Abstract
An unexpected outbreak of boll weevils, Anthonomus grandis, an insect pest of cotton, across the Southern Rolling Plains (SRP) eradication zone of west-central Texas, USA, was detected soon after passage of Tropical Storm Erin through the Winter Garden district to the south on 16 August 2007. The synchrony and broad geographic distribution of the captured weevils suggest that long-distance dispersal was responsible for the reinvasion. We integrated three types of assessment to reconstruct the geographic origin of the immigrants: (i) DNA fingerprinting; (ii) pollen fingerprinting; and (iii) atmospheric trajectory analysis. We hypothesized the boll weevils originated in the Southern Blacklands zone near Cameron, or in the Winter Garden district near Uvalde, the nearest regions with substantial populations. Genetic tests broadly agree that the immigrants originated southeast of the SRP zone, probably in regions represented by Uvalde or Weslaco. The SRP pollen profile from weevils matched that of Uvalde better than that of Cameron. Wind trajectories supported daily wind-aided dispersal of weevils from the Uvalde region to the SRP from 17 to 24 August, but failed to support migration from the Cameron region. Taken together the forensic evidence strongly implicates the Winter Garden district near Uvalde as the source of reinvading boll weevils.
Keywords: boll weevil, cotton, invasive, population genetics, pollen, atmospheric trajectory
1. Introduction
Successful colonization of non-native habitat by an invasive species can significantly disrupt ecological equilibria within the recipient ecosystem (Suarez & Tsutsui 2008; Kenis et al. 2009). An important sub-type of invasion is that of an insect herbivore into agroecosystems harbouring an abundance of vulnerable, economically important host plants arrayed as monoculture crops (Pimentel et al. 2005). The boll weevil, Anthonomus grandis Boheman (Coleoptera: Curculionidae), is an invasive and ruinous pest of cotton that began a rapid northward expansion out of its native range in southern Mexico in the nineteenth century (Burke et al. 1986). It entered Texas from Mexico in 1892 and within three decades had successfully invaded the southeastern USA, leaving economic devastation and social chaos in its wake (Haney 2001). Although it essentially completed its invasion of the USA almost nine decades ago (Culin et al. 1990), its invasive potential is once again of immediate and widespread societal concern. At huge public expense, a large-scale eradication programme has eliminated the boll weevil from much of the USA over the last 30 years (Smith 1998; Carter et al. 2001; El-Lissy & Grefenstette 2006), but substantial populations remain in eastern and southern Texas and northern Mexico. Throughout cotton growing areas that have been cleared of the boll weevil, extensive arrays of pheromone traps are deployed to monitor for reintroductions, and various mitigation protocols may be implemented to eliminate colonizing populations depending on the context and presumed origin of the migrants.
Although the geographic range of the boll weevil currently is contracting through eradication efforts, reinvasion of weevil-free areas through long-range dispersal of migrants originating in the remaining reservoirs of infested habitat is a constant concern (Allen et al. 2008; Kiser & Catanach 2008). During a range expansion, the rate of geographic spread of an invading organism is partly a function of short-range dispersal into adjacent habitat and population growth along the leading edge of an invasion front. However, long-distance dispersal and colonization ahead of an invasion front act to accelerate range expansion (Shigesada et al. 1995). The boll weevil exhibits this pattern of both short- and long-distance movement, called stratified dispersal, which enhances invasion potential (Liebhold & Tobin 2008). Most boll weevils during most of the year move relatively short distances, of the order of about 15 km or less (Johnson et al. 1975, 1976; Rummel et al. 1977; Raulston et al. 1996). However, trapping studies and estimates of gene flow indicate that natural dispersal of boll weevils over hundreds of kilometres is not uncommon (Guerra 1988; Spurgeon et al. 1997; Kim & Sappington 2004b,c, 2006; Kim et al. 2006).
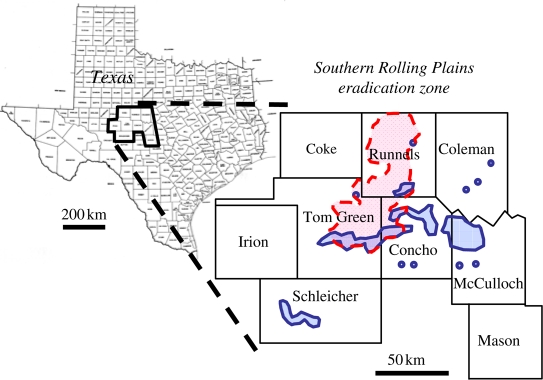
In late August and early September 2007, more than 150 boll weevil adults were unexpectedly captured in traps across an extensive area of the Southern Rolling Plains (SRP) eradication zone of west-central Texas (figure 1), which had been essentially weevil-free since 2003. Thirteen weevils were captured in traps at 12 different fields during the week ending 1 September, 30 weevils at 26 different fields during the week ending 8 September, and 109 weevils at 82 different fields during the week ending 15 September. By the end of 2007, more than 6000 boll weevils had been captured in this zone triggering treatment of 158 000 ha with malathion to combat the reinvasion (Allen et al. 2008), resulting in more than $1.4 million in direct insecticide application costs alone.
Figure 1.
Geographic pattern of boll weevil captures in Texas counties comprising the SRP eradication zone, September 2007. Major concentrations of boll weevil captures are outlined in blue, with disjunct captures indicated with blue circles. Main cotton-growing area in the SRP is outlined with dashed red line.
This outbreak was detected about two weeks after the passage of Tropical Storm (TS) Erin through the still-infested Winter Garden district about 180 km south of the SRP on the night of 16 August. The Winter Garden district is located southwest of San Antonio in the western half of the South Texas/Winter Garden eradication zone, and is characterized by high-input irrigated agriculture including about 4000 ha of cotton (Cleveland et al. 2006). It is comprised of Zavala, Frio, Dimmitt and LaSalle counties with parts of Uvalde and Medina counties forming the northern boundary. The relative synchrony and broad geographic distribution of large numbers of captured weevils around the southeastern perimeter of the SRP (figure 1) suggest that long-distance dispersal was responsible for the reinvasion. Long-distance dispersal by many insects, including boll weevils, is wind-aided (Westbrook et al. 2007). Hurricanes and tropical storms seem to have accelerated the initial range expansion of the boll weevil through the southern USA (Hinds 1916; Culin et al. 1990), and it is likely that winds generated by TS Erin were responsible for the transport of large numbers of boll weevils into the SRP. However, the meteorological effects of the storm were geographically widespread, so the source of the insects transported by its winds is not obvious.
