Abstract
Therian mammals have an extremely conserved XX/XY sex determination system. A limited number of mammal species have, however, evolved to escape convention and present aberrant sex chromosome complements. In this study, we identified a new case of atypical sex determination in the African pygmy mouse Mus minutoides, a close evolutionary relative of the house mouse. The pygmy mouse is characterized by a very high proportion of XY females (74%, n = 27) from geographically widespread Southern and Eastern African populations. Sequencing of the high mobility group domain of the mammalian sex determining gene Sry, and karyological analyses using fluorescence in situ hybridization and G-banding data, suggest that the sex reversal is most probably not owing to a mutation of Sry, but rather to a chromosomal rearrangement on the X chromosome. In effect, two morphologically different X chromosomes were identified, one of which, designated X*, is invariably associated with sex-reversed females. The asterisk designates the still unknown mutation converting X*Y individuals into females. Although relatively still unexplored, such an atypical sex chromosome system offers a unique opportunity to unravel new genetic interactions involved in the initiation of sex determination in mammals.
Keywords: African pygmy mouse, Mus minutoides, atypical sex determination system; sex-reversed females, Sry gene, X* chromosome
1. Introduction
Sex poses some of the most fascinating questions in evolutionary biology. Many eukaryotic organisms have separate males and females, but sex is determined by many different strategies, even among closely related species (for a review in fish, see Volff & Schartl 2001; vertebrates, Ezaz et al. 2006; Diptera, Saccone et al. 2002). By contrast, mammals, or at least therians (i.e. placentals and marsupials) have an extremely conserved sex determination system (for the egg-laying monotremes see Veyrunes et al. 2008). Therian sex determination has a unique origin and is syngamic, meaning that sex is fixed by the presence of the sex chromosome complement at fertilization, and not influenced by environmental factors (e.g. temperature, hormones, social structure). In effect, females have two X chromosomes (XX) and males have a single X and a Y chromosome (XY), and variation of this XX/XY dichotomy generally leads to sterility. Mammalian sex is in fact regulated by a single gene on the Y chromosome: SRY (Sex-determining Region of the Y chromosome). The SRY gene is at the top of the gene activity cascade which triggers testis formation and thus male development (Sinclair et al. 1990). In the absence of this gene, gonads develop into ovaries. It has generally been assumed that heteromorphic sex chromosomes with a highly degraded and specialized Y chromosome, such as that found in mammals, are a strong barrier to transitions between sex-determining systems. This is owing to formation of lethal genotypes (such as YY) and the potential adverse effects of Y-chromosome genes on the feminization process of neo-XY females or, conversely, the lack of necessary Y-chromosome genes in neo-XX males (Bull 1983; Marin & Baker 1998). A limited number of mammal species do, however, escape this constraint and present atypical sex chromosome systems (see excellent reviews by Fredga 1983, 1988, 1994). So far, these unusual sex-determining systems have been unambiguously identified by molecular or cytogenomic methods in seven genera, all rodents, which group in four categories: (i) XX or XY females and XY males in the lemmings Myopus schisticolor and Dicrostonyx torquatus (Fredga et al. 1976; Fredga 1983, 1988, 1994), as well as several species of the South-American field mouse Akodon sp. (Hoekstra & Edwards 2000; Bianchi 2002; Ortiz et al. 2009); (ii) XO females (only one X) and XY males in the vole Microtus oregoni (Ohno et al. 1963, 1966; Fredga 1983); (iii) females and males with an identical XO karyotype (loss of the second X chromosome in females and of the Y in males) in the Japanese spiny rats Tokudaia osimensis and Tokudaia tokunoshimensis (Soullier et al. 1998; Sutou et al. 2001; Arakawa et al. 2002), and the mole vole Ellobius lutescens (Matthey 1953; Just et al. 1995, 2007), and finally; (iv) XX males and females in two other species of Ellobius, E. tancrei and E. talpinus (Just et al. 1995, 2007). Categories (iii) and (iv) are remarkable since sex determination occurs in the absence of a Y chromosome, and even without the Sry gene.
In this study we report a new atypical sex determination system, this time in the African pygmy mouse Mus minutoides. The pygmy mouse is one of the smallest African mammals (adult weight = 4–6 g), but with one of the widest sub-Saharan distributions (Veyrunes et al. 2005). It is included in the same genus as the house mouse, Mus musculus, but is assigned to a different subgenus, Nannomys (Musser & Carleton 2005; Veyrunes et al. 2005, 2006). Mus minutoides presents an extreme example of karyotypic diversity owing largely to Robertsonian fusion variation, but more unexpectedly to rare chromosomal changes involving the sex chromosomes (Veyrunes et al. 2004, 2007). Here, we show that this species possesses a very high proportion of sex-reversed females, that is, individuals that are phenotypically female and fertile, but with a male karyotype. Thus, females are either XX or XY, males being XY. This represents the fourth report of such an unusual sex chromosome system within mammals (see above, category (i)), and given its close phylogenetic association to the emblematic M. musculus, may provide valuable insights into the complex initiation of sex determination in therian mammals.
