Abstract
We previously identified a novel nuclear RNA species derived from the preproenkephalin (PPE) gene. This transcript, which we have named PPEIA-3′ RNA, hybridizes with probes directed at a region of PPE intron A downstream of an alternative germ-cell transcription start site, but does not contain PPE protein coding sequences. We now report that estrogen treatment of ovariectomized rats increases the expression of conventional PPE heteronuclear RNA, and also induces the expression of PPEIA-3′ RNA, apparently in separate cell populations within the ventromedial nucleus of the hypothalamus. Further, we show that cells expressing PPEIA-3′ are found in several neuronal groups in the rat forebrain and brainstem, with a distinct topographical distribution. High densities of PPEIA-3′ containing cells are found in the reticular thalamic nucleus, the basal forebrain, the vestibular complex, the deep cerebellar nuclei, and the trapezoid body, a pattern that parallels the distribution of atypical nuclear RNAs described by other groups. These results suggest that this diverse neuronal population shares a common set of nuclear factors responsible for the expression and retention of this atypical RNA transcript. The implication of these results for cell-specific gene transcription and regulation in the brain and the possible relationship of PPEIA-3′ RNA and other atypical nuclear RNAs is discussed.
Keywords: transcription termination, RNA processing, intron, vestibular complex, cerebellar nuclei
Studies carried out over 20 years concluded that a large fraction of RNA turns over exclusively within the cell nucleus (1). Subsequently, it has been shown that the nuclear RNA population is made up of a number of different classes of RNA molecules including intronic sequences derived from pre-mRNAs(2), RNA transcribed from repetitive DNA sequences (e.g., LINE sequences) (3), and small nuclear and nucleolar RNAs involved in the pre-mRNA and pre-rRNA processing, respectively (4). Further, the discovery of the XIST, a large nuclear RNA expressed exclusively from the inactive X-chromosome(5, 6), suggests the possibility that nuclear RNAs may also be involved in the control of gene expression or chromatin structure. These findings indicate that heterogeneous nuclear (hn) RNA represents a complex population of RNA molecules with diverse functional roles.
We have previously identified a novel nuclear RNA species derived from the preproenkephalin (PPE) gene (7). This abundant transcript, named PPEIA-3′ RNA, hybridizes with probes directed at a region of PPE intron A downstream of an alternative transcription start site originally discovered in testicular germ cells (8) but does not contain PPE protein coding exon sequences. PPEIA-3′ RNA was originally discovered in a population of neurons in the reticular nucleus of the thalamus and basal forebrain. We now report that estrogen treatment of ovariectomized rats increases levels of the conventional PPE hn RNA, and also dramatically induces the expression of PPEIA-3′ RNA in the ventromedial nucleus of the hypothalamus. In addition, we further describe the distribution of cells expressing PPEIA-3′ in the rat brain, with emphasis on the brainstem.
METHODS
Animals.
Experiments were carried out on adult female Sprague–Dawley rats obtained from Charles River Breeding Laboratories. For the estrogen regulation studies, ovariectomized (OVX) rats were obtained from the supplier. Three groups of OVX rats were studied (n = 5 per group). One group was injected with 10 μg estradiol (E) benzoate 18 hr prior to sacrifice. A second group was given subcutaneous implants of silastic capsules containing crystalline E (1 mm long) that were left in place for 2 weeks. The third group of animals was implanted with empty capsules and served as a control. Tissues were obtained and processed for in situ hybridization as described (7, 9).
In Situ Hybridization Using Radiolabeled Probes.
In situ hybridization using radiolabeled (32P or 3H) probes was carried out as described (7, 9). Single-stranded DNA probes were produced by amplified primer extension labeling as described (9).
Quantitative Analysis of In Situ Hybridization.
For quantitation of PPE hnRNA in the ventromedial nucleus (VMN), matched sections from the VMN of control, 18 hr, and 2 week E benzoate-treated animals (n = 5 per group) were hybridized with probe I-A-labeled with 3H. Sections were dipped in NTB-2 emulsion, exposed for 6 weeks, then photographically processed, lightly counterstained with cresyl violet, and coverslipped. Sections were then coded, and manual grain counting was carried out over cells in the VMN. Statistical analysis of between-group comparisons was made using the Kolmogorov–Smirnov technique.
