Abstract
Bio-electrospray, the direct jet-based cell handling approach, is able to handle a wide range of cells (spanning immortalized, primary to stem cells). Studies at the genomic, genetic and the physiological levels have shown that, post-treatment, cellular integrity is unperturbed and a high percentage (more than 70%, compared with control) of cells remain viable. Although, these results are impressive, it may be argued that cell-based systems are oversimplistic. Therefore, it is important to evaluate the bio-electrospray technology using sensitive and dynamically developing multi-cellular organisms that share, at least some, similarities with multi-cell microenviorments encountered with tissues and organs. This study addressed this issue by using a well-characterized model organism, the non-parasitic nematode Caenorhabditis elegans. Nematode cultures were subjected to bio-electrospraying and compared with positive (heat shock) and negative controls (appropriate laboratory culture controls). Overall, bio-electrospraying did not modulate the reproductive output or induce significant changes in in vivo stress-responsive biomarkers (heat shock proteins). Likewise, whole-genome transcriptomics could not identify any biological processes, cellular components or molecular functions (gene ontology terms) that were significantly enriched in response to bio-electrospraying. This demonstrates that bio-electrosprays can be safely applied directly to nematodes and underlines its potential future use in the creation of multi-cellular environments within clinical applications.
Keywords: bio-electrosprays, Caenorhabditis elegans, gene chip micro-array, heat shock proteins, life cycle
1. Introduction
The current demand for tissues and organs required in transplantation medicine is over threefold and steeply rising (Inverardi & Camillo 2001; Emond 2002). In order to circumvent incompatibilities and minimize rejection responses, patients are routinely administered suppressive medications. This has given rise to the research field of tissue engineering and regenerative medicine (Langer & Tirrell 2004; Jayasinghe 2008a), which aspires towards designing and developing synthetic tissue, possibly even organs, to aid in the repair and replacement of damaged microenvironments.
Two main avenues are currently explored for the handling of a wide range of advanced materials needed for the design and development of functional microenvironments, namely non-jet-based and jet-based approaches. Non-jet-based approaches include lithography-driven approaches (Xia & Whitesides 1998; Mirkin 2007) that are capable of forming fine nanometre-scaled structures from functional materials which invite cells to form cell patterns. However, several drawbacks limit their wider exploration, including the inability to simultaneously handle cells and matrices for the controlled seeding of cell layers. In contrast, jet techniques have advanced significantly and have seen the development of ink-jet printing (IJP) (Jayasinghe 2008b; Nair et al. 2009) and laser-guided deposition (LGCD) (Nahmias & Odde 2006). It is important to note that jet-based techniques are (unlike non-jet-based approaches) not limited to handling cells directly with a biocompatible matrix but can also add advanced materials during the fabrication of a microenvironment, thus making the processes more economical. However, IJP, by means of either thermal or piezoelectric technology, faces process-driven obstacles that limit the handling capacity of materials and cells. Handling of heterogeneous, highly loaded cell suspensions increases cellular mortality significantly with the reduction in the inner bore diameter of ink jets, thus limiting resolution-dependent fabrication of a fully functional microenvironment. Similarly, although fine-tuned laser approaches are capable of handling single cells, this unique approach cages its exploration as tissue fabrication necessitates the creation of a multi-cellular, complex and three-dimensional microenvironment.
Although ink jet and LGCD of cells have been investigated for over a decade, true cell viability screening has, to date, not been studied in detail, thus the promise of clinical exploration is at this stage unpredictable. The limitations of IJP and LGCD have spurred recent developments giving rise to the technology now referred to as ‘bio-electrosprays’ (Jayasinghe et al. 2006a,b). A unique attribute of bio-electrosprays is that it is able to handle highly concentrated heterogeneous cell populations using large-bore needles while possessing the ability to form fine cell-bearing residues at least an order of magnitude smaller than those generated via IJP and LGCD. Furthermore, using large-bore needles allows the reduction in shear rates on cells, or indeed organisms, travelling within these needles, thus maintaining the viability.
