Abstract
Historic museum specimens are increasingly used to answer a wide variety of questions in scientific research. Nevertheless, the scientific value of these specimens depends on the authenticity of the data associated with them. Here we use individual-based genetic analyses to demonstrate erroneous locality information for archive specimens from the late nineteenth century. Specifically, using 10 microsatellite markers, we analysed 350 contemporary and 43 historic yellow-eyed penguin (Megadyptes antipodes) specimens from New Zealand's South Island and sub-Antarctic regions. Factorial correspondence analysis and an assignment test strongly suggest that eight of the historic specimens purportedly of sub-Antarctic origin were in fact collected from the South Island. Interestingly, all eight specimens were obtained by the same collector, and all are currently held in the same museum collection. Further inspection of the specimen labels and evaluation of sub-Antarctic voyages did not reveal whether the erroneous data are caused by incorrect labelling or whether deliberate falsification was at play. This study highlights a promising extension to the well-known applications of assignment tests in molecular ecology, which can complement methods that are currently being applied for error detection in specimen data. Our results also serve as a warning to all who use archive specimens to invest time in the verification of collection information.
Keywords: historic specimens, Megadyptes antipodes, New Zealand, error, fraud, assignment test
1. Introduction
Museum collections, archived in natural history museums worldwide, provide invaluable resources of materials and knowledge that are of utmost importance to science and society (Suarez & Tsutsui 2004). These collections currently hold an estimated total of three billion specimens, of which 7–10 million are bird skins (Brooke 2000; Pennisi 2000). The crucial role of museum collections in defining species and their ranges started with the diligent efforts of nineteenth century collectors, and even today this wealth of information still plays a vital role in the documentation of species decline and conservation status assessment (Shaffer et al. 1998; Collar & Rudyanto 2003). During the last two decades, the use of museum specimens as sources of DNA samples has been facilitated by advances in molecular techniques, initiating a vast increase in the use of such archived specimens in population and evolutionary genetic studies (reviewed in Wandeler et al. 2007). Comparative studies now frequently compare levels of genetic diversity over time, thereby inferring changes in population size and population connectivity (e.g. Miller & Waits 2003; Johnson et al. 2004; Larsson et al. 2008; Taylor et al. 2008).
The potential problems arising from working with low-quality DNA from historic specimens are well known and can be addressed using clear laboratory guidelines (Sefc et al. 2003, 2007; Pääbo et al. 2004), but any additional pitfalls arising from errors in specimen data are potentially much more complex (reviewed in Rasmussen & Prŷs-Jones 2003). Specimen data are recorded on attached labels (and collectors' notes when available) and the scientific value of specimens ultimately depends on the accuracy of these data. The minimum information typically associated with a specimen includes the identity, location, collection date and the name of the collector, but one or more of these entries may be missing. Additionally, these labels are also the most prone to error in specimen collection (Winker 2000). Most errors are found in identity and location, which can lead to false representation of a species' distribution (Graham et al. 2004). Causes of inaccuracy in museum specimen data vary from simple mistakes or carelessness during collection or post-collection to serious cases of fraud (Rasmussen & Prŷs-Jones 2003). Neither inadvertent mistakes nor cases of deliberate fraud have received much attention in the literature, with a few exceptions such as the case of deceit by British Colonel Richard Meinertzhagen and the data falsification by Joseph H. Batty (Knox 1993; Rasmussen & Prŷs-Jones 2003; Dalton 2005; Olson 2008).
Detecting errors in specimen data can be extremely challenging. Museum staff typically use collectors' field notes, information related to the voyages and travels of collectors and thorough examination of preparatory techniques (including X-rays) to identify errors (Knox 1993; Rasmussen & Collar 1999; Rasmussen & Prŷs-Jones 2003). An approach using geo-referencing of temporally collected samples was introduced to detect specimens with high probability of error without a priori suspicion (Peterson et al. 2004). Nevertheless, the above methods are limited by their focus on: (i) specimens from suspicious collectors; (ii) specimens that form outliers with respect to the species' natural range; or (iii) specimens that form outliers with respect to collection date (e.g. collected years after a species was reported extinct or collected on dates that clash with collectors' itineraries). When specimens do not fall into any of the above categories, error or fraud detection becomes nearly impossible. In the current study we present an unforeseen case in which individual-based genetic analyses reveal previously unsuspected inaccuracies in the geographical origin of museum material. Specifically, our data suggest a case of mislabelling or even possibly fraud involving eight yellow-eyed penguin (Megadyptes antipodes) specimens purportedly from New Zealand's sub-Antarctic islands. This detection not only highlights a promising approach to detecting errors in archive specimen data, but additionally implies that errors in museum collections may be more common than previously anticipated.