2. Material and methods
(a). Approach
In the 1960s, several reports described the occurrence of heteromorphic X chromosomes in populations of Nannomys, i.e. in M. minutoides from Ivory Coast and the Central African Republic and in M. triton from Congo (Matthey 1967, 1970; Jotterand 1972). Some females possessed one normal-sized X chromosome, while the other was two-thirds smaller and entirely heterochromatic, being similar in size and staining properties to a typical Y chromosome. The authors considered this chromosome as a partially deleted X, named Xd. Recently, Castiglia et al. (2002) and Veyrunes et al. (2004), respectively, described in M. minutoides, two females (from Mutanda, Zambia) and one female (from Kuruman, South Africa) with heteromorphic sex chromosomes, designated as XXd in accordance with the previous literature. To further critically investigate the nature of the Xd chromosome, an analysis of the M. minutoides sex chromosomes was performed using cytogenetic (G-banding), cytogenomic (fluorescence in situ hybridization) and molecular (DNA sequencing) analyses.
(b). Sampling
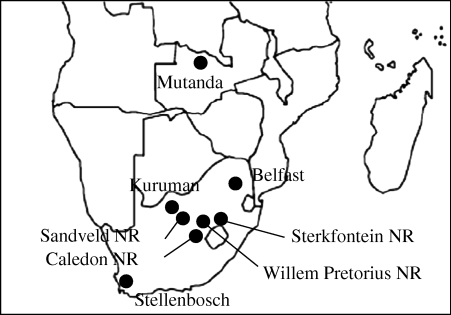
A total of 46 wild-caught specimens from eight localities of Southern and Eastern Africa were used in this study: Caledon Nature Reserve (NR), Stellenbosch, Sandveld NR, Sterkfontein NR, Kuruman, Belfast and Willem Pretorius NR from South Africa, and Mutanda from Zambia (table 1). The geographical distribution of the localities is indicated in figure 1. Since several species of African pygmy mice may coexist in sympatry, the assignation of all specimens to M. minutoides was established by comparison with published karyotypes and/or cytochrome b sequences (Veyrunes et al. 2004, 2005). Additionally, we expanded our sample to include specimens of Mus mattheyi, Mus famulus and Mus crociduroides for the analysis of Sry (see below).
Table 1.
List of the sampling localities with the number of males and females of each sex chromosome complement.
| females |
||||
|---|---|---|---|---|
| localities | X*Y | XX* | XX | males XY |
| Belfast | 0 | 0 | 1 | 0 |
| Caledon NR | 11 | 0 | 0 | 10 |
| Kuruman | 1 | 0 | 0 | 1 |
| Mutanda | 2 | 0 | 0 | 1 |
| Sandveld NR | 1 | 1 | 0 | 3 |
| Stellenbosch | 3 | 2 | 2 | 3 |
| Sterkfontein NR | 2 | 0 | 0 | 1 |
| Willem Pretorius NR | 0 | 0 | 1 | 0 |
| total | 20 | 3 | 4 | 19 |
Figure 1.
Map of Southern Africa showing the geographical origin of the samples studied.
(c). Karyotype and chromosome identifications
Chromosome preparations of all specimens were made either from bone marrow of yeast-stimulated animals (Lee & Elder 1980) or fibroblast cell-cultures established from skin biopsy following standard procedures. Identification of chromosomes was accomplished by G-banding (Seabright 1971). Several karyotypes of specimens used in this study have previously been published (Castiglia et al. 2002; Veyrunes et al. 2004, 2007).
(d). Fluorescence in situ hybridization
The commercial (Cambio) Y chromosome-specific painting probe of the house mouse M. musculus was hybridized to metaphase spreads of M. minutoides females with heteromorphic sex chromosomes from Kuruman (1 specimen) and Caledon NR (2 specimens). Hybridization and detection followed a slightly modified procedure from that described in Robinson et al. (2004). The Y chromosome probe (50 ng) was made up to 11 µl with hybridization buffer (50% deionized formamide, 10% dextran sulfate, 2 × SSC, 0.5 mol l−1 phosphate buffer, pH 7.3, and 1 × Denhardt's solution). The slides were pretreated with 10 mM HCl containing 0.01 per cent pepsin at 37°C for 5 min. The probe was denaturated at 65°C for 10 min and then preannealed at 37°C for 30 min. Metaphase slides were denaturated by incubation in 70 per cent formamide/30 per cent 2 × SSC solution at 65°C for 2 min, quenched in ice-cold 70 per cent ethanol, and dehydrated through a 70, 80, 90 and 100 per cent ethanol series. The pre-annealed paint was applied to slides, and incubated for 24 h at 37°C. Post-hybridization washes involved two successive 5 min incubations at 42°C in 50 per cent formamide/1 × SSC, 2 × SSC, and one in 4XT (250 µl Tween in 50 ml 4 × SSC). Biotin-labelled probe was visualized using Cy3-avidin (1 : 500 dilution, Amersham). Slides were mounted in Vectashield mounting medium with DAPI (Vector Laboratories), and images captured using the Genus software (Applied Imaging).
