Abstract
Subunit assembly governs regulation of AMPA receptor (AMPA-R) synaptic delivery and determines biophysical parameters of the ion channel. However, little is known about the molecular pathways of this process. Here, we present single-particle EM three-dimensional structures of dimeric biosynthetic intermediates of the GluA2 subunit of AMPA-Rs. Consistent with the structures of intact tetramers, the N-terminal domains of the biosynthetic intermediates form dimers. Transmembrane domains also dimerize despite the two ligand-binding domains (LBDs) being separated. A significant difference was detected between the dimeric structures of the wild type and the L504Y mutant, a point mutation that blocks receptor trafficking and desensitization. In contrast to the wild type, whose LBD is separated, the LBD of the L504Y mutant was detected as a single density. Our results provide direct structural evidence that separation of the LBD within the intact dimeric subunits is critical for efficient tetramerization in the endoplasmic reticulum and further trafficking of AMPA-Rs. The contribution of stargazin on the subunit assembly of AMPA-R was examined. Our data suggest that stargazin affects AMPA-R trafficking at a later stage of receptor maturation.
Introduction
The majority of fast excitatory synaptic transmission in the brain is mediated by AMPA receptors (AMPA-Rs), a subset of ligand-gated ion channels of the glutamate receptor family. Trafficking, anchoring, and gating of AMPA-Rs form the molecular basis for certain types of synaptic plasticity involved in learning and memory (Barry and Ziff, 2002; Malinow and Malenka, 2002; Nicoll et al., 2006). Dysfunction of AMPA-Rs is implicated in a variety of neurological and psychiatric disorders, including X-linked mental retardation, Alzheimer's disease, amyotrophic lateral sclerosis, and Rasmussen's encephalitis (Rogers et al., 1994; Shepherd and Huganir, 2007; Wu et al., 2007).
AMPA-R subunits are encoded by four different genes (GluA1-4) (Hollmann et al., 1989; Keinänen et al., 1990; Nakanishi et al., 1990). Each subunit consists of four domains (see Fig. 1A). The N-terminal domain (NTD) and ligand-binding domain (LBD) are both extracellular. The LBD, made of S1 and S2 subdomains, undergoes conformational changes resulting in channel gating upon glutamate binding. The polypeptide chain forming the LBD is interrupted by the channel pore-forming transmembrane domain (TMD), which consists of three membrane-spanning segments (M1, M3, and M4) and one re-entrant loop (M2) (Hollmann et al., 1994). A small C-terminal domain (CTD) extends into the cytoplasm, interacting with cytosolic proteins that regulate receptor anchoring and trafficking (Scannevin and Huganir, 2000; Sheng and Lee, 2001; Malinow and Malenka, 2002; Ziff, 2007). Cumulative evidence suggests an AMPA-R subunit assembly model in which two dimers come together to form a tetramer; hence, a dimer-of-dimer organization for mature tetrameric AMPA-Rs. (Armstrong et al., 1998; Gouaux, 2004; Mayer, 2006). In the brain, AMPA-R auxiliary subunits of the stargazin/TARP (transmembrane AMPA-R regulatory protein) family and the cornichon family are physically associated with the channel and regulate their trafficking and gating (Chen et al., 2000; Nakagawa et al., 2005; Nicoll et al., 2006; Ziff, 2007; Schwenk et al., 2009).
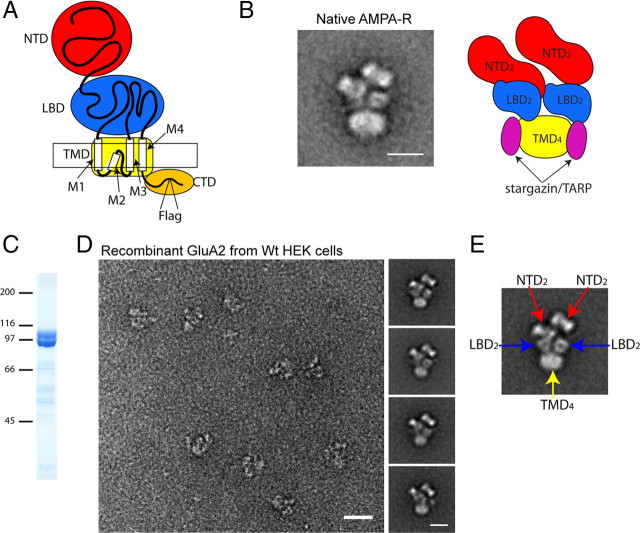
Figure 1.
Characterization of recombinant GluA2 homotetramers. A, Diagram depicting domain organization of an AMPA-R subunit. The box represents the lipid bilayer. M1–4 indicate the subdomains within the TMD. B, Representative class average of negative-stained native heterotetrameric AMPA-R particles purified from rat brain with schematic representation. TMD density includes stargazin/TARP protein(s). Scale bar, 10 nm. C, 7.5% SDS-PAGE gel of GluA2 doublet purified from constitutively expressing HEK cells (Coomassie brilliant blue-stained gel). Numbers on left indicate positions of the molecular weight marker in kDa. D, Left panel, Raw particle images of negative-stained recombinant GluA2 homotetramers purified from stably expressing HEK cells. Scale bar, 20 nm. Right small panels, Representative class averages. Scale bar, 10 nm. E, Class average of recombinant tetrameric GluA2 particle labeled with domain designations.
Trafficking of both newly synthesized and recycling AMPA-Rs is a critical component of synaptic plasticity (Malinow and Malenka, 2002; Ju et al., 2004; Park et al., 2004; Matsuo et al., 2008). The molecular anatomy of early phases of AMPA-R trafficking, including biosynthesis, is largely unknown. Studies have identified point mutations and splice variants that alter receptor trafficking and, together with the crystal structures of the mutated S1S2 domains, have provided insight into what might be happening at the ultrastructural level of the intact subunits during receptor assembly (Greger et al., 2002, 2003, 2006; Coleman et al., 2006). It is not clear how the domains are organized during the normal assembly of full-length subunits and how mutant subunits interfere with this process. In addition, how the auxiliary subunit stargazin influences biosynthesis of AMPA-R remains controversial.
Here, we investigate AMPA-R subunit assembly and report single-particle EM structures of newly synthesized AMPA-Rs in dimeric states. The study reveals that efficient subunit assembly requires a preferred conformation of AMPA-R biosynthetic intermediates and that stargazin affects AMPA-R trafficking at the later stages of receptor maturation.
Materials and Methods
Recombinant DNA.
The GluA2 flop splice variant was used for all experiments. The L504Y mutation was introduced by in vitro mutagenesis using Quick change kit (Stratagene). The green fluorescent protein (GFP)–GluA2 fragment was a gift from Y. Hayashi (RIKEN, Wako, Japan), and GFP was inserted immediately after the signal peptide, following the exact design as previously described (Hayashi et al., 2000). The FLAG epitope tag was inserted in the C-terminal domain of GluA2 (FATDYKDDDDKEGYNVYGIESVKI, where bold case indicates FLAG epitope), and placement preserves the original anti-GluA2CT epitope.
Generation of stable HEK cell line.
Wild-type HEK cells, GnTI(-)HEK cells, and the transformants created were maintained in a base media that consisted of high-glucose DMEM, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum. To isolate stable clones, we cotransfected a plasmid vector that expresses GluA2 under the CMV promoter and another plasmid vector that expresses a hygromycin-resistant gene. Transfection was performed by calcium phosphate methods, and the selection of clones was performed over 2 weeks in the presence of 160 μg/ml hygromycin. Isolated colonies were cultured until morphologically homogeneous cultures were established. Expression of GluA2-FLAG was tested for each clone using Western blotting of the whole-cell lysate by probing with custom-made antibodies raised against the C-terminal peptide of GluA2 (EGYNVYGIESVKI) (Nakagawa et al., 2005). Through screening ∼200 colonies, we identified several clones that met the criteria of optimal growth speed and expression. There was a tendency for highly expressing clones to be slow growing, consistent with toxicity to the host cell caused by overexpressing an ion channel. To assess stability, we kept culturing the established clones for 7 months and detected by immunofluorescence microscopy that 65% of the cells maintained expression of GluA2 (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Thus, the stable cell line we established can be used for large-scale culture to produce recombinant GluA2 in large quantities. Typically, a 1 L culture of HEK cells was used for each purification in this study.