Determining the origin of immigrant insects in general is challenging, but several methods are applicable to boll weevils. In late summer and early autumn, there tend to be more boll weevils flying high above the canopy (Glick 1939; Taft & Jernigan 1964; Rummel et al. 1977) where the effect of wind on dispersal is greater than near the surface. Atmospheric dispersion models can help in determining whether wind events were a likely transport mechanism, and if so, to reconstruct the incoming trajectory of immigrants (Westbrook et al. 2007). Genetic population assignment analyses based on neutral microsatellite DNA markers have provided important clues to the origin of boll weevils captured unexpectedly in or near eradication zones (Kim et al. 2006, 2008). Finally, the profile of pollen species in the gut or on the surface of insects can serve as another natural marker to help identify origins of dispersed insects based on plant species distributions (Jones & Jones 2001). Boll weevil adults readily pick up pollen grains when feeding (Jones & Coppedge 1999).
However, any of these methods alone are seldom sensitive enough to assign immigrants to their probable source region at the desired levels of geographic resolution and confidence. In this study we integrated all three types of assessment to reconstruct the most likely geographic origin of the immigrant boll weevils captured in the SRP in autumn 2007. In particular, we hypothesized that they originated in the Southern Blacklands zone near Cameron, or in the Winter Garden district near Uvalde, the nearest regions still harbouring substantial populations of boll weevils.
2. Material and methods
2.1. Collections
As with all eradication zones from which boll weevil populations have been eliminated, the SRP zone is monitored with traps (Smith 1998; Allen et al. 2008) baited with a synthetic aggregation pheromone that attracts both sexes (Tumlinson et al. 1971). The traps also contained dichlorvos insecticide strips to kill any weevils that entered the trap and reduce the chance for escape (Suh et al. 2003; Armstrong & Greenberg 2008). The SRP traps were checked once per week by Texas Boll Weevil Eradication Foundation (TBWEF) personnel. The 20 boll weevils from the SRP used in the genetics portion of this study were collected on 4 (n = 1), 10 (n = 17), 17 (n = 1) or 24 (n = 1) September, and were shipped to the USDA-Agricultural Research Service, Corn Insects and Crop Genetics Research Unit in Ames, IA (figure 1). All 16 boll weevils from the SRP used for pollen analysis were collected on 10 September. These were not the same individuals used for genetic analyses, but were captured in the same area, primarily from Concho County with a few specimens from neighbouring counties. The weevils were arbitrarily allocated for genetic or pollen analysis by sorting them numerically according to TBWEF work unit followed by alternate assignment. Additional boll weevils for both genetic and pollen analyses were collected near Uvalde and Cameron, TX, in late September 2007, areas targeted a priori as candidate source locations of the SRP immigrants because they represented the nearest areas harbouring substantial populations.
2.2. Genetic fingerprinting
Genomic DNA was extracted from individuals using the BioRad (Hercules, CA, USA) Aqua Pure kit according to the manufacturer's instructions. Ten microsatellite loci (AG-D1 to AG-D7 and AG-10 to AG-D12) were amplified by polymerase chain reaction in two multiplex reactions, and genotyped using a Beckman-Coulter CEQ 8000 Genetic Analysis System, following methods in Kim & Sappington (2004a).
Individual multilocus genotypes of the SRP weevils were screened in population assignment and exclusion tests against the new profiles from Uvalde and Cameron, as well as against profiles from a database of 18 additional USA and four northern Mexico populations (figure 2), where sample sizes generally ranged from 30 to 50 (Kim & Sappington 2006; Kim et al. 2006, 2008). Individual assignment likelihood values were calculated using both Bayesian and frequency methods (Paetkau et al. 1995; Rannala & Mountain 1997; Cornuet et al. 1999). Based on similarities and differences in their microsatellite profiles, each SRP weevil was assigned a per cent likelihood of having originated in any of the 24 reference (potential source) populations at a threshold of α = 0.05 using the program GeneClass2 (Piry et al. 2004). In the exclusion tests (Cornuet et al. 1999), which also employed the Bayesian statistical approach of Rannala & Mountain (1997), the resampling method of Paetkau et al. (2004) was used to determine the distribution of multilocus genotypes in each source population based on Monte Carlo simulations of 1000 independent individuals from that population. Only those populations where p > 0.01 are considered statistically possible sources of the SRP weevils. Each SRP weevil also was screened against the other SRP weevils to determine how similar the subject weevils are to one another.
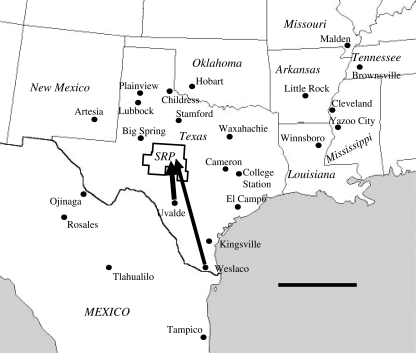
Figure 2.
Locations of potential boll weevil source populations whose microsatellite genotype profiles were compared with those of immigrants to the SRP eradication zone. Uvalde and Weslaco, TX, in order of likelihood, represent the most likely origins of reinvading boll weevils based on population assignment, population exclusion and population differentiation evidence. Scale bar, 400 km.
Further analyses were conducted with the SRP individuals pooled and considered a single population, under the assumption that they originated from the same source region as part of a single dispersal event. Thus, mean assignment likelihoods and exclusion tests were determined for SRP boll weevils as a group. Pairwise FST values, a measure of genetic differentiation (Weir & Cockerham 1984), were calculated using FSTAT v. 2.9.3 (Goudet 1995) between pooled SRP weevils and each potential source population.
2.3. Pollen fingerprinting
Sixteen boll weevils captured in the SRP during the week ending 10 September 2007 were processed for pollen analysis. Although pollen profiles were characterized previously from boll weevils collected in the Uvalde area (Jones & Coppedge 1999), the insects in that study were collected in April through July, and the profiles changed monthly as the temporal profile of blooming plant species changed. To obtain expected profiles for Uvalde and Cameron, five boll weevils were sampled from each of eight sites near these two municipalities in the last half of September 2007. Each set of five weevils was pooled for pollen extraction.
Pollen was recovered by acetolyzation of insect tissue (Cate & Skinner 1978) and stained (Jones & Coppedge 1999). Pollen grains were identified and counted via compound light microscopy to the lowest taxonomic level possible based on comparisons with pollen from the Areawide Pest Management Research Unit Pollen Reference Collections. Chenopodiaceae and Amaranthus (Amaranthaceae) pollens are morphologically too similar to distinguish and were pooled as ‘Cheno-Am’ (Martin 1963; Benedict et al. 1991; Hardee et al. 1999; Jones & Coppedge 1999).
