Abstract
Cell surface receptors acquire information from the extracellular environment and coordinate intracellular responses. Evidence from biochemical and structural studies indicates that many receptors do not operate as individual entities, but rather as part of higher-order complexes (e.g. dimers and oligomers). Coupling the functions of multiple receptors may endow signaling pathways with the sensitivity and malleability required to govern cellular responses. Moreover, multireceptor signaling complexes may provide a means of spatially segregating otherwise degenerate signaling cascades. Despite the proposed importance of receptor-receptor processes in cellular signaling, questions concerning the mechanisms, extent, and consequences of receptor co-localization and inter-receptor communication remain unanswered.
Chemical synthesis can provide a variety of compounds with which to address the role of receptor assembly in signal transduction. The focus of this review is one such approach — the use of synthetic multivalent ligands to characterize receptor function. Multivalent ligands can be generated that possess a variety of sizes, shapes, valencies, orientations, and densities of binding elements. Their unique architectures imbue multivalent ligands with the ability to access binding modes not available to monovalent compounds. Multivalent ligands, therefore, are capable of illuminating aspects of inter-receptor processes that are not readily probed using conventional approaches. We suggest that, as focus shifts from investigations of the function of individual proteins and toward the analysis of multi-receptor signaling complexes, multivalent ligands will become even more valuable tools with which to ask sophisticated mechanistic questions. Further, multivalent ligands may provide new opportunities for manipulating receptor systems for the deconvolution of pathways, diagnosis, and, ultimately, the treatment of disease.
Keywords: multivalent, oligomerization, receptors, signal transduction, synapses, biomaterials
1. Introduction to Receptor Function
To achieve success in the competition for favorable chemical and physical conditions, organisms from bacteria to worms to humans must rapidly and accurately sense changes in their environments. Likewise, the complex orchestrations that govern developmental patterning or immunological responses rely on communication between cells and their environs. The task of monitoring extracellular conditions falls to the cell-surface receptors. These proteins coordinate the cell's internal machinery by collecting, compiling, and translating external information. The pressure for survival demands that responses to stimuli be sensitive and accurate; synergistic and competitive signals must be amplified and integrated. In the face of these requirements, cells have developed sophisticated and elegant methods for achieving sensitive yet malleable receptor-mediated responses.
One important mechanism for efficient and sensitive signaling is to couple the functions of multiple cell surface receptors.[1-9] Cell surface receptors do not typically operate as individual entities; rather, they function in concert. An ermerging paradigm is that receptors within multi-receptor signaling complexes communicate with each other.[7] Although the evidence for inter-receptor communication is mounting, an understanding of the underlying mechanisms is elusive. This review focuses on the role of synthetic multivalent ligands as agents for the investigation of receptor collaboration and the exploitation of its consequences.
2. Direct and Indirect Receptor-Receptor Interactions
2.1. Signaling Complexes
Receptors transmit information on extracellular signals to the internal machinery of the cell. Because different receptors often share common cytoplasmic components, information transfer must be constrained to prevent unwanted exchange between disparate pathways.[7, 10-12] To understand how signals are transmitted, how cells can respond distinctly to different stimuli using a set of common components must be unraveled.[7] Both chemists and biologists are poised to contribute.
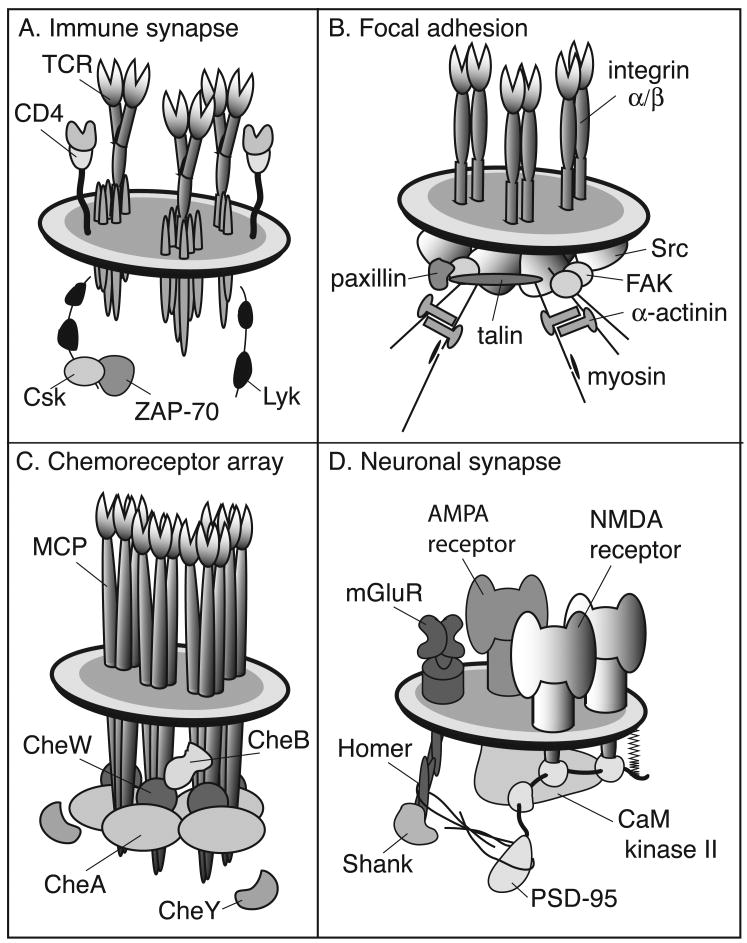
The subcellular localization of receptors into signaling complexes is proposed to be one mechanism by which cells achieve spatial and temporal regulation.[13-19] Multiprotein signaling complexes constitute the principle signaling units of neuronal synapses,[20, 21] immune synapses,[13, 22, 23] focal adhesions, [24] and bacterial chemoreceptor arrays (Figure 1).[25, 26] These ensembles are composed of receptors, signaling proteins, adapter proteins, and cytoskeletal components.[27-29] In eukaryotic cells, many signaling complexes appear to be associated with detergent insoluble lipid microdomains that provide unique physical environments for the concentration of signaling components.[30-35] Often it is not known whether cells use microdomains or other mechanisms to organize and assemble signaling components.
Figure 1.
Examples complexes of involved in signal transduction: (A) eukaryotic immune recognition; (B) eukaryotic cell adhesion; (C) prokaryotic chemotaxis; and (D) neuronal signaling. Although these schematic depictions are necessarily simplified, they have been designed to illustrate the complexity and elegance of biological multi-protein signaling complexes.
The formation of signaling complexes is not restricted to any particular receptor classes. Data from microscopy, covalent cross-linking, and x-ray crystallography experiments have revealed that cell surface receptors from many structural classes assemble into multi-receptor complexes; these include some heptahelical G-protein coupled receptors (GPCRs),[36-38] methyl-accepting chemotaxis proteins (MCPs),[39] gated ion channels,[40] receptor protein tyrosine kinases (RPTKs),[41, 42] and multichain immune recognition receptors (MIRRs).[22, 33, 35, 43-45] The size of these ensembles varies: Some complexes are composed of two receptors while others contain thousands. Some receptors,[46-48] including the ryanodine receptor[40] and MCPs,[39] are so highly concentrated that they dominate certain subcellular regions. Unfortunately, little is known about the structures of these assemblies, where they are localized within the cells, and how their localization influences signaling. Understanding these issues, however, could lead to new strategies to precisely control receptor function and therefore cellular responses. Although there remains a need to explore the role of receptor localization and assembly, the data acquired to date suggest some general modes for inter-receptor communication.
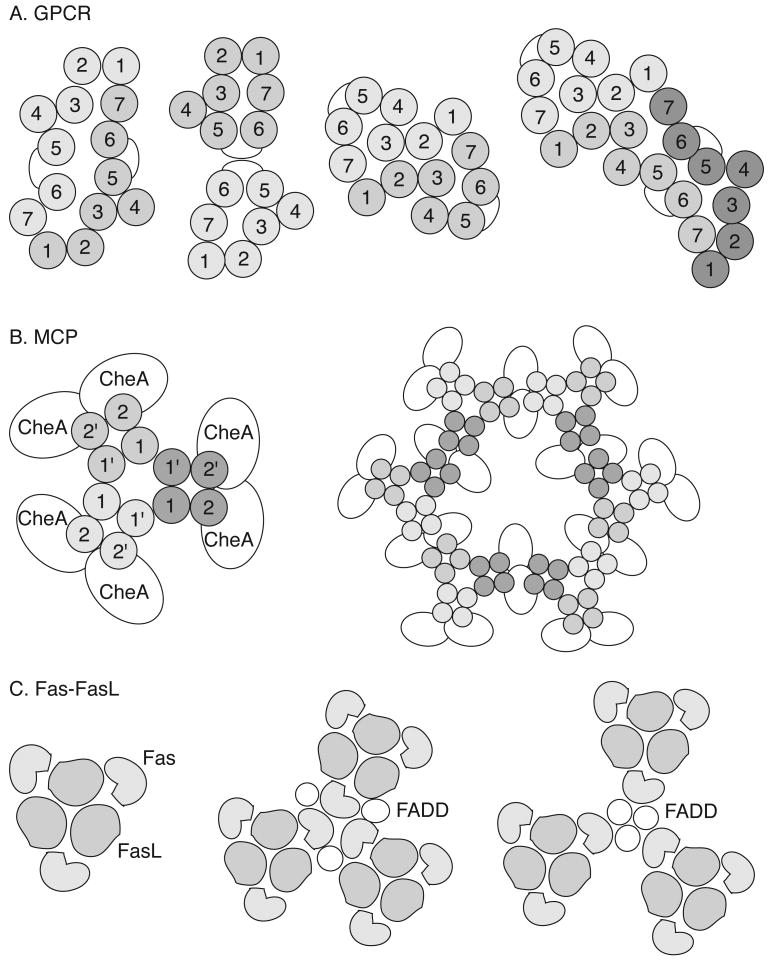
Receptors can exchange information via direct protein – protein contacts or through intermediary proteins. Direct receptor contact typically involves interactions between specific protein regions.[49] For example, helix-mediated multimerization is proposed to mediate the dimerization and oligomerization of GPCRs (Figure 2).[38, 50, 51] Evidence for the functional importance of these interactions is suggested by engineering disrupting mutations in the proposed contact sites or by adding isolated transmembrane helices to GPCRs. [37, 52, 53] Although their effects on receptor oligomerization are not yet well-established, these manipulations modulate signaling; presumably, they disrupt receptor-receptor contacts. As mentioned above, an alternative strategy by which receptors exchange information is through intermediary proteins. These scaffolding proteins can organize multi-receptor complexes, thereby, acting as frameworks for protein assembly. Examples of receptors localized via this mechanism include the MCPs and members of the tumor necrosis factor (TNF) receptor family (Figure 2b-c). [54-56]
Figure 2.
Proposed multi-receptor assemblies for some receptors. (A) Models of heptahelical GPCR dimers and a trimer based on mutagenesis results and the structure of bacteriorhodopsin. Each numbered circle represents a specific transmembrane helix. A variety of 1-7, 5-6, and 2-3 dimers and multimers have been proposed. (B) Bacterial chemoreceptors (methyl-accepting chemotaxis proteins or MCPs) are dimeric; and these can assemble into a trimer-of-dimers; this trimer of MCP dimers can further interact with signaling proteins, including the kinase CheA. Each MCP passes through the membrane twice (1 and 2) and coiled-coil interactions between these transmembrane domains (1 and 1′) mediate dimerization. A lattice model of MCP organization constructed of six of these trimers-of-dimers and 24 copies of CheA is shown. (C) The Fas-FasL interaction has been modeled using protein interfaces suggested by mutagenesis and crosslinking studies. Both the receptor Fas and its corresponding ligand, FasL, are trimers. The corresponding trimeric complex may be employed as a unit in the lattice stabilized by the adapter protein, FADD. Two models are shown with either Fas- or FADD-centered symmetries. See the text for the appropriate references.
Ligand binding can transduce signals by directly stabilizing or destabilizing protein assemblies.[57-59] For example, data collected from diverse experiments suggest that attractant binding to a single MCP can impact signaling from nearby MCP partners. [46] [60-64] Similarly, cross-phosphorylation of some growth factor receptors is facilitated by ligand-induced contacts. [65] In addition to homodimeric (or oligomeric) interactions, interactions between structurally dissimilar receptors also have been demonstrated. [66] The neuronal dopamine and gamma-aminobutyric receptors, a gated ion channel and GPCR, have been shown to exchange information by direct receptor-receptor contact. [67]
An alternative means by which ligands can induce receptor crosstalk and co-localization is by facilitating the recruitment of intermediary proteins to the target receptors; these intermediary proteins are called adapters or scaffolding proteins. [68-73] Specialized modules within intermediary proteins act as information conduits; examples include SH2, SH3, and PDZ domains. These domains participate in a series of protein–protein interactions that transfer information and localize receptors. The simplest adapters directly link one receptor to another. Additionally, many intermediary proteins contain multiple domains that facilitate communication between a receptor and a variety of membrane-associated and soluble signaling proteins.