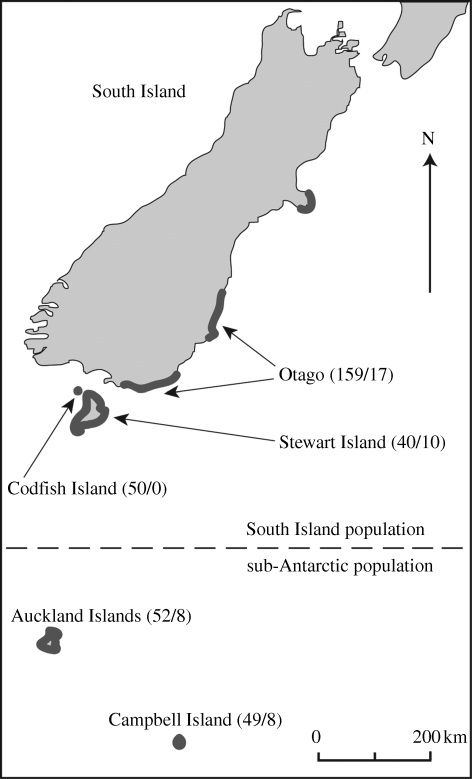
Megadyptes antipodes was first described from an Auckland Island's specimen in 1841 (Hombron & Jacquinot 1841). The species is endemic to the New Zealand region, where it inhabits the sub-Antarctic Auckland and Campbell Islands, along with the southeast coast of South Island (Marchant & Higgins 1990; McKinlay 2001; figure 1). Genetic and morphological analyses of sub-fossil and historic specimens have shown that M. antipodes probably expanded its range from the sub-Antarctic islands to the South Island of New Zealand after approximately AD 1500, following the anthropogenic extinction of its sister species M. waitaha (Boessenkool et al. 2009a). Despite the recent expansion event, the presence of significant microsatellite DNA structuring—and inferred low migration rates—among contemporary breeding sites support the genetic recognition of two separate populations, one on South Island and the other in the sub-Antarctic (Boessenkool et al. 2009b; figure 1). In the present study we analysed contemporary and historic museum specimens from both the South and sub-Antarctic Islands using 10 microsatellite markers. Our data strongly suggest that eight of the historic specimens believed to be of sub-Antarctic origin were in fact collected from the South Island.
Figure 1.
Map of the South and sub-Antarctic islands of New Zealand. The dark grey line represents the current breeding range of Megadyptes antipodes. Arrows point to the geographical locations where samples were collected. Sample sizes for contemporary/historic samples are given in brackets. The dashed line refers to the split between the South Island and the sub-Antarctic population.
2. Material and methods
(a). Sample collection, DNA extraction and genotyping
Yellow-eyed penguin blood samples were collected in 2005–2007 on the South Island (n = 249) and sub-Antarctic Auckland and Campbell Islands (n = 101) of New Zealand (figure 1) as described in Boessenkool et al. (2009b). DNA was extracted and purified using 40 µg proteinase K in 5 per cent Chelex (Biorad; Walsh et al. 1991).
Historic toe pad samples were obtained from 55 specimens collected between 1840 and 1944 on the South Island (n = 35) and sub-Antarctic Auckland and Campbell Islands (n = 20). These specimens are held in 15 museum collections around the world (for specimen details see table S1 in the electronic supplementary material). Following rehydration, toe pad samples were finely chopped and DNA was extracted using the Chargeswitch Forensic DNA Purification Kit (Invitrogen) or the DNeasy Tissue Kit (Qiagen) following manufacturers' instructions.
All samples were genotyped at 10 microsatellite loci previously developed for yellow-eyed penguins (Man03, Man08, Man13, Man21, Man39, Man47, Man50, Man51, Man54, Man55; Boessenkool et al. 2008). Microsatellite primer sequences and polymerase chain reaction (PCR) conditions for modern samples are described in Boessenkool et al. (2008). PCR reactions for historic samples were performed in 10 µl volumes containing 2 µl DNA, 0.5 µM of each primer, 0.5 U Taq DNA polymerase (Mango Taq, Bioline), 1× Taq buffer, 0.8 µM dNTP and 1.5 µM MgCl2, with the addition of betaine and dimethylsulphoxide (1.1 M and 2%, respectively) if necessary (see Boessenkool et al. 2008). The amplification profile was 2 min at 94°C, 35–50 cycles of 15 s at 96°C, 15 s at 45–50°C and 30 s at 72°C, followed by a 4 min final extension at 72°C.
Strict guidelines were followed in order to prevent contamination of historic DNA and to minimize the risk of erroneous genotypes owing to allelic dropout and the amplification of false alleles (Taberlet et al. 1996; Sefc et al. 2003). DNA extractions and PCR set-up of historic samples were performed inside an ultraviolet hood in a separate laboratory where no contemporary yellow-eyed penguin DNA or vertebrate PCR products have ever been present. Historic samples were extracted in small batches of nine samples and potential contamination was monitored by negative extraction and PCR controls. A subset of historic samples (n = 4) from the sub-Antarctic was re-extracted and genotyped to validate results. Secondary extract genotypes from three of these re-extracted samples agreed with those from primary extracts. The fourth re-extracted sample had failed to amplify successfully for the primary extraction. For all samples, two to seven successful amplifications (i.e. resulting in bands visible on a gel) were obtained before a genotype was scored, and genotypes were only confirmed once every allele was observed at least twice. Samples were always re-amplified if genotypes were either not clearly visible on the gel, or if inconsistencies in genotypes suggested allelic dropout or amplification of a false allele.
(b). Genetic analyses
Deviations from Hardy–Weinberg proportions and linkage equilibrium were assessed separately for modern South Island and modern sub-Antarctic samples using Genepop v. 4.0 (Rousset 2008). Markov chain parameters employed 10 000 dememorizations, 1000 batches and 10 000 iterations. Significance levels were adjusted for multiple comparisons using Bonferroni corrections (Rice 1989). Allele frequencies and unique alleles were evaluated using Genetix v. 4.05.2 (Belkhir et al. 1996–2004).