2.4. Atmospheric trajectory
Wind trajectories were estimated using the Windows PC v. 4.8 of the HYSPLIT Transport and Dispersion model created by the National Oceanic and Atmospheric Administration/Air Resources Laboratory (NOAA/ARL; Draxler & Rolph 2003; Rolph 2003; Westbrook et al. 2007). Boll weevils are diurnal fliers and have been captured in aircraft tow-nets at altitudes ranging from 30 to 600 m (Glick 1939; Rummel et al. 1977). Therefore, trajectories were estimated every day from 13 August to 10 September 2007 for 6 h of transport at a representative altitude of 500 m above ground level. This range of dates encompasses the period immediately before the arrival of TS Erin until the date when an accumulation of more than 40 weevils had been captured in the SRP. The trajectories originated at 14:00 h Central Daylight Time (CDT) in Concho County in the SRP zone and extended backward in time to 08:00 h CDT on the same date. Weather information for calculating the trajectories was obtained from the (40 km resolution) EDAS atmospheric data files archived at the NOAA/ARL site.
3. Results
3.1. Genetic fingerprinting
Based on microsatellite genotype data, each of the 20 boll weevils captured in the SRP eradication zone was assigned a per cent likelihood of having originated in any of the 24 reference (potential source) populations (table 1). Uvalde was ranked as the most likely source for 10 of the 20 weevils. Five of the weevils were most likely from Weslaco, four were most likely from Kingsville, and one was most likely from Tampico. All of these locations are to the southeast of the SRP (figure 2). Uvalde and Weslaco are always ranked among the three most likely source populations for weevils captured in the SRP. Kingsville was usually, and Tampico occasionally, ranked among the four most likely source populations.
Table 1.
Summary of individual assignment and exclusion tests for 20 individual boll weevils captured in the Southern Rolling Plains (SRP) eradication zone, September 2007, based on genotypes from 10 microsatellite loci. (Each individual was assigned to the most likely of 24 possible source populations (per cent likelihood in parentheses; figure 2). Any population that could not be statistically excluded (i.e. p > 0.01) as a potential source of an SRP immigrant is listed (p-value in parentheses). Thus, not-excluded populations are those considered statistically possible sources, and the higher the value, the more probable it is the source. Values from screens of individuals against other SRP weevils are provided separately as an indication of similarity to one another. Other assignment and not-excluded values are from screenings without other SRP weevils included. Abbreviations: Uvl, Uvalde; Wes, Weslaco; Tam, Tampico; Kgv, Kingsville; ElC, El Campo; Stm, Stamford.)
| SRP not included |
||||||
|---|---|---|---|---|---|---|
| individual | methoda | SRP only | rank 1 | rank 2 | rank 3 | rank 4 |
| SRP1 | assignment | 27.4 | Uvl (67.9) | Wes (26.2) | Kgv (5.9) | — |
| not excluded | 0.545 | Uvl (0.623) | Wes (0.612) | Tam (0.186) | Kgv (0.140) | |
| SRP2 | assignment | 42 | Wes (76.9) | Uvl (20) | Kgv (3.1) | — |
| not excluded | 0.332 | Wes (0.445) | Uvl (0.183) | Tam (0.119) | Kgv (0.048) | |
| SRP3 | assignment | 32.9 | Uvl (82.6) | Wes (17.1) | Kgv (0.1) | Tam (0.1) |
| not excluded | 0.310 | Uvl (0.361) | Wes (0.331) | Tam (0.152) | Kgv (0.013) | |
| SRP4 | assignment | 0 | Wes (80.1) | Kgv (14.9) | Uvl (4.7) | Tam (0.1) |
| not excluded | 0.011 | Wes (0.470) | Tam (0.133) | Uvl (0.098) | Kgv (0.087) | |
| SRP5 | assignment | 37.1 | Kgv (56.1) | Wes (22.8) | Uvl (21.1) | — |
| not excluded | 0.653 | Uvl (0.468) | Wes (0.605) | Kgv (0.302) | Tam (0.102) | |
| SRP6 | assignment | 12.3 | Wes (56.2) | Kgv (39.8) | Uvl (4.0) | — |
| not excluded | 0.207 | Wes (0.486) | Uvl (0.106) | Kgv (0.149) | — | |
| SRP7 | assignment | 13.4 | Tam (59.2) | Wes (40.7) | Uvl (0.1) | — |
| not excluded | 0.193 | Tam (0.698) | Wes (0.367) | Uvl (0.015) | — | |
| SRP8 | assignment | 42.1 | Kgv (60.7) | Uvl (32.2) | Wes (7.1) | — |
| not excluded | 0.848 | Uvl (0.703) | Wes (0.601) | Kgv (0.405) | Tam (0.030) | |
| SRP9 | assignment | 29.7 | Uvl (80.9) | Wes (19.1) | — | — |
| not excluded | 0.280 | Uvl (0.343) | Wes (0.331) | Tam (0.049) | — | |
| SRP10 | assignment | 64.3 | Wes (51.4) | Kgv (30.7) | Uvl (17.9) | — |
| not excluded | 0.550 | Wes (0.489) | Uvl (0.234) | Kgv (0.140) | Tam (0.065) | |
| SRP11 | assignment | 8.6 | Kgv (50.6) | Wes (29.9) | Uvl (19.5) | — |
| not excluded | 0.184 | Wes (0.408) | Uvl (0.234) | Kgv (0.166) | — | |
| SRP13 | assignment | 93.8 | Uvl (54.1) | Wes (42.6) | Kgv (3.3) | — |
| not excluded | 0.902 | Wes (0.518) | Uvl (0.402) | Kgv (0.078) | Tam (0.022) | |
| SRP14 | assignment | 78.7 | Uvl (98.6) | Wes (0.9) | Tam (0.5) | — |
| not excluded | 0.308 | Uvl (0.146) | Tam (0.093) | Wes (0.047) | — | |
| SRP15 | assignment | 37.7 | Wes (44.0) | Uvl (39.2) | Tam (16.8) | — |
| not excluded | 0.042 | Tam (0.171) | Wes (0.085) | Uvl (0.035) | — | |
| SRP16 | assignment | 55.3 | Uvl (99.1) | Wes (0.7) | Kgv (0.2) | — |
| not excluded | 0.406 | Uvl (0.339) | Wes (0.079) | Tam (0.061) | Kgv (0.012) | |
| SRP18 | assignment | 34.3 | Uvl (71.6) | Wes (24.5) | Kgv (3.8) | — |
| not excluded | 0.448 | Uvl (0.471) | Wes (0.461) | Kgv (0.084) | Tam (0.013) | |
| SRP19 | assignment | 83 | Uvl (99.4) | Wes (0.6) | — | — |
| not excluded | 0.487 | Uvl (0.252) | Wes (0.059) | — | — | |
| SRP20 | assignment | 85.2 | Kgv (51.0) | Uvl (23.1) | Wes (22.4) | ElC (3.5) |
| not excluded | 0.598 | Uvl (0.184) | Wes (0.309) | Kgv (0.125) | — | |
| SRP21 | assignment | 98 | Uvl (50) | Wes (40) | Kgv (10) | — |
| not excluded | 0.358 | Wes (0.093) | Tam (0.068) | Uvl (0.047) | Kgv (0.014) | |
| SRP22 | assignment | 96.5 | Uvl (68.6) | Wes (14.3) | Kgv (14.3) | Stm (2.8) |
| not excluded | 0.086 | Wes (0.018) | Uvl (0.014) | — | — | |
aAssignment test was carried out using the direct approach without probability computation, and the exclusion test was carried out using a simulation method (Cornuet et al. 1999). Both tests employed the Bayesian statistical approach of Rannala & Mountain (1997). The simulation method of Paetkau et al. (2004) was used in the exclusion test.