2.2. Factors that Influence Inter-Receptor Communication
Defining the features of a multi-receptor signaling complex is an important first step in investigating and manipulating its function. However, the potential diversity created by the presence of multiple receptors and modular adapters raises challenges for those that seek to understand them. The features of the immune synapse illustrate some of the issues relevant for elucidating the function of signaling complexes.
T lymphocytes are critical mediators of effective immune responses to invading pathogens. The contribution of T cells to the immune response is largely controlled by signaling through the T cell receptor (TCR). Like many other important cell surface receptors, the TCR can be assembled into a multi-receptor complex. When a T cell contacts an antigen-presenting cell (i.e., a dendritic or B cell), the cytoskeleton reorganizes, and adhesion receptors and the TCR undergo changes in localization at the interface. This interface has been referred to as the “immune synapse” (Figure 1a).
The features of the immune synapse assembled when T cells interact with APCs can vary. Its structure is influenced by the type of APC involved, the type and activation state of the T cell, the duration of the T cell–APC interaction, and the local physiological environment in which it forms.[74] Powerful new imaging methods have been used to visualize immune synapses formed in vitro and in vivo; these studies reveal that dynamic changes in protein localization and organization can occur. At a molecular level, the role protein organization within the synapse not yet been established: It is not known whether the observed organization of the receptors on the T cell influences signaling.
It is clear that the TCR[75] and co-receptors, such as CD4,[76, 77] are localized at the cell–cell interface. Some of these co-receptors (e.g., CD4) augment the signal generated by the TCR; others attenuate it. The balance and orientation of these co-receptors can therefore tune the level of T cell activation. This tuning is achieved by changes in the phosphorylation of intracellular protein domains and receptors; the immune synapse also contains kinases such as Lck and ZAP-70 that catalyze the phosphorylation of ligated receptors and other proteins.[77, 78] Phosphorylation can either positively or negatively impact the subsequent recruitment and/or the enzymatic activity of various components of the signaling complex.
Evidence from microscopy and signaling experiments suggests that the immune synapse can be highly organized.[76] Because the cytoskeleton plays a critical role in organizing some cell surface proteins, the findings that cytoskeletal components are critical for TCR signaling supports a role for receptor organization. [13, 79, 80] The positioning of proteins within the immune synapse also is highly dynamic. For example, when a T cell engages with a cell presenting an antigen, the negative regulator CD45 moves to the periphery of the synapse. [76, 81-83] Under these circumstances, the TCR is recruited to the inner portion of the synapse. These changes in receptor positioning suggest that synapse organization affects signal transmission: These intricate, orchestrated[76] movements are believed to affect the transfer and propagation of the activating signal. The position of the TCR in relation to the various positive and negative regulators of its function appears to set signal intensity.
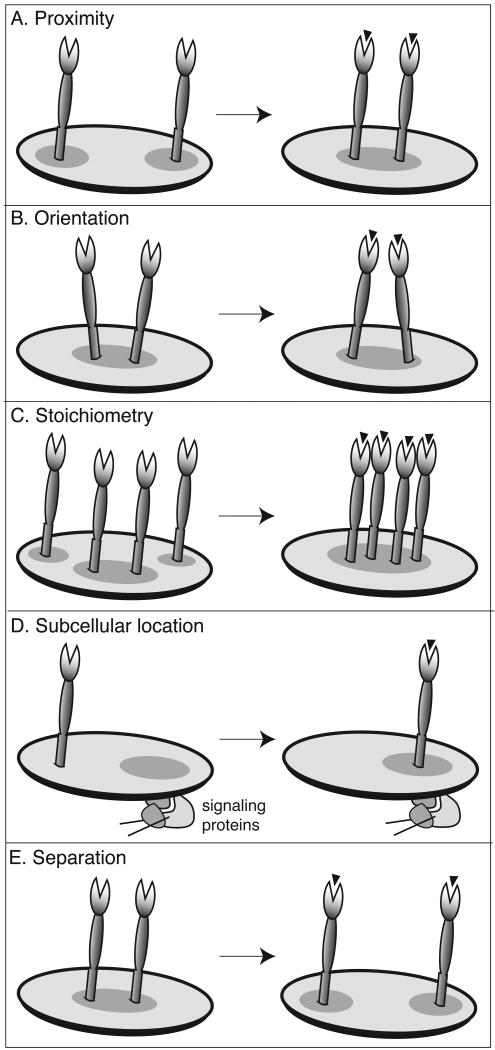
Dissecting the contribution of different factors to T cell signaling is complicated; there are many parameters that can affect signal output. Based on observations made of the immune synapse and other signaling complexes, [20] factors that influence signal output have been proposed. These include the number of activated receptors, their identity, and relative stoichiometry (Figure 3).[4, 84, 85] The stoichiometric composition of a signaling complex (i.e. the number of activating versus dampening receptors) can influence the amount of signal required to overcome thresholds. Additionally, the number and types of receptors will determine the opportunity for crosstalk. In addition to the identity and stoichiometry of receptors in the complex, signaling can also be influenced by the proximity and subcellular compartmentalization of receptors. Determining the relative contribution of all of the aforementioned variables remains a major challenge. To isolate and dissect such a complex signaling system, a variety of biological and chemical methods must be brought to bear. Multivalent ligands are valuable tools in the armamentarium.
Figure 3.
Possible methods for regulating inter-receptor communication. (A) The distance between receptors can influence the transfer of information between receptors or other proteins. (B) The relative orientation of two receptors can influence the alignment of enzyme active sites and govern the rates of covalent modifications that result in signal generation. (C) The number of receptors in a complex can influence the intensity of a signal. Additionally, the likelihood that receptors will come into contact increases when the numbers of localized receptors is greater. (D) The subcellular location of a receptor controls the access of the receptor to some intracellular signaling proteins. Changes in position can govern the flow of information through a receptor or cluster of receptors. (E) When co-receptors act as negative regulators, ligand binding can lead to activation of receptors by separation. This receptor mechanism is conceptually related to proximity-induced activation, but the underlying molecular interactions are quite different.
2.3. Challenges to Studying Receptor Function
Historically, cellular functions have been investigated by detailed examination of the structure and activity of a single protein component within a pathway. Yet, signal transduction involves the coordinated interactions of many different proteins. Thus, understanding cellular functions requires uncovering how heterogeneous collections of proteins interact at a supramolecular level. Advances in genomics, transgenic animal technologies, and chemical biology, provide the means to identify components of a pathway. Elucidating the functional and structural relationships between these components remains a major challenge. [86-88] Even after evidence for physical association is obtained by techniques such as the two-hybrid system, the order in which these interactions occur and their kinetics must be explored. Strategies that provide insight into complex cellular processes are needed.
Understanding multi-receptor assemblies requires the use of methods that reveal both molecular and supramolecular detail. For example, immunoprecipitation, confocal microscopy, and Förster resonance energy transfer (FRET) experiments have been influential in exploring changes in subcellular protein distributions.[89, 90] Moreover, fluorescent proteins and advanced imaging techniques have revolutionized the ability to follow proteins in live cells in real time. [91-93] Although these approaches provide insight into the machinations of multiprotein complexes, the molecular details that underlie inter-receptor processes often are obscure. This low resolution view is largely due to an old problem: the details revealed by an experiment are limited by the resolution of the investigative methods. For example, the optical resolution of light microscopy is approximately 200 nm. Fluorescence microscopy experiments, therefore, can be used to discern whether receptors are moving from one side of a cell to another but information concerning the orientation of two adjacent receptors is more difficult to obtain.
Methods with superior resolution are required for investigating receptor-receptor interactions that occur over sub-nanometer distances. While FRET can be used to investigate receptor assembly at this level, the application of FRET typically requires that cell lines be engineered to produce fluorescent proteins. Electron microscopy, which does not require transfected cell lines, has a limit of resolution of approximately 0.1 nm; thus, it reveals features that are 1000-fold smaller than those observed by optical microscopy. More recently, advances in single particle methods have begun to provide molecular detail for some signaling complexes. [45, 92-94] These advances in experimental methodology provide a framework from which to develop reagents for studying the function of multireceptor complexes. Still, new strategies are needed.
New methods for exploring receptor signaling are emerging from synthetic organic chemistry and chemical biology. Chemical synthesis provides access to novel compounds that can be used to dissect the role of molecular interactions; in concert with the new imaging techniques, these ligands can illuminate the roles of protein assemblies. This review focuses on the application of synthetic multivalent ligands to the analysis of signaling complexes.[9, 84] Experiments with multivalent ligands can address the next generation of questions concerning the function of receptors and signaling systems.
3. Multivalent Ligands as Probes of Inter-Receptor Processes
3.1. Definitions of Multivalent Ligand Structure and Function
Multivalent ligands present multiple copies of a recognition element (RE) from a central scaffold.[84, 95, 96, 97, 98] The REs of a multivalent ligand can be a carbohydrate, peptide, protein, or small molecule — any moiety that binds to a receptor. The nature of the scaffold chosen to present the REs determines the structural features of the multivalent ligand; it also dictates how easily they can be varied. These issues are relevant because the architecture of a multivalent ligand -- its shape, orientation of REs, flexibility, size, valency -- can influence its biological activity and its mechanism of action.[99-101] For instance, in systems that are activated by receptor clustering, the most potent ligands may be those with many, closely-spaced REs. Such defined ligand features may be attained using chemical synthesis. Thus, synthetic chemists are uniquely positioned to create ligands with tailored biological activities.
From the prospective of an organic chemist, there is an important concept that underlies the design of multivalent ligands that activate cellular signaling – many cell-surface receptors are modular. For example, some receptors possess intracellular catalytic domains, which are distinct from their extracellular binding domains; the RE-binding and signal-generating regions of receptors are therefore structurally and spatially distinct. Thus, multivalent binding to sites at the surface of the cell can be used to assemble and thereby influence interactions of distal intracellular (cytoplasmic) components. This situation is distinct from that encountered using typical organic catalysts. In these systems, the formation of the catalyst-substrate complex results in a chemical transformation. Thus, substrate binding and catalytic activity are inextricably linked. Because binding and catalytic activity are separated for most cell surface receptors, multivalent ligands can be used to manipulate the localization of intracellular catalytic domains. Ligand binding can promote changes in receptor and therefore catalyst proximity; these proximity changes can enhance or restrict access of a catalytic domain to its substrate. In this way, a multivalent ligand can influence cellular responses.
Because of the way in which multivalent ligands function, the nomenclature used to describe the activities monovalent compounds can lead to confusion.[102] Specifically, bioactive compounds are often referred to as agonists or antagonists: The former activates a response; the latter inhibits it. Still, there is often a mechanistic implication in the use of these terms (i.e., agonists are believed to induce a conformational change similar to that of the natural ligand but antagonists induce an inactive conformation). It is these mechanistic implications that complicate the use of this nomenclature to describe multivalent ligands. An individual RE, for example, can be an agonist, partial agonist, or antagonist. Unlike a single RE, however, the ability of a multivalent ligand to activate or inhibit a biological response will be influenced not only by the RE itself but also by the architecture of the scaffold upon which it is displayed. A multivalent display of an RE that serves as a monovalent antagonist, for example, may cluster receptors; thus, the multivalent ligand could serve as an agonist.[103] Likewise, a multivalent ligand composed of REs that bind to a site remote from that of the physiological ligand can induce activation of some systems solely by decreasing inter-receptor distances.[104, 105] Conversely, it has been proposed that multiple agonistic REs displayed in an orientation that holds receptors in unproductive arrangements for signaling can result in a multivalent antagonist.[106, 107] Ligand-induced sequestering of key signaling proteins also can cause antagonist- or agonist-like effects.[108, 109] Thus, rather than terming multivalent ligands agonists or antagonists, we refer to them as either multivalent effectors or inhibitors.[84] Inhibitors block receptor function and multivalent effectors activate cellular processes. This terminology does not imply any particular activity for an individual recognition element nor a specific binding mode; it is a function of both the RE identity and the architecture of the multivalent ligand.
3.2. Classes of Multivalent Ligand Architectures
A variety of scaffolds have been used to probe receptor function (see Figure 4 for examples). These scaffolds have natural, synthetic and semi-synthetic origins; they vary in size, shape, and physical characteristics. As alluded to previously, distinct scaffolds display REs differently; therefore, the structure of a scaffold can have a significant effect on its activity. For instance, a globular scaffold, such as a protein or dendrimer, may not be capable of spanning the large distances needed to cluster multiple proteins. However, such a ligand it may effectively occupy multiple binding sites on an oligomeric receptor. Thus, the architecture of a ligand influences its ability to form different types of macromolecular receptor complexes. [84, 100, 110]
Figure 4.