To evaluate the provenance of the historic museum specimens we employed two different methods. First, a two-dimensional factorial correspondence analysis (FCA) was performed using Genetix v. 4.05.2 (Belkhir et al. 1996–2004). An FCA visualizes genetic (dis)similarity of individual genotypes without grouping individuals a priori. Second, the assignment test implemented in Structure v. 2.2 (Pritchard et al. 2000) was used to infer the probability that historic museum specimens originated from the South Island or the sub-Antarctic populations, respectively. This assignment is a fully Bayesian method that uses geographical sampling location of individuals with confirmed geographical origin as prior information (Pritchard et al. 2000). The method assumes that all source populations have been sampled. In our analysis we specified the origin of the contemporary samples to be known, and the proportional membership coefficient (Q) of the historic samples to either of the two populations to be estimated by the program. An important assumption when applying this analysis to historic data is that the allele frequencies of the modern samples are representative of the allele frequencies of the historic populations (see §4 for further comments on this assumption). In the model, allele frequencies were assumed to be correlated among populations and parameters for priors of λ and FST were left at default values. For the historic samples we applied the admixture model with a uniform prior for α, bounded by a maximum of 10, and we set ALPHAPROPSD to 0.025. The migration prior (v) for the assignment test was set to 0.01, but to account for uncertainty in v we ran replicate analyses using v = 0.05 and v = 0.1. The outcome of the analyses was unaffected by the migration prior and we only present the results from runs with v = 0.01. The Markov chain Monte Carlo simulation was performed with a burn-in of 100 000 followed by 500 000 iterations. We also performed a clustering analysis without specifying any a priori geographical sampling information as implemented in Structure, using the same parameter settings as for the historic samples described above. This analysis revealed the same pattern as the analysis in which geographical information of contemporary samples was incorporated in the model (data not shown).
3. Results
DNA was successfully extracted (i.e. resulting in positive amplification of M. antipodes microsatellite markers) from 43 of the 55 historic samples. Twenty-seven of the successful extractions were from ‘South Island’ specimens, whereas 16 were from purportedly ‘sub-Antarctic’ specimens (Campbell and Auckland Islands; figure 1). Thirteen historic samples had missing genotypes at one (six samples), two (one sample), three (two samples), four (two samples), five (one sample) or six loci (one sample). The amplification of a false allele was encountered in one out of a total of 1066 successful PCRs. Allelic dropout was detected in 36 out of 420 PCR amplifications of confirmed heterozygous historic samples. In 26 out of the 36 cases dropout occurred in multiple replicate amplifications of samples at specific loci.
All 350 modern samples amplified at all microsatellite loci with the exception of six samples from the South Island, which had missing genotypes for one (three samples), three (two samples) or five loci (one sample), respectively. The dataset revealed no evidence for linkage disequilibrium between any pairs of loci and no significant departure from Hardy–Weinberg proportions in the modern M. antipodes South Island and sub-Antarctic populations (see table S2 in the electronic supplementary material for diversity indices per locus per population).
(a). Evaluation of the origin of historic samples
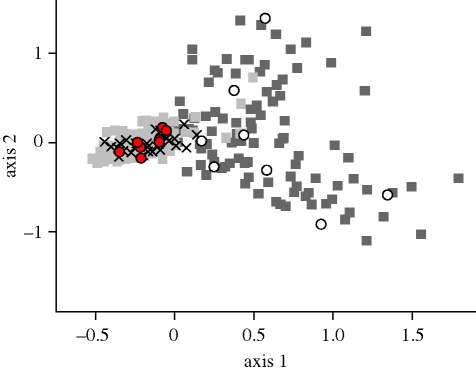
The two-dimensional FCA illustrates the genetic distinction between modern South Island versus sub-Antarctic populations (figure 2). To evaluate the origin of the historic samples, we superimposed their genotypes over the modern samples. All historic samples that were reportedly collected on the South Island clustered genetically among the modern South Island samples, consistent with their geographical origins. By contrast, only 8 of the 16 historic ‘sub-Antarctic’ samples were grouped with the modern sub-Antarctic samples, whereas the remaining eight samples clustered among the modern South Island samples (figure 2).
Figure 2.
Plot of the two-dimensional FCA based on genotypic variation at 10 microsatellite loci of modern and historic M. antipodes samples. The axes explain 9.26 per cent (x-axis) and 5.47 per cent (y-axis) of the total variation, respectively. Eight of the 16 samples with purported sub-Antarctic origin group with the modern sub-Antarctic samples (historic sub-Antarctic 1, open white circles), whereas the remaining eight samples cluster among the modern South Island samples (historic sub-Antarctic 2, filled red circles). Light grey squares, modern South Island; dark grey squares, modern sub-Antarctic; crosses, historic South Island.
