Abstract
Distinctive groups of fungi are involved in the diverse mycorrhizal associations of land plants. All previously known mycorrhiza-forming Basidiomycota associated with trees, ericads, liverworts or orchids are hosted in Agaricomycetes, Agaricomycotina. Here we demonstrate for the first time that Atractiellomycetes, members of the ‘rust’ lineage (Pucciniomycotina), are mycobionts of orchids. The mycobionts of 103 terrestrial and epiphytic orchid individuals, sampled in the tropical mountain rainforest of Southern Ecuador, were identified by sequencing the whole ITS1-5.8S-ITS2 region and part of 28S rDNA. Mycorrhizae of 13 orchid individuals were investigated by transmission electron microscopy. Simple septal pores and symplechosomes in the hyphal coils of mycorrhizae from four orchid individuals indicated members of Atractiellomycetes. Molecular phylogeny of sequences from mycobionts of 32 orchid individuals out of 103 samples confirmed Atractiellomycetes and the placement in Pucciniomycotina, previously known to comprise only parasitic and saprophytic fungi. Thus, our finding reveals these fungi, frequently associated to neotropical orchids, as the most basal living basidiomycetes involved in mycorrhizal associations of land plants.
Keywords: Orchid mycorrhiza, simple-septate Basidiomycota, Pucciniomycotina, Atractiellales, Helicogloea, neotropical mountain rainforest
1. Introduction
The tropical mountain rainforest of the Northern Andes is one of the hottest hotspots of biodiversity in the world (Beck et al. 2008). Orchidaceae are the most species-rich plant group in these forests (Homeier & Werner 2008) and cover tree stems and branches as well as the forest floor when light is sufficiently available, but orchids are also main elements of early succession on frequent natural and man-made landslides (Ohl & Bussmann 2004). The extraordinary successful orchid life strategy in the tropical mountain rainforest area is bewildering because orchids depend on distinct fungi for germination of their tiny seeds and for nutrition of the heterotrophic early stage of seedlings (protocorms; Bernard 1909; Smith & Read 2008). The tropical orchids maintain the fungal associations in the root cortical cells during their adult, photosynthetic phase. Molecular phylogenetic and transmission electron microscopic (TEM) studies, congruently, revealed only few, distinct fungal lineages as mycobionts of green orchids worldwide and mycorrhiza forming capabilities were only shown for selected fungal isolates by re-infection of protocorms (Smith & Read 2008). Orchids that are autotrophic in the adult stage were found to form mycorrhizae with Tulasnellales, Sebacinales and Ceratobasidiales (see Kottke & Suárez 2009; Yukawa et al. 2009), the former ‘binucleate Rhizoctonias’ (Taylor et al. 2002), hosted in Agaricomycetes in Agaricomycotina of Basidiomycota (Hibbett 2006). Fungi of further lineages of Agaricomycetes were found associated with mixotrophic and heterotrophic orchids linked to ectomycorrhiza-forming trees or living as saprophytes (see Smith & Read 2008; Kottke & Suárez 2009; Martos et al. 2009; Ogura-Tsujita et al. 2009; Roy et al. 2009). Thus, all so far known mycorrhiza-forming Basidiomycota are hosted in Agaricomycotina, while Ustilaginomycotina and Pucciniomycotina, the two other subphyla within the Basidiomycota (Bauer et al. 2006; AFTOL), to the best of our knowledge, comprise mainly parasites and to less extent presumed saprophytes (Weiß et al. 2004).
While our previous mycorrhizal studies in the tropical mountain rainforest combining molecular analysis with ultrastructural studies confirmed Tulasnellales and Sebacinales in epiphytic orchids (Suárez et al. 2006, 2008), large-scale sampling of terrestrial and epiphytic orchid roots in primary and regenerating forests and open, recovering landslides revealed, additionally and quite frequently, Atractiellomycetes, simple-septate Basidiomycota, not observed as mycorrhiza fungi previously. Here we present the fungal and host ultrastructure in the mycorrhizal association and the phylogenetic placement of the fungi.
2. Material and methods
(a). Field sites and root sampling
Roots of orchid individuals were collected on 56 permanent plots established in 2007 on four sites in the tropical mountain rainforest area of Reserva Biológica San Francisco (Beck et al. 2008) on the eastern slope of the Cordillera El Consuelo in the Andes of southern Ecuador bordering the Podocarpus National Park half way between Loja and Zamora, Zamora-Chinchipe Province (3°58′S, 79°04′W). The plots comprised 1 m2 each and included at least five orchid individuals of different species. Eight terrestrial plots and eight plots on tree stems between 1 and 2 m above ground were established in two pristine forests at about 2000 m (site 1) and 2170 m (site 4), and in a neighbouring 40 year old, regenerating forest at 2170 m (site 3). Eight terrestrial plots were installed on a 40-year-old manmade landslide at about 1900 m, regenerating mainly by orchids and some ericaceous shrubs (site 2). Plots were established randomly according to the highly variable conditions within the sites at least 10 m apart.