Localization of Cells Expressing PPEIA-3′ RNA.
To identify additional populations of cells expressing PPEIA-3′ (PPE intron A 3′) RNA, we hybridized probes described (7): I-A, I-A5′, and I-A3′ (see Fig. 1). Initially, sagittal sections through the rat brain were hybridized with probe I-A, washed, and exposed to x-ray film. Areas of the brain showing a strong signal with probe I-A were identified in this manner. In subsequent studies, sections containing such regions were hybridized with probes I-A5′ and I-A3′. Brain areas containing major collections of cells expressing PPEIA-l were identified by showing strong hybridization with probe I-A3′, but not I-A5′ using x-ray film detection. Sections containing such cells were dipped in Kodak NTB-3 emulsion, exposed for 2 weeks, then processed for autoradiography to identify cells expressing PPEIA-3′ RNA. Additional coronal sections were then examined with the same probes to further confirm the location of major PPEIA-3′ expressing cells.
Figure 1.
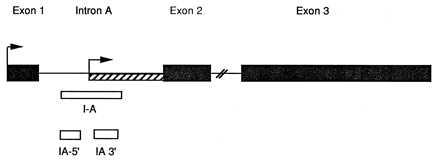
Schematic drawing of the PPE gene showing the location of the probes I-A, I-A5′, and I-A3′ used in the present study. The hatched region indicates the portion of intron A that is retained in the germ cell-specific PPE transcript (8). Single stranded probes corresponding to the regions shown were generated as described (7, 9).
Negative controls for all the experiments shown include hybridization with probe I-A in the sense orientation, as well as pretreatment of the sections with DNAse-free ribonuclease (50 μg/ml) for 1 hr at 37°C prior to hybridization with antisense probes.
RESULTS
Estrogen Regulation of PPE hnRNA and PPEIA-3′ RNA in the VMN of the Hypothalamus.
Previous work from this lab (10) has shown that estrogen treatment of OVX female rats increases PPE mRNA in the VMN of the hypothalamus. Because steroids can increase mRNA levels by stabilization of mRNA (11) independent of changes in gene transcription, we sought to determine whether E would also increase PPE hnRNA, which would indicate an E induced change in PPE gene transcription. Roberts and colleagues (12) have demonstrated the utility of in situ hybridization with intron probes for analyzing rapid changes in gene transcription in brain neurons.
In situ hybridization was carried out on sections through the VMN of control (OVX) rats, or OVX rats treated with E for either 18 hr or 2 weeks. In control OVX rats, hybridization with probe I-A showed a very light signal over the nuclei of cells in the VMN (Fig. 2). In contrast, sections from the E-treated animals showed stronger hybridization signals (Fig. 2). In addition, a population of cells was present in the E-treated animals that showed very strong hybridization signals, with foci of silver grains over the cell nucleus similar to the appearance of PPEIA-3′ RNA in other brain areas (7) (Fig. 2 Upper).
Figure 2.
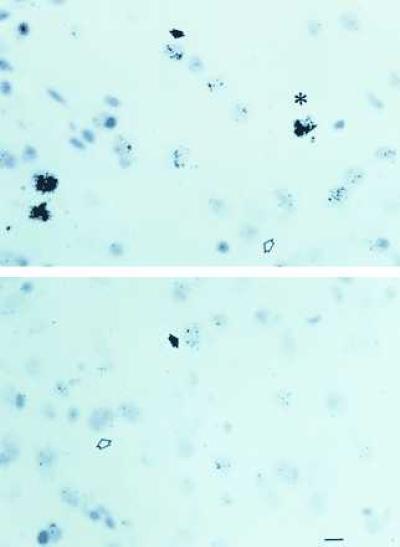
Photomicrographs of sections through the VMN of ovariectomized rats treated with E for 18 hr (Upper) or control ovariectomized female rats (Lower) hybridized with 3H-labeled antisense probe I-A. The open arrowheads point to unlabeled cells, the filled arrows point to labeled cells, and the asterisk indicates a highly labeled cell containing a focus of silver grains. (Bar = 20 μ.)