Bio-electrospraying works on the principle of accelerating media towards a grounded electrode, flowing within a conducting needle held in an electric field. Over the past 5 years, the technology has been shown to be gentle to cells with no major genomic, genetic or physiological effects on immortalized, primary and stem cells (Jayasinghe et al. 2006a,b; Odenwälder et al. 2007; Abeyewickreme et al. 2009; Mongkoldhumrongkul et al. 2009). This study aims to further characterize the suitability of the technology by exploring the whole transcriptional response, viability and life cycle parameters of the model organism Caenorhabditis elegans subjected to these electric field-driven jets.
2. Material and methods
2.1. Bio-electrospray equipment
The bio-electrospraying of C. elegans larvae was carried out with a stainless steel needle with an internal and external bore diameter of approximately 6 and 8 mm, respectively. The mid-section of the needle, held firmly in resin, was connected to a high voltage power supply (FP-30, Glassman Europe Ltd, Tadley, UK) capable of providing a voltage of up to ±30 kV with a drawing current of approximately 4 mA with a high resolution. The inlet of the needle was connected to medical grade silicone tubing with a similar internal bore diameter. The inlet end of the silicone tube was connected to a reservoir of M9 buffer containing age synchronous C. elegans larvae (L1s). The flow of suspended nematodes was controlled by way of gravitational flow and could be varied from approximately 10−5 to 10−8 m3 s−1. Centrally below the needle exit was a ring-shaped ground electrode with an internal diameter of approximately 18 mm (figure 1). This needle and ground electrode configuration allowed the distance between the electrodes to be varied as required. The bio-electrospray set-up was housed within a class II safety cabinet.
Figure 1.
Digital image of the equipment set-up explored in these investigations which was operated within a laminar flow class II safety hood.
2.2. Nematode cultivation and synchronization
C. elegans wild-type (Bristol N2) and hsp-4::GFP (green fluorescent protein) (BC11945) were obtained from the Caenorhabditis Genetics Center (CGC) at the University of Minnesota. C. elegans were maintained according to standard procedures (Brenner 1974; Sulston & Hodgkin 1988; Lewis & Fleming 1995) using culture dishes containing nematode growth media (NGM) seeded with Escherichia coli strain OP50 (a uracil-requiring E. coli strain to prevent overgrowth of the bacterial culture) as a food source. Gravid hermaphrodites were bleached to release the eggs and maintained overnight in standard M9 buffer. The resultant age synchronized L1 larvae were used for further experimentation. The following conditions were investigated: (i) a culture control, CC (untreated nematodes maintained under standard control conditions in a laboratory incubator at 20°C), (ii) a travel control, TC (untreated nematodes maintained as CCs but transported between laboratories within London, UK), (iii) a needle control, NC (nematodes maintained as in CCs, but transported between laboratories, and passed through the needle in the absence of an applied voltage to the needle), (iv) a bio-electrospray sample, BES (maintained as NC nematodes but passed with an applied voltage) and (v) transgenic heat shock protein tagged nematodes (HSP), namely the hsp-4::GFP expressing strain BC11945 (maintained as CC, TC, NC, BES or treated at 30°C for 24 h to induce heat shock). In every scenario, at least 200 nematodes were used and maintained in sealed sterile 15 ml tubes. Nematodes were observed using standard upright and inverted light microscopes with fluorescent attachments (Leica MZ10F, DMIL, DM2500 and Nikon SMZ1500 and TE2000S).