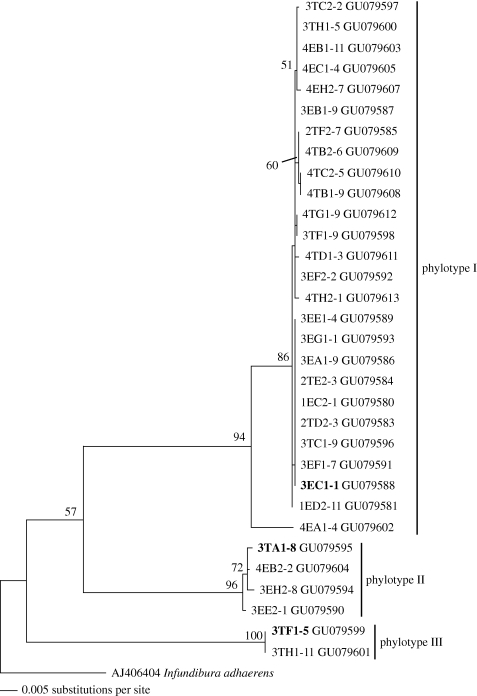
Orchid roots were sampled during February to May 2008 to extract fungal DNA. Three roots were collected for this study from two to three plant individuals per plot, 351 roots on total from 117 orchid individuals. The orchid individuals were identified by vegetative characters on genus level. Species could not be identified because of lack of flowering during the sampling period; several orchids displayed only insufficient vegetative characters for safe discrimination in the field. Because of conservational reasons in the protected area orchids were not removed to be sent to experts. Molecular barcoding was applied using pieces of orchid leaves but very few good sequences were obtained. It is, however, apparent from the genus list given in electronic supplementary material table S1e that all the identified orchids belong to subfamily Epidendroideae. Only roots in contact with the tree bark were collected from epiphytic orchids. Terrestrial orchids sampled in the forest sites had their roots in the pure humus layer while on the landslide roots were collected from the mineral soil. Preliminary observation had shown mycorrhizae in these microhabitats. Roots were screened for fungal colonization of the cortical tissue the day of sampling by microscopic observation of freehand sections stained using methyl blue (0.05% in lactic acid, Merck C.I. 42780). Well-colonized parts of roots were selected for DNA extraction. DNA was conserved at −20°C at Cellular and Molecular Biology, UTPL, Loja, Ecuador. Sequences of Atractiellomycetes are deposited in GenBank; accession numbers are given in figure 3.
Figure 3.
ML analysis of Atractiellomycetes sequences (ITS + nucLSU rDNA, 1550 bp) obtained from orchid mycorrhizae with I. adhaerens as outgroup. Bootstrap values are given for 1000 replicates, values below 50 per cent are omitted. The names of the sequences correspond to the orchid IDs giving site (1–4), habitat (Epiphytic or Terrestrial), plot (A–H), plant individual (1–5) and clone (corresponding to electronic supplementary material, table S1e). Sequences in bold were selected as representatives of the phylotypes.
Roots of orchids were sampled on sites 2, 3 and 4 from 22 of the epiphytic and terrestrial individuals as given above, in April 2008 and April 2009, to be examined by TEM. Mycorrhizal state was pre-examined and well-colonized root slices of 5 mm length were fixed in 2.5 per cent glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) the day of sampling.
(b). Transmission electron microscopy
Samples were post-fixed in 1 per cent osmium tetroxide and conventionally embedded in Spurr's plastic according to Bauer et al. (2006) but prolonging the infiltration steps up to 12 h. Semi-thin sections of 36 root samples were stained by crystal violet and observed for fungal colonization of the cortical tissue using light microscopy. Healthy looking colonized parts were selected for ultrathin sectioning. Serial ultrathin sections of 21 samples, six from site 2 terrestrial plots, four from site 3 terrestrial plots, six from site 3 epiphytic plots, and five from site 4 epiphytic plots were finally examined using a ZEISS TEM at 80 kV. Vouchers were deposited as plastic-embedded samples in the herbarium of Organismic Botany, Eberhard-Karls-University Tübingen.
(c). DNA isolation, polymerase chain reaction, cloning and sequencing
A 1–2 cm long piece was cut from each of the three selected colonized roots per plant for one DNA extraction per plant individual. The root pieces were rinsed in sterile water and freed from the velamen. Genomic DNA was recovered using a Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturers' instructions. The whole ITS1-5.8S-ITS2 region and part of the 28S rDNA were amplified with the universal primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′; White et al. 1990) and TW14 (5′-GCTATCCTGAGGGAAACTTC-3′; Cullings 1994) using the Phusion High-Fidelity PCR Mastermix (Finnzymes, Espoo, Finland). Success of PCR amplification was tested in 0.7 per cent agarose stained in ethidium bromide solution (0.5 µg ml−1). PCR products were cloned with the Zero Blunt TOPO PCR Cloning Kit (Invitrogen) according to manufacturer's protocol and Stockinger et al. (2009). Twelve colonies per individual were selected for PCR amplification using modified M13F and M13R primers (Krüger et al. 2009). Success of PCR was tested in 1 per cent agarose stained in a solution of ethidium bromide (0.5 µg ml−1). Eight colonies per orchid individual showing correct fragment size were grown in liquid LB Broth, MILLER (Difco) and purified with S.N.A.P. miniprep kit (Invitrogen) according to manufacturers' instructions. Clones were sequenced by Macrogen (Seoul, Korea) using universal primers M13F and M13R.