Generation of stable HEK cell lines that express GluA2-FLAG by doxycycline induction.
A neomycin (G418)-resistant TetON-HEK cell line (Clontech) has in its genome the expression module to produce reverse Tet transcriptional activator (see Fig. 2A). GluA2-FLAG, GluA2L504Y-FLAG, GFP-GluA2-FLAG, and GFP-GluA2L504Y-FLAG were subcloned into pTREtight vector (Clontech). TetON-HEK cells were cotransfected with a plasmid that expresses a hygromycin-resistant gene and a GluA2 construct in pTREtight described above. Transfection was performed using calcium phosphate, and the selection of clones was performed over 2 weeks in the presence of 120 μg/ml hygromycin. The remaining procedure follows the generation of the stable HEK cell lines described above, except that we detected the expression of GluA2 using Western blotting after inducing the isolated clones with 5 μg/ml doxycycline (DOX) for 24 h.
Figure 2.

Purification and EM imaging of GluA2 dimers. A, Schematic of TetON system of protein induction in which the addition of DOX promotes GluA2 production. B, Western blot depicting the time course of GluA2 protein expression after induction. Note the formation of double band after 24 h. C, Gel filtration chromatograph for GluA2 expressed and purified at 20 h (solid line) and 24 h (dotted line) after induction. Positions of tetramer and dimer are indicated. D, Raw particle images of GluA2 dimers (top) and representative class averages (bottom small panels) purified from TetONGluA2 HEK cells. Scale bar, 10 nm. E, Diagram of Fab fragment labeled GluA2 subunit (left). Raw particle images GluA2 dimer labeled with Fab fragment. Under each class average is a representation to facilitate interpretation. Receptor complex is in white, and Fabs are in gray. Fab labels C-terminal portion. Scale bar, 10 nm. F, Diagram of GFP-tagged GluA2 subunit (left). Class averages of GFP-GluA2 dimer particles (right top). Under each class average is a representation to facilitate interpretation. The receptor complex is in white, and GFP is in green. GFP labels the N-terminal portion. Scale bar, 10 nm. G, Summary of domain labeling. Class average of a dimeric AMPA-R particle labeled with domain designations.
Generation of TetOnGluA2-stg stable HEK cells.
Stargazin-IRES-mCherry cassette was subcloned into pBOSS vector (a gift from Shigekazu Nagata and Hideki Sakahira, Osaka University, Osaka, Japan) downstream of the elongation factor promoter. pBOSS-stg-IRES-mCherry vector and pCMVZeocin (Invitrogen) were cotransfected into the parental TetONGluA2 stable HEK cell and stable clones were isolated by selecting with antibiotics 125 μg/ml zeocin, 150 μg/ml hygromycin, and 125 μg/ml neomycin (G418). mCherry-positive colonies were visually identified using an epifluorescent microscope, isolated, and subcultured. Eighty percent of the mCherry-positive clones also expressed stargazin, as determined by Western blotting. DOX-inducible GluA2 expression was also reconfirmed in all of the isolated cell lines.
Harvesting HEK cell monolayer from a large number of plates.
Five confluent 15 cm dishes were taken from the CO2 incubator at a time. Media was aspirated off, and 6 ml of ice-cold PBS was added to each plate. The plates were tapped strongly from the side about 10–15 times to dislodge all cells from the bottom of the dish. Cells were pooled in a 250 ml centrifuge tube on ice. The dishes were further rinsed with 3 ml of PBS twice to collect all remaining cells. Cells collected from 20 plates filled up the 250 ml centrifuge tube. We centrifuged the cells at 1000 rpm at 4°C for 10 min. After discarding the supernatant, the pellet was resuspended in 50 ml of PBS and further centrifuged. After discarding the supernatant, the cell pellet was flash frozen in liquid nitrogen and stored at −80°C until use.
Purification of recombinant GluA2.
All purification procedures were conducted on ice or in the cold room to maintain a specimen temperature of <4°C. One liter of HEK cell culture (6 ml of cell pellet) was resuspended in 50 ml of buffer containing 50 mm K-HEPES, pH 7.4, 100 mm NaCl, 1 mm Kynurenic acid, protease inhibitors (1 mm PMSF, 10 μg/ml leupeptin, atropinin, benzamidine, and pepstatin A). The cells were extracted with the detergent dodecyl maltoside (DDM) (0.25%) at 4°C for 3 h. Solubilization yield was >90%, which is unsurprising given that AMPA-Rs from the brain solubilize efficiently when synaptosomal fractions are extracted with mild detergents (Leonard et al., 1998). After clearing the lysate by ultracentrifugation (Beckman 45 Ti) at 45,000 rpm for 1 h at 4°C, the supernatant was applied to a column made of protein A Sepharose beads (GE Healthcare) crosslinked using DMP (Pierce) with anti-FLAG M2 monoclonal antibody (Sigma) at a concentration of 2 mg/ml. Following washing, bound proteins were released from the column using a buffer containing 0.5 mg/ml FLAG epitope peptide. The peak fraction from the peptide elution was further separated by Superdex 200 gel filtration column (GE Healthcare) in a buffer that contains 50 mm K-HEPES, pH 7.4, 100 mm NaCl, and 0.1% DDM.
Detection of stargazin from the preparation of recombinant GluA2.
Purified GluA2 was resolved in 7.5% SDS-PAGE, and Western blotting was performed using the custom-made anti-panTARP antibody as described previously (Nakagawa et al., 2005). In the case of the mass spectrometry, the gel was stained with CBB. All the bands between 30 and 60 kDa were cut out and digested with trypsin. The identity of each band was determined by liquid chromatography followed by tandem mass spectrometry (LC/MS/MS) at the University of California, San Diego, mass spectrometry facility.
Negative staining of purified proteins and EM.
Four hundred-mesh copper grids were coated with carbon to create a substrate for proteins to bind. Four microliters of protein solution was applied to a glow-discharged grid and left for 30 s to 5 min to allow the proteins to bind. The excess water was blotted on filter paper, and the specimen was washed twice in water droplets to remove excess detergent. Purified proteins were negative stained with 0.75% (w/v) uranyl formate, as described previously (Ohi et al., 2004). Images were recorded using a FEI Sphera electron microscope equipped with an LaB6 filament operated at an acceleration voltage of 200 kiloelectron volts. Images were taken at a magnification of 50,000 × and defocus value = −1.5 μm. Specimens were imaged at 0° and 60° tilt for random conical tilt three-dimensional (3D) reconstruction; the defocus value for 0° was −1.5 to approximately −1.8 μm, and for 60° tilt it was −2.0 to approximately −2.2 μm. All images were recorded using SO-163 film and developed with Kodak D-19 developer at full strength for 12 min at 20°C. Particle images were taken at room temperature and under low-dose conditions (20 e/Å2) to minimize radiation damage.
Antigen-binding fragment labeling.
Antigen-binding fragments (Fabs) were purified using the Immunopure IgG1 F(ab′) and the F(ab′)2 Fab purification kit (Pierce) followed by gel filtration on a Superdex 200 column (Pharmacia). Anti-FLAG M2 monoclonal antibody (Sigma) was used as the source. Labeling was performed by incubating dimeric AMPA-Rs with Fab fragments at a molar ratio of 1:4–1:8 overnight at 4°C in 50 mm HEPES, pH 7.4, 100 mm NaCl, and 0.1% DDM.
Image processing.