Similarly, an exclusion probability for each SRP weevil was determined for each population (table 1). For ease of interpretation, we present those populations that cannot be statistically ruled out as potential sources, and these are indicated as ‘not excluded’. The results are generally consistent with those of the assignment tests, with Uvalde, Weslaco, Kingsville and Tampico the only locations that are included as potential source areas for the weevils captured in the SRP. All other populations, including Cameron, were always excluded as possible sources (p < 0.01). Only Uvalde and Weslaco were never excluded.
Each weevil was screened against the other weevils from the SRP to reveal how similar the subject weevils are to one another. The assignment and exclusion probability values for SRP were consistently high (table 1), with the lone exception of weevil SRP4, which appears genetically dissimilar to the other SRP weevils as a group. These data along with the unexpected and synchronous capture of these insects in the SRP, suggest that the influx of weevils was part of a single immigration event from the same source region. Under this assumption, we pooled the weevils from the SRP samples and analysed them as a group. The mean assignment likelihoods for SRP are presented for both the Bayesian and frequency methods (Cornuet et al. 1999) of calculation (table 2), which are in good agreement with one another. The data are on a negative log scale, so the lowest value indicates the most likely source population. By these criteria, Uvalde is the most likely source of SRP immigrants, although the Lower Rio Grande Valley represented by Weslaco, and the Lower Coastal Bend represented by Kingsville also have relatively low values.
Table 2.
Mean individual assignment likelihood (Li→j) calculated using Bayesian and frequency methods, and pairwise FST values for 20 boll weevils captured in the Southern Rolling Plains (SRP) eradication zone in September 2007 and 24 potential source populations. (The number of SRP individuals, of 20 tested, assigned to and not excluded from potential source areas are indicated by relative ranking. Based on genotypes from 10 microsatellite loci.)
| SRP population |
no. individuals assigned to population |
no. individuals not excluded |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mean likelihoodb |
differentiationc | rank |
rank |
||||||||
| potential source (reference) populationa | Bayesian | frequency | FST | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 |
| Cameron, TX | 16.24 | 13.77 | 0.319 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Uvalde, TX | 8.05 | 7.82 | 0.016ns | 10 | 4 | 6 | 0 | 10 | 6 | 4 | 0 |
| Big Spring, TX | 16.06 | 15.47 | 0.242 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Childress, TX | 15.77 | 13.99 | 0.278 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| College Station, TX | 14.36 | 13.78 | 0.247 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| El Campo, TX | 13.27 | 13 | 0.258 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Kingsville, TX | 9.27 | 8.97 | 0.079 | 4 | 3 | 8 | 0 | 0 | 0 | 8 | 6 |
| Lubbock, TX | 15.59 | 13.56 | 0.281 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Plainview, TX | 16.44 | 14.17 | 0.296 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Stamford, TX | 14.56 | 14.04 | 0.245 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Waxahachie, TX | 14.49 | 13.97 | 0.291 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Weslaco, TX | 8.25 | 8.19 | 0.034ns | 5 | 13 | 2 | 0 | 8 | 11 | 1 | 0 |
| Little Rock, AR | 19.12 | 16.44 | 0.384 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Winnsboro, LA | 19.07 | 16.28 | 0.386 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cleveland, MS | 19.14 | 16.01 | 0.415 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Yazoo City, MS | 19.87 | 16.61 | 0.401 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Malden, MO | 17.74 | 15.08 | 0.329 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Artesia, NM | 17.99 | 15.59 | 0.311 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hobart, OK | 17.49 | 15.40 | 0.299 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Brownsville, TN | 19.59 | 16.44 | 0.403 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rosales, Mexico | 16.98 | 14.40 | 0.263 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ojinaga, Mexico | 22.71 | 17.61 | 0.357 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tampico, Mexico | 11.67 | 11.69 | 0.156 | 1 | 0 | 2 | 2 | 2 | 3 | 5 | 5 |
| Tlahualilo, Mexico | 17.65 | 15.22 | 0.278 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
aSample locations are indicated in figure 2.
bMean likelihood values are expressed on a negative log scale.
cns, not significantly greater than zero (α = 0.05); all other values are significant.
Pairwise FST values were calculated between pooled SRP weevils and each potential source population (table 2). The higher the FST value, the more genetically distinct the two populations are from one another. Low and non-significant FST values indicate that the two populations are genetically indistinguishable. There was no significant genetic differentiation between SRP and Uvalde, or between SRP and Weslaco, and these FST values were the lowest of any comparison. The Kingsville FST value is also rather low, but is significant. Pairwise comparisons with all other populations indicate moderate to high FST values, suggesting that they are all unlikely sources of the SRP immigrants.
Together, results from the variety of population genetics tests applied in this study are in fundamental and broad agreement that the boll weevils captured in the SRP in autumn of 2007 most likely originated in the Uvalde area, with the Lower Rio Grande Valley around Weslaco coming in a close second as a likely source (figure 2). Kingsville is another possibility, but not as likely. The geographic positions of these three leading candidates suggest movement of weevils into the SRP from the south or southeast. The hypothesis that the Uvalde area is the source of boll weevil immigrants into the SRP, posited on its proximity to the SRP and the presence of still substantial infestation levels, is therefore supported by the genetic evidence. There is no genetic evidence in support of Cameron as the source of the SRP boll weevils. Other populations to the east, as well as those to the southwest, west and north are very unlikely sources of the SRP reintroduction event as well.