(d). Sequence editing, sequence identity and phylogenetic analysis
Sequences were edited and consensuses were generated using Sequencher 4.6 software (Gene Codes, Ann Arbor, MI, USA). BLAST (Altschul et al. 1997) against the NCBI nucleotide database (GenBank; http://www.ncbi.nlm.nih.gov/) was used to find published sequences with high similarity. The search yielded Tulasnella, Helicogloea/Infundibura, Sebacina, some few further basidiomycetes and few ascomycetes as close to our sequences. Only sequences showing high similarities to Atractiellomycetes (Helicogloea/Infundibura) were further analysed. These sequences were aligned using MAFFT v. 5.667 (Katoh et al. 2005) under the E-INS-i option. Maximum-likelihood (ML) analysis of the 32 new Atractiellomycetes sequences (ITS and part of the LSU) and Infundibura adhaerens as outgroup was performed using RAxML software v. 7.0.3 (Stamatakis 2006) under the GTRMIX model of DNA substitution with 1000 rapid bootstrap replicates (Felsenstein 1985). Two additional sequences were not included in the analysis because they were too short, but are given in electronic supplementary material, table S1e. Subsequently, one sequence from each resulting cluster was selected and aligned with sequences from a representative subphyla sampling across Pucciniomycotina, Ustilagomycotina and Agaricomycotina to confirm the phylogenetic placement within Basidiomycota. Six of the currently available 10 LSU sequences of Atractiellomycetes were included in the analysis. The taxon Taphrina deformans was used as outgroup. Highly divergent portions of the Basidiomycota alignment were eliminated using the Gblocks program v. 0.91b (Castresana 2000) with the following options: ‘Minimum Number of Sequences for a Conserved Position’ to 23, ‘Minimum Number of Sequences for a Flank Position’ to 37, ‘Maximum Number of Contiguous Non-conserved Positions’ to 8, ‘Minimum Length of a Block’ to 10 and ‘Allowed Gap Positions’ to ‘With half’. Phylogenetic trees and bootstrap replicates from the resulting Gblocks alignment were estimated as indicated above. Graphical processing of the trees with best likelihood and bootstrapping were generated using TreeViewPPC v. 1.6.6 (Page 1996) and PAUP* 4.0b10 (Swofford 2002).
3. Results
(a). Light and transmission electron microscopy
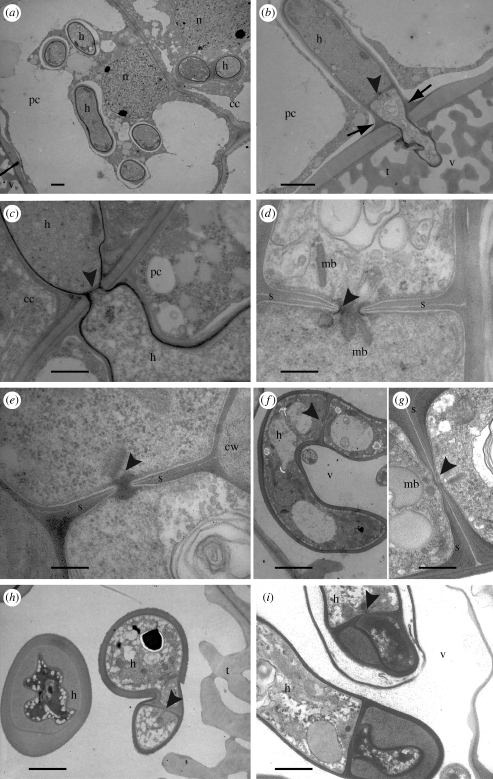
Light microscopic studies revealed the typical structures of orchid mycorrhizae in all the investigated 36 samples. Roots of the neotropical orchids consist of a stele surrounded by several layers of cortical cells, a suberized exodermal layer with intermingled non-suberized passage cells, covered by a mostly multilayered velamen of dead cells of which the innermost cell wall displays tilosomes, lignified wall protuberances (figure 1a,c). A multitude of fungi colonized the velamen (figure 1a,b). Coils (pelotons) of living or moribund hyphae were observed in the cortical cell layers at random distribution (figure 1a–c). TEM revealed that host cytosol was increased in cells colonized by living fungi and activity of hyphae and host cells was indicated by dense cytosol and large amounts of mitochondria (figure 1d,e). Living hyphae were separated from the host cytosol by the host plasma membrane (perifungal membrane) and an interfacial matrix (figure 1d,e). Dead pelotons showed the typical collapsed hyphae and encasement layers (electronic supplementary material, figure S1e). Focusing on the septal pore apparatus of the fungi in the cortical tissue we discerned Tulasnellales, Sebacinales and Ceratobasidiales by their dolipores with membrane caps and slime in the cell walls of Tulasnellales (electronic supplementary material, figure S1e, a–d). Unexpectedly, we found fungi with simple septal pores and no membrane caps (figure 1f,g,i) forming the just described, typical pelotons in the cortical tissue (figure 1b). The simple pores had rounded margins and were usually surrounded by membrane-coated microbodies of different shape, elongate or ovoid to rectangular, probably according to orientation of the sections (figures 1g and 2d and electronic supplementary material, figure S1e, g). The microbodies were visible as small dark points already at low magnification of TEM, a fact that facilitated search for the respective fungi. Occasionally, one microbody was located within the pore channel and plugged the pore (figure 1f). Close to the pores the cytosol may be rather dense and homogeneous (figure 1g). The hyphae contained symplechosomes, each consisting of two or more stacked cisternae of the endoplasmic reticulum that were interconnected by filaments (figure 1h and electronic supplementary material, figure S1e, h). Hyphal branches were clampless but formed septa close to branching (figure 1i). Hyphal cells were binucleate (electronic supplementary material, figure S1e, f).