Electron micrographs were digitized with a CoolScan 9000 (Nikon) using a step size of 6.35 μm, and 3 × 3 pixels were binned so the specimen level pixel size used was 3.81 Å. Projection averages were calculated from windowed small images of 100 × 100 pixels over 10 cycles of K-means classification and multireference alignment specifying 100 classes. For 3D reconstruction of GluA2 wild-type dimer, 269 tilt pairs (a total of 538 micrographs) were recorded on film, from which 178 tilt pairs were selected based on image quality. A total of 13,345 particle pairs were interactively selected using WEB display program for SPIDER (Frank et al., 1996) and windowed, and the untilted particles were averaged into 100 classes, as was done before. In the case of the GluA2L504Y dimer, 238 tilt pairs (a total of 476 micrographs) were recorded, from which 159 tilt pairs were selected based on image quality. A total of 9000 particle pairs were selected and used for further analysis. Raw particle images were visually inspected after classification to make sure that tetramers were not mistakenly introduced into our dimer 3D reconstruction. Images of the tilted specimens for each class were used to calculate initial 3D reconstructions of individual classes by back projection, back-projection refinement, and angular refinement (implemented in SPIDER). The final volume obtained by angular refinement with SPIDER was used as the input model for FREALIGN (Grigorieff, 2007); this was used for refinement of orientation parameters of individual particles and for individual image contrast transfer function correction based on the defocus value. The tilt angles and defocus values of the center of each micrograph were determined with CTFTILT (Mindell and Grigorieff, 2003). The defocus of each particle was deduced from its position on the micrograph. Particles selected from tilted and untilted specimens were used for FREALIGN refinement (500 and 800 particles were used in the final reconstruction for wild-type and L504Y particles, respectively). To ensure that the final 3D reconstruction agreed with the raw data, the particle images were compared with reprojections.
Surface labeling of HEK cells.
Expression of GFP-GluA2 wild type and GFP-GluA2L504Y was induced with 7.5 μg/ml DOX. Thirty hours after induction cells were live labeled using an anti-GluA2 NTD monoclonal antibody for 15 min (MAB397, Millipore). Cells were washed with warm DMEM and fixed with 4% formaldehyde in 0.1 m phosphate buffer, pH 7.4. Surface GluA2 was detected using Alexa 568-conjugated anti-mouse IgG secondary antibody (Invitrogen). Total GluA2 was detected by the GFP fluorescence signal. Imaging was performed using an Olympus Fluoview confocal microscope using the 60× objective lens.
Subcellular colocalization of total GFP-GluA2 and internalized GFP-GluA2.
GFP-GluA2 expression was induced for 30 h with 7.5 μg/ml DOX followed by labeling with 10 μg/ml anti-GluA2NTD monoclonal antibody (MAB397, Millipore) in the CO2 incubator for 1 h. During the 1 h incubation period, GFP-GluA2 labeled with the antibody undergoes endocytosis. Unbound antibodies were washed with DMEM (37°C), and the cells were fixed with 4% formaldehyde. Alexa 568-conjugated anti-mouse IgG antibody (Invitrogen) was used as a secondary antibody to label the primary antibodies. All the images were taken on an Olympus Fluoview confocal microscope, as described previously in this section.
Time-lapse imaging.
HEK cells were grown on a glass-bottom dish for 24 h before 7.5 μg/ml DOX was added. Time-lapse imaging occurred 30 h after induction using an Olympus Fluoview confocal microscope mounted with a temperature- (37°C), CO2- (5%), and humidity-controlled chamber and equipped with a 60× objective lens [Olympus PLAPON; numerical aperture (NA), 1.42]. One-half-micrometer-thick optical sections were obtained up to 4 μm from the bottom of the cell. Recording was done continuously, resulting in 20 s time intervals between each Z-stack.
Neuron culture and immunostaining.
Primary cultures of rat hippocampal neurons were prepared from embryonic day 18 rat embryos, as previously described (Sala et al., 2003). HA-tagged GluA2 constructs were introduced into neurons at DIV14 using calcium phosphate transfection. Surface HA-GluA2 was stained while neurons were alive. Specifically, after labeling the DIV 18 neurons with anti-HA monoclonal antibody (HA.11, Covance), neurons were washed in DMEM to remove unbound antibodies and fixed with 4% formaldehyde in 0.1 m phosphate buffer, pH 7.4. After washing with PBS, internal HA-GluA2 was labeled using anti-HA polyclonal antibody (Y-11, Santa Cruz Biotechnology) diluted in 1XGBD (0.2% gelatin, 0.6% Triton X-100, 33 mm phosphate buffer, pH 7.4, and 0.9 m NaCl). Alexa 488-conjugated anti-mouse IgG (Invitrogen) and Alexa 568-conjugated anti-rabbit IgG (Invitrogen) were used as secondary antibodies. Neurons were imaged using an Olympus Fluoview confocal microscope using the 60× objective lens (Olympus PLAPON; NA, 1.42). Z-projections of confocal stacks are shown in Figure 3B.
Figure 3.

Contrasting dynamics of GFP-GluA2 wild type and L504Y. A, Diagram showing position of the L504Y mutation in GluA2 subunit. B, Fluorescent confocal images of primary rat hippocampal neurons overexpressing HA-GluA2 wild type (top) or HA-GluA2L504Y (bottom). Surface GluA2 (green) was live labeled with anti-HA monoclonal antibody. Fixed and permeablized neurons were labeled with anti-HA polyclonal antibody (red) to stain intracellular GluA2. Scale bar, 20 μm. C, Fluorescent confocal images of HEK cells expressing GFP-GluA2 wild type (top) or GFP-GluA2L504Y (bottom). Surface expression of the L504Y mutant is reduced in our inducible expression system. Surface labeling (red) was done 30 h after induction. Total GluA2 was detected by the GFP signal (green). Scale bar, 5 μm. D, Scheme of time-lapse imaging. The 4 μm between the bottom of the nucleus and the bottom of the cell attached to the coverglass (yellow volume) provided the clearest images of receptor trafficking. E, Confocal Z-stack projection of GFP-GluA2 wild type in HEK cells. Imaged 30 h after GluA2 induction. Scale bar, 4 μm. F, Confocal Z-stack projection of GFP-GluA2L504Y. Imaged 30 h after GluA2 induction. Same magnification as in E. G, Time-lapse image taken from the red square in E. Note difference in dynamics between the puncta pointed out by red and yellow arrows. Scale bar, 4 μm.
Time course of GluA2 expression in TetOnGluA2-stg cell lines.
Four clones (2, 8, 9, and 10) of TetOnGluA2-stg cell lines and the parental TetONGluA2 cell were plated on the six-well plates at a density of 0.6 × 106 cells/well and were incubated for 24 h to allow the cells to attach. At this point, the cells were near confluent, and 7.5 μg/ml DOX was added with 30 μm NBQX to induce GluA2 expression. Cells were harvested 0, 6, 12, 18, and 24 h after induction. Equal amounts of protein sample from each time point were loaded into each well of the SDS-PAGE gel (12.5% and 7.5% gels were used to resolve stargazin and GluA2, respectively). Western blotting was done using anti-panTARP and anti-GluA2C-terminal polyclonal antibodies [both generated in the study by Nakagawa et al. (2005)].
Immunostaining of HEK cells expressing stargazin and GluA2.
TetOnGluA2-stg clone 10 cells were fixed, as described previously in this section, 24 h after DOX induction. Anti-FLAG monoclonal (Sigma) and anti-panTARP polyclonal antibodies (Nakagawa et al., 2005) were used to detect GluA2 and stargazin, respectively. Cy5-conjugated anti-mouse IgG (Jackson ImmunoResearch) and Alexa 488-conjugated anti rabbit (Invitrogen) were used as secondary antibodies. Confocal Z-stack images of the cells were recorded as above.
Results
Simplified biochemical preparation to study AMPA receptor assembly
To obtain pure intact AMPA-Rs for EM analyses, we stably expressed FLAG epitope-tagged GluA2 flop (Fig. 1A) in HEK cells. Recombinant GluA2 was solubilized in DDM in the presence of 1 mm kynurenic acid and purified by affinity chromatography using a Sepharose column conjugated with anti-FLAG M2 monoclonal antibody (Fig. 1C). The purity was further improved by gel filtration chromatography. The majority of purified GluA2 existed as tetramers (supplemental Fig. 1C, available at www.jneurosci.org as supplemental material). Because of glycosylation, the purified protein migrated in the SDS-PAGE as a doublet (Fig. 1C). Only the tetrameric species were glycosylated (supplemental Fig. 1B–D, available at www.jneurosci.org as supplemental material). Based on the yield of purification we estimate that at least 15,000∼20,000 GluA2 tetramers are expressed per cell.