2.3. Microarray sample generation and execution
A synchronized nematode suspension (approx. 10 000 worms) was centrifuged and the supernatant discarded. The worm pellet was snap frozen in liquid nitrogen and stored at −80°C until use. Total RNA was isolated using Trizol (Invitrogen) as recommended by the manufacturer. An additional purification step was performed using the RNeasy mini kit (Qiagen). Total RNA integrity and quantity were evaluated by agarose gel electrophoresis and spectrophotometry (Nanodrop-1000, Thermo Fisher Scientific), respectively; 200 ng of total RNA was subjected to RNA amplification and biotin labelling as described in the MessageAmp Premier RNA Amplification Kit (Ambion) for microarray analysis. Amplified RNA (aRNA) clean-up was achieved using magnetic beads (Ambion), and 10 µg of biotin end-labelled aRNA was fragmented and hybridized to the Affymetrix C. elegans chips as recommended by Affymetrix. The quality and quantity of the RNA probe preparation were assessed before hybridization using an Agilent 2100 Bioanalyzer and Nanodrop-1000 spectrophotometer, respectively. Affymetrix GeneChip Command Console (AGCC) software was used for the control of array washing (GeneChip Fluidics Station-450), data acquisition (Scanner 3000-7G) as well as for the preliminary data analysis. Standard Affymetrix recommended quality control parameters were used during the entire experiment.
2.4. Microarray data analysis
Pre-processing of microarray raw data included probe-specific background correction, summarization of probe set values and normalization using the GCRMA algorithm with CARMAweb 1.4, an R- and Bioconductor-based web service for microarray data analysis (Rainer et al. 2006). The quality of the normalization was accessed by boxplot, histogram and MA graph analyses. Differences between treatments were visualized by principal component analysis (PCA) plotting with MeV/TM4 (Saeed et al. 2003). Data were initially filtered out for missing values and then subjected to a CLEAR test that combines differential expression and variability using the GEPAS web server at http://www.gepas.org. In our case, the false discovery rate was set to a stringent level of 5 per cent. Differentially exposed genes (DEGs) showing a fold change of two were further analysed with regard to their molecular functions, biological processes and biological pathways using information provided by Wormbase (www.wormbase.org) with high throughput GoMiner software (Zeeberg et al. 2005). CateGOrizer software was used to summarize gene ontology terms (Hu et al. 2008).
2.5. Brood size
The daily and total reproductive output (brood size) was determined from nematodes subjected to the different culturing conditions (CC, TC, NC and BES). In detail, at least 30 synchronized L1 larvae were plated individually onto 12-well culture dishes containing NGM seeded with OP50 as a food source. Once nematodes reached adulthood, they were transferred to new plates every 24 h. The number of offspring was determined by counting hatched eggs the following day.
3. Results and discussion
3.1. Bio-electrospraying Caenorhabditis elegans larvae
The equipment arrangement explored within the class II safety cabinet allowed the variation of several bio-electrospray set-up configurations (figure 1). A wide operational parametric window was traversed, namely approximately 1–30 kV, 10−5–10−8 m3 s−1 and 5–20 mm for the applied voltage, flow rate to the needle and the distance between the electrodes, respectively. We were unable to achieve stable jetting and jet continuity conditions, which is not surprising given the media properties, in particular the electrical conductivity and the viscosity which were high and low, respectively, a finding that was analogous to previous reports (Jayasinghe et al. 2006a,b; Clark & Jayasinghe 2008; Abeyewickreme et al. 2009; Geach et al. 2009). The observed instability and jet discontinuity are a likely consequence of the suspension properties of the living larvae. The high concentration of ions within the suspension and the low viscosity provides a higher electrical relaxation time with regards to the hydrodynamic time, giving rise to unstable jetting (figure 2). In general, stable jetting requires a hydrodynamic time that is smaller than the electrical relaxation time, unless the viscosity is above a threshold value which results in the manifestation of the process referred to as ‘electrically forced jetting’ (Jayasinghe & Edirisinghe 2004a,b). However, in the bio-electrospray-based studies described here, stable jetting with jet continuity is not of paramount importance as the primary goal was to determine whether this technique induces significant molecular-based perturbations on the nematode. Indeed, if stable jetting with jet continuity is desired, coaxial bio-electrosprays would have facilitated stable conditions (Jayasinghe & Townsend-Nicholson 2006a,b).
Figure 2.
Representative high-speed image sequence (a–f) depicting the unstable jetting of the living C. elegans.