Quantification of the hybridization signal was carried out by an observer unaware of the treatment conditions. Cells that were so highly labeled that individual grains could not be counted (see Fig. 2) with simply classified as highly labeled (50–100 grains) or very highly labeled (>100 grains). Fig. 3 shows the results of this analysis. Compared with OVX controls, histograms of the 18 hr and 2 week estrogen-treated animals were shifted to the right, indicating greater numbers of cells expressing higher levels of PPE hnRNA. The shape of both the 18 hr and 2 week groups was significantly different that the OVX group (Kolmogorov–Smirnov test, P < 0.01 for OVX vs. both E-treated groups). Further analysis of the grain counts (in cells where grains could be accurately counted), showed that cells in the VMN of both E-treated groups had ≈2-fold higher grain densities than VMN cells in OVX animals (mean grain counts: OVX = 12 ± 2.3, 18 hr = 23 ± 4.1; 2 week = 21 ± 2.0; P > 0.01).
Figure 3.
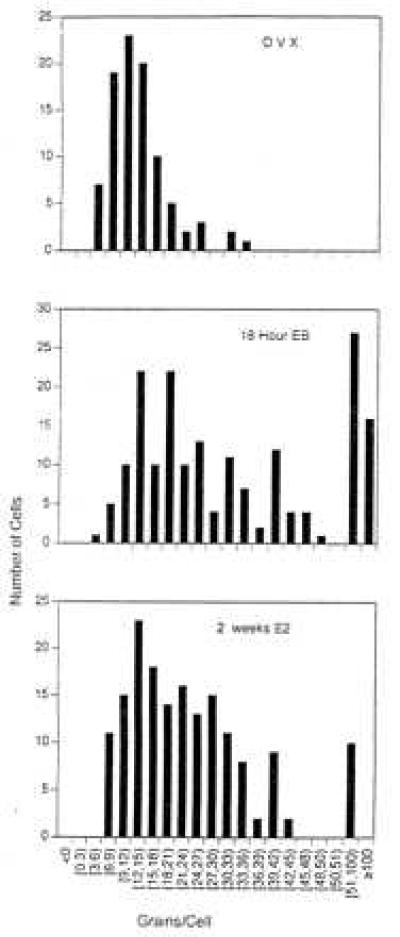
Histograms showing the distribution of labeled cells in the VMN of control OVX rats (Top), OVX rats treated with E for 18 hr (Middle) or 2 weeks (Bottom). Data are pooled from five animals per group. The highly labeled (51-100 grains per cell) and very highly labeled (>100 grains per cell) classifications are estimates as the grains could not be counted accurately in these cells. All quantification was carried out without knowledge of the treatment group. The shape of the 18 hr and 2 week distributions are significantly different from the OVX group (P < 0.01, Kolmogorov–Smirnov analysis).
The number of highly labeled cells in the VMN was dramatically affected by E treatment. Highly labeled cells with nuclear foci of silver grains were never seen in sections from the OVX animals, but were detected in both the 18 hr and 2 week E group, with higher values in the 18 hr group (Figs. 2 and 3).
Given the appearance and intensity of the hybridization signal in the highly labeled VMN cells following estrogen treatment, we hypothesized that the highly labeled cells may represent the induction of PPEIA-3′ RNA by estrogen. To test this, we took advantage of the fact that expression of PPEIA-3′ can easily be detected at the x-ray film level by hybridizing adjacent sections with probe I-A3′ and I-A5′. Sections through the VMN from an additional group of OVX and 18 hr E-treated rats were hybridized to 32P-labeled probes I-A3′ and I-A5′, followed by exposure to x-ray film. As shown in Fig. 4, E treatment of OVX animal produced a dramatic increase in the hybridization signal over the VMN detected with probe I-A3′. In contrast, use of the probe I-A5′ showed no difference in VMN hybridization signal following E treatment, presumably because PPE hnRNA was present at too low abundance to be detected. Long exposure of the sections hybridized with probe I-A5′ revealed that this probe does hybridize to the striatum present in the same sections, indicating that the probe is capable of detecting conventional PPE hnRNA.
Figure 4.