Recombinant GluA2 tetramers were homogeneous in shape and size when imaged by negative stain EM (Fig. 1D, left panel). The projection structures (class averages) of GluA2 tetramers were very similar to those of native AMPA-Rs from rat brain (Fig. 1, compare B, D). The NTD, LBD, and TMD were clearly identified as distinct features (Fig. 1E). The TMD of native AMPA-Rs is larger than that of recombinant GluA2, a difference caused by the presence or absence of auxiliary stargazin/TARP subunits (Chen et al., 2000). Consistently, we did not detect stargazin/TARP protein in the purified recombinant GluA2 fraction when examined by Western blotting or by protein identification using LC/MS/MS (data not shown). Glycosylation did not affect the overall structure of tetrameric GluA2. The shapes of negatively stained glycosylated GluA2 tetramers were very similar to GluA2 tetramers purified from the GnTI(-) HEK cell line, a line defective in complex mannose glycosylation (Reeves et al., 2002) (supplemental Fig. 1E, available at www.jneurosci.org as supplemental material). Altogether, our stable HEK cell line is a simple, robust, and highly reproducible system to study AMPA-Rs using single-particle EM.
Isolation of dimer intermediates of newly synthesized AMPA receptors
To enrich biosynthetic intermediates that have a shorter lifetime than mature and targeted proteins, we induced expression of GluA2-FLAG in HEK cells and purified them at an early time point. In this system, HEK cells stably express a reverse Tet transcriptional activator that enhances transcription from the minimal CMV promoter upstream of the GluA2-FLAG transgene only in the presence of DOX (Fig. 2A). We named this cell line TetONGluA2. GluA2 protein expression reached a maximum 24 h after DOX application (Fig. 2B). The magnitude of GluA2 expression in DOX inducible system was similar to that of the constitutive expression when the expression was induced for 24 h at 7.5 μg/ml DOX. Enrichment of the glycosylated band became prominent 12–24 h after transcription initiation (emergence of the upper band in Fig. 2B).
When GluA2 was purified 20 h after addition of DOX and resolved by gel filtration, equal amounts of tetramers and dimers were detected (Fig. 2C, solid line). The tetramer peak increased relative to the dimer peak 20 to 24 h after DOX induction. At earlier time points (15 h after induction) the majority of GluA2 existed as dimers (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Collectively, this suggests that the dimer population precedes the formation of the tetramer population, and thus represents a biosynthetic intermediate of preassembled GluA2 tetramers.
Interestingly, even at 15 h after DOX induction we were unable to detect a distinct population that represents the monomeric subunits (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Because the appearance of the monomers should in theory precede the dimers, we interpret that at a given moment the monomer population is much lower relative to the dimer population. It is thus difficult to detect monomers in our experimental system. Such an interpretation is consistent with an idea that the dimer-to-tetramer transition is the rate-limiting step compared with the monomer-to-dimer transition.
Projection structures of the GluA2 wild-type dimers
When GluA2 biosynthetic dimer intermediates were imaged by negative stain EM, the particles were homogeneous in size but existed in a variety of shapes (Fig. 2D, top). Approximately 7000 particles were analyzed by multivariate statistical analysis, classification and multi reference alignment. Representative class averages are shown in Figure 2D, bottom. There is an elongated bipartite density at the top of the particle, and a featureless globular density at the bottom of the particle. In some class averages the upper bipartite density appears as a squarish density with a weaker density in the center (Fig. 2D). Between these structures two small round densities are positioned at both sides. The arrangement of the bipartite density relative to the small round densities on the sides differs significantly between class averages, and as a result, the heights of the particles vary between class averages.
The projection structures were interpreted by molecular labeling. To immunolabel the C-terminal FLAG epitope, Fabs were generated by proteolytic digest from IgG of anti-FLAG monoclonal antibody (Fig. 2E, left). Anti-FLAG Fab consistently labeled the bottom featureless density of the particles (Fig. 2E, right panels). Consistent with our interpretation that these particles are dimers, some particles were labeled with two Fabs (Fig. 2E, far right). Because the CTD is small and attached to the TMD, we interpret that the bottom round density represents the TMD and CTD (Fig. 2G).
To label the NTD with GFP, we purified particles from a DOX-inducible HEK cell line that expresses a GluA2 construct whose N-terminal end of the NTD is fused to GFP (Fig. 2F). The time course of GFP-GluA2 expression after DOX induction and the elution profile of purified GFP-GluA2 in gel filtration chromatography were similar to those without the GFP tag (data not shown), suggesting that GFP does not affect overall processing. In the class averages of GFP-GluA2 dimers, two GFP densities were always attached to each side of the bipartite density (Fig. 2F). The small round densities between the NTD and the TMD were neither labeled by anti-FLAG Fab or GFP, so we interpret that they are the LBDs (Fig. 2G).
Contrasting vesicle trafficking of GluA2 wild type and L504Y
Maturation and trafficking are tightly coupled processes in membrane proteins. To understand the functional significance of the dimer structure, we decided to study the structure of the L504Y mutant, which has trafficking and maturation deficits (Fig. 3A). This mutation within the LBD was originally identified in the GluR3 subunit because it blocks receptor desensitization (Stern-Bach et al., 1998). L504Y mutation is located in helix D in the S1 subdomain and the mutated tyrosine interacts with helix J in the S2 subdomain of the adjacent subunit (Sun et al., 2002). This interaction stabilizes the LBD dimer interface and, thus, blocks the receptor from entering into the desensitized state. Recently, it was found that surface expression of the GluA2L504Y mutant is impaired (Greger et al., 2006). Consistently, we observe significantly reduced surface expression of GluA2L504Y compared with GluA2 wild type when expressed for 4 d starting from DIV14 in primary rat hippocampal neurons (Fig. 3B).
We created stable HEK cell lines in which expression of GFP-GluA2 wild type or GFP-GluA2L504Y is DOX inducible. In both cell lines, the time courses of protein expression after DOX induction were similar to those without the GFP tag (data not shown), suggesting that the GFP has minimal effect on protein turnover. At 30 h after DOX induction, there was significantly less GFP-GluA2L504Y on the cell surface compared with GFP-GluA2 wild type, while the total expression levels of each protein were the same (Fig. 3C). This suggests that the defect in surface expression of GluA2L504Y can be replicated in our simplified HEK cell system.
To gain further insight into the differential dynamics of newly synthesized GFP-tagged GluA2 wild type and L504Y, we conducted time-lapse confocal imaging of HEK cells. We imaged a thin layer of cytoplasm between the bottom of the nucleus and the bottom of the cell that provides clear images of receptor trafficking (Fig. 3D, yellow volume).
For the first 24 h following DOX induction, GluA2 wild type and GluA2L504Y showed similar subcellular distribution, localized predominantly in the endoplasmic reticulum (ER) (data not shown). After 24 h, dynamic punctate structures emerged in the GluA2 wild-type cells but not in the GluA2L504Y cells (Fig. 3, compare E, F). The vesicles appeared at time points after the majority of receptor maturation was complete. More than 50% of the punctate structures in GluA2 wild-type cells were dynamic and translocated rapidly (Fig. 3G; supplemental movies, available at www.jneurosci.org as supplemental material). These punctate structures were absent from GluA2L504Y cells even 48 h after DOX induction, suggestive of differential vesicle trafficking patterns caused by the mutation (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Double staining GFP-GluA2 puncta with the known organelle markers revealed no colocalization with EEA1 (an early endosomal marker), transferrin (a recycling endosomal marker), or lysotracker (a lysosomal marker). However, a subpopulation of GFP-GluA2 vesicles colocalized with rab6 (a small GTPase localized to a subset of the post-Golgi vesicles), GM130 (a Golgi marker), and PDI (an ER marker) (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). In addition, the GFP-GluA2 puncta partially colocalized with a population of GFP-GluA2 that underwent endocytosis while they were live labeled for 1 h with anti-GluA2NTD antibody. These data suggest that the GFP-GluA2 puncta are a mixture of vesicles that belong to the ER or Golgi apparatus, or are involved in post-Golgi trafficking and endocytic pathways (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). The time lag between tetramer formation and the appearance of puncta of GFP-GluA2 wild type may also suggest the possibility that tetrameric GFP-GluA2 is not readily competent to exit the ER. Collectively, these results suggest that vesicle trafficking is severely reduced in GluA2L504Y cells compared with wild-type cells. Next, we tested whether there is also a maturation deficit in this mutant.