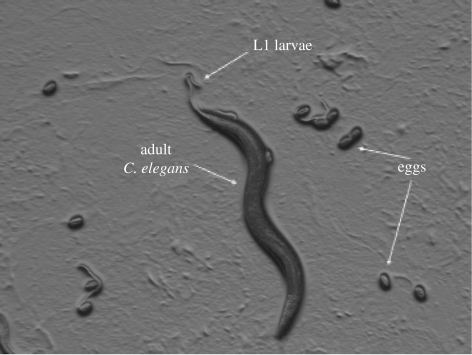
A fully grown adult nematode is approximately 1 mm in length and a hatched L1 larva is approximately 250 µm in length (figure 3). During bio-electrospraying (operational conditions were approx. 10 kV, 10−6 m3 s−1 and 15 mm for the applied voltage and flow rate to the needle, and the distance between the electrodes, respectively), we observed that the jetting process was similar to the previously observed and reported microdripping (figure 2). However, we noticed that droplet break-up essentially took place not via unstable jetting by way of a formed instantaneous jet but by means of the external electric field (figure 2b–d). At this stage, we speculate that this is directly due to the suspension properties having either extremes for the parameters electrical conductivity and viscosity of the nematode suspension. We also noted that upon elevating the applied voltage (approx. 15 kV) for a constant flow rate (approx. 10−6 m3 s−1), the resulting nematode-containing suspension was seen to undergo an unstable jetting behaviour that resembled multi-spindle-type morphologies at the needle exit. If the reverse was carried out, namely keeping the applied voltage constant for an increasing flow rate, the resulting droplet generation process would still take place, but with the formation of larger nematode-bearing residues.
Figure 3.
Representative image of a mixed population of C. elegans containing a gravid adult, eggs and freshly hatched L1 larvae. Size of the adult: approximately 1 mm.
3.2. Genomic, genetic and reproductive effects of bio-electrospraying
The C. elegans genome was the first multi-cellular organism that was fully sequenced and therefore Affymetrix microarrays have been manufactured to contain all 22 500 transcripts predicted from intricate bioinformatic analyses. This resource allows gene profiling at a whole-genome level. Here, we compared the transcriptional profile of control, heat exposed and bio-electrosprayed nematodes. The complete dataset can be viewed in the National Center for Biotechnology Information's (NCBI) Gene Expression Omnibus (GEO) database (accession number GSE16915). Overall, when taking all transcripts that pass the quality threshold into account, PCA identified that heat shock induced a very distinctive response, while bio-electrospray was closely clustered with control nematodes (figure 4a). Stringent downstream bioinformatics frequently include false discovery rate (FDR) algorithms to account for false positives (type I error) and false negatives (type II error). A good indicator of variance and error estimation can be made when two biologically independent, but otherwise identical, samples are analysed. Previous data from our laboratory consisting of successive hybridizations of any two replicate arrays under each individual treatment group and deep bioinformatics (to minimize errors without excessive data loss) reproducibly yielded a coefficient of correlation (R) > 0.9954 with a type I error contribution of 10–12 genes (unpublished data from 45 Affymetrix chips). In this context, the 16 transcripts highlighted to be differentially regulated by at least a factor of two can be stipulated to be only marginally above what should be expected by random type I error (figure 4b) and is in stark contrast to the 806 genes shown to be differentially regulated by heat shock.
Figure 4.
(a) PCA of whole-genome microarray data. Note how the transcriptome of control nematodes clusters closely with nematodes passed through a needle passed with an applied voltage (bio-electrosprayed), both of which are distinct from nematodes heat shocked at 30°C for 24 h. (b) Only 16 transcripts were identified to be differentially expressed (FDR p < 0.05, cut-off twofold up/downregulation) owing to electrospraying, a number that is significantly lower than the 806 heat stress responsive genes.