(e). Sry gene sequencing and phylogenetic reconstruction
Total genomic DNA was extracted from tissues preserved in ethanol using a QiaAmp DNA extraction kit (Qiagen). A 472-bp region of the Sry gene, beginning 103 bp 5′ to the highly conserved 79-amino-acid high mobility group (HMG) box, was amplified from genomic DNA using two oligonucleotide primers (SRY1: 5′-AGATCTTGATTTTTAGTGTTC-3′ and SRY2: 5′-GAGTACAGGTGTGCAGCTCTA-3′) as described in Lundrigan & Tucker (1994). Polymerase chain reaction (PCR) parameters were: one step at 94°C for 5 min, followed by 35 cycles (30 s at 94°C, 30 s at 50°C, 1 min at 72°C). The final extension at the end of the profile was at 72°C for 10 min. Double-stranded PCR products were purified from agarose gel using Amicon Ultrafree-DNA columns (Millipore), and both strands were sequenced directly using an automatic sequencer ABI 310 (PE Applied Biosystems) or sent to Cogenics for sequencing. New Sry sequences obtained for M. minutoides (27), M. mattheyi, M. famulus and M. crociduroides were deposited in the EMBL data bank under the accession numbers FN568 495 to FN568 524. Additional sequences were retrieved from GenBank and added to the alignment: M. minutoides (AY057 745), M. musculus (MMU03 645), Mus cervicolor (L29 548) and Mus pahari (L29 543). Sequences were aligned using SeaView (Galtier et al. 1996). The maximum likelihood (ML) phylogeny was reconstructed with PHYML (Guindon & Gascuel 2003) under a GTR + G + I model of sequence evolution as specified by the Akaike Information Criterion criteria implemented in Modeltest v. 3.06 (Posada & Crandall 1998), and 100 ML bootstrap replicates were performed on the resulting topology.
3. Results and discussion
(a). Phenotypic versus karyotypic sex
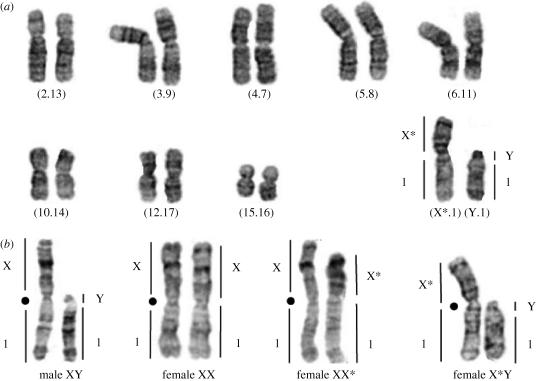
Phenotypic sex of the 46 specimens analysed herein (table 1) was identified by examination of gonad morphology, yielding 27 females and 19 males. All specimens were then karyotyped by G-banding. Our analysis included published data from Mutanda (2n = 24–25; Castiglia et al. 2002), Caledon NR (2n = 18; Veyrunes et al. 2004, 2007), Stellenbosch (2n = 18; Veyrunes et al. 2004) and Kuruman (2n = 34; Veyrunes et al. 2004), that we expanded with additional samples from Caledon NR, Stellenbosch and new localities where G-banded karyotypes were determined for the first time: Belfast and Sandveld NR (2n = 34, similar to the karyotype of Kuruman), Sterkfontein NR (2n = 18, similar to the one of Caledon NR), and finally, Willem Pretorius NR (2n = 26). The difference among karyotypes results from variation in the number and combination of Robertsonian fusions (i.e. fusion by the centromere of two non-homologous chromosomes), but all specimens have two fusions in common, those involving the sex chromosomes and pair 1, (X.1) and (Y.1), which represent a diagnostic signature for this species (Veyrunes et al. 2004, 2005).
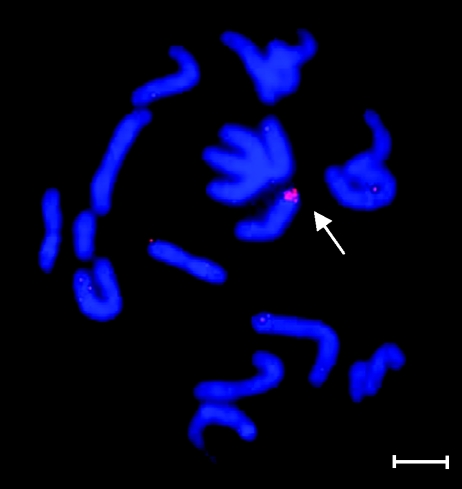
The karyotypic analyses identified the presence of a small, atypical heterochromatic X chromosome in 20 of the 27 females, named Xd according to Matthey (1967) and subsequently followed in recent studies (Castiglia et al. 2002; Veyrunes et al. 2004). The Xd chromosome varied from medium-sized in the Mutanda and Kuruman localities to minute in the Caledon NR and Stellenbosch localities, but was invariant within populations. The observation that the Xd arm always matched the size of the Y arm in a given population (see Veyrunes et al. 2004), prompted our efforts to critically re-examine the derivation of this chromosome. Fluorescence in situ hybridization of the mouse Y chromosome painting probe on metaphase spreads resulted in a strong signal covering the entire Xd arm (figure 2), indicating unambiguously that Xd is in fact a Y chromosome and had been incorrectly assigned to an X chromosome in previous studies. The females carrying this derivative chromosome are therefore XY females (figure 3a). Remarkably, all the females (n = 11) from the Caledon NR population were diagnosed as being sex-reversed (table 1).
Figure 2.
Fluorescence in situ hybridization using a house mouse Y chromosome painting probe on a Mus minutoides XXd female from Caledon NR. The strong signal on the Xd arm (arrow) indicates that this female is in fact XY. Scale bar, 10 µm.
Figure 3.
(a) G-banded karyotype of an X*Y female from Stellenbosch, the numbers in parentheses designate the chromosome pairs involved in the Robertsonian fusions. (b) The different sex chromosome complements (X, X* and Y chromosomes fused to autosome pair 1); black dots indicate centromere position.