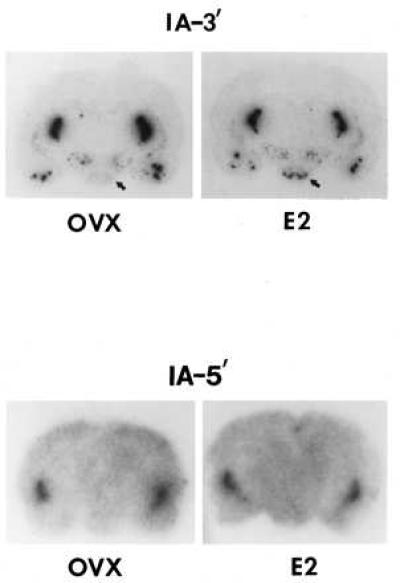
Representative film autoradiographs of brain sections of an OVX and 18 hr E-treated female rat hybridized with probe I-A3′ and I-A5′-labeled with 32P. Note that in the VMN (identified by arrow) of the E2-treated animal, a hybridization signal is detectable with probe I-A of similar intensity as in the reticular thalamic nucleus, indicating the appearance of PPEIA-3′ RNA in the VMN following E treatment. Exposure time: Upper; 1 day; Lower, 5 days. The Lower panel was exposed for a longer period to show that probe I-A5′ does label the striatum as expected.
Taken together, these data indicate that E increases PPE hnRNA, and also induces the expression of PPEIA-3′ RNA, in the VMN.
Major Groups of PPEIA-3′ Expressing Cells in the Hindbrain Are Found in the Vestibular Complex, Deep Cerebellar Nuclei, and Trapezoid Complex.
While the main focus of this work was on the regulation of PPE hnRNAs by estrogen in the VMN, we were also interested in further examining the expression of PPEIA-3′ RNA in other brain areas. We previously identified cells expressing PPEIA-3′ RNA in the basal forebrain and reticular thalamic nucleus (7). To screen for additional brain regions containing PPEIA-3′ cells, sagittal sections of the rat brain were hybridized with probe I-A (Fig. 1), labeled with 32P, and sites of hybridization detected with x-ray film. The high abundance of PPEIA-3′ facilitates this approach. Examination of sagittal sections hybridized with probe I-A showed a strong signal over the dorsal and ventral hindbrain (not shown). Additional hybridization experiments were then carried out on coronal sections taken from those areas showing the highest signal in the sagittal sections. These areas are the deep cerebellar nuclei, the vestibular complex (especially the medial vestibular nucleus) and the trapezoid complex (Fig. 5). Cells in all three areas showed a strong signal with probe I-A in the antisense but not the sense orientation, and strongly hybridized with probe I-A3′, but not with I-A5′ (Fig. 5). In each case, the signal was abolished by pretreating the tissue with DNAse free-RNAse prior to hybridization, indicating hybridization to RNA. Examination of emulsion autoradiographs indicated that the signal appeared to be concentrated over the nucleus (not shown). Taken together, these data indicate that the deep cerebellar nuclei, vestibular complex, and trapezoid complex all contain large numbers of cells that express PPEIA-3′ RNA.
Figure 5.
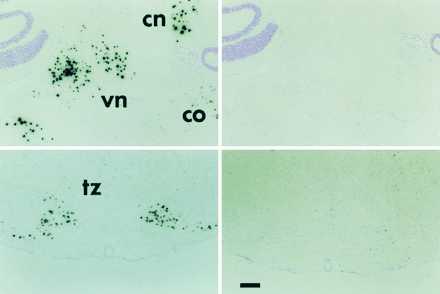
Low power photomicrographs showing the hybridization of probe I-A in the brainstem. The left panels show hybridization with the antisense probe, the right panels show hybridization with the sense strand probe. cn, Cerebellar nuclei; vn, vestibular nuclei; co, cochlear nucleus; tz, trapezoid body. (Bar = 200 μ.)
In addition to the above mentioned areas, cells expressing PPEIA-3′ were also seen in a number of other midbrain and brainstem areas. These include the ventral lateral lemniscus, anterior tegmental nucleus, the nucleus abducens, the cochlear nucleus, scattered cells in the ventral pontine and medullary reticular formation, and cells in the medial laminae of the spinal cord (not shown).