Scaled diagram of the classes of multivalent ligand scaffolds. Although the actual size of ligands that are based on these scaffolds varies, representative examples of each class are pictured. To illustrate the importance of resolution, the sizes are shown relative to a mammalian lymphocyte. Size bars are as follows: cell and bead 1 nm; liposome and polymer 0.5 nm; antibody, dendrimer, and albumin 0.05 nm.
To understand how to design synthetic multivalent ligands, it is useful to examine the structural complexity of natural multivalent ligands. Natural ligands possess an enormous diversity of architectures. For example, a TNF family member, sTALL, was found to be an ensemble of 60 monomer units interacting non-covalently to generate a symmetric “virus-like” complex. [94, 111, 112] It is hypothesized that geometric complexity of sTALL is required to organize multiple copies of its cell surface receptor into an active signaling complex. Other natural ligands, such as lipopolysaccharides, mucins, and glycosaminoglycans, present a heterogeneous display of potential receptor binding sites, even within the same molecule. [113, 114] A casual visual comparison between the architecture of sTALL and a typical glycosaminoglycan will reveal very few similarities in either size, shape, or RE density. Despite their dramatic structural differences, both sTALL and glycosaminoglycans are involved in clustering cell surface receptors. This simple observation can lead one to begin formulating hypotheses about the differences in scaffold architecture that might impact the underlying signaling. Often, however, determining which of the many possible ligand features influence activity is difficult. For example, naturally-occurring polysaccharides vary in length, branching, and monosaccharide composition. Tracing the specific feature of a physiological multivalent ligand that is critical for activity can be arduous.
Synthetic organic chemistry can provide a wide range of defined multivalent ligand architectures. Because of this diversity, synthetic ligands can be used to dissect the features of physiological ligands responsible for their activity. For example, unlike natural ligands, the valency of a synthetic ligand can be systematically altered by varying the length or size of the scaffold. Similarly, by generating polymers of defined lengths, the effect of valency on activity can be assessed. For example, the effect of dendrimer valency on activity can be explored by testing different generations. By examining the impact of these changes on the biological response, the mechanisms of natural receptor-ligand function can be illuminated. In the following sections, we briefly introduce some structural classes of multivalent ligands and the synthetic challenges and opportunities available. Finally, we discuss how synthetic multivalent ligands have been used to elucidate and exploit the mechanisms by which key receptors function.
3.3. Synthesis of Multivalent Ligands
The most versatile methods for synthesizing multivalent ligands involve adding REs to a preformed synthetic or naturally occurring scaffold. The types of scaffolds used in the generation of multivalent ligands using this approach vary; proteins, dendrimers, polymers, and solid supports have all been used. [115] With an appropriately functionalized scaffold (or RE), multiple copies of an RE can be appended. This approach provides straightforward access to ligands with variable valency and density of REs, because the mole fraction of functionalized RE used in the conjugation reaction can be readily controlled. [115] An alternative to this strategy is the approach typically used in creating polymer-based multivalent ligands: A single step can be used to assemble RE-bearing monomers into a multivalent product. In most of the examples presented herein, the REs have been added to a preformed scaffold to generate the multivalent ligands; for those with little expertise in organic synthesis, this assembly method is the simplest to perform. The examples that we discuss can guide the selection of scaffold; the corresponding literature references can be used to design the most efficient synthetic route.
3.3a. Low Molecular Weight Ligands
We use the descriptor “low molecular weight ligands” to refer to compounds that present fewer than ten REs and typically possess molecular masses less than 1000 Da. The synthesis of such compounds can be quite challenging; however, some general strategies have been developed. [116-119] Although methods for the direct dimerization of individual REs are known,[119] low molecular weight multivalent ligands are typically generated by conjugation of REs to a core scaffold. An advantage of these ligands is that the core scaffold can be rigid to give rise to a specific RE orientation. Additionally, low molecular weight ligands are amenable to combinatorial approaches. [120, 121] Low molecular weight ligands can be generated by combinatorial methods; therefore, their shape, flexibility and other structural features can be varied. Despite the opportunities to exert precise control over ligand features, most low molecular weight ligands are dimeric; dimeric ligands can be more readily synthesized. Because dimeric ligands can interact with only two cell surface receptors, their utility for investigating the role of higher order receptor organization in signaling is limited.
3.3b. Protein Conjugates
Semi-synthetic routes to multivalent ligands often involve the incorporation of REs onto well-characterized carrier proteins such as streptavidin, bovine serum albumin (BSA) and keyhole limpet hemocyanin (KLH). [122, 123] The size and shape of the scaffolds vary: streptavidin, a tetramer, has a rectangular structure with dimensions of approximately 6 by 10 nm, while human serum albumin has a globular shape with a diameter of approximately 10 nm. The drawback of protein conjugates and other multivalent compounds generated by semi-synthesis is their relative heterogeneity compared to scaffolds generated by chemical synthesis. Protein conjugation reactions typically depend on the presence and accessibility of specific endogenous amino acid side chains. Thus, the opportunities for controlling epitope orientation are limited. Interestingly, this limitation has been partially overcome recently by introducing functionality to viral coat proteins. Viral particles bearing these modified proteins serve to organize the functionality in defined patterns. [124-126] This is a promising strategy that is just beginning to be explored.
3.3c. Solid Supports
Receptor binding, activation, and endocytosis have been studied using functionalized beads and surfaces. [127-130] The beads employed in these investigations vary in composition; they can be derived polystyrene, latex, polysaccharides, or other insoluble materials. The typical reactions used to conjugate binding epitopes to beads are straightforward and general, though rarely are these processes chemoselective or regioselective. A variety of small and large, synthetic and natural REs have been incorporated. The number of potential sites that can be functionalized on the surface varies with the bead composition. Although beads are widely used, the orientation and availability of their binding sites has not been characterized; generally, the distribution of sites on the bead (and perhaps on the RE) is assumed to be random. Bead size is another variable that can influence bead activity. Indeed, the diameter of beads can vary widely. For example, Sepharose beads, which are used for size exclusion chromatography, have diameters of 30 to 300 μm; latex beads, at 0.2 to 2 μm across, are much smaller. The number of receptors that a functionalized bead can occupy will depend on many variables: the number of functional REs presented, the density of RE sites; and the size of the beads. The only variable that can be controlled readily is the latter. As with protein scaffolds, therefore, a drawback of beads is the lack of control they offer.
3.3d Liposomes
Liposomes are typically non-covalent assemblies that can be used to present multiple REs. It is the lipid-lipid interactions that give rise to the array;[131-133] thus, liposome composition can be controlled by varying the ratio of RE-bearing and unmodified lipids. Liposomes can be generated in a wide range of sizes. For example, liposomes generated by treatment with BioBeads affords species with diameters of 0.05 to 0.5 μm. [134] Large liposomes (termed giant vesicles) with a diameter of 5-200 μm also can be produced. [135] The size of liposomes can be nearly homogeneous; however, the arrangement of REs within each liposome is difficult to regulate. Fluctuations in RE presentation within a liposome can be an advantage in generating RE displays that are highly active. The orientation and density of REs can change as the lipid components undergo 2-dimensional diffusion; thus, the most active arrangement of REs can be found. For mechanistic studies this dynamic behavior is detrimental. Another potential drawback of liposomes is that individual components can partition into biological membranes; therefore, the use of liposomes can be problematic for mechanistic studies involving organisms or even cells.
3.3e. Dendrimers
Dendrimers are often used as multivalent ligand scaffolds. [96, 136-141] An advantage of these ligands is that they can be fairly homogeneous; [96] this quality can aid in relating ligand features to biological activity. With beads or carrier protein conjugates, the sites of RE conjugation often are unknown; therefore, the population of conjugates is heterogeneous. In contrast, the architectural features of a dendrimeric ligand are defined by the choice of scaffold and the methods used for its synthesis. For example, Starburst (PAMAM) dendrimers of generation 0 have diameters of 1.5 nm and valencies of four. [142] Each increase in generation enhances the diameter by 0.7 to 1.6 nm while also doubling the maximum valency. Thus, as the size of the dendrimer increases, so does the density of REs. The REs of such highly functionalized dendrimers are often inaccessible to proteins.
3.3f. Polymers
Modern polymer chemistry is providing new opportunities for the synthesis of tailored, biologically active polymeric (oligomeric) ligands. The multivalent ligands derived from polymers typically are composed of a central backbone that presents multiple copies of a RE; ligands of this type can be produced using a variety of synthetic methods. Polymers are generally assembled in a single step by methods such as radical, ionic, or ring-opening metathesis polymerization of RE-bearing monomers. [101, 143-149] Alternatively, REs can be appended to a pre-formed polymeric scaffold. [101, 147] Certain polymerization reactions are more tolerant of biologically active functionality than others; thus, the type of epitopes to be incorporated will determine the most effective synthetic strategy.
With select polymerization reactions, the biologically-active ligand can be synthesized with controlled valency. [84] Living polymerization reactions, in which the rates of chain termination are low, are powerful methods for generating well-defined multivalent ligands. When these polymerizations have fast initiation and slow propagation rates, polymers of narrow polydispersity can be generated. [150] Such polymerization reactions can be used to synthesize multivalent ligands with a variety of sizes. For example, linear polymers with molecular weights of approximately 3,000 generated via ring-opening metathesis polymerization (ROMP) can span 100 nm.[151] Polymers displaying dendrimeric appendages can span 10 to 100 nm.[152] In addition to allowing control over the degree of polymerization, living polymerization reactions can be used to synthesize block copolymers that display different REs (or simply different functionality) within each block.[153]
Polymers generated by ROMP have found increasing use as biologically active multivalent ligands. [101, 149, 154-158] Ruthenium carbene catalysts for ROMP can be used to generate materials of distinct valency.[150, 159-161] These ligands have been generated as inhibitors of saccharide-protein interactions, [151, 162-164] as ligands for combating vancomycin-resistant bacteria,[156] and as effectors of biological responses. [46, 165] Importantly, this polymerization method allows control over the RE display and density.[166, 167] The differences in activity between these compounds can, therefore, be attributed to specific aspects of ligand structure.
3.3g. Combinatorial Multivalent Ligand Synthesis
One new frontier in multivalent ligand synthesis is the application of combinatorial methods. Combinatorial and diversity-oriented synthesis of monovalent molecules has facilitated the discovery of low molecular weight ligands that are optimally designed to bind to a target receptor. [168-173] Similarly, libraries of multivalent ligands might be expected to contain compounds that possess diverse activities.[119-121, 166, 167, 174] However, the potential diversity of multivalent materials is great and strategies to synthesize diverse collections of multivalent ligands are only beginning to emerge.
The technical hurdles to overcome in the synthesis of small molecule libraries versus multivalent ligand libraries are distinct. For example, multivalent ligands bearing different REs can have different physical properties; therefore, they will require different purification methods. Combinatorial approaches to multivalent ligand synthesis are in their infancy: Only a few synthetic strategies for generating diverse multivalent ligands have been described. [119-121, 166, 167, 174] [153] Future advances in synthetic methods and purification technologies will be instrumental in providing multivalent materials with broad architectural diversity.
3.4. Mechanisms of Multivalent Ligand Binding
Multivalent ligands can access receptor-binding modes inaccessible to monovalent compounds (Figure 5).[100, 115, 175] For example, a ligand with multiple copies of a RE can bind to multiple binding sites on a single oligomeric receptor; examples of such receptors are immunoglobulins and some lectins. When a multivalent ligand binds to an oligomeric receptor, it can occupy multiple binding sites. The cost for translational entropy is paid with the first receptor–ligand contact, and subsequent binding interactions proceed without additional translational entropy penalties.[176] Excellent examples of this chelation binding mode are presented in some recent studies directed at devising multivalent inhibitors of pentavalent toxins.[177-179] Similarly, certain receptors possess binding subsites in addition to their primary site of interaction. Unlike monovalent ligands, which can only access subsites adjacent to their primary binding pocket, multivalent ligands may gain binding energy from contacting these secondary sites.[180] Either a RE or another component of the scaffold can contact these subsites. In situations in which the multivalent ligand occupies more than 1 binding site -- whether it is a subsite or a primary site within an oligomeric receptor -- the ligand typically will bind with a high functional affinity. [181] Another mechanism by which a multivalent ligand can act is through steric stabilization: The steric bulk of the multivalent ligand may preclude the engagement of a bound receptor with an opposing viral particle or cell.[182] This aspect of multivalent ligand activity is particularly relevant for inhibitors of binding interactions at the cellular or viral surface. Collectively, these binding modes are unique to multivalent ligands and have led to numerous applications of these reagents as inhibitors of macromolecular interactions.