Although bio-electrospray has to be considered to be statistically indistinguishable from control nematodes, it is worth highlighting that, among the 16 genes identified, there are 11 uncharacterized transcripts, one p-glycoprotein (Wormbase ID C05A9.1), one C-type lectin (T09F5.9) and notably three dehydrogenase, an acyl-CoA dehydrogenase (C17C3.12), a short-chain alcohol dehydrogenase (F09E10.3) and an iron-containing alcohol dehydrogenase (Y38F1A.6) (see electronic supplementary material, file 1). At this stage, it is currently not possible to stipulate whether the abundance of dehydrogenases was identified by chance or represents a finding of biological importance. As expected, many genes involved in direct and indirect responses, notably the heat shock proteins hsp-16.41, hsp-70, hsp-16A and hsp-16.49, were identified in the positive control (see electronic supplementary material, file 1). Similarly, downstream gene ontology analysis returned no significant biological processes, molecular functions or cellular components that were significantly enriched upon bio-electrospraying. In contrast, a complex response was observed upon heat shock (figure 4b; for details, see electronic supplementary material, file 2).
Given that heat stress was chosen as a positive control, we explored the use of the transgenic nematode expressing a GFP driven by the hsp-4. HSP-4 is a member of the HSP-70 superfamily and encodes a predicted homologue of the mammalian ER chaperone BiP. It is expressed at low levels under control conditions but is readily activated upon stress in general; in particular, it is an endoplasmic reticulum unfolded protein response (Kapulkin et al. 2005; Olsen et al. 2006). While exposure to heat (30°C for 24 h) induced a strong expression of hsp-4 in the intestinal cells and reproductive organs (in particular the distal tip cell), the levels of hsp-4 remained low (at background levels) in control and bio-electrosprayed animals (figure 5). This highlights further that the transcriptional response upon bio-electrospray is likely to be negligible.
Figure 5.
Comparing the effect of heat shock and bio-electrospray on transgenic nematodes expressing an extrachromosomal heat shock protein (hsp-4::GFP). (a) A superimposed image using Differential Interference Contrast (DIC; Nomaski) optics and a fluorescent image of an adult nematode maintained under heat shocked conditions (30°C for 24 h). Note expression levels of hsp-4 in the intestinal cells and reproductive organs (in particular the distal tip cell). (b) Representative (non-superimposed) fluorescent images of heat-shocked nematodes, nematodes maintained under control conditions at 20°C and transported between laboratories, (c) the TC and the (d) bio-electrospray-treated nematodes. Note, while heat shock induced a pronounced effect on hsp-4 expression, no significant difference was observed between control and bio-electrosprayed nematodes. The photographs were taken using invariant settings (1/40 s exposure time with a gain setting of 13 using a 60× magnification).
In nematodes, one of the most sensitive life cycle endpoints is the brood size. Previous reports have identified that stress and toxicosis can have a marked effect on egg laying (time to first egg), duration of reproductive period or total offspring number (Calafato et al. 2008; Hughes et al. 2009; Lagido et al. 2009). CC, TC, NC and bio-electrosprayed animals (BES) all developed synchronous production of between 250 and 300 eggs, a value that is in agreement with the published literature. No statistically identifiable differences were identified in terms of offspring produced per day or over the entire brood period (figure 6).
Figure 6.
Brood size of CC (white bar), TC (black bar), NC (grey bar) and bio-BES (striped bar) nematodes over a 4-day time period. Every condition consists of 30 replicates and the error bars represent s.d. of the mean. Note that the respective culturing conditions and notably the bio-electrospray induce no time delay in reproductive performance, nor a difference in total reproductive output.
4. Conclusions
These studies demonstrate that bio-electrosprays are not lethal to nematodes. The reproductive behaviour (a sensitive life cycle trait) of nematodes subjected to bio-electrospray was indistinguishable from nematodes maintained under optimal laboratory conditions, nematodes transported between laboratories and nematodes passed through a needle in the absence of an applied voltage. Likewise, neither in vivo stress responsive biomarkers nor whole-genome transcriptomics could identify significant differences between bio-electrosprayed animals and appropriate negative controls. This baseline study demonstrates the utility of applying bio-electrosprays to multi-cellular organisms and driving this technology towards possible future clinical applications, such as the fabrication of fully functional microenvironments which could be explored for tissue repair and replacement.