DISCUSSION
Estrogen Regulation of PPE hnRNA and PPEIA-3′ RNA in the Ventromedial Hypothalamic Nucleus.
The initial aim of these studies was to examine the effects of E on PPE hnRNA. Given the well-studied effect of E on PPE mRNA (10), our finding that E increases PPE hnRNA in the VMN was not unexpected, but does provide additional evidence that E increases PPE gene transcription. Surprisingly, in addition to E regulation of PPE hnRNA, we also found that E could stimulate the expression of PPEIA-3′ RNA in a subset of VMN neurons. The magnitude of the effect is quite striking, in that we did not see any PPEIA-3′ cells in the VMN of OVX animals. This result indicates that the expression of PPEIA-3′ in VMN neurons in completely dependent upon E. Even low physiological concentrations of E appear to be sufficient to maintain expression of this transcript, however, since we have observed constant expression of PPEIA-3′ throughout the estrous cycle in intact female rats (T.F. et al., unpublished manuscript).
Given that E increases both PPE hnRNA and PPEIA-3′ RNA in the VMN, an important question concerns the possibility that both transcripts are expressed in the same cell. The data shown in the histograms in Fig. 3 indicate two populations of cells in VMN of E-treated animals. We believe that the population of highly labeled cells to the right of the histogram represents cells expressing PPEIA-3′ RNA, and the remaining population of cells corresponds to cells expressing traditional PPE hnRNA for the following reasons. (i) The two-fold increase in silver grain counts (in those cells where the grains could be counted accurately) after 18 hr of E treatment closely parallels the 2-3 fold increase in PPE mRNA following E treatment (10). (ii) The similarity in grain counts between the 18 hr and 2 week E animals shown here is consistent with data showing that PPE mRNA in the VMN remains elevated for at least 1 month of E exposure (10). Finally the intensity and focal appearance of the silver grains over the nucleus of the highly labeled cells is characteristic of appearance of PPEIA-3′ RNA (7). Further, in additional studies combining radiolabeled probes for PPE mRNA and dig-labeled probes for PPEIA-3′, we have not identified cells expressing both PPEIA-3′ and PPE mRNA in the VMN of estrogen-treated animals (not shown). Therefore, our interpretation of these data is that E increases PPE hnRNA, and also induces the expression of PPEIA-3′ RNA in separate populations of cells in the rat VMN, consistent with the divergent expression of these two transcripts in other brain areas (7).
Widespread Expression of PPEIA-3′ RNA in the Rat Central Nervous System.
In our initial work (7) we identified cells expressing PPEIA-3′ RNA in the basal forebrain and reticular thalamic nucleus. In the present work, we have extended these findings in showing that cells expressing PPEIA-3′ RNA are not limited to the forebrain, but are found throughout the central nervous system, including the spinal cord. In addition to the basal forebrain, reticular thalamus, and other forebrain sites described previously, major collections of cells expressing PPEIA-l RNA are found in the deep cerebellar nuclei, the vestibular complex, and the trapezoid body, with smaller numbers of PPEIA-3′ cells found throughout the pons, medulla, and spinal cord. Thus cells expressing PPEIA-3′ RNA are widely distributed throughout the rat central nervous system.
The distribution of cells expressing PPEIA-3′ RNA is remarkable in that it is not coextensive with the distribution of any known neurotransmitter substances, including PPE. As we showed previously, in the forebrain, PPE mRNA and PPEIA-3′ are clearly not colocalized; the striatum contains many PPE neurons but no PPEIA-3′ neurons, whereas the reticular thalamic nucleus contains many PPEIA-3′ neurons but no PPE neurons. In hindbrain regions, while some PPE neurons have been reported (13) in the regions where we see PPEIA-3′, the details of the two distributions are different. For example, we see large numbers of PPEIA-3′ cells in the medial trapezoid (Fig. 2) whereas the few PPE neurons reported in the trapezoid were mainly located in the lateral subdivision (13). Thus in the hindbrain as well as the forebrain, PPEIA-3′ appears to be expressed in a different cell population than PPE mRNA.