Inefficient tetramerization of GluA2L504Y mutant
The time course of GluA2L504Y protein expression following DOX induction was similar to that of GluA2 wild type. However, unlike GluA2 wild type, GluA2L504Y was detected as a single band by SDS-PAGE when purified 24 h after induction (Fig. 4A,B). Furthermore, the mobility of GluA2L504Y was identical to that of GluA2 wild type purified from GnTI(-)HEK cells deficient in complex mannose glycosylation (data not shown), indicating that the majority of GluA2L504Y is not glycosylated within 24 h of expression.
Figure 4.

Inefficient tetramerization of GluA2 L504Y. A, Western blot depicting time course of GluA2L504Y protein expression after induction. B, Coomassie brilliant blue stained 7.5% SDS-PAGE of GluA2 purified from TetOnGluA2L504Y HEK cells. Note only a single band is present. C, Gel filtration chromatograph of GluA2 wild type (red) and L504Y (blue) purified from TetONGluA2 and TetONGluA2L504Y HEK cells, respectively. Proteins were harvested 24 h after induction. Peaks corresponding to tetramer and dimer are indicated. D, Peak fractions from gel filtration in C were resolved and processed for Western blotting using an anti-GluA2 C-terminal antibody. The arrows indicate the glycosylated (upper) and nonglycosylated (lower) bands. Note the absence of any glycosylated GluA2L504Y in the peaks corresponding to tetramers.
Next, we tested whether tetramerization was affected by the L504Y mutation. Interestingly, even 24 h after induction, the majority of GluA2L504Y remained as dimers when resolved by gel filtration chromatography (Fig. 4C, blue). In contrast, GluA2 wild type exists primarily as tetramers 24 h after DOX induction (Fig. 4C, compare red and blue). Western blotting of the gel filtration fractions suggests that the glycosylated species of GluA2 wild type are tetramers, whereas the nonglycosylated species are primarily enriched as dimers (Fig. 4D). In contrast to GluA2 wild type, GluA2L504Y exists mostly as dimers, although a small quantity is detected as tetramers. These results are in keeping with the observation that GluA2L504Y mutant has high degree of aberrant multimerization of subunits including limited tetramer formation in HEK cells (Penn et al., 2008).
The projection structure of the negative-stained tetrameric GluA2L504Y is very similar to that of GluA2 wild type (supplemental Fig. 5A,B, available at www.jneurosci.org as supplemental material). The yield of these particles was extremely low, so, despite our efforts, it was not feasible to obtain 3D structures of GluA2L504Y tetramers. We considered the possibility that the yield was low because tetramers in the non-desensitizing state might be less stable in detergent. However, this is unlikely because our previous study showed that brain-derived AMPA-Rs locked in the non-desensitized state by treatment with 330 μm cyclothiazide and 1 mm glutamate had similar structures to the untreated receptors (Nakagawa et al., 2005). Collectively, these results suggest that the GluA2L504Y mutation causes defects in the dimer-to-tetramer transition.
Projection structures of the GluA2L504Y dimer
To gain further insight into the difference in tetramerization between GluA2 wild type and L504Y, EM images were taken from negative-stained GluA2L504Y dimers. GluA2L504Y dimer particles were homogeneous in size, but had less conformational heterogeneity and were more elongated than wild-type dimers (Fig. 5A). Representative class averages show three layers of domains stacked on top of each other. Similar to the GluA2 wild-type dimer structures, the top domain of GluA2L504Y was bipartite and elongated or appeared squarish with a weaker density in the center in some class averages. (Fig. 5A, bottom right panels). The bottom domain was a featureless globular density. In contrast to the wild-type dimer, in most class averages, the middle portion existed as a single density, and was split into two in only a minority of the class averages (Fig. 5A, bottom row, second from right).
Figure 5.

EM imaging of GluA2L504Y dimers. A, Raw particle images of the GluA2L504Y dimer (upper large panel) and representative class averages (lower small panels). Scale bar, 10 nm. B, Diagram of Fab fragment-labeled GluR subunit (left). Raw particle images of GluA2L504Y dimer labeled with Fab fragment (right top). Under each class average is a representation to facilitate interpretation. Receptor complex is in white, and Fabs are in gray. Fabs label the C terminus. Scale bar, 10 nm. C, Diagram of GFP-tagged GluA2L504Y subunit (left). Class averages of GFP-GluA2L504Y dimer particles (right top). Under each class average is a representation to facilitate interpretation. Receptor complex is in white, and GFP is in green. GFP labels the N terminus. Scale bar, 10 nm. D, Summary of domain labeling. Class average of dimeric GluA2L540Y particle labeled with domain designations.
Fab labeling and GFP tagging were used for domain assignment. Similar to GluA2 wild-type dimers, anti-FLAG epitope Fabs consistently decorated the bottom round featureless density of the particles (Fig. 5B) and two Fabs sometimes bound to a single particle (Fig. 5B, right panel). Moreover, two extra densities representing N-terminal GFP tags were detected at the top of the bipartite density of GFP-GluA2L504Y dimers. From these experiments we interpret the bottom density as the TMD and CTD dimer, the middle density as the LBD dimer, and the top elongated bipartite domain as the NTD dimer (Fig. 5D).
The particles were quantified based on LBD separation. Approximately 7000 particles from negative-stained EM images were classified into 100 classes by multivariate statistical analysis and multireference alignment. We assigned each class average (and the particles in these classes) either to “LBD separated” or “LBD fused”. All classes that could not unambiguously be assigned to either were termed “unclassifiable.” In GluA2 wild-type dimers, 71.3% had a separated LBD, 2.3% had a fused LBD, and 26.4% were unclassifiable. In contrast, of GluA2L504Y dimers, 66% had a fused LBD, 20% had a separated LBD, and 14% were unclassifiable. Among the 66% of GluA2L504Y dimers that adopted fused LBD, 33.3% had shapes exhibiting twofold symmetry, while 66.7% had asymmetric shapes. Among the classifiable particles of GluA2 wild-type and L504Y dimers, 7.8% and 12.1% adopted squarish NTDs, respectively, indicating that the particles that adopt squarish NTDs are the minority. These results demonstrate a robust difference in the molecular shapes between dimers of GluA2 wild-type and L504Y (Figs. 2D, 5A, 6). Furthermore, the difference in molecular shapes is correlated with their contrasting ability to transition from dimer-to-tetramer (Fig. 4).
Figure 6.

3D structures of GluA2 wild-type and L504Y dimers. A, Structures of GluA2 wild-type dimers are shown. Left box, Class average of untilted particles used for random conical tilt reconstruction. Right, Four different views of 3D density map calculated from particles classified into the class average shown in the left box. B, Placement of known crystal structures into the EM density map for the GluA2 wild-type dimer. Red: NTD of GluA2 (PDB:3H5W); blue: LBD of GluA2 S1S2 wild type (PDB:1FTJ). Scale bar, 7.3 nm. C, Structures of GluA2L504Y dimers are shown. Left box, Class average of untilted particles used for random conical tilt reconstruction. Right, Four different views of the 3D density map calculated from particles classified into the class average shown in the left box. D, Placement of known crystal structures into the EM density map for GluA2L504Y dimer. Red, NTD of GluA2 (PDB:3H5W); blue, LBD of GluA2 S1S2 L483Y (PDB:1LB8). Scale bar, 7.3 nm. 3D maps in A–D are shown at the same scale. E, Juxtaposition of GluA2 wild-type dimer and brain-derived tetrameric AMPA-R devoid of stargazin/TARPs (Nakagawa et al., 2006). Two different views are shown. Note that the TMD is smaller in the dimer than in the tetramer when the EM density map is viewed from the bottom. Scale bar, 7.3 nm.