3.2. Pollen fingerprinting
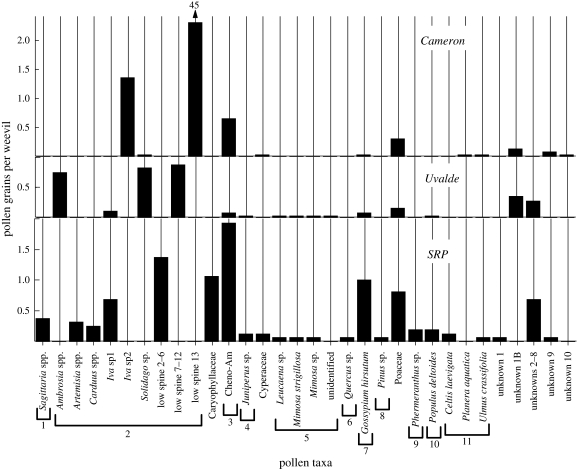
Overall, 158 pollen grains representing at least 33 species from 15 families were associated with the 16 boll weevils examined from the SRP (figure 3). The pollen fingerprint of the SRP weevils included 14 taxa not recovered in the Cameron and Uvalde samples, notably arrowhead (Sagittaria spp.), sagebrush (Artemisia spp.), plumeless thistle (Carduus spp.), five unidentified low-spine species of Asteraceae, pinks (Caryophyllaceae), flameflower (Phemeranthus spp., Sy = Talinum) and sugarberry (Celtis laevigata C. von Willdenow).
Figure 3.
Pollen fingerprints from the guts and surface of boll weevil adults captured in the SRP eradication zone, compared with those of boll weevils collected near Cameron and Uvalde, TX, September 2007. Families of taxa not already indicated: 1, Alismataceae; 2, Asteraceae; 3, Chenopodiaceae+Amaranthus; 4, Cupressaceae; 5, Fabaceae; 6, Fagaceae; 7, Malvaceae; 8, Pinaceae; 9, Portulacaceae; 10, Salicaceae; 11, Ulmaceae.
The diversity of pollen taxa associated with boll weevils collected in Uvalde and Cameron was not as great as that of boll weevils collected from the SRP (figure 3). The Uvalde profile was characterized by greater species diversity than that of Cameron, each with 26 and 12 taxa, respectively. The pollen fingerprint of Uvalde boll weevils included nine taxa not shared with SRP or Cameron including ragweed (Ambrosia spp.), six unidentified species of low-spine Asteraceae, a species of Fabaceae and a species from an unidentified family. The pollen fingerprint of boll weevils captured near Cameron was dominated by an abundant low-spine of Asteraceae (low spine 13) present in samples from all sites. Its complete absence in SRP samples (figure 3) effectively rules out Cameron as the source. Although the Uvalde pollen profile did not perfectly match that of SRP weevils, 48.5 per cent of SRP pollen taxa were shared with Uvalde versus only 18.2 per cent shared with Cameron. If the weevils picked up pollen from the SRP after immigration, it is expected that they might have pollen taxa in their profile not present in Uvalde or Cameron, so the lack of a perfect match of profiles does not in itself rule out Uvalde as a potential source.
3.3. Atmospheric trajectory
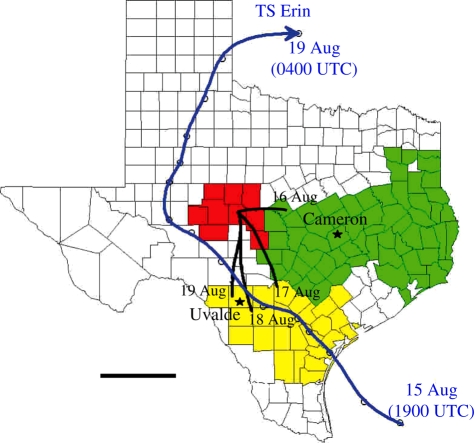
TS Erin made landfall near Lamar, TX, on the morning of 16 August 2007. Its centreline trajectory passed through the Winter Garden district on the night of 16 August, and continued west and north around the western perimeter of the SRP zone, entering southwestern Oklahoma on 19 August (figure 4). Copious rainfall was associated in the northeastern sector of Erin as it moved across central Texas, Oklahoma and southern Missouri, and was primarily limited to the eastern side of Erin's centreline trajectory.
Figure 4.
Atmospheric trajectory analysis of potential boll weevil transport (black lines) into Concho County in the SRP eradication zone (red) from the Winter Garden district of the South Texas/Winter Garden zone (yellow), and from the Southern Blacklands eradication zone (green). Path of Tropical Storm (TS) Erin through Texas in August 2007 is indicated with blue arrow and labelled in Universal Coordinated Time (UTC). Scale bar, 200 km.
Wind trajectories calculated using the HYSPLIT model (Draxler & Rolph 2003; Rolph 2003) for the 3 days before the passage of TS Erin showed no significant transport potential into the SRP. On 16 August there was a potential for transport from about 125 km east near Star in Mills County in the Southern Blacklands eradication zone (figure 4). After 16 August, counterclockwise atmospheric circulation associated with TS Erin generated strong southerly winds between the Winter Garden district near Uvalde and the SRP zone. Calculated trajectories supported wind-aided dispersal of weevils from the Uvalde region to the SRP daily from 16 to 24 August, but provided no support for migration of boll weevils from areas near Cameron in the Southern Blacklands zone through mid-September.
4. Discussion
All three methods used in this study have inherent strengths and weaknesses. Both the genetic and pollen fingerprinting approaches are based on natural marking of individual insects suspected of long-distance movement, and both rely on matching profiles of marks associated with the immigrants to profiles characteristic of potential source areas. The nature of these marks and of their information content is very different, however. The information contained in pollen fingerprints reflects the profile of blooming plants in the landscape inhabited by the boll weevil. Cameron is in the Blackland Prairie Vegetational Zone, while Uvalde is in the South Texas Plains Vegetational Zone (Correll & Johnston 1979; Diggs et al. 1999), each characterized by its own unique blend of plant species. However, pollen profiles are temporally dynamic, not only for the source landscape where the array of blooming plants changes continuously through the year, but also for the individuals themselves (Jones 1997). The marking of a boll weevil with pollen is not a one-time discrete event. Boll weevil adults commonly ingest pollen, but most, if not all, of the grains are cleared from the gut within 24 h (Cate & Skinner 1978). Thus, if there is a lag of more than a day between a boll weevil immigration event and capture in a trap, pollen recovered from the gut will reflect only the plant species profile of its new surroundings. The SRP boll weevils were captured in the Rolling Plains Vegetational Zone and would be expected to carry pollen typical of this area if they foraged locally before being trapped. However, pollen deposited on the external surface of the insect is expected to have a longer potential residence time, and thus the surface profile will reflect a composite of regions in which the weevil has been foraging, including the source area.