Figure 1.
(a–c) Light micrographs of transverse section through root of Pleurothallis sp. 3TC1 displaying fungal colonization of velamen and cortical cells. (a) Overview (scale bar, 30 µm). (b) Cortical cell with coils (pelotons) of living hyphae (scale bar, 10 µm). (c) Cortical cell with collapsed, lysed hyphae and pycnoid nucleus (scale bar, 10 µm). (d–i) TEM of orchid-mycorrhiza structures and details of fungal features in unidentified orchids 3EB1 (d,e) and 3EE2 (f), Maxillaria sp. 4EC1 (g) and Pleurothallis sp. 3TC1 (h,i); (d) living hyphae embedded in dense root cytosol separated by plasma membrane and interfacial matrix (scale bar, 3 µm); (e) enlargement of hypha with well-preserved interfacial matrix (arrows; scale bar, 1 µm). (f) Hypha in root cortical cell displaying simple-pored septum without membrane caps, the pore plugged by a microbody (arrow head; scale bar, 1 µm); (g) enlargement of simple septal pore (arrow head) surrounded by several membrane-coated microbodies in dense cytosol (scale bar, 0.5 µm); (h) symplechosome in hypha of simple-septate basidiomycete (scale bar, 0.2 µm); (i) hypha branching without clamp formation displaying simple-pored septum (scale bar, 1 µm). cc, Cortical cell; cv, cortical cell vacuole; cy, cortical cell cytosol; dp, degenerating hyphal peloton; ex, exodermis; h, hypha; m, mitochondrion; mb, microbody; n, nucleus; p, living hyphal peloton; pc, passage cell; s, septum; st, stele; t, tilosomes; v, velamen.
Figure 2.
TEM showing series of root colonization in Pleurothallis sp. 3TC1 by the simple-septate basidiomycetes. (a) Overview of colonization of passage cell and adjacent cortical cell, note velamen with tilosomes at lower left corner (scale bar, 3 µm); (b) simple-septate basidiomycete (arrow head) passing from velamen among tilosomes into the passage cell, note narrowing of hyphal diameter, formation of septum and host cell wall apposition at the entry point (arrows; scale bar, 3 µm); (c) fungus passing from passage cell into cortical cell, note narrowing of hyphal diameter and formation of septum (scale bar, 2 µm); (d) enlargement of simple-pored septum with microbodies of micrograph (b) (scale bar, 0.5 µm); (e) enlargement of simple-pored septum of micrograph (c), pore plugged by electron dark material (scale bar, 0.5 µm); (f–i) simple-septate basidiomycete in the velamen: (f) large hypha in velamen (scale bar, 5 µm); (g) enlargement of the simple-pored septum with microbodies of micrograph (f) (scale bar, 1 µm); (h) square section through thick-walled hypha and hypha branching with septum formation (scale bar, 2 µm); (i) hypha with partial cell wall thickening and simple-pored septum, indicating that the thick-walled hyphae belong to the respective fungus (scale bar, 2 µm). For abbreviations see figure 1.
We observed the simple-septate basidiomycetes forming coils in the thin-walled passage cells (figure 2a) passing in from the velamen between the wall protuberances (tilosomes; figure 2b). The hyphal diameter was narrowed at the penetration point. Host plasma membrane and a kind of host cell wall apposition layer encased the penetrating hyphae (figure 2b). A simple-pored septum with adjoining microbodies was observed close to the entry point of the hyphae (figure 2b,d). The basidiomycetes further invaded the cortical cell layer forming narrow hyphal segments and septa at the penetration points that were transversed by simple pores (figure 1c,e). No signs of plant cell damage were noticed.
The fungus with simple septa was also observed in the velamen (figure 2f–i). Hyphal diameter was enlarged (5 µm) compared with hyphae in the cortical pelotons (3.5 µm) and showed local, prominent cell wall thickenings (figure 2h,i). Figure 2i displays the wall thickenings in a simple-septate hypha proving that both features belong to the same fungi. Wall thickenings were well visible in square sections of hyphae facilitating recognition of the respective fungi even by light microscopy.