3D EM maps of GluA2 wild type and L504Y dimer
To confirm that the structural differences seen in the projection structures indeed reflect structural differences in 3D, the 3D density maps of the GluA2 wild type and L504Y dimers were calculated using random conical tilt reconstruction (Frank, 1996; Frank et al., 1996). For this purpose, particle images were recorded as tilt pairs at specimen tilt angles of 0° and 60°. Raw particle images recorded at 0° were analyzed by multivariate statistical analysis, classification and multi reference alignment. Well aligned and highly represented class averages were chosen (Fig. 6A,C, left box) and corresponding tilted images of particles in the selected class were used to calculate the 3D structure. Refinement was done in three steps using back-projection refinement, angular refinement (implemented in SPIDER) (Frank et al., 1996) and FREALIGN refinement (Grigorieff, 2007). Three different views of the final reconstruction of wild-type and L504Y dimers are shown in Figure 6, A and C, respectively. The resolution of the final reconstruction was 35Å (wild type) and 34 Å (L504Y) at FSC = 0.5, and with the less conservative criteria 29Å (wild type) and 28Å (L504Y) (FSC = 0.142) (Rosenthal and Henderson, 2003). The images shown in Figure 6 are filtered at the resolution determined by FSC = 0.5, the more conservative resolution criteria. In addition, to demonstrate the structural contrast between immature and mature receptors, the 3D maps of GluA2 wild-type dimer and AMPA-R tetramer are compared side-by-side in Figure 6E.
The 3D maps of wild-type and L504Y dimers were significantly different. The projection structures and the 3D EM maps viewed from the front correspond well for both wild-type and L504Y dimers (Fig. 6A,C). We assigned each globular feature in the 3D maps to individual domains of the subunit based on the domain labeling experiments (Figs. 2G, 5D). The wild-type dimer has the bipartite NTD dimer at the top and two smaller globular LBD densities attached at both sides of the NTD dimer. The following dimensions are the maximal distances. The height of the particle is 14.5 nm and the width is 14.6 nm. The central empty cavity of the particle suggests a clear physical separation of the two LBDs. The dimensions of the cavity are 3.7 nm in height and 5.6 nm in width. In contrast, the height and width of the L504Y dimer is 17.0 nm and 8.7 nm, and the L504Y dimer has no LBD separation.
Similar to the class averages (Fig. 6A,C), the overall 3D structures did not reveal global two fold symmetry, despite that they represent homodimeric subunits of GluA2 wild type or L504Y. The linker between the NTD and LBD consists of 16 amino acids, and we predict it has structural flexibility to accommodate twist that may exist between the NTD and LBD dimers. Consistently, the NTD was observed as an elongated bipartite density in some class averages, whereas in the others it appeared as a squarish density (Figs. 2D, 5A). We believe that the different appearance of the NTD in different class averages results from viewing the NTD dimer from different angles. A variety of orientations of the NTD relative to LBD can be explained by the structural flexibility present in the linker connecting these domains.
To interpret our 3D density map, we used known crystal structures of AMPA-R subunit domains. The crystal structure of the NTD of GluA2 (Jin et al., 2009) fits nicely in the NTD density of our EM structure (Fig. 6B,D). Additionally, the two separated small globular densities of the wild-type dimer can each accommodate the crystal structure of the GluA2 LBD monomer (Armstrong et al., 1998), while the density that corresponds to the LBD in the L504Y dimer is consistent with the dimeric crystal structure of the LBD (Fig. 6B,D) (Sun et al., 2002). Because of the limited resolution of our EM map, we did not refine the position of the crystal structures using computational algorithms. However, placing crystal structures into our 3D reconstruction demonstrates that the size and shape of the densities of the extracellular domains of dimeric GluA2 wild type and L504Y are compatible with known crystal structures. Thus, we conclude that the structural differences of the class averages between GluA2 wild type and GluA2L504Y do not reflect different views of an identical 3D structure but represent true structural differences. Specifically, the structural difference between the wild-type and L504Y dimers can be attributed to the different global arrangement of the LBDs.
Interaction of GluA2 and stargazin during the subunit assembly of AMPA-Rs
Stargazin/TARPs are auxiliary subunits of native AMPA-Rs in the brain. Because endogenous stargazin/TARPs were not detected in HEK cells, we investigated the effect of stargazin on subunit assembly of GluA2 by introducing stargazin into the parental stable HEK cell that DOX-dependently expresses FLAG-tagged GluA2 flop (TetONGluA2 cell) (Fig. 2A). The new stable HEK cell lines, which we named TetONGluA2-stg cells, constitutively express stargazin-IRES-mCherry using the elongation factor promoter and DOX-dependently express GluA2 flop (Fig. 7A,B). The mCherry was coexpressed (not as a fusion protein) with stargazin to facilitate the visual isolation of stable clones by fluorescent microscopy.
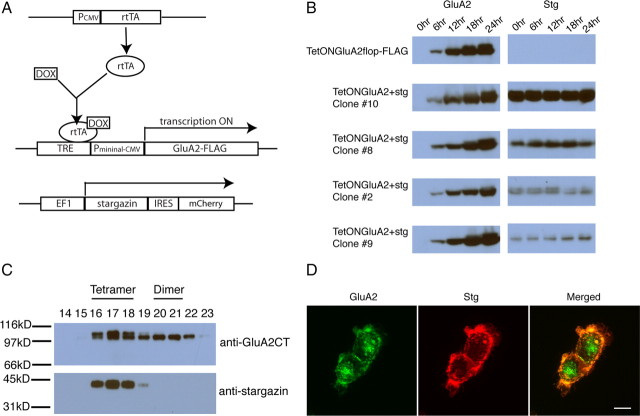
Figure 7.
Stargazin forms a stable complex with GluA2 tetramers, but not with dimers. A, Schematic of our strategy used to coexpress GluA2 and stargazin in HEK cells that we named TetONGluA2-stg cells. EF1, Elongation factor, B, Western blotting showing time course of GluA2 (left) and stargazin (Stg; right) expression in the cell lines indicated on the left of each row. C, Western blotting of the fractions eluted from Superdex200 gel filtration column. Membranes were probed with anti-GluA2C-terminal antibody (top) and anti-stargazin (bottom) antibodies. Fractions corresponding to the tetramers and dimers are indicated. D, Confocal immunofluorescent Z-stack projections of clone 10 24 h after induction. Red is stargazin and green is GluA2. Superposition of the two colors is shown in merged image on the right. Scale bar, 15 μm.
The four stable TetONGluA2-stg cell lines (clone #2, 8, 9, and 10), express stargazin at different levels. Interestingly, these cells started to die after 24 h of DOX induction, suggesting that the expression of GluA2-induced cytotoxicity in the presence of stargazin. The cytotoxicity was stronger in the clones that express higher levels of stargazin and, as a result, all the cells died within 48 h in clone 10 and 8. The cytotoxicity was suppressed when the cells were cultured with 30 μm NBQX, an antagonist of AMPA-Rs. Because the same level of GluA2 expression was not cytotoxic in TetONGluA2 cells, we interpret that the glutamate in the media caused cell death by activating AMPA-Rs whose function was enhanced by stargazin. These observations suggest that both GluA2 and stargazin are functional in these cells.
First, we investigated whether stargazin alters the biosynthesis of GluA2. All the TetONGluA2-stg cell lines expressed similar levels of GluA2 24 h after DOX induction. To assess the effect of stargazin on GluA2 maturation, we induced GluA2 expression in various TetONGluA2-stg cells and compared the time course of expression with the parental TetONGluA2 cells that lack stargazin. We were unable to detect any significant difference in the rate of GluA2 expression (Fig. 7B). Thus, in HEK cells stargazin is not the rate-limiting molecular chaperone for GluA2 biosynthesis.
When GluA2 was purified from clone 10, 12 h after DOX application, no detectable amount of stargazin was copurified. However, when purification was done 24 h after DOX induction, stargazin copurified with GluA2 and associated only with the tetrameric forms of GluA2, as determined by Superdex 200 gel filtration (Fig. 7C). Consistently, both proteins colocalized at the cell periphery when clone 10 cells were double stained 24 h after induction (Fig. 7D).
Discussion
This study establishes a new approach to investigate subunit assembly of AMPA-Rs using a combination of genetic engineering and single-particle EM. We use recombinant GluA2 tetramers obtained from HEK cells whose structures (at the resolution of our current study) are indistinguishable from those purified from rat brain (Nakagawa et al., 2005, 2006) (Fig. 1). This system has several advantages because the subunit composition can be precisely controlled, genetic manipulation is feasible, and the protocol is simple and highly reproducible. While AMPA-Rs form heterotetramers in the brain (Wenthold et al., 1996), it has been established that homotetrameric receptors are functional ion channels when expressed in HEK cells (Swanson et al., 1997). Many structural studies of AMPA-Rs are done using the GluA2 subunit. In addition, this subunit renders AMPA-Rs impermeable to calcium; therefore, they are less toxic when overexpressed in cells. Insights obtained from homomeric GluA2 may therefore also apply to the heteromeric AMPA-Rs.