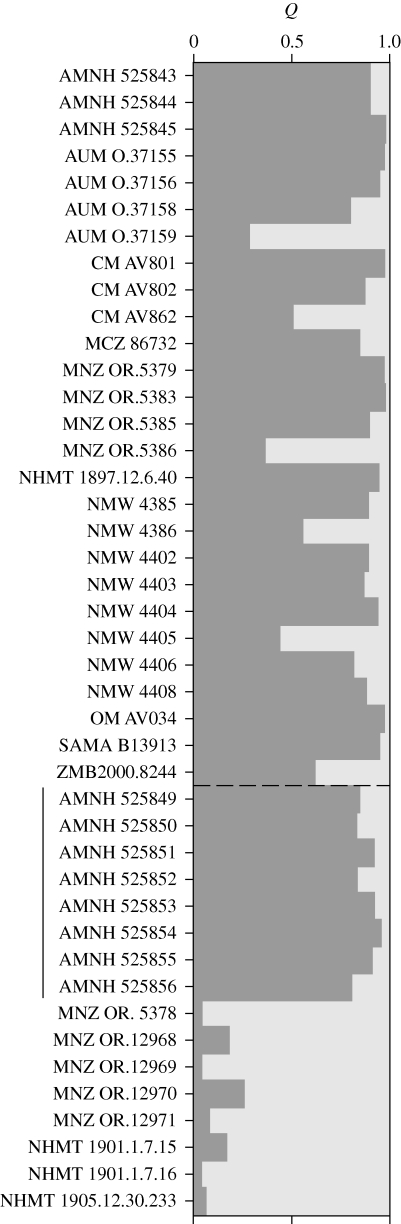
Using the assignment test implemented in Structure we estimated the proportional membership coefficient (Q) for each of the historic samples to both the South Island and the sub-Antarctic populations (figure 3). Of the 27 historic samples with South Island origin, 20 have high Q (i.e. Q > 0.80) to the South Island population. The other seven samples do not assign strongly to either of the populations; their Q-values lie between 0.20–0.80 indicating that they potentially have admixed ancestry (Lecis et al. 2006; Bergl & Vigilant 2007). Note that of these individuals with admixed ancestry, two have missing data at four loci. Of the historic samples with purported sub-Antarctic origins, eight have high Q to the sub-Antarctic population (Q > 0.80), whereas seven show strong membership to the South Island population (Q > 0.9 for four samples and 0.80 < Q > 0.90 for three samples, respectively). One sample has weak evidence for mixed ancestry (Q = 0.744 to the South Island population), but this individual lacked genotypic data at five of ten loci. The eight sub-Antarctic samples that have strong membership to the South Island population are the same eight samples that were placed among the modern South Island samples in the FCA. Four of these eight specimens were reportedly collected on the Auckland Islands (three in 1893, one in 1894) and the other four were reportedly collected on Campbell Island (two in 1893, two in 1894). Interestingly, these eight specimens were all collected by the same collector, namely Henry Hamersley Travers (1844–1928), and are currently held in the same museum collection (American Museum of Natural History).
Figure 3.
Proportional membership (Q) of historic M. antipodes specimens to the South Island (dark grey) and sub-Antarctic (light grey) populations as estimated using the assignment test in Structure. Each horizontal bar represents a single specimen identified by its museum accession number and specimens are ordered by reported geographical sampling location and museum collection. Specimens above the dashed line were purportedly collected on the South Island, those under the dashed line on the sub-Antarctic Auckland and Campbell Islands. The vertical line borders the eight specimens with suspect geographical origin. Museum abbreviations can be found in table S1 in the electronic supplementary material.
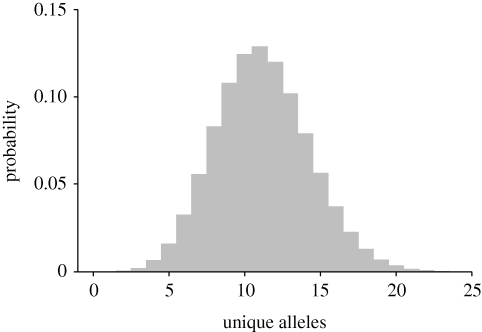
The modern sub-Antarctic population has 18 unique alleles, whereas only one unique allele was detected in the modern South Island population (see also Boessenkool et al. 2009b). It is particularly noteworthy, therefore, that the eight historic samples supposedly collected from the sub-Antarctic, but genetically categorized as South Island specimens (figures 2 and 3), do not possess any of the alleles unique to the sub-Antarctic. By contrast, five of the eight historic samples with confirmed sub-Antarctic origin possess a total of nine unique sub-Antarctic alleles. The probability of recovering zero unique alleles (by chance) in our sample of eight suspect individuals would appear to be very low. We investigated this probability further by calculating the probability distribution of sampling unique alleles (figure 4), assuming these eight suspect individuals were randomly sampled from the sub-Antarctic population. We simulated random resampling of the genotypes (100 million iterations and allowing for the missing data present in the original sample) of the suspect eight individuals using probabilities based on the allele-frequency vectors of the modern sub-Antarctic population and recorded the number of unique alleles in our sets of resampled genotypes. Our simulation confirmed that the probability of having zero unique alleles is extremely low (figure 4) and over 99.99 per cent of our random draws resulted in genotype sets with one or more unique alleles.
Figure 4.
Probability distribution of sampling unique alleles in the genotype sets of eight individuals from the modern sub-Antarctic population. The genotypes of the eight individuals were resampled (100 million iterations) based on the allele-frequency distribution of the current sub-Antarctic population, and the number of alleles that are unique to this population was recorded each iteration.
4. Discussion
(a). Erroneous origins of historic Megadyptes specimens?