The simple-septate basidiomycetes were observed in six of the 21 well-preserved orchid mycorrhizae, while dolipores were found in 14 samples, 11 of Tulasnella, two of Sebacina (electronic supplementary material, table S1e), and one of the Ceratobasidium parenthesome structure (electronic supplementary material, figure S1e, a–d). The simple-septate basidiomycetes were recorded from site 3 (regenerating forest at 2170 m) in two orchid individuals, an epiphytic and a humus terrestrial orchid (orchid ID: 3EE2 and 3TC1) and from site 4 (pristine forest at 2170 m) in roots of two epiphytic orchid individuals (orchid ID: 4EB2 and 4EC1). No simple-septate basidiomycetes were encountered by TEM in samples collected on site 2.
(b). Molecular phylogeny
Sequences of Tulasnellales were obtained from 89 orchid individuals (86%), Atractiellomycetes from 32 orchid individuals (31%) and Sebacinales in 19 orchid individuals (18%) from successful sequencing of mycobionts from 103 orchid individuals on total (electronic supplementary material, table S1e). The 32 sequences of Atractiellomycetes fell in three clusters supported by bootstrap values of 94, 96 and 100, respectively, indicating three phylotypes (figure 3 I–III). Sequences of phylotype I showed 90 per cent identity to phylotype II and 88 per cent identity to phylotype III, phylotype II and III showed 88 per cent identity (ITS1-TW14 fragment, 1500 bp). Proportional differences within the phylotypes were at most 0.4 per cent, with exception of sequence 4EA1_4 in phylotype I with 2.5 per cent difference. Phylotype I was the most frequent type, detected in 28 orchid individuals, phylotypes II and III were more rare, proven in four, respectively, two orchid individuals (figure 3; electronic supplementary material, table S1e). In two orchid individuals (3TF1, 3TH1) phylotypes I and III were both verified (electronic supplementary material, table S1e).
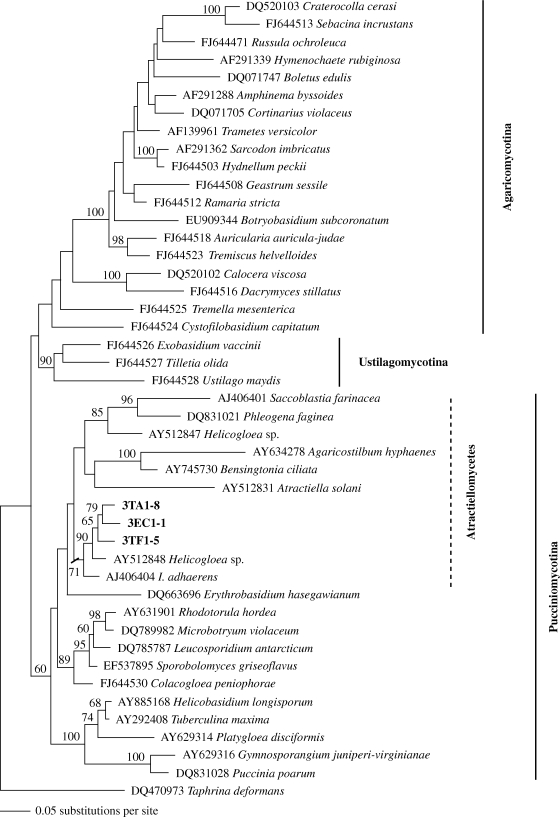
The Basidiomycota tree confirmed clustering of our new phylotypes in Atractiellomycetes, Pucciniomycotina. All three phylotypes clustered together with bootstrap 65 and form a supported cluster with Helicogloea sp. AY512848 (bootstrap 90) and I. adhaerens AJ406404 (bootstrap 71) (figure 4). Saccoblastia farinacea, Phleogena faginea and Helicogloea sp. AY512847 clustered in a separate clade of Atractiellomycetes (figure 4).
Figure 4.
Phylogenetic placement of the fungal representatives of phylotypes I–III associated with orchids (figure 3) within Basidiomycota based on ML analysis from an alignment of partial nuclear large subunit rDNA sequences. Bootstrap values are given for 1000 replicates; values below 50 per cent are omitted.
4. Discussion
(a). Indication of mycorrhizal state
The roles of mycorrhizal fungi in tropical, green orchids have been hitherto neglected; however, the intracellular hyphal coils (pelotons) in root cortical cells are seen as the major defining characteristics of orchid mycorrhizae (Smith & Read 2008; Rasmussen & Rasmussen 2009). We, therefore, argue for an orchid–mycorrhizal interaction of the simple-septate basidiomycetes as described on the basis of the ultrastructure here for the first time. No ultrastructural differences among mycorrhizal state of Tulasnellales, Sebacinales and the simple-septate basidiomycetes were observed in our material (compare Suárez et al. 2006, 2008; Kottke & Suárez 2009). The interface between living hyphae and host cytosol consisting of the plant plasma membrane and an encasement of most likely plant cell wall material correspond to the situation in other orchid mycorrhizae (Barroso & Pais 1985; Peterson et al. 1996). To confirm this conclusion isolating these fungi and establishing well-functioning experimental systems is required. Physiology and gene activities of the mycobionts need to be studied to clarify substrate access and interaction processes with orchid protocorms and roots.
