi:10.2307/2261605) [Google Scholar]
- Wildman D. E., et al. 2007Genomics, biogeography, and the diversification of placental mammals. Proc. Natl Acad. Sci. USA 104, 14395–14400 (doi:10.1073/pnas.0704342104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders C. P., Anderson S. A.2006The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7, 687–696 (doi:10.1038/nrn1954) [DOI] [PubMed] [Google Scholar]
- Yang Z.1997PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 [DOI] [PubMed] [Google Scholar]
- Zaitsev A. V., Povysheva N. V., Gonzalez-Burgos G., Rotaru D., Fish K. N., Krimer L. S., Lewis D. A.2009Interneuron diversity in layers 2–3 of monkey prefrontal cortex. Cereb. Cortex 19, 1597–1615 (doi:10.1093/cercor/bhn198) [DOI] [PMC free article] [PubMed] [Google Scholar]