Individual-based genetic analyses and evaluation of unique alleles for 10 microsatellite loci indicate that eight historic M. antipodes museum specimens have incorrect specimen data with respect to geographical collection location. We show that these eight specimens were not collected on the sub-Antarctic Auckland and Campbell Islands, as is stated on their specimen labels, but were in fact collected on the South Island of New Zealand. We have previously analysed these specimens for mitochondrial DNA (mtDNA) variation (Boessenkool et al. 2009a,b), and these results do not conflict with our microsatellite data. However, M. antipodes mtDNA lacks the phylogeographic structure required to reliably discriminate between sub-Antarctic versus South Island (Boessenkool et al. 2009b).
The historic expansion of M. antipodes (Boessenkool et al. 2009a) and the current pattern of migration (Boessenkool et al. 2009b) both indicate an asymmetric pattern of rare dispersal in this species, involving migration northwards from the sub-Antarctic to the South Island. It is not surprising, therefore, that a few South Island individuals show admixed genetic ancestry (figure 3; see also Boessenkool et al. 2009b). On the other hand, the single batch of eight purportedly ‘sub-Antarctic’ birds that genetically have South Island origins, clearly conflicts with observed dispersal patterns for M. antipodes. Perhaps most intriguingly, all eight of these birds are attributed to the same collector who purportedly obtained them within two years of one another.
The eight incorrectly labelled specimens were obtained by the American Museum of Natural History (AMNH) from Lionel Walter Rothschild the second Baron Rothschild (in 1932; Rothschild 1983), who in turn seems to have obtained the specimens from the collector Henry Hamersley Travers. The specimens have Rothschild collection labels as well as labels from H. H. Travers, which state the collection location (i.e. Auckland or Campbell Islands) and date (i.e. 1893 or 1894; M. LeCroy 2009, personal communication). An assessment of the sub-Antarctic voyages documented from the 1890s (Anonymous 1895; Headland 1989) revealed that it is unlikely that Travers or any other collector visited the Campbell and/or Auckland Islands in 1893. It therefore seems that, in addition to incorrect collection localities, the collection dates for at least five of the eight incorrectly labelled specimens are also incorrect.
Whether the erroneous collection data for the eight penguin specimens is attributable to incorrect labelling or to deliberate fraudulence remains unclear. Monetary gain is considered the most evident motivation for the deliberate falsification of specimen information and cases within ornithology provide some well-known examples of major specimen fraud (Rasmussen & Prŷs-Jones 2003). While M. antipodes is more abundant on the sub-Antarctic islands than on the South Island, specimens from geographically remote locations, such as the sub-Antarctic islands would nonetheless have been more valuable than readily available specimens from the South Island. The path from initial suspicion of specimen fraud to conclusive proof is, however, a long one (Rasmussen & Prŷs-Jones 2003). H. H. Travers was a well-known professional collector and taxidermist in nineteenth century New Zealand (Cyclopedia Company Limited 1897) and although some have questioned the veracity of some of his collection data (i.e. R.A. Falla in Murphy & Pennoyer 1952; Scofield 2005), no substantive proof has been found of any incorrect labelling until now. Alternatively, it is possible that Travers may have bought the penguin specimens from someone else: indeed, he apparently had a standing agreement with crew members of the New Zealand government steamer to collect specimens on sub-Antarctic islands for him (Warham & Bell 1979). It is therefore currently not possible to trace the exact origin or cause of the erroneous specimen labels. Nevertheless, further investigation of Travers' specimens as well as others that have been collected on sub-Antarctic New Zealand Islands in the late twentieth century may shed more light on the possibility of dishonesty.
(b). Genetic analysis as a means to detect errors in specimen locality data
The detection of errors in archive museum specimen information can be an arduous and time-consuming process that often relies on the availability of historical data such as collectors' field notes. The development of molecular techniques has now facilitated the use of DNA in this process. Genetic data have successfully been used to detect sexing errors in museum bird skins (Lee & Griffiths 2003; Bantock et al. 2008) and to verify the identity of species (e.g. Hennache et al. 2003; Olson et al. 2005) or even eggs (Lee & Prŷs-Jones 2008). In this study we present an extension of this approach by using individual-based population genetic analysis to detect errors in the locality data of archive museum specimens.
The use of individual-based genetic analyses, including assignment tests, to identify an organism's geographical origin is well known (Waser & Strobeck 1998; Manel et al. 2005). The extension of these approaches to the verification of locality data of archive specimens is a promising new direction in museum science. This method, however, is not without its limitations. First, the reliability of the method depends on the availability of specimens with confirmed geographical origin that can be used to calculate population allele frequencies. Often such historic samples are not available, so one relies on the assumption that allele frequencies of modern samples are representative of the allele frequencies of the historic populations. In our study we fortunately had good knowledge of the study system, including the colonization history and dispersal patterns of M. antipodes, and we can therefore be confident that this temporal assumption was not violated. Nevertheless, when populations are known to have suffered severe bottlenecks, for example, verifying the validity of this assumption will be challenging. Second, successful assignment of specimens is contingent on the existence of sufficient genetic structuring among populations and the statistical power of the applied markers to detect such a pattern. If limited levels of dispersal exist, the method can still be applied to detect errors in a sample of specimens that were collected by the same collector, or for example on the same voyage, but the detection of single misinformed specimens will probably be unreliable. If researchers aim to verify the origin of a single specimen, the guidelines already developed for the use of assignment tests in the detection of wildlife poaching should be applicable (Manel et al. 2002). Importantly, high thresholds (e.g. a probability of 0.999 that an individual belongs to a specific population) would have to be satisfied before a specimen can be confidently assigned to a specific population (Manel et al. 2002). Furthermore, care has to be taken when choosing an assignment method, as some techniques require sampling of all potential source populations (Pritchard et al. 2000; Manel et al. 2002; Piry et al. 2004). The present study underlines the value of combining distinct genetic approaches to improve the reliability of error detection.