(b). Sry sequence in M. minutoides
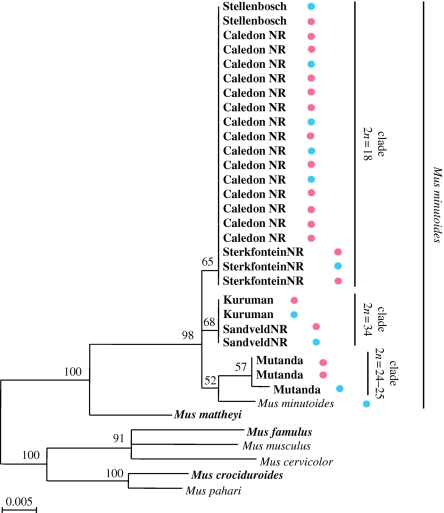
An obvious explanation for the pattern of sex-reversal outlined above would involve the male-promoting gene, Sry. In effect, since the Y chromosome is present in the females, we hypothesized that the Sry gene was either deleted in these specimens or, if present, was not expressed. Given that the window for Sry expression in the mouse is extremely narrow (a few hours during embryogenesis; Hacker et al. 1995; Bullejos & Koopman 2001), we chose an indirect approach to test the integrity/functionality of the Sry gene in sex-reversed females. Our decision to target 472 bp of the Sry gene (whole HMG domain and part of the flanking regions) in 27 individuals (18 XY females and nine males), was based on the observation that most SRY mutations leading to sex-reversal in humans have fallen within the HMG box (O'Neill & O'Neill 1999). Surprisingly, the sequencing results revealed that the gene was present in XY females (but not in XX females), and that there was no evidence of deletion or nucleotide substitution that would result in frameshift or stop codon mutations. In fact, the sequences were identical between males and XY females of a given population (with the exception of Mutanda; figure 4). The Sry sequences were clustered into four haplotypes corresponding to the different cytotypes: one shared by all specimens analysed with 18 chromosomes, i.e. Stellenbosch, Sterkfontein NR and Caledon NR (14 females; six males); one shared by all specimens with 34 chromosomes, i.e. Kuruman and Sandveld NR (two females; two males); one for the two females from Mutanda (2n = 24–25); and one specific to the male of Mutanda (2n = 25). Within M. minutoides, the bootstrap values supporting clades are extremely low since only one or two nucleotide substitutions characterize each haplotype (figure 4). Although Nagamine (1994) reported the presence of several Y-specific Sry copies in Mus, including M. minutoides, no ambiguities were recorded in the PCR products or sequences in our study, suggesting that either the gene copies were identical over the sequenced region, or the PCR primers were specific for only the functional copy (see also Lundrigan & Tucker 1994; Lundrigan et al. 2002). These results suggest that the Sry gene, or at least the HMG box is intact (mutations outside the analysed region, such as promoter sequences, have not been considered), and is thus potentially functional in both sexes. In light of these results, therefore, the sex reversal mechanism in M. minutoides does not appear to involve an Sry mutation.
Figure 4.
Maximum likelihood phylogeny using the 472 bp sequences of the Sry gene. The samples in bold are those sequenced in this study. Bootstrap values supporting each clade are indicated on nodes. Within Mus minutoides, specimens are characterized by their locality of capture. Blue and pink dots represent males and X*Y females, respectively. 2n = diploid number.
(c). Sex-reversal mutation in M. minutoides
In addition to the presence of a Y chromosome (and Sry gene) in some of the females, the G-banding data detected two X chromosome morphs that differ in length and banding pattern: one that is similar to other pygmy mouse species (Veyrunes et al. 2004), designated as X herein and which is always associated with XY males, and a second, always associated with XY females that we termed X* (figure 3b). The difference in banding pattern and size between the two morphs suggests that a complex rearrangement has occurred possibly involving a deletion and an inversion. When the phenotypic sex was compared to the genotypic sex established from karyotypes, three types of females were identified: XX, XX* and X*Y; whereas all males were XY (figure 3b). Of the 27 females examined in our study, four were XX, three were XX* and 20 were X*Y females from geographically widespread populations (table 1). There was one exception to this: both the male and sex-reversed female from Kuruman possessed the same X chromosome morph, which was slightly longer, and differed in banding pattern from the X and X* chromosomes of the other specimens (data not shown). This inconsistency could be attributed to an independent event in this lineage, or to a further rearrangement on the X chromosome. We also identified a pregnant X*Y female (from Stellenbosch) carrying five embryos, all but one of which were karyotyped, and sexed according to their sex chromosome complement. This pregnancy comprised two X*Y females, one XX* female, and one XY male, suggesting that X*Y females are fertile in marked contrast to pathological sex-reversal in humans, where the affected individuals are invariably sterile (e.g. McElreavey & Fellous 1999).