Although these factors make the pollen fingerprint of a weevil difficult to interpret, the amount of information contained is potentially great, and is enhanced by abundance data. For example, the absence of the low spine 13 pollen taxon from the SRP profile would not be enough by itself to discount Cameron as a possible source region. Indeed, there are three other taxa present in the Cameron profiles that are missing in the SRP profiles, including marsh elder (Iva sp2), water elm (Planera aquatica) and an unknown, but their absence in SRP could simply be owing to rarity in the source region. However, the absence of low spine 13 among SRP weevils is important in disqualifying Cameron as the source because of its overwhelming abundance in the latter location, averaging 45 grains per individual (figure 3).
Although it did not happen in this study, the presence of pollen from a taxon of restricted geographic range in the profile of an immigrant can be diagnostic (Mikkola 1971; Hendrix et al. 1987; Lingren et al. 1994; Jones & Jones 2001). For example, the presence of pollen from Chloroleucon spp. (SY = Pithecellobium spp.) and Calliandra spp. (both Fabaceae) on the surface of black cutworm (Agrotis ipsilon Hufnagel) moths captured in the spring in Iowa demonstrated that these individuals originated at least 1800 km to the south in the Lower Rio Grande Valley of Texas or northeastern Mexico, the northernmost limit of these species' ranges (Hendrix & Showers 1992).
Unlike pollen profiles, the genetic profile of an individual is unchanging, and the profile of selectively neutral genetic markers for a resident population is relatively stable over several generations if the population size is large compared with numbers of immigrants. This relative stability makes sophisticated population genetics analyses possible, such as population assignment and exclusion tests. On the other hand, the sensitivity of these analyses depends on the genetic diversity of the markers in the populations and the degree of genetic structuring over the geographic scale of interest. If movement and gene flow are high across the spatial scale separating two potential source populations, their genetic profiles will be indistinguishable and immigrants from either source will have the same profile. The level of gene flow among boll weevil populations is restricted enough for marginal structuring to occur at the spatial resolution of 200–300 km separating the SRP, Uvalde and Cameron (Kim & Sappington 2006). However, sensitivity of the assignment tests is differentially affected by a progressive south to north loss in boll weevil genetic diversity (Roehrdanz 2001; Kim & Sappington 2004b,c, 2006), a legacy of the founder effects of the range expansion out of Mexico.
Atmospheric trajectory analysis can provide clues to the source of immigrants over a specified time window under the assumption that wind was a major determinant of direction and distance of a dispersal event. Boll weevils clearly are capable of long-distance movement as discussed previously, but even light winds can interfere with local boll weevil flight (Hardee et al. 1969; Sappington & Spurgeon 2000), and flight mill experiments suggest they cannot fly much faster than approximately 5 km h−1 under their own power (McKibben et al. 1991). Thus, movement over great distances is wind-aided, and analysis of wind trajectories has been a powerful tool in modelling boll weevil dispersal (Culin et al. 1990; Westbrook et al. 2000, 2007). Uncertainties arise in not knowing the precise date of boll weevil arrival in an eradication zone, which can predate capture in a trap. Even the precise date of capture of the SRP weevils is unknown because traps were checked only weekly. It is also possible that immigrants to the SRP arrived after two or more days of travel from a more distant ultimate source, and that the calculated trajectories into Concho County represent only the final leg of a longer journey.
Because of their inherent limitations, application of any of these methods alone is not sufficient to ascertain the origin of the boll weevils captured during late summer in the SRP with tolerable certainty or within reasonably narrow geographic limits. However, by taking advantage of their independent strengths, these complementary multidisciplinary approaches can largely compensate for one another's weaknesses. Thus, taken together, the forensic evidence from genetic, palynological and atmospheric data strongly implicates the Winter Garden district near Uvalde as the source.
Such a multidisciplinary approach is widely applicable to similar problems associated with other invasive and quarantine insects. Our study demonstrates that through the use of multiple, independent methods, likely source areas of dispersing insects may be identified or ruled out with a greater level of confidence than is usually possible with any single method. Furthermore, this multidisciplinary forensic analysis portends the incorporation of similar, different or additional methods that can refine estimates of likely source areas of other dispersing insect species of interest, whether or not they are invasive.
Acknowledgements
We thank S. Esquivel, R. Eyster, L. Fraser, M. Minner and E. Wilson for technical assistance, and C. Allen for boll weevil specimens. We gratefully acknowledge the NOAA Air Resources Laboratory for providing the HYSPLIT model and READY website (http://www.arl.noaa.gov/ready.html).