(b). Systematic implications
Among the kingdom Fungi and the organisms in general, Atractiellomycetes (Pucciniomycotina, Basidiomycota) are unique in having symplechosomes (Bauer et al. 2006), cell organelles consisting of stacked cisternae of the endoplasmic reticulum, which are interconnected by hexagonally arranged filaments (Bauer & Oberwinkler 1991). The simple-septate orchid mycobionts share this characteristic feature which unambiguously makes clear that the simple-septate orchid mycobionts are members of the Atractiellomycetes. The single order Atractiellales of Atractiellomycetes comprises the teleomorphic genera Helicogloea, Saccoblastia, Phleogena, Atractiella and Basidiopycnis and the anamorphic genera Infundibura (anamorph of Helicogloea), Hobsonia, Leucogloea (anamorph of Helicogloea) and Proceropycnis (Oberwinkler & Bauer 1989; Kirschner 2004; Weiß et al. 2004; Bauer et al. 2006; Oberwinkler et al. 2006). Two types of septal pore apparatus were observed in these species, the atractielloid type in which the simple septal pores are surrounded by atractosomes and the puccinialean type in which the simple pores are surrounded by microbodies (Weiß et al. 2004; Bauer et al. 2006). The orchid mycobionts have the puccinialean septal pore apparatus.
Molecular analysis confirmed phylogenetic placement of our sequences in Atractiellomycetes of Pucciniomycotina. Representative sequences of the three phylotypes were retrieved in a clade together with Helicogloea sp. AY512848 and I. adhaerens, while Saccoblastia clustered with P. faginea and Helicogloea sp. AY512847. Atractiellomycetes do not appear as a monophylum in our LSU tree as was found by Bauer et al. (2006) and Aime et al. (2007). This incongruence needs future clarification but is probably not relevant to our conclusion. The three well-supported phylotypes of our Atractiellomycetes, separated by differences of 10–12%, argue for at least three different distant taxa.
Prominent wall thickenings are known from Tulasnellales, but these appeared fibrillar or strongly osmiophilic and were observed in pelotons of root cortical cells, in the velamen and in cultures (electronic supplementary material, figure S1e, b; Kottke & Suárez 2009). The wall thickenings of the simple-septate basidiomycetes are of homogeneous material appearing less osmiophilic than the cell wall layers and were only observed in hyphae colonizing the velamen.
(c). Ecological and evolutionary aspects
TEM observation and DNA sequencing found Atractiellomycetes in material from the same plant individual, but molecular approach revealed these fungi in many more samples, often together with Tulasnella or Sebacina sequences (electronic supplementary material, table S1e). Atractiellomycetes occurred on all the four sites, in primary and regenerating tropical mountain rainforest and on the regenerating landslide, in terrestrial orchids with roots in the mineral soil and those growing on pure humus layer and in roots of stem epiphytic orchids. Orchid individuals comprised diverse orchid genera (electronic supplementary material, table S1e) and most likely a multitude of species. The ecological amplitude of Atractiellomycetes, thus, appeared as broad as that of Tulasnellales mycobionts in the research area (electronic supplementary material, table S1e). Atractiellomycetes were the second most frequent orchid mycobionts implying potential importance for orchid conservation in the area.
Pucciniomycotina so far comprised parasitic and saprophytic fungi and Atractiellales have been suspected to be saprophytic (Bauer et al. 2006). Our finding is the first evidence of mycorrhizal fungi in this subphylum. The position of the mycobiont among potential saprophytes may indicate physiological flexibility from saprophytism to mutualism which is required for orchid mycobionts (Rasmussen & Rasmussen 2009).
It is puzzling that Atractiellomycetes were not reported from orchid mycorrhizae of other areas so far. At the current stage it is risky to speculate about these fungi as a peculiarity of the area under study, the extraordinary species-rich mountain rainforest of the northern Andes. The fungi may have been just overlooked because few studies used TEM and restricted number of studies identified orchid mycobionts by molecular tools.
It is generally accepted that mycorrhizae evolved several times independently in the fungal and plant kingdoms (Smith & Read 2008; Hibbett & Matheny 2009). In Basidiomycota, mycorrhiza-forming fungi were previously only known from Agaricomycetes of Agaricomycotina and therein Sebacinales were so far the most basal lineage known to form mycorrhiza associations (Weiß & Oberwinkler 2001; Hibbett 2006). We now encountered a mycorrhizal fungal group in Orchidaceae even more basal in Basidiomycota. Orchidaceae is an ancient plant group (Chase 2001). The tiny seeds with very few reserves and the mycorrhizal habit are almost certainly interdependent and could not have evolved without the appropriate fungi (Smith & Read 2008). It is tempting to speculate about the finding of a fungus in the Pucciniomycotina as an early evolutionary event in orchid mycorrhiza formation which was preserved in the tropical mountain rainforest, and would then probably predate the switch to Tulasnella and Ceratobasidium by the basal subfamily Apostasioideae (Yukawa et al. 2009). However, all the orchid individuals sampled for this study and by far the majority of orchids in the investigation area (Homeier & Werner 2008) belong to subfamily Epidendroideae, the most derived orchid subfamily (Cameron et al. 1999; Cameron 2004). The northern Andean mountain rainforest is a rather young ecotype sharing the most floral elements with the paleoflora from Meso- and North America (Taylor 1995). A rather recent local switch by orchids to Atratiellomycetes appears more reasonable, therefore. Currently, we can only encourage attention to Atractiellomycetes during future studies of orchid and other mycorrhizal types in tropical mountain and lowland forests worldwide in order to clarify their role in ecology and evolution of orchid mycorrhizae and mycorrhizae in general.