These data allowed us to infer the presence of a mutation on the X* chromosome that prevents masculinisation of X*Y specimens. Although, it might appear counterintuitive that the mutation causing male-to-female sex reversal is on the X chromosome and not the Y, the X is known to have accumulated a disproportionate number of genes controlling sex and reproductive traits, and even genes expressed in spermatogonia (Saifi & Chandra 1999; Wang et al. 2001; Graves et al. 2002). For example, over-expression of the DAX1 gene on the X chromosome appears to over-ride SRY, and is associated with male-to-female sex reversal in humans (Bardoni et al. 1994). It is noteworthy that similar, atypical sex determination systems have been described in three other rodent genera (see §1). The genetic bases of these modifications are all unknown, but are associated with an X-linked mutation in Myopus (deletion of Xp21–23; Liu et al. 1998), and possibly so too in Dicrostonyx and Akodon (Fredga 1988; Ortiz et al. 2009). Although these mutations have appeared independently in the four different lineages (even the two lemmings are not closely related; Buzan et al. 2008), they are all X-linked, and may therefore involve the same gene(s). Clearly, further molecular/genomic analyses are required in M. minutoides to identify the mutational event changing the normal X chromosome into the derivative X*. Once the rearranged synteny block is identified, it should be possible to pinpoint sex-reversal candidate genes by comparison to the mouse genome assembly.
(d). Evolution of sex determination in M. minutoides
The question remains of the selective pressures favouring the establishment of the atypical sexual system in pygmy mice given the reproductive cost associated with X*Y females (loss of YY embryos, i.e. 25% lethality). Several evolutionary mechanisms for compensating for the loss of the YY embryos have been identified in the literature. In effect, the X*Y females may bypass this reproductive disadvantage by meiotic drive (i.e. preferential transmission of the X* in gametes). This is particularly evident with the meiotic double non-disjunction that occurs in M. schisticolor in which only X*-carrying ovules are formed, producing only XX* and X*Y females, thus avoiding embryo loss (Fredga 1983). Additionally X*Y females may compensate for foetal loss through higher reproductive efficiency, as is the case in Akodon azarae where X*Y females have both a longer reproductive lifespan and higher rates of preimplantation embryonic development compared to XX females (Espinosa & Vitullo 2001). Although constrained by sample size, the pregnant X*Y female in our analysis provided a preliminary clue to its ability to establish: (i) no litter size reduction was evident, since five embryos were formed, marginally higher than the documented mean litter size of four (Willan & Meester 1978); and (ii) the presence of a XY male offspring does not support a Myopus-like double non-disjunction mechanism. An additional feature of the X*Y female sex system is an anticipated biased sex-ratio since the XX* and X*Y females are theoretically expected to produce 75 and 66 per cent female offspring, respectively. Skewed sex-ratios such as these may be under selection and contribute to evolutionary changes in sex determination systems (e.g. Werren & Beukeboom 1998; Uller et al. 2007; Vuilleumier et al. 2007). For example, in the two gregarious and polygamous lemming species M. schisticolor and D. torquatus, cycles of high densities followed by population crashes (boom and bust scenario) have been recorded, suggesting that the production of an excess of females may represent an adaptive strategy for rapid recovery from low densities. Coincidently, surveys of M. minutoides populations have shown similarly seasonal population cycles (Monadjem 1999; Fichet-Calvet et al. 2009).
(e). How pervasive is this sex determining system within African pygmy mice?
By extension, our results suggest that all females previously described with heteromorphic X chromosomes in M. minutoides from Ivory Coast, the Central African Republic and even in a population of M. triton from Congo may in fact be X*Y females (Matthey 1967, 1970; Jotterand 1972). If confirmed, M. minutoides X*Y females would not be restricted to Southern and Eastern Africa, but would be widespread throughout sub-Saharan Africa, and would extend to related species. Additionally, the apparently unique situation in mice collected from the Caledon NR population deserves closer scrutiny. All females analysed from this population (n = 11) were identified as X*Y (table 1). If this unexpected result is confirmed by additional sampling, it would mean that the XX and X*X females are no longer produced or viable (and that half of the embryos produced are lethal). A sex chromosome system such as this, where all females are X*Y and males XY, has never been described, and would lead to a unique mechanism in which the sex determining switch would no longer be determined by the Y chromosome, but by the presence of a modified X chromosome (either X or X*). Interestingly, Jotterand-Bellomo (1988) reported a further sex chromosome system in a population of M. triton from Burundi, where both males and females are XO, mimicking the situation in E. lutescens and T. osimensis (see §1). Consequently, there are at least two sex determining systems in the African pygmy mice: the standard XX/XY and the one described in the present study, and maybe two more if the extraordinary systems of M. minutoides from Caledon NR and M. triton, respectively X*Y/XY and XO/XO, are confirmed. Whereas modification of sex determination is extremely rare in mammals, and is usually associated with a significant decrease in fitness, our study confirms that the African pygmy mice have an unprecedented predisposition to accumulate chromosomal rearrangements involving sex chromosomes. This makes them an excellent model for investigating the evolution of mammalian sex determination and the evolutionary modification of sex chromosomes (Veyrunes et al. 2007).
4. Conclusions
Notwithstanding the accumulation of karyotypic data and the considerable advances that have been made in genomic/cytogenetic methods, the present study reports, to our knowledge, the first instance of a new atypical sex determination system recorded in a therian mammal in almost 30 years. Mus minutoides is one of a few species that escapes the constraints of the conventional mammalian XX/XY system. It shows a widespread occurrence and high prevalence (74%, n = 27) of fertile sex-reversed females. Atypical sex determination systems in mammals are poorly studied. The genes involved in the mutations, the mechanisms of expression and regulation of the genes, and/or organization of the genome are completely unknown, owing in a large part to a lack of knowledge on the complexity of the standard sex determination pathway in mammals itself. For example, still remarkably little is known about the mode of action and identity of the downstream genes involved in sex determination (see reviews by Brennan & Capel 2004; Sekido & Lovell-Badge 2009). However, in return, the study of the atypical sex determination systems discussed herein, and evidenced by the pygmy mice, may provide valuable clues to better understanding standard mammalian sex determination. In effect, most of the major advances in the field (including the identification of SRY) have come from the analysis of variant sex determination systems and pathological sex reversal cases in humans and mice (McElreavey & Fellous 1999; Vaiman & Pailhoux 2000; Camerino et al. 2006). As a result, the finding of an unusual sex determining system in a close phylogenetic relative of the house mouse, an index species for biomedical research, raises the possibility that comparative genomic approaches may provide novel insights into sex determination in mammals in general, and the identification of candidate genes involved in the pathological reversal of sex in humans in particular.