Using individual-based genetic analyses to detect erroneous specimen information should be regarded as complementary to the methods that are currently being applied for error detection in specimen data (see Knox 1993; Rasmussen & Collar 1999; Rasmussen & Prŷs-Jones 2003; Peterson et al. 2004; Olson 2008). The strength of the genetic method, however, is that potential errors can be detected without a priori suspicion. With the continuously increasing number of population and evolutionary genetic studies that use archive specimens (Wandeler et al. 2007), there is considerable scope to apply these methods.
(c). Consequences of erroneous specimen data
Historical material is a limited resource and studies employing historical specimens are typically constrained by low sample sizes. The detection of errors in specimen data consequently imposes a dilemma: should specimens with erroneous information be retained in the study or should they be removed from the dataset? If samples are retained in cases where errors were detected in locality data, then all subsequent analyses could be performed with samples re-allocated to their true geographical origin. Although this appears to be a valid solution, such an approach can result in some form of circularity, as the genetic data used to verify specimen location will probably be the same data used for subsequent analysis. Alternatively, if specimens found to be incorrectly labelled are completely removed from a dataset, this may severely limit sample sizes for subsequent analyses. In the case of yellow-eyed penguins, for example, we removed the eight specimens with erroneous specimen data from calculations of temporal effective population size (Ne; see Boessenkool et al. in press). As a result, the sample size of the sub-Antarctic population was too low to estimate the population's temporal Ne, but we still had sufficient data to estimate the temporal Ne for the South Island population (Boessenkool et al. in press). Whether samples should be retained or removed from the dataset clearly has to be decided on a case-by-case basis, as determined by the availability of additional samples, the specific analysis to be undertaken, and the type of error encountered.
The results of the present study serve as a warning to all those who use museum specimens in population genetic studies. Especially when working on endangered species or populations, the reliability of specimen locality data is essential. Mistakes in locality data can potentially confound inferences of historical population connectivity, identification of conservation units, effective population size estimates and associated management strategies. Although it was recently argued that physical specimens provide the most reliable evidence for assessing species ranges (McKelvey et al. 2008), our analysis shows that specimen data are only as reliable as the associated collection details. Researchers are encouraged to invest in the verification of specimen data to ensure that archive specimens remain a valuable resource for many years to come.
Acknowledgements
Samples were collected under Department of Conservation permits SO-17933-FAU and OT-19097-RES and University of Otago Animal Ethics Approval 69/06.
We are very grateful to the Auckland Museum, American Museum of Natural History, Australian Museum, Museum of Comparative Zoology, Natural History Museum Geneva, Natural History Museum Tring, Natural History Museum Paris, Museum of New Zealand Te Papa Tongarewa, Natural History Museum Vienna, Swedish Museum of Natural History, Otago Museum, South Australian Museum, Smithsonian Institution, Museum für Naturkunde Berlin and C. Millar for supplying tissue samples of historic specimens. We are greatly indebted to Mary LeCroy and Paul Sweet from the American Natural history Museum. We thank the New Zealand Department of Conservation for help with collecting contemporary samples, and Tania King for guidance in the laboratory. This research was supported by the Department of Zoology, University of Otago, including PBRF Research Enhancement Grants to P.J.S. and J.M.W.
References
- Anonymous 1895Appendixes to the Journals of the House of Representatives for 1894 H25, 1–4 [Google Scholar]
- Bantock T. M., Prŷs-Jones R. P., Lee P. L. M.2008New and improved molecular sexing methods for museum bird specimens. Mol. Ecol. Resour. 8, 519–528 (doi:10.1111/j.1471-8286.2007.01999.x) [DOI] [PubMed] [Google Scholar]
- Belkhir K., Borsa P., Chikhi L., Raufaste N., Bonhomme F.1996–2004Genetix 4.05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Laboratoire Génome, Populations, Interactions, Université de Montpellier II [Google Scholar]
- Bergl R. A., Vigilant L.2007Genetic analysis reveals population structure and recent migration within the highly fragmented range of the Cross River gorilla (Gorilla gorilla diehli). Mol. Ecol. 16, 501–516 (doi:10.1111/j.1365-294X.2006.03159.x) [DOI] [PubMed] [Google Scholar]