Acknowledgements
N.M. wishes to gratefully thank the Royal Thai Government for providing a fully funded PhD scholarship for her studies in the BioPhysics Group. S.J. wishes to thank the Royal Society for funding the BioPhysics Group and S.R.S. thanks the Royal Society and the BBSRC for continuous support. We wish to acknowledge Dr Estibaliz Aldecoa-otalora Astarloa and Dr Matthew Arno (KCL Genomics Center) for invaluable help with the arrays and the CGC, which is funded by the National Institutes of Health National Center for Research Resources, for the supply of C. elegans strains and E. coli.
References
- Abeyewickreme A., Kwok A., McEwan J. R., Jayasinghe S. N. 2009. Bio-electrospraying embryonic stem cells: interrogating cellular viability and pluripotency. Integr. Biol. 1, 260–266. ( 10.1039/b819889f) [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafato S., Swain S. C., Hughes S., Kille P., Stürzenbaum S. R. 2008. Knock down of Caenorhabditis elegans cutc-l exacerbates the sensitivity toward high levels of copper. Toxicol. Sci. 106, 384–391. ( 10.1093/toxsci/kfn180) [DOI] [PubMed] [Google Scholar]
- Clark J. D. W., Jayasinghe S. N. 2008. Bio-electrosprayed multicellular zebrafish embryos are viable and develop normally. Biomed. Mater. 3, 011001 ( 10.1088/1748-6041/3/1/011001) [DOI] [PubMed] [Google Scholar]
- Emond J. C. 2002. What's new in transplantation. J. Am. Coll. Surg. 194, 636–641. ( 10.1016/S1072-7515(02)01161-4) [DOI] [PubMed] [Google Scholar]
- Geach T., Mongkoldhumrongkul N., Zimmerman L. B., Jayasinghe S. N. 2009. Assessing viability and development of bio-electrosprayed Xenopus laevis embryos. Analyst 134, 743–747. ( 10.1039/b817827e) [DOI] [PubMed] [Google Scholar]
- Hu Z.-L., Bao J., Reecy J. M. 2008. CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J. Bioinformatics 9, 108–112. [Google Scholar]
- Hughes S. L., Bundy J. G., Want E. J., Kille P., Stürzenbaum S. R. 2009. The metabolomic responses of Caenorhabditis elegans to cadmium are largely independent of metallothionein status, but dominated by changes in cystathionine and phytochelatins. J. Proteome Res. 8, 3512–3519. ( 10.1021/pr9001806) [DOI] [PubMed] [Google Scholar]
- Inverardi L., Camillo R. 2001. Tolerance and pancreatic islet transplantation. Phil. Trans. R. Soc. Lond. B 356, 759–765. ( 10.1098/rstb.2001.0849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe S. N. 2008a. Cell engineering: spearheading the next generation in healthcare. Biomed. Mater. 3, 034004. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S. N. 2008b. Advanced jet-protocols for directly engineering living cells: a genesis to alternative bio-handling approaches for the life sciences. Regener. Med. 3, 49–61. ( 10.2217/17460751.3.1.49) [DOI] [PubMed] [Google Scholar]
- Jayasinghe S. N., Edirisinghe M. J. 2004a. Electric-field driven jetting from dielectric liquids. Appl. Phys. Lett. 85, 4243–4245. ( 10.1063/1.1812574) [DOI] [Google Scholar]
- Jayasinghe S. N., Edirisinghe M. J. 2004b. Electrically forced jets and microthreads of high viscosity dielectric liquids. J. Aerosol Sci. 35, 233–243. ( 10.1016/j.jaerosci.2003.08.004) [DOI] [Google Scholar]
- Jayasinghe S. N., Townsend-Nicholson A. 2006a. Stable electric-field driven cone-jetting of concentrated biosuspensions. Lab Chip 6, 1086–1090. ( 10.1039/b606508m) [DOI] [PubMed] [Google Scholar]
- Jayasinghe S. N., Townsend-Nicholson A. 2006b. Bio-electrosprays: the next generation of electrified jets. Biotechnol. J. 9, 1018–1022. [DOI] [PubMed] [Google Scholar]