Acknowledgements
The specimens were collected under permit AAA004-00295-0035.
We are grateful to N. Avenant, F. Catzeflis, C. Gilbert, B. & D. Hayter, J. A. J. Nel and C. Newberry for assistance with various aspects of this investigation. This study was supported by CNRS-UM II grants to UMR 5554, the Plateforme Cytogénomique IFR 119, an Ellerman Fellowship from the University of Stellenbosch to J.B.D. and three collaborative grants: CNRS-NRF (nos. 13293 and 15439), PICS (no. 3196) and GDRI (no. 191). This is publication ISEM no. 2009-117.
References
- Arakawa Y., Nishida-Umehara C., Matsuda Y., Sutou S., Suzuki H.2002X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet. Genome Res. 99, 303–309 (doi:10.1159/000071608) [DOI] [PubMed] [Google Scholar]
- Bardoni B., et al. 1994A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat. Genet. 7, 497–501 (doi:10.1038/ng0894-497) [DOI] [PubMed] [Google Scholar]
- Bianchi N. O.2002Akodon sex reversed females: the never ending story. Cytogenet. Genome Res. 96, 60–65 (doi:10.1159/000063029) [DOI] [PubMed] [Google Scholar]
- Brennan J., Capel B.2004One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat. Rev. Genet. 5, 509–521 (doi:10.1038/nrg1381) [DOI] [PubMed] [Google Scholar]
- Bull J. J.1983Evolution of sex determining mechanisms San Francisco, CA: Benjamin/Cummings [Google Scholar]
- Bullejos M., Koopman P.2001Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn. 221, 201–205 (doi:10.1002/dvdy.1134) [DOI] [PubMed] [Google Scholar]
- Buzan E. V., Krystufek B., Hanfling B., Hutchinson W. F.2008Mitochondrial phylogeny of Arvicolinae using comprehensive taxonomic sampling yields new insights. Biol. J. Linn. Soc. 94, 825–835 (doi:10.1111/j.1095-8312.2008.01024.x) [Google Scholar]
- Camerino G., Parma P., Radi O., Valentini S.2006Sex determination and sex reversal. Curr. Opin. Genet. Dev. 16, 289–292 (doi:10.1016/j.gde.2006.04.014) [DOI] [PubMed] [Google Scholar]
- Castiglia R., Gormung E., Corti M.2002Cytogenetic analyses of chromosomal rearrangements in Mus minutoides/musculoides from North-West Zambia through mapping of the telomeric sequence (TTAGGG)n and banding techniques. Chromosome Res. 10, 399–406 (doi:10.1023/A:1016853719616) [DOI] [PubMed] [Google Scholar]
- Espinosa M., Vitullo A.2001Fast-developing preimplantation embryo progeny from heterogametic females in mammals. Zygote 9, 289–292 [DOI] [PubMed] [Google Scholar]
- Ezaz T., Stiglec R., Veyrunes F., Marshall Graves J. A.2006Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 16, 736–743 [DOI] [PubMed] [Google Scholar]
- Fichet-Calvet E., Lecompte E., Veyrunes F., Barrière P., Nicolas V., Koulemou K.2009Diversity and dynamics in a small mammal community in coastal Guinea, West Africa. Belg. J. Zool. 139, 93–102 [Google Scholar]
- Fredga K.1983Aberrant sex chromosomes mechanisms in mammals. Differentiation 23, 23–30 [DOI] [PubMed] [Google Scholar]
- Fredga K.1988Aberrant chromosomal sex-determining mechanisms in mammals, with special reference to species with XY-females. Phil. Trans. R. Soc. Lond. B 322, 83–94 (doi:10.1098/rstb.1988.0116) [DOI] [PubMed] [Google Scholar]
- Fredga K.1994Bizarre mammalian sex-determining mechanisms. In The differences between the sexes (eds Short R. V., Balaban E.), pp. 419–431 Cambridge, UK: Cambridge University Press [Google Scholar]
- Fredga K., Gropp A., Winking H., Frank F.1976Fertile XX- and XY-type females in the wood lemming Myopus schisticolor. Nature 261, 225–227 (doi:10.1038/261225a0) [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy M., Gautier C.1996SeaView and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12, 543–548 [DOI] [PubMed] [Google Scholar]
- Graves J. A. M., Gecz J., Hameister H.2002Evolution of the human X: a smart and sexy chromosome that controls speciation and development. Cytogenet. Genome Res. 99, 141–145 (doi:10.1159/000071585) [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O.2003A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- Hacker A., Capel B., Goodfellow P., Lovell-Badge R.1995Expression of Sry, the mouse sex determining gene. Development 121, 1603–1614 [DOI] [PubMed] [Google Scholar]