Cell Populations that Express PPEIA-3′ RNA Also Express Other Atypical Nuclear Transcripts.
The distribution of cells expressing PPEIA-3′ RNA in the rat brain is strikingly similar to the distribution of cells expressing atypical nuclear RNAs described by other groups. Using an oligonucleotide probe directed to the alpha 2 adrenergic receptor, Nicholas et al. (22) reported the detection of intense hybridization signals located over the nucleus of cells located in the reticular thalamic nucleus, vestibular complex, trapezoid body, and deep cerebellar nuclei, as well as other brain areas. The same authors found a similar pattern with one beta-1 adrenergic receptor oligonucleotide probe (23). In both cases, as in the present work, the nuclear hybridization signal was strand and probe specific. The sequence of the probes used in these two studies did not show significant identity with each other, or with the probes used in the present study (P.B., unpublished observation). These data indicate that the expression of atypical nuclear transcripts in a subset of cells in the basal forebrain, reticular thalamus, deep cerebellar nuclei, vestibular complex, and trapezoid body (as well as scattered cells in other brain regions) is not limited to the PPE gene, but includes transcripts from other genomic loci as well. These findings lend further support to the hypothesis that these cells share a common set of factors that influence the production of atypical, nuclear localized transcripts from several different genes.
Generation of PPEIA-3′ RNA.
PPEIA-3′ RNA could be produced either by atypical transcription initiation and termination, or by an RNA processing mechanism. In our previous paper, we proposed that in cells expressing PPEIA-3′ RNA, transcription initiates from within intron A, but does not extend beyond the exon 2 boundary. Since nuclear run-on assays reflect RNA polymerase activity over different genomic regions in vivo, our hypothesis would predict that nuclear run-on assays in tissues containing large numbers of cells expressing PPEIA-3′ would detect higher levels of transcription over intron A than over exons 2 and 3. Independently, G. Weisinger et al. (unpublished manuscript) have carried out nuclear run-on assays using rat tissues, and found higher levels of transcription over intron A than over exon 3 in the cerebellum. This result is consistent with our data showing high levels of expression of PPEIA-3′ RNA in cells in the deep cerebellar nuclei. Our hypothesis (7) would explain both our results and those of Weisinger et al. in the cerebellum, and by extension, other cells in which PPEIA-3′ is expressed. Weisinger and coworkers (14, 15) proposed that a transcriptional down-regulation event in the region of intron A can influence PPE gene transcription initiated from exon 1 under certain conditions.
Control of transcription during the elongation phase has been shown to be a regulatory mechanism in several eukaryotic genes (16, 17). In cases where the RNA products of premature termination can be observed, they are typically heterogeneous (see e.g., refs. 18 and 19) that may explain our inability to identify PPEIA-3′ by biochemical methods. A parallel to our results with the PPE gene may be found in work with the c-Myc gene. Transcription from one promoter in c-Myc gives rise to full-length transcripts, while transcription from an alternate promoter gives rise to short transcripts that terminate heterogeneously (18, 20). Transcripts from the c-myc and N-myc genes have also been reported to show anomalous accumulation in the nucleus, apparently over nucleoli (21).
Functional Role of PPEIA-3′ RNA in the Nucleus.
The functional importance of PPEIA-3′ RNA, as well as the other aberrant nuclear RNAs expressed in this cell population remains to be determined. At a minimum, PPEIA-3′ RNA is a marker for a specific cell population in the rat brain, reflecting an underlying molecular similarity of cells within functionally diverse brain regions. The possibility that these atypical nuclear RNA transcripts may play a functional role in cells should also be considered, in view of recent work demonstrating that nontranslated nuclear RNAs can regulate gene expression by associating with specific chromosomal regions (24–27). Other data indicates a role for nontranslated RNA in processing other RNAs (28, 29), in DNA (30) and RNA (31) methylation, and in response to oxidative stress (32). Further studies of chromatin structure, gene expression and RNA processing mechanisms in the cell population that expresses PPEIA-3′ RNA are therefore warranted.