- Boessenkool S., King T. M., Seddon P. J., Waters J. M.2008Isolation and characterization of microsatellite loci from the yellow-eyed penguin (Megadyptes antipodes). Mol. Ecol. Resour. 8, 1043–1045 (doi:10.1111/j.1755-0998.2008.02149.x) [DOI] [PubMed] [Google Scholar]
- Boessenkool S., Austin J. J., Worthy T. H., Scofield P., Cooper A., Seddon P. J., Waters J. M.2009aRelict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proc. R. Soc. B 276, 815–821 (doi:10.1098/rspb.2008.1246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boessenkool S., Star B., Waters J. M., Seddon P. J.2009bMultilocus assignment analyses reveal multiple units and rare migration events in the recently expanded yellow-eyed penguin (Megadyptes antipodes). Mol. Ecol. 18, 2390–2400 (doi:10.1111/j.1365-294X.2009.04203.x) [DOI] [PubMed] [Google Scholar]
- Boessenkool S., Star B., Seddon P. J., Waters J. M.In press Temporal genetic samples indicate small effective population size of the endangered yellow-eyed penguin. Conserv. Genet. (doi:10.1007/s10592-009.9988.8) [Google Scholar]
- Brooke M. D.2000Why museums matter. Trends Ecol. Evol. 15, 136–137 (doi:10.1016/S0169-5347(99)01802-9) [Google Scholar]
- Collar N. J., Rudyanto2003The archive and the ark: bird specimen data in conservation status assessment. In Why museums matter: avian archives in the age of extinction. Bulletin of the British Ornithologist's Club 123A (eds Collar N. J., Fisher C. T., Feare C. J.), pp. 95–113 The British Ornithologist's Club [Google Scholar]
- Cyclopedia Company Limited 1897The cyclopedia of New Zealand (Wellington Provincial District) Wellington, New Zealand: The Cyclopedia Company Limited [Google Scholar]
- Dalton R.2005Ornithologists stunned by bird collector's deceit. Nature 437, 302–303 (doi:10.1038/437302a) [DOI] [PubMed] [Google Scholar]
- Graham C. H., Ferrier S., Huettman F., Moritz C., Peterson A. T.2004New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol. Evol. 19, 497–503 (doi:10.1016/j.tree.2004.07.006) [DOI] [PubMed] [Google Scholar]
- Headland R. K.1989Chronological list of Antarctic expeditions and related historical events Cambridge, UK: Cambridge University Press [Google Scholar]
- Hennache A., Rasmussen P., Lucchini V., Rimondi S., Randi E.2003Hybrid origin of the imperial pheasant Lophura imperialis (Delacour and Jabouille, 1924) demonstrated by morphology, hybrid experiments, and DNA analyses. Biol. J. Linn. Soc. 80, 573–600 (doi:10.1111/j.1095-8312.2003.00251.x) [Google Scholar]
- Hombron J. B., Jacquinot H.1841Description de plusieurs oiseaux nouveaux ou peu connus, provenant de l'expédition autour du monde faite sur les corvettes l'Astolabe et la Zélée. Annales Des Sciences Naturelles, Zoologie 16, 312–320 [Google Scholar]
- Johnson J. A., Bellinger M. R., Toepfer J. E., Dunn P.2004Temporal changes in allele frequencies and low effective population size in greater prairie-chickens. Mol. Ecol. 13, 2617–2630 (doi:10.1111/j.1365-294X.2004.02264.x) [DOI] [PubMed] [Google Scholar]
- Knox A. G.1993Richard Meinertzhagen: a case of fraud examined. Ibis 135, 320–325 (doi:10.1111/j.1474-919X.1993.tb02851.x) [Google Scholar]
- Larsson J. K., Jansman H. A. H., Segelbacher G., Hoglund J., Koelewijn H. P.2008Genetic impoverishment of the last black grouse (Tetrao tetrix) population in the Netherlands: detectable only with a reference from the past. Mol. Ecol. 17, 1897–1904 (doi:10.1111/j.1365-294X.2008.03717.x) [DOI] [PubMed] [Google Scholar]
- Lecis R., Pierpaoli M., Biro Z. S., Szemethy L., Ragni B., Vercillo F., Randi E.2006Bayesian analyses of admixture in wild and domestic cats (Felis silvestris) using linked microsatellite loci. Mol. Ecol. 15, 119–131 (doi:10.1111/j.1365-294X.2005.02812.x) [DOI] [PubMed] [Google Scholar]
- Lee P. L. M., Griffiths R.2003Sexing errors among museum skins of a sexually monomorphic bird, the moorhen Gallinula chloropus. Ibis 145, 695–698 (doi:10.1046/j.1474-919X.2003.00201.x) [Google Scholar]
- Lee P. L. M., Prŷs-Jones R. P.2008Extracting DNA from museum bird eggs, and whole genome amplification of archive DNA. Mol. Ecol. Resour. 8, 551–560 (doi:10.1111/j.1471-8286.2007.02042.x) [DOI] [PubMed] [Google Scholar]
- Manel S., Berthier P., Luikart G.2002Detecting wildlife poaching: identifying the origin of individuals with Bayesian assignment tests and multilocus genotypes. Conserv. Biol. 16, 650–659 (doi:10.1046/j.1523-1739.2002.00576.x) [Google Scholar]
- Manel S., Gaggiotti O. E., Waples R. S.2005Assignment methods: matching biological questions with appropriate techniques. Trends Ecol. Evol. 20, 136–142 (doi:10.1016/j.tree.2004.12.004) [DOI] [PubMed] [Google Scholar]
- Marchant S., Higgins P. J.1990Handbook of Australian, New Zealand and Antarctic Birds Melbourne, Australia: Oxford University Press [Google Scholar]
- McKelvey K. S., Aubry K. B., Schwartz M. K.2008Using anecdotal occurrence data for rare or elusive species: the illusion of reality and a call for evidentiary standards. Bioscience 58, 549–555 (doi:10.1641/B580611) [Google Scholar]