- Hoekstra H. E., Edwards S. V.2000Multiple origins of XY female mice (genus Akodon): phylogenetic and chromosomal evidence. Proc. R. Soc. Lond. B 267, 1825–1831 (doi:10.1098/rspb.2000.1217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jotterand M.1972Le polymorphisme chromosomique des Mus (Leggadas) africains. Cytogénétique, zoogéographie, évolution. Rev. Suisse Zool. 79, 287–359 [PubMed] [Google Scholar]
- Jotterand-Bellomo M.1988Chromosome analysis of five specimens of Mus bufo-triton (Muridae) from Burundi (Africa): three cytogenetic entities, a special type of chromosomal sex determination, taxonomy, and phylogeny. Cytogenet. Cell Genet. 48, 88–91 (doi:10.1159/000132596) [Google Scholar]
- Just W., Rau W., Vogel W., Akhverdian M., Fredga K., Graves J. A. M., Lyapunova E.1995Absence of Sry in species of the vole Ellobius. Nat. Genet. 11, 117–118 (doi:10.1038/ng1095-117) [DOI] [PubMed] [Google Scholar]
- Just W., Baumstark A., Süb A., Graphodatsky A., Rens W., Schäfer N., Bakloushinskaya I., Hameister H., Vogel W.2007Ellobius lutescens: sex determination and sex chromosome. Sex. Dev. 1, 211–221 (doi:10.1159/000104771) [DOI] [PubMed] [Google Scholar]
- Lee M. R., Elder F. F. B.1980Yeast stimulation of bone marrow mitosis for cytogenetic investigations. Cytogenet. Cell Genet. 26, 36–40 (doi:10.1159/000131419) [DOI] [PubMed] [Google Scholar]
- Liu W. S., Eriksson L., Fredga K.1998XY sex reversal in the wood lemming is associated with deletion of Xp21–23 as revealed by chromosome microdissection and fluorescence in situ hybridization. Chromosome Res. 6, 379–383 (doi:10.1023/A:1009273205788) [DOI] [PubMed] [Google Scholar]
- Lundrigan B. L., Tucker P. K.1994Tracing paternal ancestry in mice, using the Y-linked, sex-determining locus, Sry. Mol. Biol. Evol. 11, 483–492 [DOI] [PubMed] [Google Scholar]
- Lundrigan B. L., Jansa S. A., Tucker P. K.2002Phylogenetic relationships in the genus Mus, based on paternally, maternally, and biparentally inherited characters. Syst. Biol. 51, 410–431 (doi:10.1080/10635150290069878) [DOI] [PubMed] [Google Scholar]
- Marin I., Baker B. S.1998The evolutionary dynamics of sex determination. Science 281, 1990–1994 [DOI] [PubMed] [Google Scholar]
- Matthey R.1953La formule chromosomique et le problème de la détermination sexuelle chez Ellobius lutescens (Rodentia-Muridae-Microtinae). Arch. Julius Klaus-Stift Vererb. Forsch 28, 65–73 [Google Scholar]
- Matthey R.1967Cytogénétique des Leggada: (1) la formule chromosomique de Mus (Leggada) bufo, (2) Nouvelles données sur la délétion portant sur le bras court d'un X chez Mus (Leggada) triton. Experientia 23, 133–134 [DOI] [PubMed] [Google Scholar]
- Matthey R.1970Nouvelles données sur la cytogénétique et la spéciation des Leggada (Mammalia—Rodentia—Muridae). Experientia 26, 102–103 (doi:10.1007/BF01900420) [DOI] [PubMed] [Google Scholar]
- McElreavey K., Fellous M.1999Sex determination and the Y chromosome. Am. J. Med. Genet. 89, 176–185 (doi:10.1002/(SICI)1096-8628(19991229)89:4<176::AID-AJMG2>3.0.CO;2-B) [DOI] [PubMed] [Google Scholar]
- Monadjem A.1999Population dynamics of Mus minutoides and Steatomys pratensis (Muridae: Rodentia) in a subtropical grassland in Swaziland. Afr. J. Ecol. 37, 202–210 (doi:10.1046/j.1365-2028.1999.00169.x) [Google Scholar]
- Musser G. G., Carleton M. D.2005Superfamily Muroidea. In Mammal species of the world, vol. 2 (eds Wilson D. E., Reeder D. M.), pp. 894–1531 Baltimore, MD: John Hopkins University Press [Google Scholar]
- Nagamine C. M.1994The testis-determining gene, SRY, exists in multiple copies in Old World rodents. Genet. Res. 64, 151–159 (doi:10.1017/S001667230003281X) [DOI] [PubMed] [Google Scholar]
- Ohno S., Jainchill J., Stenius C.1963The creeping vole (Microtus oregoni) as a gonosomic mosaic. I. The OY/XY constitution of the male. Cytogenetics 2, 232–239 (doi:10.1159/000129781) [DOI] [PubMed] [Google Scholar]
- Ohno S., Stenius C., Christian L.1966The XO as the normal female of the creeping vole (Microtus oregoni). In Chromosome today, vol. 1 (eds Darlington C. D., Lewis K. R.), pp. 182–187 Edingburgh, UK: Oliver and Boyd [Google Scholar]
- O'Neill M. J., O'Neill R. J. W.1999Whatever happened to SRY? Cell. Mol. Life Sci. 56, 883–893 (doi:10.1007/s000180050481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. I., Pinna-Senn E., Dalmasso G., Lisanti J. A.2009Chromosomal aspects and inheritance of the XY female condition in Akodon azarae (Rodentia, Sigmodontinae). Mamm. Biol. 74, 125–129 (doi:10.1016/j.mambio.2008.03.001) [Google Scholar]