Recombinant AMPA-Rs were structurally more homogeneous than the AMPA-R particles purified from the brain (Nakagawa et al., 2005, 2006). Because application of glutamate to the purified AMPA-R induces conformational changes of the NTD, we speculate that the structural heterogeneity observed in brain-derived AMPA-Rs is caused by exposure of the particles to the endogenous glutamate during purification. Consistently, it is critical to include antagonist kynurenic acid (or NBQX) during the detergent solubilization and subsequent immunoaffinity chromatography to obtain structurally homogeneous recombinant AMPA-R tetramers.
The AMPA-R auxiliary subunit stargazin/TARP was not detected in HEK cells, nor from fractions of purified GluA2 from HEK cells that should in principal enrich endogenous stargazin/TARPs if they are present [note, however, that two proteins copurified when stargazin was introduced into the system (Fig. 7C)]. We estimate that there is very little, if any, and that possible endogenous stargazin/TARPs in HEK cells contribute little to tetrameric assembly of AMPA-R subunits. Consistently, stargazin/TARPs form a stable complex with GluA2 tetramers but not with the dimer intermediates during biogenesis (Fig. 7C). In addition, the time course of expression of the newly synthesized GluA2 was not affected by the presence of stargazin in these cell lines. Stargazin functions as a molecular chaperone for AMPA-Rs in the presence of the ER stress response (Vandenberghe et al., 2005a). Because our expression level of GluA2 was modest, it is unlikely that the ER stress response was elicited. Taken together, it is likely that stargazin functions as a chaperone only when there is a high demand to synthesize AMPA-Rs. The lack of accumulation of AMPA-R dimers in the stargazer mutant mice is likely due to the fact that the ER is unlikely to be in a stress state (Vandenberghe et al., 2005b). Cornichons are newly identified auxiliary subunits of AMPA-Rs that modulate receptor trafficking (Schwenk et al., 2009). This suggests the possibility that they are also involved in the assembly process of AMPA-Rs. However, no definitive data are currently available.
Trafficking intermediates of membrane receptors are typically not easily accessible. By harvesting GluA2 within a short period after induction of transcription, we were able to enrich the biosynthetic intermediates. GluA2 protein expression reached a plateau >24 h after DOX induction in HEK cells. Interestingly, the half-life of an AMPA-R subunit is 18 h as determined by radioisotope metabolic labeling in cortical neurons (O'Brien et al., 1998). As the duration of protein turnover and molecular structures are consistent between AMPA-Rs expressed in HEK cells and endogenous receptors in neurons, HEK cells likely have the necessary molecular chaperones to correctly assemble AMPA-Rs.
The ratio of tetramer to dimer increased as a function of time after GluA2 induction by DOX. When determined by negative stain EM, purified AMPA-R tetramers remained in tetrameric form up to 5 d in buffer containing 0.1% DDM, suggesting that the tetrameric complexes are relatively stable (data not shown). Thus, it is unlikely that the dimers we purified are an artifact of detergent solubilization. Dimeric GluA2, a biogenic trafficking intermediate, is a previously uncharacterized molecular species of AMPA-Rs from a structural point of view. We speculate that stable dimers are common biosynthetic intermediates of AMPA-Rs, regardless of subunit composition. Such an idea is consistent with the preferential interaction of GluA1 with GluA1 NTD relative to GluA2 NTD (Leuschner and Hoch, 1999).
The 3D structure of dimeric GluA2 revealed that the LBDs are spatially separated, a novel feature that was not previously observed in AMPA-R structures. The NTD and TMD form dimers but the LBD remains separated. This observation provides a structural basis for a previous model derived from electrophysiological experiments in which the NTD and TMD are critical determinants for the formation of functional channels (Ayalon and Stern-Bach, 2001). Our structural finding suggests that dimerization of both the NTD and TMD precedes tetramerization. This agrees with the interruption of the polypeptide sequence of the LBD by the TMD, which implies that folding of the TMD coordinates with that of the LBD during translation.
To study the functional significance of the structure of GluA2 wild-type dimers, we identified GluA2L504Y as a mutant defective in tetramerization, determined the structure of the dimers of this mutant, and compared the structure with the wild type. GluA2L504Y and analogous mutants in other AMPA-R subunits have been studied extensively as non-desensitizing mutants (Rosenmund et al., 1998; Stern-Bach et al., 1998; Robert et al., 2001; Sun et al., 2002). The tetramerization defect, however, was not observed when GluA2L504Y was exogenously expressed in neurons (Greger et al., 2006). This can be explained by the possible coassembly of exogenous mutant GluA2L504Y subunits with endogenous wild-type AMPA-R subunits. However, it remains unclear why heterotetramers that contain GluA2L504Y did not exit the ER. Additional mechanisms such as sampling of gating motion in the ER may regulate this process (Penn et al., 2008).
In the dimers of GluA2L504Y, all three domains (NTD, LBD, and TMD) appear to be dimerized or at least in close proximity. The dimeric modules are arranged linearly, thus making the overall shape of the complex elongated compared with wild-type structures (compare Figs. 2, 5, 6). The compact structure of the LBD in dimeric GluA2L504Y is consistent with the high-affinity dimer formed by the S1S2 construct of the LBD that carries the same mutation (Sun et al., 2002). Our data are also consistent with previous results indicating that native tetrameric AMPA-Rs treated with 1 mm glutamate have more compact structures in the presence of 330 μm cyclothiazide, an allosteric inhibitor of desensitization (Nakagawa et al., 2005).
The two LBDs are separated in the GluA2 wild-type dimers and, therefore, intermolecular dimerization of the LBDs can potentially occur between two GluA2 wild-type dimers during assembly of a tetramer. In contrast, the LBDs are fused in the GluA2L504Y dimers. Since this mutation causes a defect in dimer-to-tetramer transition, it is conceivable that the separation of LBD dimers is required to drive tetramerization (Fig. 8A). Thus, our results support a new model for the subunit assembly pathway of AMPA-Rs in which the dimer-to-tetramer transition accompanies formation of two new LBD dimers between the two molecular dimers of subunits (Fig. 8B). This suggests an unexpected domain arrangement in tetrameric AMPA-Rs, in which the NTD and LBD of each subunit forms a dimer with a different neighboring subunit.
Figure 8.

Working model of AMPA-R tetramerization and trafficking. A, In the ER, dimers of AMPA-R subunits are formed and transition into tetramers. With dimeric wild-type subunits, the NTD and TMD form dimers but the LBD is separated. In contrast, the NTD, LBD, and TMD are all compactly dimerized in the GluA2L504Y mutant, which transition into tetramers much less efficiently. The domain organization seen in the structure of the wild-type dimer is critical for efficient tetramerization. A small quantity of GluA2L504Y tetramers is formed and reaches the cell surface by an unknown mechanism. The majority of GluA2L504Y is not complex mannose glycosylated, suggesting that it did not receive modification in the Golgi apparatus. It is likely that most GluA2L504Y cannot even exit the ER or reach the cis-Golgi. Stargazin/TARPs are associated with the mature AMPA-Rs. Stargazin preferentially forms a stable complex with GluA2 tetramers, but not with dimers. Cornichons are another subset of AMPA-R auxiliary proteins in the membrane. The precise timing of incorporation into the AMPA-R complex remains to be determined. B, Proposed AMPA-R subunit assembly pathways are shown. In the wild-type subunit dimers, the LBDs are not dimerized. We propose that the LBD dimers in wild-type subunits are formed during the dimer-to-tetramer transition. On the other hand, the LBDs in the L504Y mutant subunits form intradimer dimers that may prevent efficient dimer-to-tetramer transition. NTD, red and orange; LBD, blue and purple; TMD, yellow and green.
As with any structural approaches, technical artifacts must always be considered. The carbon support and the negative stain can potentially introduce a small distortion in the molecular structure. Even if we assume the presence of a small distortion, the robust structural differences between the GluA2 wild-type dimer and L504Y dimer were detected when both specimens were prepared under identical experimental procedures. Therefore, the observed structural differences of the particles on EM grid should reflect the structural differences in solution. In addition, when carefully inspected, the peak elution volumes of wild-type and L504Y dimers are different (Fig. 4C,D). Because the molecular weights of the wild-type dimer and the L504Y mutant dimer are nearly identical, this small difference in the peak elution volume suggests a structural difference between the two molecular forms. The direct structural inspection by single-particle EM and the hydrodynamic properties obtained from quantitative gel filtration chromatography both support the presence of a structural difference between the wild-type and L504Y dimers, thus experimental artifacts are an unlikely explanation for the observed gross structural differences.