Figure 5.
Receptor binding mechanisms that are unique to multivalent ligands.
As mentioned previously, there are applications of multivalent ligands that do not involve inhibiting a process; an alternative and complementary use is signal transduction activators. Multivalent ligands can bind avidly to multiple receptors on the cell surface, a process that is facilitated in the fluid lipid bilayer by the two dimensional diffusion of receptors. If multivalent ligands cluster signaling receptors (Figure 5c), they can activate signaling pathways. It is this mechanism, which is uniquely available to multivalent ligands, that is the focus of the remainder of this review.
We have outlined the three major concepts that are critical for the application of multivalent ligands as probes of signal transduction. First, signal transduction cascades are mediated by receptor–receptor interactions, and promoting receptor assembly is critical for signaling (Figure 3). Second, multivalent ligands can interact with the target receptors via multiple binding modes (Figure 5). Third, the structural architecture of a multivalent ligand will determine the favored binding modes (Figure 4). Thus, the structure of the ligand can be optimized to elicit the desired biological response.[84] In the next sections, we provide some examples that illustrate the utility of multivalent ligands as mechanistic probes. Insights into receptor function gleaned from these studies and possibilities for further advances are presented.
4. Using Multivalent Ligands to Gain Insights into Receptor Function
As mentioned earlier, the binding of a multivalent ligand to a cell surface can assemble a multiprotein complex with distinct features. The ligand valency, orientation of REs, and stability can be used to control features of the complex: its stoichiometry; its size; the orientation of the receptor within it; and its lifetime. Thus, a multivalent ligand can have a marked influence on the output response of a signaling cascade. The following examples describe how specific multivalent ligands can be used to illuminate signaling.
4.1. Receptor Proximity
The binding of multivalent ligands can result in receptor clustering in the membrane.[84] Here, we discuss some of the many examples in which the ability of multivalent ligands to promote clustering has been used to reveal aspects of receptor function.
4.1a. Integrins
Integrins are cell-surface receptors that mediate cell adhesion; they also are the central component of the focal adhesion signaling complex. [24, 183-185] Integrin function is important in multicellular organisms, and targeted deletion of integrin subunits can strongly impair tissue formation.[186] Given their fundamental physiological roles, it is not surpising integrin-mediated adhesion and signaling events are carefully regulated.
A key feature of integrins is their ability to switch between low- and high-affinity binding modes.[187] These two modes are thought to allow rapid and reversible adjustments to the strength of cell adhesion. This switch likely involves changes in integrin conformation and/or proximity.[188] Integrins are proposed to adopt at least two conformational states: a bent inactive state and an extended active state.[187] In addition, they can undergo changes in receptor proximity, which can be induced either by interaction with ligand (an “outside-in” change) or by cellular activation (an “inside-out” change). Although there is evidence that both proximity and conformation influence signaling and adhesive strength, more specific and useful therapeutics may result from a deeper understanding of the relative contributions of these mechanisms. Insight into the influence of receptor proximity in focal adhesion function is emerging from experiments utilizing synthetic multivalent ligands.
One type of synthetic ligand used to probe integrin clustering is a chemical inducer of dimerization (CID). CIDs are low-molecular weight, cell-permeable, bivalent ligands that can mediate the clustering of two receptors.[4, 116, 189-191] A key feature of CIDs is that they can oligomerize receptors fused to a specific ligand-binding protein (Figure 6). In its first incarnation, this strategy utilized fusions to FKBP, a protein that binds to the immunosuppressant FK506.[189] FK506 is a small molecule that can be functionalized and converted to a dimer, FK1012; the latter is capable of dimerizing FKBP. FKBP-fusion proteins can be transfected or otherwise introduced into cells permitting examination of the functional consequences of FK1012-mediated dimerization of the target fusion proteins. If multiple copies of FKBP are fused to the target protein of interest, ligand addition can induce higher order assemblages. [192] Such strategies have been expanded to include heterodimerizers and dimerizers that avoid endogenous proteins (e.g., FKBP) and specifically target engineered fusion proteins. [192-194]
Figure 6.
Depiction of the intracellular-mediated alteration of receptor proximity by CID. A receptor fused to a binding protein is expressed in a target cell. Addition of a small molecule dimerizer (FK1012) induces the clustering of the receptor.
As discussed above, both integrin proximity and conformation contribute to the switch between low- and high-affinity integrin binding. To dissect the role of conformational change from that of receptor proximity, a CID strategy was employed.[195] A gene encoding two copies in tandem of FKBP was fused to that encoding αIIb; the latter is a partner in the integrin complex αIIb-β3. In cells producing the resulting fusion protein, the addition of a dimerizer, AP1510, elicits integrin clustering. If proximity alone is important for the low- to high-affinity switch, one would expect that the addition of AP1510 would fully replace the need for a natural binding partner. As a measure of the switch to the high-affinity binding mode, an antibody (PAC1) was used that binds specifically to the high-affinity form of the integrin. When the FKBP dimerizer AP1510 was added to cells, a dose-dependent increase in binding of PAC1 was observed. To investigate the role of receptor conformational change, these authors used a monovalent antibody fragment, termed LIBS, that binds and potently activates integrins. The increase in binding observed in the present of the dimerizer AP1510 was modest; the effect from LIBS treatment was more dramatic. These studies with CIDs indicate that, although there is a contribution from receptor proximity, other factors, such has adopting the high affinity conformational state, contribute significantly to integrin-mediated cell adhesion.
As described, integrins are cell surface proteins that function not only in adhesion but also in intracellular signaling. Signaling from integrins involves the activation of multiple pathways. As is the case for most complex signaling systems, efforts to deconvolute the contributions of each pathway are ongoing. In integrin signaling, two kinases, Syk and FAK, are implicated in transducing signals through different pathways. Integrin clustering may favor information transfer through one of these pathways (Syk or FAK) over others. To test this hypothesis, the role of receptor proximity in signaling by Syk and FAK was explored with the dimeric ligand AP1510. When cells were treated with AP1510 alone to cluster the integrin αIIb-β3, an increase in tyrosine phosphorylation by Syk was observed. The requirements for activation of FAK, however, include both high affinity receptor occupancy (treatment with LIBS) and oligomerization (treatment with AP1510). These results demonstrate the complex interplay between signaling, adhesion, and cellular response that can be achieved by changes in integrin occupancy, proximity, or both. These investigations illustrate the utility of the CID strategy for deconvoluting these contributions. One minor disadvantage of the CID approach, however, is that the cells be genetically engineered to express the fusion proteins.
Multivalent surfaces can be used to probe integrin function directly. Integrins can facilitate cell adhesion to extracellular matrix components as well as other cells. [185, 196] The extracellular matrix, a naturally occurring multivalent display, may therefore also influence integrin activity.[197] In one study, surfaces were modified with peptide REs of different valencies for use in cell adhesion studies. [198] Lymphoblastoid B cells were allowed to adhere to microtiter wells coated with naturally monomeric (fibrinogen) or multimeric (fibrin) REs (Figure 7A). Multivalent fibrin was able to support cell adhesion but unactivated cells did not adhere to immobilized monovalent fibrinogen.
Figure 7.
The density of REs presented influences the adhesion of cells to integrin-ligand-bearing surfaces. (A) Surfaces coated with multivalent ligands have a greater functional affinity for B cells. (B) Star polymers are depicted that display variable copies of the pentapeptide YGRGD, an integrin-binding RE. The relative potency of each surface is indicated by the number of cells bound.
In a series of related experiments, a ligand specific for the integrin αvβ3, adenovirus PB, was coated in wells in either its monomeric or pentameric form (Figure 7b). As in the fibrin experiment, the B cells attached selectively to the wells coated with multivalent ligand. To understand the mechanisms responsible for the differences in binding, the number of possible integrin binding sites in the wells displaying monomeric (800 sites/mm2) or pentameric ligand (125 sites/mm2) was determined via radioimmunoassay. Although it had 6-fold fewer sites per mm2, the surface displaying pentameric integrin ligand was more adhesive. These results suggest that the local arrangement of binding sites, and not the overall number of sites, is critical for integrin-mediated adhesion.
These results are supported by additional data that indicate local RE density is important for integrin adhesion strength.[127] Maheshwari et al. used star polymers that display variable numbers of copies of the minimal integrin ligand YGRGD to investigate the impact of binding site density on integrin-mediated cell migration and adhesion of αvβ3-bearing cells. The integrin αvβ3 binds RGD sequences, and surfaces with more YGRGD were more effective at interacting with cells (Figure 7B). Interestingly, however, when surfaces with equal numbers of REs but different local densities were compared, those with closely packed REs were more efficient at mediating cell migration and adhesion. These results suggest that clustering of the integrins improves their adhesive strength, likely through both enhanced functional affinity for ligand and by activation of signaling pathways.
Together, experiments using synthetic multivalent surfaces have illuminated a role for receptor proximity in integrin function. These studies also provide information about the most effective types of natural integrin ligands. Unfortunately, these multivalent surfaces cannot be used to explore integrin function in whole organisms. Experiments utilizing soluble multivalent ligands could be used to investigate the consequences of integrin clustering in physiological settings.
4.1b. G-Protein Coupled Receptors (GPCRs)
GPCRs are one of the largest families of mammalian cell surface receptors. They participate in many important physiological processes. Their significance is underscored by their importance to medicine: over 50% of therapeutics on the market act through GPCRs.[199] Mounting evidence implicates GPCR homo- and heterodimerization is involved in the regulation of signaling via these receptors.[117, 200] Therefore, to understand GPCR signaling and to exert control over it, it would be valuable to elucidate the contribution of GPCR oligomerization. The role of ligand in modulating the clustering of GPCRs is unknown, but experiments using synthetic multivalent ligands are beginning to address this issue.
Low molecular weight divalent ligands have been used to with the goal of assessing the consequences of GPCR dimerization.[103, 117, 186, 201-207] An illustration of this approach is the use small molecule dimer of GPCR ligand, which has been used to activate neutrophil chemotaxis. Neutrophils are attracted to sites of bacterial infection by byproducts of bacterial protein synthesis, such as N-formyl methionine-containing peptide fragments. Perhaps the most widely studied neutrophil chemoeffector, is N-formylmethionine-leucine-phenylalanine (fMLF), which is recognized by the formyl peptide receptor (FPR), a GPCR. Dimers presenting two copies of fMLF, therefore, were employed to explore the role of receptor proximity in the regulation of FPR signaling.
Mono-, di-, and triethylene glycol-linked dimers of fMLF were generated by coupling a linker to the C-terminal end of the peptide. The monoethylene glycol dimer would not be expected to be capable of dimerizing the FPR; consequently, any enhanced chemotactic activity relative to fMLF is likely due to an increase in avidity. In contrast, the authors suggest that the triethylene glycol-linked dimer may facilitate receptor dimerization, although the linker length required to simultaneously occupy two copies of the FPR is not known. The authors generated a series of divalent ligands, hoping that those with longer linkers might cluster the FPR.
Two assays were used to explore FPR function: chemotaxis and superoxide production.[208] In vivo, neutrophils are recruited to the location of the invading pathogen through chemotaxis; the release of superoxide serves a killing role. These two cellular responses must occur with strict spatial and temporal control to minimize damage to healthy tissue. Specifically, chemotaxis towards a site of bacterial infection must precede superoxide formation or neutrophils could prematurely release toxic oxygen species. Both of these responses can be elicited in response to ligand binding to FPR. It is important, therefore, to understand what factors influence the triggering of these distinct cellular responses.
The synthetic dimers were added to cells expressing FPR, and the chemotaxis and superoxide levels were monitored. The triethylene glycol linked-dimer was the most active chemoattractant, while the monoethylene glycol dimer elicited the highest level of superoxide production. Although FPR dimerization was not tested directly, these results suggest that receptor dimerization may influence chemotactic responses. The increased ability of the monoethylene glycol dimer to elicit superoxide release was attributed to it having higher affinity for the FPR. While the specific mechanisms of action of these divalent agents were not probed further, these initial results suggest that synthetic multivalent ligands will be useful in illuminating the role of receptor clustering in GPCR signaling, in general, and chemotactic signaling in neutrophils, in particular.
Receptor heterooligomerization can provide a means of integrating information from multiple pathways into a coherent cellular response. The consequences of GPCR heterodimer formation have been probed by creating heterodimeric ligands.[103] This approach was taken to study the effects of co-clustering a GPCR that responds to enkephalin with one that is activated by neurotensin. Both natural ligands are important regulatory neuropeptides that function as synergistic activators of GPCR signaling.[209] The goal of the studies was to determine whether there might be a direct potentiation of signal via assembly of a heteroligomeric GPCR complex.