- Posada D., Crandall K. A.1998ModelTest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- Robinson T. J., Fu B., Ferguson-Smith M. A., Yang F.2004Cross-species chromosome painting in the golden mole and elephant-shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc. R. Soc. Lond. B 271, 1477–1484 (doi:10.1098/rspb.2004.2754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone G., Pane A., Polito L. C.2002Sex determination in flies, fruitflies and butterflies. Genetica 116, 15–23 (doi:10.1023/A:1020903523907) [DOI] [PubMed] [Google Scholar]
- Saifi G. M., Chandra H. S.1999An apparent excess of sex- and reproduction-related genes on the human X chromosome. Proc. R. Soc. Lond. B 266, 203–209 (doi:10.1098/rspb.1999.0623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright J.1971A rapid technique for human chromosomes. Lancet II, 971–972 [DOI] [PubMed] [Google Scholar]
- Sekido R., Lovell-Badge R.2009Sex determination and SRY: down to a wink and a nudge? Trends Genet. 25, 19–29 (doi:10.1016/j.tig.2008.10.008) [DOI] [PubMed] [Google Scholar]
- Sinclair A. H., et al. 1990A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244 (doi:10.1038/346240a0) [DOI] [PubMed] [Google Scholar]
- Soullier S., Hanni C., Catzeflis F., Berta P., Laudet V.1998Male sex determination in the spiny rat Tokudaia osimensis (Rodentia: Muridae) is not Sry dependent. Mamm. Genome 9, 590–592 (doi:10.1007/s003359900823) [DOI] [PubMed] [Google Scholar]
- Sutou S., Mitsui Y., Tsuchiya K.2001Sex determination without the Y chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm. Genome 12, 17–21 (doi:10.1007/s003350010228) [DOI] [PubMed] [Google Scholar]
- Uller T., Pen I., Wapstra E., Beukeboom L., Komdeur J.2007The evolution of sex ratios and sex determining systems. Trends Ecol. Evol. 22, 292–297 (doi:10.1016/j.tree.2007.03.008) [DOI] [PubMed] [Google Scholar]
- Vaiman D., Pailhoux E.2000Mammalian sex reversal and intersexuality: deciphering the sex-determination cascade. Trends Genet. 16, 488–494 (doi:10.1016/S0168-9525(00)02126-0) [DOI] [PubMed] [Google Scholar]
- Veyrunes F., Catalan J., Sicard B., Robinson T. J., Duplantier J. M., Granjon L., Dobigny G., Britton-Davidian J.2004Autosome and sex chromosome diversity among the African pygmy mice, subgenus Nannomys (Muridae; Mus). Chromosome Res. 12, 369–382 (doi:10.1023/B:CHRO.0000034098.09885.e6) [DOI] [PubMed] [Google Scholar]
- Veyrunes F., Britton-Davidian J., Robinson T. J., Calvet E., Denys C., Chevret P.2005Molecular phylogeny of the African pygmy mice, subgenus Nannomys (Rodentia, Murinae, Mus): implications for chromosomal evolution. Mol. Phylogenet. Evol. 36, 358–369 (doi:10.1016/j.ympev.2005.02.011) [DOI] [PubMed] [Google Scholar]
- Veyrunes F., Dobigny G., Yang F., O'Brien P. C. M., Catalan J., Robinson T. J., Britton-Davidian J.2006Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse. Proc. R. Soc. B 273, 2925–2934 (doi:10.1098/rspb.2006.3670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrunes F., Watson J., Robinson T. J., Britton-Davidian J.2007Accumulation of rare sex chromosome rearrangements in the African pygmy mouse, Mus (Nannomys) minutoides: a whole-arm reciprocal translocation (WART) involving an X-autosome fusion. Chromosome Res. 15, 223–230 (doi:10.1007/s10577-006-1116-8) [DOI] [PubMed] [Google Scholar]
- Veyrunes F., et al. 2008Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18, 965–973 (doi:10.1101/gr.7101908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff J. N., Schartl M.2001Variability of genetic sex determination in poeciliid fishes. Genetica 111, 101–110 (doi:10.1023/A:1013795415808) [DOI] [PubMed] [Google Scholar]
- Vuilleumier S., Lande R., Van Alphens J., Seehausen O.2007Invasion and fixation of sex-reversal genes. J. Evol. Biol. 20, 913–920 (doi:10.1111/j.1420-9101.2007.01311.x) [DOI] [PubMed] [Google Scholar]
- Wang P. J., McCarrey J. R., Yang F., Page D. C.2001An abundance of X-linked genes expressed in spermatogonia. Nature Genet. 27, 422–426 (doi:10.1038/86927) [DOI] [PubMed] [Google Scholar]
- Werren J. H., Beukeboom L. W.1998Sex determination, sex ratios, and genetic conflict. Annu. Rev. Ecol. Syst. 29, 233–261 (doi:10.1146/annurev.ecolsys.29.1.233) [Google Scholar]
- Willan K., Meester J.1978Breeding biology and postnatal development of the African dwarf mouse. Acta Theriol. 23, 55–73 [Google Scholar]