References
- Allen C. T., Patton L. W., Smith L. E., Newman R. O. 2008. Progress report—Texas boll weevil eradication program. Proc. Beltwide Cotton Conf., Nashville, TN, 8–11 January 2008, pp. 1147–1153. [Google Scholar]
- Armstrong J. S., Greenberg S. M. 2008. Evaluation of extended-life pheromone formulations used with and without dichlorvos for boll weevil (Coleoptera: Curculionidae) trapping. J. Econ. Entomol. 101, 399–403. ( 10.1603/0022-0493(2008)101[399:EOEPFU]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- Benedict J. H., Wolfenbarger D. A., Bryant V. M., Jr, George D. M. 1991. Pollens ingested by boll weevils (Coleoptera: Curculionidae) in southern Texas and northeastern Mexico. J. Econ. Entomol. 84, 126–131. [Google Scholar]
- Burke H. R., Clark W. E., Cate J. R., Fryxell P. A. 1986. Origin and dispersal of the boll weevil. Bull. Entomol. Soc. Am. 32, 228–238. [Google Scholar]
- Carter F. L., Nelson T. C., Jordan A. G., Smith J. R. 2001. U.S. cotton declares war on the boll weevil. In Boll weevil eradication in the United States through 1999 (eds Dickerson W. A., Brashear A. L., Brumley J. T., Carter F. L., Grefenstette W. J., Harris F. A.), pp. 25–54. Memphis, TN: The Cotton Foundation Publisher. [Google Scholar]
- Cate J. R., Skinner J. L. 1978. Fate and identification of pollen in the alimentary canal of the boll weevil Anthonomus grandis. Southwest. Entomol. 3, 263–265. [Google Scholar]
- Cleveland C. J., et al. 2006. Economic value of the pest control service provided by Brazilian free-tailed bats in south-central Texas. Front. Ecol. Environ. 4, 238–243. ( 10.1890/1540-9295(2006)004[0238:EVOTPC]2.0.CO;2) [DOI] [Google Scholar]
- Cornuet J.-M., Piry S., Luikart G., Estoup A., Solignac M. 1999. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153, 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll D. S., Johnston M. C. 1979. Manual of the vascular plants of Texas. Richardson, TX: The University of Texas at Dallas. [Google Scholar]
- Culin J., Brown S., Rogers J., Scarborough D., Swift A., Cotterill B., Kovach J. 1990. A simulation model examining boll weevil dispersal: historical and current situations. Environ. Entomol. 19, 195–208. [Google Scholar]
- Diggs G. M., Jr, Lipscomb B. L., O'Kennon R. J. 1999. Shinners and Mahler's flora of north-central Texas. Fort Worth, TX: Botanical Research Institute of Texas. [Google Scholar]
- Draxler R. R., Rolph G. D. 2003. HYSPLIT (HYbrid Single-Particle Lagrangian Integrated Trajectory) Model. See NOAA ARL READY website http://www.arl.noaa.gov/ready/hysplit4.html. NOAA Air Resources Laboratory, Silver Spring, MD. [Google Scholar]
- El-Lissy O. A., Grefenstette W. J. 2006. Progress of boll weevil eradication in the U.S. In Proc. Beltwide Cotton Conf., San Antonio, TX, 3–6 January 2006, pp. 1266–1276. [Google Scholar]
- Glick P. A. 1939. The distribution of insects, spiders and mites in the air. USDA Technical Bulletin, no. 673, US Government Printing Office, Washington, DC. [Google Scholar]
- Goudet J. 1995. FSTAT (version 1.2): a computer program to calculate F statistics (version 2.9.03). J. Hered. 86, 485–486. [Google Scholar]
- Guerra A. A. 1988. Seasonal boll weevil movement between northeastern Mexico and the Rio Grande Valley of Texas, USA. Southwest. Entomol. 13, 261–271. [Google Scholar]
- Haney P. B. 2001. The cotton boll weevil in the United States: impact on cotton production and the people of the Cotton Belt. In Boll weevil eradication in the United States through 1999 (eds Dickerson W. A., Brashear A. L., Brumley J. T., Carter F. L., Grefenstette W. J., Harris F. A.), pp. 7–24. Memphis, TN: The Cotton Foundation Publisher. [Google Scholar]
- Hardee D. D., Cross W. H., Mitchell E. B., Huddleston P. M., Mitchell H. C., Merkl M. E., Davich T. B. 1969. Biological factors influencing responses of the female boll weevil to the male sex pheromone in field and large-cage tests. J. Econ. Entomol. 62, 161–165. [Google Scholar]
- Hardee D. D., Jones G. D., Adams L. C. 1999. Emergence, movement, and host plants of boll weevils (Coleoptera: Curculionidae) in the Delta of Mississippi. J. Econ. Entomol. 92, 130–139. [Google Scholar]
- Hendrix W. H., III, Showers W. B. 1992. Tracing black cutworm and armyworm (Lepidoptera: Noctuidae) northward migration using Pithecellobium and Calliandra pollen. Environ. Entomol. 21, 1092–1096. [Google Scholar]
- Hendrix W. H., III, Mueller T. F., Phillips J. R., Davis O. K. 1987. Pollen as an indicator of long-distance movement of Heliothis zea (Lepidoptera: Noctuidae). Environ. Entomol. 16, 1148–1151. [Google Scholar]
- Hinds W. E. 1916. Boll weevil in Alabama. AAES Bulletin, no. 188, Alabama Polytechnic Institute, Auburn, AL. [Google Scholar]
- Johnson W. L., Cross W. H., Leggett J. E., McGovern W. L., Mitchell H. C., Mitchell E. B. 1975. Dispersal of marked boll weevil: 1970–1973 studies. Ann. Entomol. Soc. Am. 68, 1018–1022. [Google Scholar]
- Johnson W. L., Cross W. H., McGovern W. L. 1976. Long-range dispersal of marked boll weevils in Mississippi during 1974. Ann. Entomol. Soc. Am. 69, 421–422. [Google Scholar]
- Jones R. W. 1997. Pollen feeding by the boll weevil (Coleoptera: Curculionidae) following cotton harvest in East Central Texas. Southwest. Entomol. 22, 419–429. [Google Scholar]
- Jones G. D., Coppedge J. R. 1999. Foraging resources of boll weevils (Coleoptera: Curculionidae). J. Econ. Entomol. 92, 860–869. [Google Scholar]
- Jones G. D., Jones S. D. 2001. The uses of pollen and its implication for entomology. Neotrop. Entomol. 30, 341–350. [Google Scholar]
- Kenis M., Auger-Rozenberg M.-A., Roques A., Timms L., Péré C., Cock M. J. W., Settele J., Augustin S., Lopez-Vaamonde C. 2009. Ecological effects of invasive alien insects. Biol. Invasions 11, 21–45. ( 10.1007/s10530-008-9318-y) [DOI] [Google Scholar]
- Kim K. S., Sappington T. W. 2004a. Isolation and characterization of polymorphic microsatellite loci in the boll weevil, Anthonomus grandis Boheman (Coleoptera: Curculionidae). Mol. Ecol. Notes 4, 701–703. ( 10.1111/j.1471-8286.2004.00765.x) [DOI] [Google Scholar]
- Kim K. S., Sappington T. W. 2004b. Genetic structuring of boll weevil populations in the U.S. based on RAPD markers. Insect Mol. Biol. 13, 293–303. ( 10.1111/j.0962-1075.2004.00487.x) [DOI] [PubMed] [Google Scholar]