Cells expressing both receptors were treated with neurotensin, a mixture of neurotensin and enkephalin, or a bifunctional neurotensin-enkephalin conjugate [209] The cytosolic cyclic GMP (cGMP) concentration was measured at a variety of ligand doses. When both hormones were added separately only a subtle increase in cGMP production was observed. In contrast, the covalent heterodimer was a potent inducer. These data suggest that the forced proximity of the two receptors is the primary cause for the increased level of cGMP. As in the previous example, however, direct data in support of heterodimerization is lacking.
Further experimentation with ligands bearing higher valencies or more diverse architectures may afford more definitive insights into the effects of receptor proximity on GPCR-mediated signaling. It will be especially useful to couple signaling studies with those that address the influence of ligands on GPCR assembly. Because so many drugs target GPCRs, these investigations may facilitate the design of a new generation of therapeutics.[210]
4.1c. T Cell Receptor
T lymphocytes are key mediators of mammalian immunity. [13, 22, 78, 211] These cells recognize foreign antigens via their T cell receptors (TCRs). T cell receptor engagement can result in cellular activation; this activation can precipitate the killing of tumor cells, pathogens, and virally infected cells. As discussed in the introduction, when T cells encounter antigen-presenting cells, an immune synapse can form at the cell-cell interface. This immune synapse, which contains the TCR and other co-receptors and signaling components, possesses exquisite organization. [76]
There is evidence that changes in the organization of the TCR and other signaling components are important for T cell activation; yet, the contribution of various factors to TCR signaling remains a subject of debate. Despite the large body of literature on TCR organization and signaling, questions concerning the molecular details of the TCR function remain. For example, it has been controversial whether the dimerization of two TCRs is sufficient stimulus for T cell activation or whether TCR oligomerization is required. Also unknown is the role of simple monovalent ligation of individual TCRs. [212] If individual or small groups of TCRs can activate signaling, the purpose of the synapse is unclear. Multivalent ligands will undoubtedly play prominent roles in addressing these fundamental issues.
One strategy for studying TCR function uses multivalent protein conjugates; these are formed using the high affinity interaction between biotin and the tetravalent protein streptavidin. [213] The four identical biotin-binding sites of biotin each bind with a dissociation constant of 10-15 M; therefore, biotinylated recognition elements can be readily displayed from a streptavidin scaffold. Because streptavidin is tetravalent, multivalent complexes can be generated that present 1, 2, 3, or 4 recognition epitopes per scaffold (Figure 8). Davis, McConnell, and coworkers developed streptavidin as a scaffold for the presentation of peptide-loaded major histocompatibility complexes (MHCs). [213] Because peptide-loaded MHCs can serve as ligands for the TCR, streptavidin-bound biotinylated MHC complexes can bind (in principle) up to 4 TCRs.
Figure 8.
Schematic of streptavidin-MHC-MCC complexes. Four complexes with one, two, three, or four biotinylated MHC-MCC moieties are shown on the left. A model for the activation of TCRs by multivalent engagement by the highest valency ligand is shown at the right.
To generate these multivalent presentations, a peptide derived from moth cytochrome c (MCC) was added to the MHC. The resulting MCC-MHC complex was singly biotinylated and mixed with streptavidin; this procedure should yield a tetravalent complex that can bind the TCR (Figure 8). The amount of active MHC complex in the assembly reaction was varied to favor formation of mono-, di-, tri-, or tetrameric structures. Biotinylated MHC that lacks MCC was also produced, and these proteins, which cannot bind the TCR, were added to occupy remaining streptavidin sites. A complex displaying 4 unloaded MHCs, which does not display foreign antigen, served as a control.
To test the effects of the assemblies on T cell activation, increases in extracellular acidification and the concentration of intracellular Ca2+ were measured. In both cases, the tetramer was the only ligand to elicit a significant increase in activity. Although effects due to changes in binding kinetics cannot be ruled out, these experimental results suggest that changes in TCR proximity influence T cell activation. Similar results were obtained using chemically defined peptide-based multivalent ligands, although a dimeric ligand was sufficient to activate TCR signaling. [214, 215] Together, these data suggest that valency is an important feature of natural TCR ligands and that receptor proximity is important for the amplification of TCR-mediated signals.
Factors other than TCR ligation also contribute to activation and signal modulation. Indeed, numerous co-receptors serve to positively or negatively modulate the activation signal. One limitation of using streptavidin-based scaffolds is that it is difficult to incorporate multiple REs that can engage co-receptors and TCRs simultaneously. Thus, scaffolds that can be functionalized to present multiple types of REs have been employed.
A variety of beads and surfaces have been used as multivalent displays to explore T cell signaling. [9, 129, 216-218] In this way, a study addressing the mechanism by which T cell responses are enhanced by the co-receptor CD28 was undertaken.[87] Ligand binding to the TCR is necessary and sufficient for activation of T cells; however, co-stimulation of CD28 yields a more potent response. To determine the mechanism for enhanced signaling, beads were used to mimic the physical size of the surface of an antigen-presenting cell (APC), which presents ligands on its cell surface for the TCR and CD28.
Latex beads were modified to assess the consequences of simultaneous receptor engagement. Specifically, they were designed to co-cluster the TCR and CD28. An antibody that recognizes the TCR was appended, as was another that binds the co-receptor CD28. The beads were tested for their ability to induce cell proliferation, which occurs upon lymphocyte activation. If T cell activation requires co-engagement of both the TCR and CD28, only the beads displaying REs for both receptors should activate the cells. Beads displaying only the TCR clustering element (anti-TCR) did not promote T cell proliferation. In contrast, beads displaying both anti-TCR and anti-CD28 were potent activators of proliferation.
One explanation for the data is that co-engagement of CD28 and TCR promotes the co-localization of these receptors to a region of the membrane termed a lipid microdomain. It has been suggested that lipid microdomains concentrate signaling components and facilitate formation of mature signaling complexes. In support of a role for microdomains in enhancing signaling, a fluorescent marker of lipid rafts (fluorescein-labeled cholera toxin B subunit) was concentrated into a dense fluorescent patch at the point of contact between the bead and the T cell. These results demonstrate that co-stimulation of the TCR and co-receptors can modify their distribution on the cell surface, which may modulate immune responses. An important caveat to these experiments, however, is that the REs on a bead-based multivalent ligand are immobilized. Thus, unlike cell surface REs, the REs attached to the bead cannot undergo rearrangement. When an antigen presenting cell was used as a natural “ligand”, however, similar results were obtained This comparison suggests that the ligand-decorated bead effectively mimics some aspects of an antigen-presenting cell.[219] Thus, a role for the CD28 co-receptor in regulating receptor proximity and immune function was implicated in studies using functionalized beads as multivalent ligands.
4.1d. Bacterial Chemoreceptors
Bacteria must migrate toward nutrients and away from toxins to survive. Bacterial chemotaxis is driven by a well-characterized signaling pathway that is responsible for the detection of these nutrients and toxins. [220-222] Five types of membrane-bound chemoreceptors, MCPs, mediate chemotaxis. Each MCP type is responsible for detecting and mediating chemotaxis towards a subset of small molecules or other stimuli. To initiate appropriate locomotion, bacteria require integration of signals from multiple stimuli into a coherent response. Moreover, responses are mounted against stimuli at very low concentrations, which requires a significant level of signal amplification.[223-225]
The mechanism used by bacteria to amplify and integrate chemotactic signals has long been debated. One recent hypothesis is that signal amplification is achieved through inter-receptor communication. [26, 60, 226, 227] [54] Interestingly, the MCPs are organized within the cell: They are concentrated at the poles of Escherichia coli and other bacterial species. [39, 228] This organization of chemoreceptors into an array has been proposed to be important in signal transduction. Recent experiments using synthetic multivalent ligands and other approaches support such a view; they suggest that communication between homologous and heterologous MCPs within the chemoreceptor array is responsible for chemotactic responses.[46] [60, 62, 229-231] [54]
The first goal was to generate multivalent attractants that could cluster the chemoreceptors. ROMP was used to synthesize multivalent ligands that display chemotactic carbohydrate residues in sufficient valency (∼25 monomer units) to mediate MCP clustering. As predicted, these were shown by microscopy to cluster the MCPs. This change in receptor proximity influenced signaling through the MCPs: ligands capable of clustering MCPs were also potent activators of chemotaxis (Figure 9). Using a fluorescent probe, it was shown that galactose-bearing multivalent ligands cluster the galactose-sensing receptor, Trg. Surprisingly, they also cluster the serine-sensing receptor Tsr. Tsr does not bind galactose; its presence in the cluster suggests that chemoreceptor–chemoreceptor interactions bring it into the cluster. These data indicate that there are interactions between different types of chemoreceptors.
Figure 9.
Synthetic multivalent polymer-based investigation of receptor proximity effects in bacterial chemotaxis. Addition of a multivalent ligand with sufficient valency can induce the re-organization of MCPs. This potentiates signaling through these receptors and activates bacterial locomotion.
As evidence in support of the functional significance of chemoreceptor–chemoreceptor interactions was obtained by examining the effects of receptor clustering on signal output. Specifically, a multivalent galactose derivative was introduced to cluster the chemoreceptors. When the bacteria had adapted to the multivalent attractant, the monovalent attractant serine was added. Under these conditions, the chemotactic response to serine was potentiated by 100- to 1000-fold. This result indicates that heterologous MCPs communicate to amplify and integrate chemotactic signals. The proximity between multiple types of receptors is critical for signal amplification. Thus, a single receptor is not all that is needed to sense a particular compound – all chemoreceptor types contribute to proper sensing and signal amplification.
The application of multivalent chemoattractants to examine chemoreceptor proximity in chemotaxis illustrates their power: They can be used to explore signal transduction even when receptor dimerization is known to be required. Changes in receptor organization had not been implicated in chemotactic signal transduction; the role of receptor organization might have been overlooked because the known chemoattractants are monomeric, and the MCPs are dimeric in the absence of ligands. Nevertheless, studies indicate that changes in MCP proximity influence signal amplification, and evidence supporting such a model was obtained using multivalent ligands. Moreover, the use of these ligands did not require genetic manipulation of the bacteria; thus, the behavioral response elicited by the multivalent ligand could be directly analyzed under physiological conditions and in wild-type genetic backgrounds. These results underscore that soluble, structurally defined multivalent ligands are valuable probes.
4.2. Receptor Orientation
Many cell surface receptors possess domains with intrinsic enzymatic activity; alternatively, some interact with proteins with catalytic domains. The substrates of these enzymatic activities are often other signaling proteins. The relative orientations of the receptor-associated enzyme active site and the substrate may influence the amount of product generated; this enzymatic efficiency, in turn, will influence signal transduction.[232][230][229][225] When a signaling complex assembles, it is often unclear whether its activity is due to a specific orientation of receptors within the assembled complex or simply the localization of signaling components.[233] Thus, questions remain about the roles of receptor orientation in signaling. As discussed above, multivalent ligands can organize receptors into specific orientations; therefore, they have the potential to serve as tools for investigating the role of protein orientation in signaling.
4.2a. G-Protein Coupled Receptors (GPCRs)
One key feature of multivalent ligands is they can occupy multiple binding sites on a single receptor and thereby exploit the chelate effect (Figure 5a).[176] Multivalent ligands that use this binding mode can interact with high functional affinities, an important determinant of agonist or antagonist potency. In a recent example, a low molecular weight dimer was constructed that can bind simultaneously to two sites on the corticotrophin-releasing factor receptor (CRFR-1), a GPCR that is a key regulator of adrenocorticotropic hormone (e.g. testosterone) release during the stress response.[234]
A series of dimers was generated, such that each component possesses REs in different orientations; the orientation of the REs was controlled by using a helical and rigid linkers. The addition of these dimers to cells expressing the CRFR-1 resulted in testosterone release. Potent activity was elicited by the dimers that maintained a trans-orientation between the terminal REs. Ligands that did not maintain this orientation were 10-fold less effective at causing testosterone release. The experiments conducted do not eliminate the possibility that the observed differences arise from ligand-induced changes in receptor oligomerization. Still, these results suggest that the orientation of REs can influence receptor activation.[117, 235]
4.2b. ZAP-70
Chemical synthesis can be used to create ligands to probe the role of protein orientation in assembled signaling complexes. In the previous example, the relative orientation of binding moieties within a multivalent ligand influence its ability to elicit signaling through a target receptor. When a multivalent ligand acts via clustering receptors, however, the geometric constraints on the binding epitopes might be relaxed. In the few examples studied to date, the localization of receptors to a signaling complex is often more important than their specific orientation within the complex. [233, 236] Consistent with this view, the ability of dimeric ligands to activate signaling via ZAP-70 highlights that different RE orientations often give rise to only subtle effects.[107]
ZAP-70 is a kinase whose function is required for the varied signaling functions of the T cell. If the orientation of ZAP-70 is important for its function, multivalent ligands that control its orientation should possess varying abilities to elicit T cell activation. To this end, a series of small molecule dimerizers was generated; these dimeric ligands possess conformationally restricted linkers between the two identical REs. To assess the activity of this series, CIDs were added to cells encoding a membrane docking protein (3 copies of FKBP fused to a myristoylation domain of v-src) and a ZAP-70 – FKBP fusion protein (Figure 10). Presumably, these CIDs can bind and cluster the fusion proteins, thereby recruiting ZAP-70 to the membrane. All of the synthetic CIDs mediated ZAP-70 recruitment; all activated signaling. While large changes in orientation may influence ZAP-70signaling, the presumed changes in orientation afforded by the CIDs had little effect.