- McKinlay B.2001Hoiho (Megadyptes antipodes) recovery plan 2000–2025 Wellington, New Zealand: Department of Conservation [Google Scholar]
- Miller C. R., Waits L. P.2003The history of effective population size and genetic diversity in the Yellowstone grizzly (Ursus arctos): implications for conservation. Proc. Natl Acad. Sci. USA 100, 4334–4339 (doi:10.1073/pnas.0735531100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. C., Pennoyer J. M.1952Larger petrels of the genus Pterodroma. Am. Mus. Novitates 1580, 1–43 [Google Scholar]
- Olson S. L.2008Falsified data associated with specimens of birds, mammals, and insects from the Veragua Archipelago, Panama, collected by J. H. Batty. Am. Mus. Novitates 3620, 1–37 (doi:10.1206/592.1) [Google Scholar]
- Olson S. L., Fleischer R. C., Fisher C. T., Bermingham E.2005Expunging the ‘Mascarene starling’ Necropsar leguati: archives, morphology and molecules topple a myth. Bull. Br. Ornithol. Club 125, 31–42 [Google Scholar]
- Pääbo S., et al. 2004Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679 (doi:10.1146/annurev.genet.37.110801.143214) [DOI] [PubMed] [Google Scholar]
- Pennisi E.2000Taxonomic revival. Science 289, 2306–2308 (doi:10.1126/science.289.5488.2306) [DOI] [PubMed] [Google Scholar]
- Peterson A. T., Navarro-Sigüenza A. G., Pereira R. S.2004Detecting errors in biodiversity data based on collector's itineraries. Bull. Br. Ornithol. Club 124, 143–151 [Google Scholar]
- Piry S., Alapetite A., Cornuet J. M., Paetkau D., Baudouin L., Estoup A.2004GeneClass2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539 (doi:10.1093/jhered/esh074) [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P.2000Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen P. C., Collar N. J.1999Major specimen fraud in the Forest Owlet Heteroglaux (Athene auct.) blewitti. Ibis 141, 11–21 [Google Scholar]
- Rasmussen P. C., Prŷs-Jones R. P.2003History vs. mystery: the reliability of museum specimen data. In Why museums matter: avian archives in the age of extinction. Bulletin of the British Ornithologist's Club 123A (eds Collar N. J., Fisher C. T., Feare C. J.), pp. 66–94 The British Ornithologist's Club [Google Scholar]
- Rice W. R.1989Analyzing tables of statistical tests. Evolution 43, 223–225 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- Rothschild M. L.1983Dear Lord Rothschild: birds, butterflies and history London, UK: Hutchinson [Google Scholar]
- Rousset F.2008Genepop ' 007: a complete re-implementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 (doi:10.1111/j.1471-8286.2007.01931.x) [DOI] [PubMed] [Google Scholar]
- Scofield P.2005The supposed Macquarie Island parakeet in the collection of the Canterbury Museum. Notornis 52, 117–120 [Google Scholar]
- Sefc K. M., Payne R. B., Sorenson M. D.2003Microsatellite amplification from museum feather samples: effects of fragment size and template concentration on genotyping errors. Auk 120, 982–989 (doi:10.1642/0004-8038(2003)120[0982:MAFMFS]2.0.CO;2) [Google Scholar]
- Sefc K. M., Payne R. B., Sorenson M. D.2007Single base errors in PCR products from avian museum specimens and their effect on estimates of historical genetic diversity. Conserv. Genet. 8, 879–884 (doi:10.1007/s10592-006-9240-8) [Google Scholar]
- Shaffer H. B., Fisher R. N., Davidson C.1998The role of natural history collections in documenting species declines. Trends Ecol. Evol. 13, 27–30 (doi:10.1016/S0169-5347(97)01177-4) [DOI] [PubMed] [Google Scholar]
- Suarez A. V., Tsutsui N. D.2004The value of museum collections for research and society. Bioscience 54, 66–74 (doi:10.1641/0006-3568(2004)054[0066:TVOMCF]2.0.CO;2) [Google Scholar]
- Taberlet P., Griffin S., Goossens B., Questiau S., Manceau V., Escaravage N., Waits L. P., Bouvet J.1996Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 24, 3189–3194 (doi:10.1093/nar/24.16.3189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Jamieson I. G., Wallis G. P.2008Ancestral and contemporary levels of genetic variation in two New Zealand passerines with different histories of decline. J. Evol. Biol. 20, 2035–2047 (doi:10.1111/j.1420-9101.2007.01362.x) [DOI] [PubMed] [Google Scholar]
- Walsh P. S., Metzger D. A., Higuchi R.1991Chelex-100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513 [PubMed] [Google Scholar]
- Wandeler P., Hoeck P. E., Keller L. F.2007Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 (doi:10.1016/j.tree.2007.08.017) [DOI] [PubMed] [Google Scholar]
- Warham J., Bell B. D.1979The birds of Antipodes Island, New Zealand. Notornis 26, 121–170 [Google Scholar]
- Waser P. M., Strobeck C.1998Genetic signatures of interpopulation dispersal. Trends Ecol. Evol. 13, 43–44 (doi:10.1016/S0169-5347(97)01255-X) [DOI] [PubMed] [Google Scholar]
- Winker K.2000Obtaining, preserving, and preparing bird specimens. J. Field Ornithol. 71, 250–297 [Google Scholar]