ABBREVIATIONS
- PPE
preproenkephalin
- hn
heterogeneous nuclear
- OVx
ovariectomized
- E
estradiol
- VMN
ventromedial nucleus
References
- 1.Lewin B. Cell. 1975;4:77–93. doi: 10.1016/0092-8674(75)90113-0. [DOI] [PubMed] [Google Scholar]
- 2.Sharp P. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz E, Mohr E, Richter D. DNA Cell Biol. 1988;10:81–91. doi: 10.1089/dna.1991.10.81. [DOI] [PubMed] [Google Scholar]
- 4.Birnsteil M. Structure and Function of the Major and Minor Small Nuclear Ribonucloproteins. London: Elsevier; 1980. [Google Scholar]
- 5.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N, Rastan S. Cell. 1992;71:510–520. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 7.Brooks P, Funabashi T, Kleopoulos S P, Mobbs C V, Pfaff D W. Mol Brain Res. 1993;19:22–30. doi: 10.1016/0169-328x(93)90144-e. [DOI] [PubMed] [Google Scholar]
- 8.Kilpatrick D, Zinn S A, Fitzgerald M, Higuchi H, Sabol S L, Meyerhardt J. Mol Cell Biol. 1990;10:3717–3726. doi: 10.1128/mcb.10.7.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks P, Kaplitt M G, Kleopoulos S P, Funabashi T, Mobbs C V, Pfaff D W. J Histochem Cytochem. 1993;41:1761–1766. doi: 10.1177/41.12.8245424. [DOI] [PubMed] [Google Scholar]
- 10.Romano G, Mobbs C V, Howells R D, Pfaff D W. Mol Brain Res. 1989;5:51–58. doi: 10.1016/0169-328x(89)90017-x. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro D J. Recent Prog Horm Res. 1989;45:29–58. doi: 10.1016/b978-0-12-571145-6.50006-6. [DOI] [PubMed] [Google Scholar]
- 12.Fremeau R, Autelitano D J, Blum M L, Wilcox J N, Roberts J L. Mol Brain Res. 1989;6:197–201. doi: 10.1016/0169-328x(89)90054-5. [DOI] [PubMed] [Google Scholar]
- 13.Harlan R E, Shivers B D, Romano G J, Howells R D, Pfaff D W. J Comp Neurol. 1987;258:159–184. doi: 10.1002/cne.902580202. [DOI] [PubMed] [Google Scholar]
- 14.Weisinger G. Biochem J. 1995;307:617–29. doi: 10.1042/bj3070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeCristofaro J D, Weisinger G, LaGamma E F. Mol Brain Res. 1993;18:133–40. doi: 10.1016/0169-328x(93)90182-o. [DOI] [PubMed] [Google Scholar]
- 16.Spencer C A, Groudine M. Oncogene. 1990;5:777–786. [PubMed] [Google Scholar]
- 17.Marshall N F, Price D H. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentley D L, Groudine M. Cell. 1988;53:245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- 19.Innis J, Kellems R E. Mol Cell Biol. 1991;11:5398–5409. doi: 10.1128/mcb.11.11.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer C A, LeStrange R C, Novak U, Hayward W S, Groudine M. Genes Dev. 1990;4:75–88. doi: 10.1101/gad.4.1.75. [DOI] [PubMed] [Google Scholar]
- 21.Bond A C, Wold B. Mol Cell Biol. 1993;13:3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholas A, Pieribone V, Hokfelt T. J Comp Neurol. 1993;328:575–594. doi: 10.1002/cne.903280409. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas A, Pieribone V, Hokfelt T. Neuroscience. 1993;4:1023–1039. doi: 10.1016/0306-4522(93)90148-9. [DOI] [PubMed] [Google Scholar]
- 24.Herzing L B K, Romer J T, Horn J M, Ashworth A. Nature (London) 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 26.Meller V H, Wu K H, Roman G, Kuroda M I, Davis R L. Cell. 1997;88:445–458. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- 27.Amrein H, Axel R. Cell. 1997;88:459–470. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 28.Sollner-Webb B. Cell. 1993;75:403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- 29.Balakin A G, Smith L, Fournier M J. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 30.Weiss A, Keshet I, Razin A, Cedar H. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 31.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–88. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 32.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]