In both AMPA-Rs and kainite receptors, point mutations that prevent receptor desensitization also result in decreased surface delivery (Greger et al., 2006; Priel et al., 2006). Accordingly, it has been proposed that glutamate receptors sense glutamate before being delivered to the cell surface, which can prevent non-desensitizing and nonfunctional subunits from going to the surface (Mah et al., 2005; Valluru et al., 2005; Penn et al., 2008). Because globular LBDs are clearly detected in immature AMPA receptors, glutamate might be used as a tool to facilitate progression through the subunit assembly pathway of AMPA-Rs as well. All in all, we have created a system that has allowed us to study molecular details of AMPA-R assembly. Comparison of wild-type and mutant receptors has provided insight into the domain arrangements of the dimeric intermediates which are conducive to the receptor transitioning into mature tetrameric form.
Note added in proof. While this manuscript was being reviewed, an article reporting the x-ray crystal structure of GluA2cryst was published (Sobolevsky et al., 2009). Despite the fact that 6 of 16 amino acids in the wild-type GluA2 were deleted from the linker that connects the NTD and LBD, the domain arrangement reported for the x-ray crystal structure of GluA2cryst is consistent with our model described in Figure 8.
Footnotes
This work is supported by grants from John Merck Fund, Hellman Foundation (to T.N.) and National Institutes of Health (NIH) Grant P41004050 (to M.H.E.). N.F.S is supported by the NIH Molecular Biophysics Training Grant (NIH GM08326). A.N.F is supported by the NIH CMG Training Grant (T32 GM007240-34). T.N. is a recipient of the National Alliance for Research on Schizophrenia and Depression Young Investigator Award. We thank Peter Seeburg and Yasunori Hayashi for rat GluA2 flop and GFP-GluA2flop cDNA clones. We acknowledge the use of the University of California, San Diego (UCSD), Cryo-Electron Microscopy Facility, which was supported by NIH Grants 1S10RR20016 and GM033050 to Dr. Timothy S. Baker (UCSD) and a gift from the Agouron Institute to UCSD. We thank Anirvan Ghosh for giving us access to their epifluorecent microscope during the initial phase of this study. We also thank Palmer Taylor for HEK wt and HEK GnTI(-) cell lines and Toni Koller for conducting LC/MS/MS. We thank Thomas Walz for discussion. We thank Hiro Hakozaki, Ohkyung Kwon, Guido Gaietta, and Tom Deerinck for their help at The National Center for Microscopy and Imaging Research. We thank Mark Elliott for transfecting and immunostaining neurons.
References
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Barry MF, Ziff EB. Receptor trafficking and the plasticity of excitatory synapses. Curr Opin Neurobiol. 2002;12:279–286. doi: 10.1016/s0959-4388(02)00329-x. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Coleman SK, Möykkynen T, Cai C, von Ossowski L, Kuismanen E, Korpi ER, Keinänen K. Isoform-specific early trafficking of AMPA receptor flip and flop variants. J Neurosci. 2006;26:11220–11229. doi: 10.1523/JNEUROSCI.2301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Three-dimensional electron microscopy of macromolecular assemblies. San Diego: Academic; 1996. [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gouaux E. Structure and function of AMPA receptors. J Physiol. 2004;554:249–253. doi: 10.1113/jphysiol.2003.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Ziff EB. RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron. 2002;34:759–772. doi: 10.1016/s0896-6273(02)00693-1. [DOI] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- Greger IH, Akamine P, Khatri L, Ziff EB. Developmentally regulated, combinatorial RNA processing modulates AMPA receptor biogenesis. Neuron. 2006;51:85–97. doi: 10.1016/j.neuron.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R. Driving AMPA receptors into synapses by LTP and CaMKII: requirement for GluR1 and PDZ domain interaction. Science. 2000;287:2262–2267. doi: 10.1126/science.287.5461.2262. [DOI] [PubMed] [Google Scholar]
- Hollmann M, O'Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Maron C, Heinemann S. N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994;13:1331–1343. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW. SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W. Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem. 1999;274:16907–16916. doi: 10.1074/jbc.274.24.16907. [DOI] [PubMed] [Google Scholar]
- Mah SJ, Cornell E, Mitchell NA, Fleck MW. Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci. 2005;25:2215–2225. doi: 10.1523/JNEUROSCI.4573-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Reijmers L, Mayford M. Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science. 2008;319:1104–1107. doi: 10.1126/science.1149967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Sheng M, Walz T. Three-dimensional structure of an AMPA receptor without associated stargazin/TARP proteins. Biol Chem. 2006;387:179–187. doi: 10.1515/BC.2006.024. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Shneider NA, Axel R. A family of glutamate receptor genes: evidence for the formation of heteromultimeric receptors with distinct channel properties. Neuron. 1990;5:569–581. doi: 10.1016/0896-6273(90)90212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science. 2006;311:1253–1256. doi: 10.1126/science.1123339. [DOI] [PubMed] [Google Scholar]
- O'Brien RJ, Kamboj S, Ehlers MD, Rosen KR, Fischbach GD, Huganir RL. Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron. 1998;21:1067–1078. doi: 10.1016/s0896-6273(00)80624-8. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification: powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH. Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 2008;27:3056–3068. doi: 10.1038/emboj.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Selak S, Lerma J, Stern-Bach Y. Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron. 2006;52:1037–1046. doi: 10.1016/j.neuron.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Irizarry SN, Hughes TE, Howe JR. Subunit interactions and AMPA receptor desensitization. J Neurosci. 2001;21:5574–5586. doi: 10.1523/JNEUROSCI.21-15-05574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SW, Andrews PI, Gahring LC, Whisenand T, Cauley K, Crain B, Hughes TE, Heinemann SF, McNamara JO. Autoantibodies to glutamate receptor GluR3 in Rasmussen's encephalitis. Science. 1994;265:648–651. doi: 10.1126/science.8036512. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci. 2003;23:6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nat Rev Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, Klöcker N. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- Sheng M, Lee SH. AMPA receptor trafficking and the control of synaptic transmission. Cell. 2001;105:825–828. doi: 10.1016/s0092-8674(01)00406-8. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Bach Y, Russo S, Neuman M, Rosenmund C. A point mutation in the glutamate binding site blocks desensitization of AMPA receptors. Neuron. 1998;21:907–918. doi: 10.1016/s0896-6273(00)80605-4. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru L, Xu J, Zhu Y, Yan S, Contractor A, Swanson GT. Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J Biol Chem. 2005;280:6085–6093. doi: 10.1074/jbc.M411549200. [DOI] [PubMed] [Google Scholar]
- Vandenberghe W, Nicoll RA, Bredt DS. Interaction with the unfolded protein response reveals a role for stargazin in biosynthetic AMPA receptor transport. J Neurosci. 2005a;25:1095–1102. doi: 10.1523/JNEUROSCI.3568-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe W, Nicoll RA, Bredt DS. Stargazin is an AMPA receptor auxiliary subunit. Proc Natl Acad Sci U S A. 2005b;102:485–490. doi: 10.1073/pnas.0408269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthold RJ, Petralia RS, Blahos J, II, Niedzielski AS. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci. 1996;16:1982–1989. doi: 10.1523/JNEUROSCI.16-06-01982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Arai AC, Rumbaugh G, Srivastava AK, Turner G, Hayashi T, Suzuki E, Jiang Y, Zhang L, Rodriguez J, Boyle J, Tarpey P, Raymond FL, Nevelsteen J, Froyen G, Stratton M, Futreal A, Gecz J, Stevenson R, Schwartz CE, et al. Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci U S A. 2007;104:18163–18168. doi: 10.1073/pnas.0708699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff EB. TARPs and the AMPA receptor trafficking paradox. Neuron. 2007;53:627–633. doi: 10.1016/j.neuron.2007.02.006. [DOI] [PubMed] [Google Scholar]