Acknowledgements
This work is dedicated to Prof. Dr. Franz Oberwinkler in honour of his 70th birthday. The authors thank the Deutsche Forschungsgemeinschaft (DFG) for funding (RU816) and Nature and Culture International (NCI) for providing research facilities in Ecuador.
References
- Aime M. C., et al. 2007An overview of the higher-level classification of Pucciniomycotina based on combined analysis of nuclear large and small subunit rDNA sequences. Mycologia 98, 896–905 (doi:10.3852/mycologia.98.6.896) [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J.1997Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (doi:10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso J., Pais M. S.1985Cytochimie—caractérisation cytochimique de l'interface hôte/endophyte des endomycorrhizes d’Ophrys lutea. Rôle de l'hôte dans la synthèse des polysaccharides. Ann. Sci. Nat., Bot. 13, 237–244 [Google Scholar]
- Bauer R., Oberwinkler F.1991The symplechosome: a unique cell organelle of some basidiomycetes. Botanica Acta 104, 93–97 [Google Scholar]
- Bauer R., Begerow D., Sampaio J. P., Weiß M., Oberwinkler F.2006The simple-septate basidiomycetes: a synopsis. Mycol. Progr. 5, 41–66 (doi:10.1007/s11557-006-0502-0) [Google Scholar]
- Beck E., Bendix J., Kottke I., Makeschin F., Mosandl R. (eds) 2008Gradients in a tropical mountain ecosystem of Ecuador In Series ecological studies, vol. 198 Berlin, Germany: Springer Verlag; ISBN 978-3-540-73525-0 [Google Scholar]
- Bernard N.1909L’évolution dans la symbiose. Les orchidées et leurs champignons commensaux. Ann. Sci. Nat. (Paris) 9, 1–196 [Google Scholar]
- Cameron K. M.2004Utility of plastid psaB gene sequences for investigating intra-familial relationships within Orchidaceae. Mol. Phyl. Evol. 31, 1157–1180 (doi:10.1016/j.ympev.2003.10.010) [DOI] [PubMed] [Google Scholar]
- Cameron K. M., Chase M. W., Whitten W. M., Kores P. J., Jarrell D. C., Albert V. A., Yukawa T., Hillis H. G., Goldman D. H.1999A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot. 86, 208–224 (doi:10.2307/2656938) [PubMed] [Google Scholar]
- Castresana J.2000Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- Chase M.2001The origin and biogeography of orchidaceae. In Genera orchidacearum, vol. 2 (ed. Pridgeon A. M.), pp. 1–5 Oxford, UK: Oxford University Press [Google Scholar]
- Cullings K. W.1994Molecular phylogeny of the Monotropoideae (Ericaceae) with a note on the placement of the Pyroloideae. J. Evol. Biol. 7, 501–516 (doi:10.1046/j.1420-9101.1994.7040501.x) [Google Scholar]
- Felsenstein J.1985Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 83–791 (doi:10.2307/2408678) [DOI] [PubMed] [Google Scholar]
- Hibbett D. S.2006A phylogenetic overview of the Agaricomycotina. Mycologia 98, 917–925 (doi:10.3852/mycologia.98.6.917) [DOI] [PubMed] [Google Scholar]
- Hibbett D. S., Matheny P. B.2009The relative ages of ectomycorrhizal mushrooms and their plant host estimated using Baysian relaxed molecular clock analyses. BMC Biol. 7, 13 (doi:10.1186/1741-7007-7-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeier J., Werner F. A.2008Spermatophyta checklist—Reserva Biológica San Francisco (Prov. Zamora-Chinchipe, S. Ecuador). In Provisional checklist of flora and fauna of San Francisco Valley and its surroundings, vol. 4 (eds Liede-Schumann S., Breckle S. W.), pp. 15–58 Ecotropical Monographs Ulm, Germany, Society for Tropical Ecology. [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T.2005MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (doi:10.1093/nar/gki198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner R.2004Sporodochial anamorphs of species of Helicogloea. In Frontiers in basidiomycote mycology (eds Agerer R., Piepenbring M., Blanz P.), pp. 165–178 Eching, Germany: IHW-Verlag [Google Scholar]
- Kottke I., Suárez J. P.2009Mutualistic, root-inhabiting fungi of orchids—identification and functional types. In Proc. Second Scientific Conf. on Andean Orchids (eds Pridgeon A. M., Suárez J. P.), pp. 84–99 Loja, Ecuador: Universidad Técnica Particular de Loja; (ISBN 978-9942-00-502-1) [Google Scholar]
- Krüger M., Stockinger H., Krüger C., Schüßler A.2009DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol. 183, 212–223 (doi:10.1111/j.1469-8137.2009.02835.x) [DOI] [PubMed] [Google Scholar]