- Kim K. S., Sappington T. W. 2004c. Boll weevil (Anthonomus grandis, Boheman) (Coleoptera: Curculionidae) dispersal in the southern United States: evidence from mitochondrial DNA variation. Environ. Entomol. 33, 457–470. [Google Scholar]
- Kim K. S., Sappington T. W. 2006. Molecular genetic variation of boll weevil populations in North America estimated with microsatellites: implications for patterns of dispersal. Genetica 127, 143–161. ( 10.1007/s10709-005-2673-z) [DOI] [PubMed] [Google Scholar]
- Kim K. S., Cano-Ríos P., Sappington T. W. 2006. Using genetic markers and population assignment techniques to infer origin of boll weevils (Coleoptera: Curculionidae) unexpectedly captured near an eradication zone in Mexico. Environ. Entomol. 35, 813–826. [Google Scholar]
- Kim K. S., Allen C. T., Sappington T. W. 2008. Genetic profiling to determine potential origins of boll weevils (Coleoptera: Curculionidae) captured in a Texas eradication zone: endemicity, immigration, or sabotage? J. Econ. Entomol. 101, 1729–1736. ( 10.1603/0022-0493-101.6.1729) [DOI] [PubMed] [Google Scholar]
- Kiser D., Catanach M. 2008. Arkansas boll weevil eradication update 2007. Proc. Beltwide Cotton Conf., Nashville, TN, 8–11 January 2008, pp. 1130–1146. Memphis, TN: National Cotton Council. [Google Scholar]
- Liebhold A. M., Tobin P. C. 2008. Population ecology of insect invasions and their management. Annu. Rev. Entomol. 53, 387–408. ( 10.1146/annurev.ento.52.110405.091401) [DOI] [PubMed] [Google Scholar]
- Lingren P. D., Westbrook J. K., Bryant V. M., Jr, Raulston J. R., Esquivel J. F., Jones G. D. 1994. Origin of corn earworm (Lepidoptera: Noctuidae) migrants as determined by Citrus pollen markers and synoptic weather systems. Environ. Entomol. 23, 562–570. [Google Scholar]
- Martin P. S. 1963. The last 10,000 years: a fossil pollen record of the American southwest. Tucson, AZ: University of Arizona Press. [Google Scholar]
- McKibben G. H., Willers J. L., Smith J. W., Wagner T. L. 1991. Stochastic model for studying boll weevil dispersal. Environ. Entomol. 20, 1327–1332. [Google Scholar]
- Mikkola K. 1971. Pollen analysis as a means of studying the migration of Lepidoptera. Ann. Entomol. Fenn. 37, 136–139. [Google Scholar]
- Paetkau D., Calvert W., Stirling I., Strobeck C. 1995. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 4, 347–354. ( 10.1111/j.1365-294X.1995.tb00227.x) [DOI] [PubMed] [Google Scholar]
- Paetkau D., Slade R., Burden M., Estoup A. 2004. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol. Ecol. 13, 55–65. ( 10.1046/j.1365-294X.2004.02008.x) [DOI] [PubMed] [Google Scholar]
- Pimentel D., Zuniga R., Morrison D. 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 52, 273–288. [Google Scholar]
- Piry S., Alapetite A., Cornuet J. M., Paetkau D., Baudouin L., Estoup A. 2004. GeneClass2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539. ( 10.1093/jhered/esh074) [DOI] [PubMed] [Google Scholar]
- Rannala B., Mountain J. L. 1997. Detecting immigration by using multilocus genotypes. Proc. Natl Acad. Sci. USA 94, 9197–9201. ( 10.1073/pnas.94.17.9197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulston J. R., Henneberry T. J., Leggett J. E., Byrne D. N., Grafton-Cardwell E., Leigh T. F. 1996. Short- and long-range movement of insects and mites. In Cotton insects and mites: characterization and management (eds King E. G., Phillips J. R., Coleman R. J.). Cotton Foundation Reference Book Series, no. 3, pp. 143–162. Memphis, TN: The Cotton Foundation Publisher. [Google Scholar]
- Roehrdanz R. L. 2001. Genetic differentiation of southeastern boll weevil and thurberia weevil populations of Anthonomus grandis (Coleoptera: Curculionidae) using mitochondrial DNA. Ann. Entomol. Soc. Am. 94, 928–935. ( 10.1603/0013-8746(2001)094[0928:GDOSBW]2.0.CO;2) [DOI] [Google Scholar]
- Rolph G. D. 2003. Real-time Environmental Applications and Display sYstem (READY). See NOAA ARL READY website (http://www.arl.noaa.gov/ready/hysplit4.html. NOAA Air Resources Laboratory, Silver Spring, MD. [Google Scholar]
- Rummel D. R., Jordan L. B., White J. R., Wade L. J. 1977. Seasonal variation in the height of boll weevil flight. Environ. Entomol. 6, 674–678. [Google Scholar]
- Sappington T. W., Spurgeon D. W. 2000. Variation in boll weevil captures in pheromone traps (Coleoptera: Curculionidae) arising from wind speed moderation by brush lines. Environ. Entomol. 29, 807–814. [Google Scholar]
- Shigesada N., Kawasaki K., Takeda Y. 1995. Modeling stratified diffusion in biological invasions. Am. Nat. 146, 229–251. ( 10.1086/285796) [DOI] [Google Scholar]
- Smith J. W. 1998. Boll weevil eradication: area-wide pest management. Ann. Entomol. Soc. Am. 91, 239–247. [Google Scholar]
- Spurgeon D. W., Raulston J. R., Zamora O. Z., Loera J. 1997. Spatial and temporal patterns of boll weevil trap captures in northeastern Mexico. Proc. Beltwide Cotton Conf., New Orleans, LA, 6–10 January 1997, pp. 984–986. [Google Scholar]
- Suarez A. V., Tsutsui N. D. 2008. The evolutionary consequences of biological invasions. Mol. Ecol. 17, 351–360. ( 10.1111/j.1365-294X.2007.03456.x) [DOI] [PubMed] [Google Scholar]
- Suh C. P.-C., Spurgeon D. W., Hagood S. 2003. Evaluation of kill strips on boll weevil (Coleoptera: Curculionidae) mortality in pheromone traps and impact on weevil escape. J. Econ. Entomol. 96, 348–351. [DOI] [PubMed] [Google Scholar]
- Taft H. M., Jernigan C. E. 1964. Elevated screens for collecting boll weevils flying between hibernation sites and cottonfields. J. Econ. Entomol. 57, 773–775. [Google Scholar]
- Tumlinson J. H., Gueldner R. C., Hardee D. D., Thompson A. C., Hedin P. A., Minyard J. P. 1971. Identification and synthesis of the four compounds comprising the boll weevil sex attractant. J. Org. Chem. 36, 2616–2621. [Google Scholar]
- Weir B. S., Cockerham C. C. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. ( 10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- Westbrook J. K., Spurgeon D. W., Eyster R. S., Schleider P. G. 2000. Wind-directed dispersal of boll weevils, Anthonomus grandis (Boh.). Proc. Beltwide Cotton Conf., San Antonio, TX, 4–8 January 2000, pp. 1261–1264. [Google Scholar]
- Westbrook J. K., Eyster R. S., Allen C. T. 2007. A model evaluation of long-distance dispersal of boll weevils. Proc. Beltwide Cotton Conf., New Orleans, LA, 9–12 January 2007, pp. 337–344. [Google Scholar]