Figure 10.
Orientation of ZAP-70 influences its kinase function. A CID strategy was used to explore the influence of ZAP-70 orientation on function of this kinase. The chemical structures of three dimerizer compounds are shown, FK1012, FK1012H2, and FK1012Z. These dimers present REs in three distinct relative orientations. The relative abilities of these compounds to induce ZAP-70 activity were similar.
The difference in the results of the CRFR-1 versus the ZAP-70 studies suggests a plasticity in signaling complex assembly: For signal activation, the REs of dimeric ligands that occupy subsites within a receptor must be more carefully aligned than those that act by inducing changes in receptor proximity. Interestingly, these findings are consistent with studies in which fusion proteins have been used to test whether different assemblies of signaling domains influence output. [233, 236] As with the ZAP-70 investigations, the orientation of different signaling domains was much less important than their recruitment to a signaling complex. It will be interesting to explore further the generality of these findings.
4.3. Receptor Stoichiometry
The number of receptors in a complex can influence its biological activity. Cell adhesion is expected to be especially sensitive to the number of receptors in a complex; the functional affinity of the array is expected to be directly related to the number of receptor-ligand complexes formed.[100] Signal transduction processes also may be sensitive to the number of receptors in the complex; however, it can be difficult to determine the quantitative role of receptor stoichiometry in signaling. To investigate either cell adhesion or cell signaling, multivalent ligands can be generated that vary in the density and valency of REs. These ligands, therefore, may have the ability to generate specific clusters of receptors that vary in the number of proteins included. This strategy has been utilized to investigate the role of stoichiometry in receptor function.
4.3a. B Cell Antigen Receptor
The B cell antigen receptor (BCR) is a complex of proteins involved in the recognition of antigens and the generation of antibodies during acquired immune responses.[23, 85, 237] The activation of BCR-mediated functions must be strictly regulated; inappropriate activation can cause autoimmune disease. Understanding the minimal requirements for BCR-mediated activity is therefore critical. Synthetic multivalent ligands have played key roles in investigating the importance of antigen stoichiometry in the activation of this system.
Pioneering work by Dintzis and coworkers in the field of BCR signaling in the 1970s and 1980s utilized synthetic multivalent ligands to explore the role of valency in B cell responses. [238-242] The binding of a multivalent antigen to the B cell receptors (BCRs) can activate B cell signaling. To test how the valency of T-cell independent antigens influences antibody production, a series of multivalent ligands were generated using polyacrylamide, dextran, carboxymethyl cellulose, and polyvinyl alcohol as scaffolds.[243] These were injected into mice, and the efficiency of antibody production was measured. In every case, independent of scaffold structure or the amount of polymer branching, the immunogenicity of the ligands strictly depended on RE valency. From these data, the authors hypothesize that the number of BCRs included in a cluster is a principle determinant of function. Moreover, they propose that occupation of approximately 20 BCRs is required for B cell activation. In this complex system, there are many steps between B cell activation and antibody production. Still, these investigations highlight the power of synthetic multivalent ligands for answering questions of fundamental biological importance.
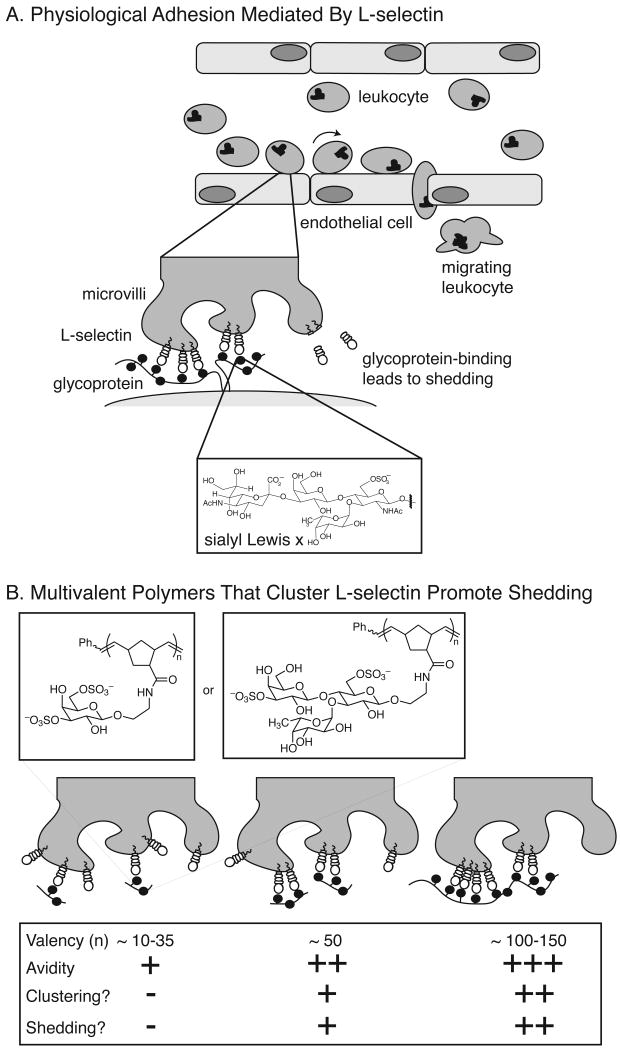
4.3b. L-Selectin
Leukocyte migration from the blood to lymphatic tissues is dependent on the function of L-selectin; L-selectin also mediates leukocyte recruitment to sites of inflammation.[244] L-selectin is displayed on the surface of leukocytes, and it typically localizes to patches at the tips of the cellular microvilli (Figure 11).[237][241-246] The natural ligands for L-selectin are glycoproteins displayed on the endothelium of the blood vessel. [247] Binding of L-selectin to these ligands slows the progress of cells through the vessel and allows tight adherence of the leukocyte to the endothelium. Physiological L-selectin ligands typically present multiple copies of derivatives of the sulfated sialyl Lewis × antigen (sLe×); synthetic multivalent ligands that display these and related carbohydrate epitopes have been shown to be effective selectin ligands.[248-253] Multivalency, therefore, may be an important determinant of L-selectin function in vivo. Experiments using synthetic multivalent ligands have begun to reveal the importance of multivalency and stoichiometry of selectin-ligand clusters for L-selectin recognition.
Figure 11.
Multivalent ligands for L-selectin mimic cell surface glycoproteins. (A) L-selectin expressed on lymphocytes binds to glycoproteins on the endothelium. This interaction slows lymphocyte progression through the vessel and triggers proteolytic release of L-selectin. (B) Multivalent polymers displaying sulfated carbohydrates also bind multiple copies of L-selectin, which leads to receptor clustering and proteolytic shedding. There is a direct relationship between the valency of the polymer, the number of L-selectin proteins bound, and the avidity of the interaction.
The role of receptor stoichiometry in regulating the adhesion function of L-selectin was investigated using synthetic multivalent ligands derived from ROMP.[254] The aforementioned properties of ROMP were exploited to generate ligands of distinct valencies. Additionally, the polymers were specifically end-labeled with a single fluorophore to visualize binding and to determine how many copies of a multivalent ligand bind L-selectin-positive cells.[255] When these compounds were tested, it was found that the number of copies of L-selectin that bind to a multivalent sulfated carbohydrate derivative depends the ligand's valency (Figure 11).[254] A related observation is that the functional affinity of a ligand for cells displaying L-selectin is directly related to its valency. Increasing the valency from 35 monomer units to 150 resulted in an increase in the relative stoichiometry from 1.1 to nearly 5 copies of L-selectin. The functional affinity of the interaction improved 10-fold. These results suggest that the adhesive strength of L-selectin is related to the stoichiometry of the selectin-ligand complex.
Intriguingly, these polymers not only bind L-selectin but also promote its proteolytic release, or shedding, from the cell surface.[256, 257] It appears that cell signaling is important for triggering L-selectin release;[165, 258] it is possible that clustering L-selectin activates the signaling cascade that leads to its cleavage; perhaps physiological L-selectin ligands also influence L-selectin levels in vivo. Together, these results suggest that the stoichiometry of L-selectin-ligand interactions is an important determinant of both the adhesive and signaling functions of this protein.
5. Summary and Outlook
Cell surface receptors mediate the essential tasks of cell adhesion and signaling. In these roles, receptors are rarely left to function as isolated entities. Rather, they collaborate as constituents of higher order macromolecular assemblies. In the case of multi-receptor signaling complexes, understanding the mechanisms underlying the function of these assemblies is critical. Multivalent ligands can serve as powerful tools for the deconvolution of complex multi-receptor networks. The use of synthetic multivalent ligands is complementary to other approaches; therefore, they provide the means to address important new questions.
In addition to utility as agents for examining receptor function, synthetic multivalent ligands have potential applications in the treatment of disease. Multivalent ligands have access to binding mechanisms not available to small molecules. For example, Whitesides, Collier and coworkers took advantage of this unique aspect of multivalent ligands to generate potent polyacrylamide-based multivalent ligands that function as inhibitors of anthrax toxin.[259] The success of these and other[260, 261] in vivo applications of multivalent ligands indicates that they may function not only as probes of biological processes but even as therapeutic agents. In addition to their potential uses as inhibitors, multivalent ligands can cluster cell surface proteins and thereby function as effectors. While many small molecule inhibitors are known, it is typically more difficult to find small molecules that serve as signal transduction activators. The ability of multivalent ligands to activate signal transduction pathways suggest that they may have unique and complementary therapeutic applications.
Multireceptor complexes are the requisite entities that convert extracellular stimuli into appropriate cellular responses. Communication and coordination between receptors provides a means to amplify, integrate and process signals. Understanding how these processes occur at a molecular level is crucial to illuminating the function of biological systems. While monovalent ligands are powerful tools for disrupting such complexes, they cannot be used to examine the importance of complex formation. Multivalent ligands have the necessary attributes direct the formation of multiprotein complexes and/or control the localization of multiple proteins within the cell. We envision that such ligands, which can be used like small molecules for temporal control, will illuminate how protein assembly and/or protein localization direct cellular responses.
Here, we have reviewed some of the uses of synthetic multivalent ligands as probes. Additional advances in target-oriented and diversity-oriented syntheses of multivalent ligands can afford novel compounds that address increasingly complex biological questions. Such investigations demand a multidisciplinary approach that melds chemical and biological concepts to yield, ultimately, a coherent understanding of the molecular mechanisms governing cellular systems.