- Martos F., Dulormne M., Pailler T., Bonfante P., Faccio A., Fournel J., Dubois M. P., Selosse M.-A.2009Independent recruitment of saprotrophic fungi as mycorrhizal partners by tropical achlorophyllous orchids. New Phytol. 184, 668–681 [DOI] [PubMed] [Google Scholar]
- Oberwinkler F., Bauer R.1989The systematics of gasteroid, auricularioid heterobasidiomycetes. Sydowia 41, 224–256 [Google Scholar]
- Oberwinkler F., Kirschner R., Arenal F., Villarreal M., Rubio V., Begerow D., Bauer R.2006Two new members of the Atractiellales: Basidiopycnis hyalina and Proceropycnis pinicola. Mycologia 98, 637–649 (doi:10.3852/mycologia.98.4.637) [DOI] [PubMed] [Google Scholar]
- Ogura-Tsujita Y., Gebauer G., Hashimoto T., Umata H., Yukawa T.2009Evidence for novel and specialized parasitism: the orchid Gastrodia confusa gains carbon from saprotrophic Mycena. Proc. R. Soc. B 276, 761–767 (doi:10.1098/rspb.2008.1225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl C., Bussmann R.2004Recolonisation of natural landslides in a tropical mountain forest of Southern Ecuador. Feddes Repertorium 115, 248–264 (doi:10.1002/fedr.200311041) [Google Scholar]
- Page R. D. M.1996TreeView: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- Peterson R. L., Bonfante P., Faccio A., Uetake Y.1996The interface between fungal hyphae and orchid protocorm cells. Can. J. Bot. 74, 1861–1870 (doi:10.1139/b96-223) [Google Scholar]
- Rasmussen H. N., Rasmussen F. N.2009Orchid mycorrhiza: implications of a mycophagous life style. Oikos 118, 334–345 (doi:10.1111/j.1600-0706.2008.17116.x) [Google Scholar]
- Roy M., Watthana S., Stier A., Richard F., Vessabutr S., Selosse A.2009Mycoheterotrophic orchids from Thailand tropical dipterocarpacean forests associate with a broad diversity of ectomycorrhizal fungi. BMC Biol. 7, 51 (doi:10.1186/1741-7007-7-51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Read D.2008Mycorrhizal symbiosis, 3rd ed.San Diego, CA: Academic Press [Google Scholar]
- Stamatakis A.2006RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- Stockinger H., Walker C., Schüßler A.2009‘Glomus intraradices DAOM197198’, a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 183, 1176–1187 (doi:10.1111/j.1469-8137.2009.02874.x) [DOI] [PubMed] [Google Scholar]
- Suárez J. P., Weiß M., Abele A., Garnica S., Oberwinkler F., Kottke I.2006Diverse tulasnelloid fungi form mycorrhizas with epiphytic orchids in an Andean cloud forest. Mycol. Res. 110, 1257–1270 (doi:10.1016/j.mycres.2006.08.004) [DOI] [PubMed] [Google Scholar]
- Suárez J. P., Weiß M., Abele A., Oberwinkler F., Kottke I.2008Members of Sebacinales subgroup B form mycorrhizae with epiphytic orchids in a neotropical mountain rain forest. Mycol. Progr. 7, 75–85 (doi:10.1007/s11557-008-0554-4) [Google Scholar]
- Swofford D. L.2002PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0b10 Sunderland, MA: Sinauer Associates [Google Scholar]
- Taylor D. W.1995Cretaceous to tertiary geologic and angiosperm paleobiogeographic history of the Andes. In Biodiversity and conservation of neotropical montane forests (eds Churchill S. P., Balslev H., Forero E., Luteyn J. L.), pp. 3–9 New York, NY: New York Botanical Garden, Bronx [Google Scholar]
- Taylor D. L., Bruns T. D., Leake J. R., Read D. J.2002Mycorrhizal specificity and function in myco-heterotrophic plants. In Mycorrhizal ecology. Ecological Studies 157 (eds van der Heijden M. G. A., Sanders I.), pp. 375–413 Berlin, Germany: Springer Verlag [Google Scholar]
- Weiß M., Oberwinkler F.2001Phylogenetic relationships in Auriculariales and related groups—hypotheses derived from nuclear ribosomal DNA sequences. Mycol. Res. 105, 403–415 (doi:10.1017/S095375620100363X) [Google Scholar]
- Weiß M., Bauer R., Begerow D.2004Spotlights on heterobasidiomycetes. In Frontiers in basidiomycote mycology (eds Agerer R., Piepenbring M., Blanz P.), pp. 7–48 Eching, Germany: IHW-Verlag [Google Scholar]
- White T. J., Bruns T. D., Lee S. B., Taylor J. W.1990Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR-protocols and applications: a laboratory manual (eds Innis M. A., Gelfand H., Sninsky J. S., White T. E.), pp. 315–322 New York, NY: Academic Press [Google Scholar]
- Yukawa T., Ogura-Tsujita Y., Shefferson R. P., Yokoyama J.2009Mycorrhizal diversity in Apostasia (Orchidaceae) indicates the origin and evolution of orchid mycorrhiza. Am. J. Bot. 96, 1997–2009 (doi:10.3732/ajb.0900101) [DOI] [PubMed] [Google Scholar]