- Jayasinghe S. N., Eagles P. A. M., Qureshi A. N. 2006a. Electric field driven jetting: an emerging approach for processing living cells. Biotechnol. J. 1, 86–94. ( 10.1002/biot.200500025) [DOI] [PubMed] [Google Scholar]
- Jayasinghe S. N., Qureshi A. N., Eagles P. A. M. 2006b. Electrohydrodynamic jet processing: an advanced electric-field-driven jetting phenomenon for processing living cells. Small 2, 216–219. ( 10.1002/smll.200500291) [DOI] [PubMed] [Google Scholar]
- Kapulkin W. J., Hiester B. G., Link C. D. 2005. Compensatory regulation among ER chaperones in C. elegans. FEBS Lett. 579, 3063–3068. ( 10.1016/j.febslet.2005.04.062) [DOI] [PubMed] [Google Scholar]
- Lagido C., McLaggan D., Flett A., Pettitt J., Glover L. A. 2009. Rapid sublethal toxicity assessment using bioluminescent Caenorhabditis elegans, a novel whole-animal metabolic biosensor. Toxicol. Sci. 109, 88–95. ( 10.1093/toxsci/kfp058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer R., Tirrell D. A. 2004. Designing materials for biology and medicine. Nature 428, 487–492. ( 10.1038/nature02388) [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Fleming J. T. 1995. Basic culture methods. In Caenorhabditis elegans: modern biological analysis of an organism (eds Epstein H. F., Shakes D. C.), pp. 4–29. San Diego, CA: Academic Press. [Google Scholar]
- Mirkin C. A. 2007. The power of the pen: development of massively parallel dip-pen nanolithography. ACS Nano 1, 79–83. ( 10.1021/nn700228m) [DOI] [PubMed] [Google Scholar]
- Mongkoldhumrongkul N., Flanagan J., Jayasinghe S. N. 2009. Direct jetting approaches for handling stem cells. Biomed. Mater. 4, 015018 ( 10.1088/1748-6041/4/1/015018) [DOI] [PubMed] [Google Scholar]
- Nahmias Y., Odde D. J. 2006. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic–endothelial sinusoid-like structures. Nat. Protoc. 1, 2288–2296. ( 10.1038/nprot.2006.386) [DOI] [PubMed] [Google Scholar]
- Nair K., Gandhi M., Khalil S., Yan K. C., Marcolongo M., Barbee K., Sun W. 2009. Characterization of cell viability during bioprinting processes. Biotechnol. J. 4, 1168–1177. ( 10.1002/biot.200900004) [DOI] [PubMed] [Google Scholar]
- Odenwälder P. K., Irvine S., McEwan J. R., Jayasinghe S. N. 2007. Bio-electrosprays: A precision ‘drop and place’ paradigm for safe handling and deployment of primary living organisms. Biotechnol. J. 2, 622–630. ( 10.1002/biot.200700031) [DOI] [PubMed] [Google Scholar]
- Olsen A., Vantipalli M. C., Lithgow G. J. 2006. Lifespan extension of Caenorhabditis elegans following repeated mild hormetic heat treatments. Biogerontology 7, 221–230. ( 10.1007/s10522-006-9018-x) [DOI] [PubMed] [Google Scholar]
- Rainer J., Sanchez-Cabo F., Stocker G., Sturn A., Trajanoski Z. 2006. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 34, W498–W503. ( 10.1093/nar/gkl038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A. I., et al. 2003. MeV /TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J. 1988. Methods. In The nematode Caenorhabditis elegans (ed. Wood W. B.), pp. 587–606. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Xia Y., Whitesides G. M. 1998. Soft lithography. Angew. Chem. Int. Edn 37, 550–575. ( 10.1002/(SICI)1521-3773(19980316)37:5%3C550::AID-ANIE550%3E3.0.CO;2-G) [DOI] [PubMed] [Google Scholar]
- Zeeberg B. R., et al. 2005. High-throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics 6, 168–176. ( 10.1186/1471-2105-6-168) [DOI] [PMC free article] [PubMed] [Google Scholar]