References
- 1.Chaneux JP, Thiéry J, Tung Y, Kittel C. Proc Natl Acad Sci USA. 1966;57:335. doi: 10.1073/pnas.57.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heldin CH. Cell. 1995;80:213. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger J. TIBS. 1988;13:443. doi: 10.1016/0968-0004(88)90219-8. [DOI] [PubMed] [Google Scholar]
- 4.Klemm JD, Schreiber SL, Crabtree GR. Annu Rev Immunol. 1998;16:569. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 5.Metzger H. J Immunol. 1992;149:1477. [PubMed] [Google Scholar]
- 6.Sacchettini JC, Baum LG, Brewer CF. Biochemistry. 2001;40:3009. doi: 10.1021/bi002544j. [DOI] [PubMed] [Google Scholar]
- 7.Smith FD, Scott JD. Curr Biol. 2002;12:R32. doi: 10.1016/s0960-9822(01)00646-7. [DOI] [PubMed] [Google Scholar]
- 8.Weiss A, Schlessinger J. Cell. 1998;94:277. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 9.Groves JT. Angewandte Chemie-International Edition. 2005;44:3524. doi: 10.1002/anie.200461014. [DOI] [PubMed] [Google Scholar]
- 10.Mellado M, Rodríguez-Frade J, Vila-Coro AJ, Fernández S, Martín de Ana A, Jones DR, Toran JL, Martínez-A C. EMBO J. 2001;20:2497. doi: 10.1093/emboj/20.10.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madhani HD. Cell. 2001;106:9. doi: 10.1016/s0092-8674(01)00422-6. [DOI] [PubMed] [Google Scholar]
- 12.Riese DJ, II, Stern DF. BioEssays. 1998;20:41. doi: 10.1002/(SICI)1521-1878(199801)20:1<41::AID-BIES7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Dustin ML, Chan AC. Cell. 2000;103:283. doi: 10.1016/s0092-8674(00)00120-3. [DOI] [PubMed] [Google Scholar]
- 14.Bray D. Annu Rev Biophys Biomol Struct. 1998;27:59. doi: 10.1146/annurev.biophys.27.1.59. [DOI] [PubMed] [Google Scholar]
- 15.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. Nature. 1997;388:243. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 16.Stein E, Lane AA, Cerretti DP, Schoecklmann HO, Schroff AD, Van Etten RL, Daniel TO. Genes Dev. 1998;12:667. doi: 10.1101/gad.12.5.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levit MN, Liu Y, Stock JB. Biochemistry. 1999;38:6651. doi: 10.1021/bi982839l. [DOI] [PubMed] [Google Scholar]
- 18.Teruel MN, Meyer T. Cell. 2000;103:181. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 19.Lauffenburger DA. Proc Natl Acad Sci USA. 2000;97:5031. doi: 10.1073/pnas.97.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy MB. Science. 2000;290:750. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, Huganir RL. Curr Opin Cell Biol. 1999;11:248. doi: 10.1016/s0955-0674(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 22.Germain RN. Curr Biol. 1997;7:R640. doi: 10.1016/s0960-9822(06)00323-x. [DOI] [PubMed] [Google Scholar]
- 23.Matsuuchi L, Gold MR. Curr Opin Immunol. 2001;13:270. doi: 10.1016/s0952-7915(00)00215-6. [DOI] [PubMed] [Google Scholar]
- 24.van der Flier A, Sonnenberg A. Cell Tissue Res. 2001;305:285. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu TS, Le Novére N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Nature Cell Biol. 2000;2:792. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 26.Bray D, Levin MD, Morton-Firth CJ. Nature. 1998;393:85. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 27.Martin PT. Glycobiology. 2002;12:1R. doi: 10.1093/glycob/12.1.1r. [DOI] [PubMed] [Google Scholar]
- 28.Damjanovich S, Gaspar R, Jr, Pieri C. Quart Rev Biophys. 1997;30:67. doi: 10.1017/s0033583596003307. [DOI] [PubMed] [Google Scholar]
- 29.Holowka D, Baird B. Annu Rev Biophys Biomol Struct. 1996;25:79. doi: 10.1146/annurev.bb.25.060196.000455. [DOI] [PubMed] [Google Scholar]
- 30.Janes PW, Ley SC, Magee AI, Kabouridis PS. Sem Immunol. 2000;12:23. doi: 10.1006/smim.2000.0204. [DOI] [PubMed] [Google Scholar]
- 31.Cherukuri A, Dykstra M, Pierce SK. Immunity. 2001;14:657. doi: 10.1016/s1074-7613(01)00156-x. [DOI] [PubMed] [Google Scholar]
- 32.Brown DA, London E. Annu Rev Cell Dev Biol. 1998;14:111. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 33.Langlet C, Bernard AM, Drevot P, He HT. Curr Opin Immunol. 2000;12:250. doi: 10.1016/s0952-7915(00)00084-4. [DOI] [PubMed] [Google Scholar]
- 34.Edidin M. Curr Opin Struct Biol. 1997;7:528. doi: 10.1016/s0959-440x(97)80117-0. [DOI] [PubMed] [Google Scholar]
- 35.Sheets ED, Holowka D, Baird B. Curr Opin Chem Biol. 1999;3:95. doi: 10.1016/s1367-5931(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 36.Hebert TE, Bouvier M. Biochem Cell Biol. 1998;76:1. doi: 10.1139/bcb-76-1-1. [DOI] [PubMed] [Google Scholar]
- 37.Bartfai T, Benovic JL, Bockaert J, Bond RA, Bouvier M, Christopoulos A, Civelli O, Devi LA, George SR, Inui A, Kobilka B, Leurs R, Neubig R, Pin JP, Quirion R, Roques BP, Sakmar TP, Seifert R, Stenkamp RE, Strange PG. Nature Reviews Drug Discovery. 2004;3:574. [Google Scholar]
- 38.Gouldson PR, Snell CR, Bywater RP, Higgs C, Reynolds CA. Prot Engineer. 1998;11:1181. doi: 10.1093/protein/11.12.1181. [DOI] [PubMed] [Google Scholar]
- 39.Maddock JR, Shapiro L. Science. 1993;259:1717. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 40.Yin CC, Lai FA. Nature Cell Biol. 2000;2:669. doi: 10.1038/35023625. [DOI] [PubMed] [Google Scholar]
- 41.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Nature. 2000;407:1029. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 42.Schlessinger J. Cell. 2000;103:211. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 43.Brümmendorf T, Lemmon V. Curr Opin Cell Biol. 2001;13:611. doi: 10.1016/s0955-0674(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 44.Chan C, George AJT, Stark J. Proc Natl Acad Sci USA. 2001;98:5758. doi: 10.1073/pnas.101113698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douglass AD, Vale RD. Cell. 2005;121:937. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gestwicki JE, Kiessling LL. Nature. 2002;415:81. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 47.Irvine DJ, Mayes AM, Griffith LG. Biomacromolecules. 2001;2:85. doi: 10.1021/bm005584b. [DOI] [PubMed] [Google Scholar]
- 48.Markovic I, Leikina E, Zhukovsky M, Zimmerberg J, Chernomordik LV. J Cell Biol. 2001;155:833. doi: 10.1083/jcb.200103005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elcock AH, McCammon JA. Proc Natl Acad Sci USA. 2001;98:2990. doi: 10.1073/pnas.061411798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hébert TE, Bouvier M. Biochem Cell Biol. 1998;76:1. doi: 10.1139/bcb-76-1-1. [DOI] [PubMed] [Google Scholar]
- 51.Dean MK, Higgs C, Smith RE, Bywater RP, Snell CR, Scott PD, Upton GJG, Howe TJ, Reynolds CA. J Med Chem. 2001;44:4595. doi: 10.1021/jm010290+. [DOI] [PubMed] [Google Scholar]
- 52.Milligan DL, Koshland DE., Jr Science. 1991;254:1651. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- 53.Terrillon S, Bouvier M. Embo Reports. 2004;5:30. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sourjik V, Berg HC. Nature. 2004;428:437. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 55.Fesik SW. Cell. 2000;103:273. doi: 10.1016/s0092-8674(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 56.Wu H. Cell Surface Receptors. 2004;68:225. [Google Scholar]
- 57.Skehel JJ, Wiley DC. Annu Rev Biochem. 2000;69:531. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 58.Yu EW, Koshland DE., Jr Proc Natl Acad Sci USA. 2001;98:9517. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blundell TL, Burke DF, Chirgadze D, Dhanaraj V, Hyvonen M, Innis CA, Parisini E, Pellegrini L, Sayed BL. Biol Chem. 2000;381:955. doi: 10.1515/BC.2000.117. [DOI] [PubMed] [Google Scholar]
- 60.Gardina PJ, Manson MD. Science. 1996;274:425. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 61.Tatsuno I, Homma M, Oosawa K, Kawagishi I. Science. 1996;274:423. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 62.Ames P, Studdert CA, Resier RH, Parkinson JS. Proc Natl Acad Sci USA. 2002;99:7060. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SH, Wang W, Kim KK. Proc Natl Acad Sci USA. 2002;99:11611. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomason PA, Wolanin PM, Stock JB. Curr Biol. 2002;12:R399. doi: 10.1016/s0960-9822(02)00885-0. [DOI] [PubMed] [Google Scholar]
- 65.Ullrich A, Schlessinger J. Cell. 1990;61:203. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 66.Alderton F, Rakhit S, Kong KC, Plamer T, Sambi B, Pyne S, Pyne NJ. J Biol Chem. 2001;276:28578. doi: 10.1074/jbc.M102771200. [DOI] [PubMed] [Google Scholar]
- 67.Liu F, Wan Q, Pristupa ZB, Yu YT, Niznik HB. Nature. 2000;403:274. doi: 10.1038/35002014. [DOI] [PubMed] [Google Scholar]
- 68.Garner CC, Nash J, Huganir RL. Trends Cell Biol. 2000;10:274. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 69.Pawson T, Nash P. Science. 2003;300:445. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 70.Sheng M, Sala C. Annu Rev Neurosci. 2001;24:1. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 71.Gomperts SN. Cell. 1996;84:659. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 72.Prehoda KE, Lim WA. Curr Opinion Cell Biol. 2002;14:149. doi: 10.1016/s0955-0674(02)00307-1. [DOI] [PubMed] [Google Scholar]
- 73.Lim WA. Curr Opinion Struct Biol. 2002;12:61. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 74.Friedl P, den Boer AT, Gunzer M. Nature Reviews Immunology. 2005;5:532. doi: 10.1038/nri1647. [DOI] [PubMed] [Google Scholar]
- 75.Krummel MF, Davis MM. Curr Opin Immunol. 2002;14:66. doi: 10.1016/s0952-7915(01)00299-0. [DOI] [PubMed] [Google Scholar]
- 76.Germain RN, Stefanova I. Annu Rev Immunol. 1999;17:467. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- 77.Long EO. Annu Rev Immunol. 1999;17:875. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 78.Hermiston ML, Xu Z, Majeti R, Weiss A. J Clin Invest. 2002;109:9. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuosto L, Parolini I, Schröder S, Sargiacomo M, Lanzavecchia A, Viola A. Eur J Immunol. 2001;31:345. doi: 10.1002/1521-4141(200102)31:2<345::aid-immu345>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 80.van der Merwe PA, Davis SJ, Shaw AS, Dustin ML. Sem Immunol. 2000;12:5. doi: 10.1006/smim.2000.0203. [DOI] [PubMed] [Google Scholar]
- 81.Viola A, Schroeder S, Sakakibara Y, Lanzavecchia A. Science. 1999;283:680. doi: 10.1126/science.283.5402.680. [DOI] [PubMed] [Google Scholar]
- 82.Bromley SK, Burack WR, Johnson KG, Somersalo K, Sims TN, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. Annu Rev Immunol. 2001;19:375. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- 83.Vyas YM, Mehta KM, Morgan M, Maniar H, Butros L, Jung S, Burkhardt JK, Dupont B. J Immunol. 2001;167:4358. doi: 10.4049/jimmunol.167.8.4358. [DOI] [PubMed] [Google Scholar]
- 84.Kiessling LL, Gestwicki JE, Strong LE. Curr Opin Chem Biol. 2000;4:696. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 85.Healy JI, Goodnow CC. Annu Rev Immunol. 1998;16:645. doi: 10.1146/annurev.immunol.16.1.645. [DOI] [PubMed] [Google Scholar]
- 86.Asthagiri AR, Lauffenburger DA. Annu Rev Biomed Eng. 2000;2:31. doi: 10.1146/annurev.bioeng.2.1.31. [DOI] [PubMed] [Google Scholar]
- 87.Ideker T, Galitski T, Hood L. Annu Rev Genomics Hum Genet. 2001;2:343. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 88.Krauss S, Brand MD. FASEB J. 2000;14:2581. doi: 10.1096/fj.00-0064com. [DOI] [PubMed] [Google Scholar]
- 89.Matko J, Edidin M. Methods Enzymol. 1997;278:444. doi: 10.1016/s0076-6879(97)78023-6. [DOI] [PubMed] [Google Scholar]
- 90.Philipona C, Chevolot Y, Léonard D, Mathieu HJ, Sigrist H, Marquis-Weible F. Bioconjugate Chem. 2001;12:332. doi: 10.1021/bc0000841. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J, Campbell RE, Ting AY, Tsien RY. Nature Reviews Molecular Cell Biology. 2002;3:906. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 92.Lippincott-Schwartz J, Snapp E, Kenworthy A. Nature Reviews Molecular Cell Biology. 2001;2:444. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 93.Wouters FS, Verveer PJ, Bastiaens PIH. Trends In Cell Biology. 2001;11:203. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 94.Liu Y, Xu L, Opalka N, Kappler J, Shu HB, Zhang G. Cell. 2002;108:383. doi: 10.1016/s0092-8674(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 95.Tully DC, Fréchet JMJ. Chem Commun. 2001:1229. [Google Scholar]
- 96.Roy R. Curr Opin Struct Biol. 1996;6:692. doi: 10.1016/s0959-440x(96)80037-6. [DOI] [PubMed] [Google Scholar]
- 97.Bovin NV. Glycoconjugate J. 1998;15:431. doi: 10.1023/a:1006963717646. [DOI] [PubMed] [Google Scholar]
- 98.Lundquist JJ, Toone EJ. Chemical Reviews. 2002;102:555. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 99.Roy R, Page D, Perez SF, Bencomo VV. Glycoconjugate J. 1998;15:251. doi: 10.1023/a:1006945028547. [DOI] [PubMed] [Google Scholar]
- 100.Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed Engl. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 101.Kiessling LL, Strong LE. Top Organomet Chem. 1998;1:199. [Google Scholar]
- 102.Kenakin T. FASEB J. 2001;15:598. doi: 10.1096/fj.00-0438rev. [DOI] [PubMed] [Google Scholar]
- 103.Carrithers MD, Lerner MR. Chem Biol. 1996;3:537. doi: 10.1016/s1074-5521(96)90144-1. [DOI] [PubMed] [Google Scholar]
- 104.Macek MB, Lopez LC, Shur BD. Dev Biol. 1991;147:440. doi: 10.1016/0012-1606(91)90301-i. [DOI] [PubMed] [Google Scholar]
- 105.Schneider H, Chaovapong W, Matthews DJ, Karkaria C, Cass RT, Zhan HJ, Boyle M, Lorenzini T, Elliot SG, Giebel LB. Blood. 1997;89:473. [PubMed] [Google Scholar]
- 106.Somjen D, Amirzaltsman Y, Gayer B, Mor G, Jaccard N, Weisman Y, Barnard G, Kohen F. J Endocrinol. 1995;145:409. doi: 10.1677/joe.0.1450409. [DOI] [PubMed] [Google Scholar]
- 107.Graef IA, Holsinger LJ, Diver S, Schreiber SL, Crabtree GR. EMBO J. 1997;16:5618. doi: 10.1093/emboj/16.18.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Torigoe C, Inman JK, Metzger H. Science. 1998;281:568. doi: 10.1126/science.281.5376.568. [DOI] [PubMed] [Google Scholar]
- 109.Nishizumi H, Horikawa K, Mlinaric-Rascan I, Yamamoto T. J Exp Med. 1998;187:1343. doi: 10.1084/jem.187.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wright D, Usher L. Curr Org Chem. 2001;5:1107. [Google Scholar]
- 111.Liu Y, Hong X, Kappler J, Jiang L, Zhang R, Xu L, Pan CH, Martin W, Murphy RC, Shu HB, Dai S, Zhang G. Nature. 2003;423:49. doi: 10.1038/nature01543. [DOI] [PubMed] [Google Scholar]
- 112.Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, Yoo OJ, Lee H, Lee JO. Nature Struct Biol. 2003;10:342. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- 113.Sasisekharan R, Venkataraman G. Curr Opin Chem Biol. 2000;4:626. doi: 10.1016/s1367-5931(00)00145-9. [DOI] [PubMed] [Google Scholar]
- 114.Esko JD, Lindahl U. J Clin Invest. 2001;108:169. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kiessling LL, Strong LE, Gestwicki JE. Annu Rep Med Chem. 2000;35:321. [Google Scholar]
- 116.Clemons PA. Curr Opin Chem Biol. 1999;3:112. doi: 10.1016/s1367-5931(99)80020-9. [DOI] [PubMed] [Google Scholar]
- 117.Portoghese PS. J Med Chem. 2001;44:2259. doi: 10.1021/jm010158+. [DOI] [PubMed] [Google Scholar]
- 118.Boger DL, Goldberg J. Bioorg Med Chem. 2001;9:557. doi: 10.1016/s0968-0896(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 119.Blackwell HE, Clemons PA, Schreiber SL. Org Lett. 2001;3:1185. doi: 10.1021/ol015647i. [DOI] [PubMed] [Google Scholar]
- 120.Pattarawarapan M, Burgess K. Angew Chem Int Ed. 2000;39:4299. doi: 10.1002/1521-3773(20001201)39:23<4299::AID-ANIE4299>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 121.Goldberg J, Jin Q, Satoh S, Desharnais J, Capps K, Boger DL. J Am Chem Soc. 2002;124:544. doi: 10.1021/ja0118789. [DOI] [PubMed] [Google Scholar]
- 122.Krantz MJ, Holtzman NA, Stowell CP, Lee YC. Biochemistry. 1976;15:3963. doi: 10.1021/bi00663a009. [DOI] [PubMed] [Google Scholar]
- 123.Lee YC, Stowell CP, Krantz MJ. Biochemistry. 1976;15:3956. doi: 10.1021/bi00663a008. [DOI] [PubMed] [Google Scholar]
- 124.Gillitzer E, Willits D, Young M, Douglas T. Chem Commun. 2002:2390. doi: 10.1039/b207853h. [DOI] [PubMed] [Google Scholar]
- 125.Wang Q, Lin T, Tang L, Johnson E, Finn MG. Angew Chem Int Ed Engl. 2002;41:459. doi: 10.1002/1521-3773(20020201)41:3<459::aid-anie459>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 126.Schlick TL, Ding Z, Kovacs EW, Francis MB. J Am Chem Soc. 2005;127:3718. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]
- 127.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. J Cell Sci. 2000;113:1677. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 128.Paran N, Geiger B, Shaul Y. EMBO J. 2001;20:4443. doi: 10.1093/emboj/20.16.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Horan N, Yan L, Isobe H, Whitesides GM, Kahne D. Proc Natl Acad Sci USA. 1999;96:11782. doi: 10.1073/pnas.96.21.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen CS, Mrsksich M, Huang S, Whitesides GM, Ingber DE. Biotechnol Prog. 1998;14:356. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 131.Houseman BT, Mrksich M. Angew Chem Int Ed Engl. 1999;38:782. doi: 10.1002/(SICI)1521-3773(19990315)38:6<782::AID-ANIE782>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 132.Mrksich M. Chem Soc Rev. 2000;29:267. [Google Scholar]
- 133.Smith EA, Thonas WD, Kiessling LL, Corn RM. J Am Chem Soc. 2003;125:6140. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 134.Lévy D, Bluzat A, Seigneuret M, R JL. Biochim Biophys Acta. 1990;1025:179. doi: 10.1016/0005-2736(90)90096-7. [DOI] [PubMed] [Google Scholar]
- 135.Menger FM, Keiper JS. Curr Opin Chem Biol. 1998;2:726. doi: 10.1016/s1367-5931(98)80110-5. [DOI] [PubMed] [Google Scholar]
- 136.Woller EK, Cloninger MJ. Org Lett. 2002;4:7. doi: 10.1021/ol016568+. [DOI] [PubMed] [Google Scholar]
- 137.Kim Y, Zimmerman SC. Curr Opin Chem Biol. 1998;2:733. doi: 10.1016/s1367-5931(98)80111-7. [DOI] [PubMed] [Google Scholar]
- 138.Aoyama Y. Chemistry-A European Journal. 2004;10:588. [Google Scholar]
- 139.Cloninger MJ. Current Opinion In Chemical Biology. 2002;6:742. doi: 10.1016/s1367-5931(02)00400-3. [DOI] [PubMed] [Google Scholar]
- 140.Turnbull WB, Kalovidouris SA, Stoddart JF. Chemistry-A European Journal. 2002;8:2988. doi: 10.1002/1521-3765(20020703)8:13<2988::AID-CHEM2988>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 141.Lindhorst TK. Host-Guest Chemistry. 2002;218:201. [Google Scholar]
- 142.Lin CH, Shen GJ, Garcia-Junceda E, Wong CH. J Am Chem Soc. 1995;117:8031. [Google Scholar]
- 143.Bovin NV, Gabius HJ. Chem Soc Rev. 1995;24:413. [Google Scholar]
- 144.Reuter JD, Myc A, Hayes MM, Gan Z, Roy R, Qin D, Yin R, Piehler LT, Esfand R, Tomalia DA, Baker JR., Jr Bioconjugate Chem. 1999;10:271. doi: 10.1021/bc980099n. [DOI] [PubMed] [Google Scholar]
- 145.Matyjaszewski K, Xia J. Chem Rev. 2001;101:2921. doi: 10.1021/cr940534g. [DOI] [PubMed] [Google Scholar]
- 146.Irvine DJ, Ruzette AVG, Mayes AM, Griffith LG. Biomacromolecules. 2001;2:545. doi: 10.1021/bm015510f. [DOI] [PubMed] [Google Scholar]
- 147.Griffith BR, Allen BL, Rapraeger AC, Kiessling LL. Journal of the American Chemical Society. 2004;126:1608. doi: 10.1021/ja037646m. [DOI] [PubMed] [Google Scholar]
- 148.Ladmiral V, Melia E, Haddleton DM. European Polymer Journal. 2004;40:431. [Google Scholar]
- 149.Maynard HD, Okada SY, Grubbs RH. Journal Of The American Chemical Society. 2001;123:1275. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]
- 150.Trnka TM, Grubbs RH. Acc Chem Res. 2001;34:18. doi: 10.1021/ar000114f. [DOI] [PubMed] [Google Scholar]
- 151.Kanai M, Mortell KH, Kiessling LL. J Am Chem Soc. 1997;119:9931. [Google Scholar]
- 152.Schlüter AD, Rabe JP. Angew Chem Int Ed Engl. 2000;39:864. doi: 10.1002/(sici)1521-3773(20000303)39:5<864::aid-anie864>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 153.Pontrello JK, Allen MJ, Underbakke ES, Kiessling LL. J Am Chem Soc. 2005;127:xxxx. doi: 10.1021/ja053931p. [DOI] [PubMed] [Google Scholar]
- 154.Strong LE, Kiessling LL. J Am Chem Soc. 1999;121:6193. [Google Scholar]
- 155.Manning DD, Strong LE, Hu X, Beck PJ, Kiessling LL. Tetrahedron. 1997;53:11937. [Google Scholar]
- 156.Arimoto H, Nishimura K, Kinumi T, Hayakawa I, Uemura D. Chem Commun. 1999;15:1361. [Google Scholar]
- 157.Gibson VC, Marshall EL, North M, Robson DA, Williams PJ. J Chem Soc, Chem Commun. 1997:1095. [Google Scholar]
- 158.Mortell KH, Weatherman RV, Kiessling LL. J Am Chem Soc. 1996;118:2297. [Google Scholar]
- 159.Lynn DM, Mohr B, Grubbs RH. J Am Chem Soc. 1998;120:1627. [Google Scholar]
- 160.Dias EL, Nguten ST, Grubbs RH. J Am Chem Soc. 1997;119:3887. [Google Scholar]
- 161.Fraser C, Grubbs RH. Macromolecules. 1995;28:7248. [Google Scholar]
- 162.Mortell KH, Gingras M, Kiessling LL. J Am Chem Soc. 1994;116:10253. [Google Scholar]
- 163.Iyer S, Rele S, Grasaa G, Nolan S, Chaikof EL. Chem Commun. 2003:1518. doi: 10.1039/b301734f. [DOI] [PubMed] [Google Scholar]
- 164.Mann DA, Kanai M, Maly DJ, Kiessling LL. J Am Chem Soc. 1998;120:10575. [Google Scholar]
- 165.Gordon EJ, Sanders WJ, Kiessling LL. Nature. 1998;392:30. doi: 10.1038/32073. [DOI] [PubMed] [Google Scholar]
- 166.Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. J Am Chem Soc. 2002;124:1615. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]
- 167.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 168.Schreiber SL. Science. 2000;287:1964. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 169.Hoekstra WJ, Poulter BL. Curr Med Chem. 1998;5:195. [PubMed] [Google Scholar]
- 170.Wilson SR, Czarnik AW. Combinatorial chemistry: synthesis and applications. Wiley; New York: 1997. [Google Scholar]
- 171.Nicolaou KC, Pfefferkorn JA. Biopolymers. 2001;60:171. doi: 10.1002/1097-0282(2001)60:3<171::AID-BIP10030>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 172.Ellman JA, Gallop MA. Curr Opin Chem Biol. 1998;2:317. doi: 10.1016/s1367-5931(98)80003-3. [DOI] [PubMed] [Google Scholar]
- 173.Shogren-Knaak MA, Alaimo PJ, Shokat KM. Annu Rev Cell Dev Biol. 2001;17:405. doi: 10.1146/annurev.cellbio.17.1.405. [DOI] [PubMed] [Google Scholar]
- 174.Wittman V, Seeberger S. Angew Chem Int Ed Engl. 2000;39:4348. doi: 10.1002/1521-3773(20001201)39:23<4348::AID-ANIE4348>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 175.Kiessling LL, Pohl NL. Chem Biol. 1996;3:71. doi: 10.1016/s1074-5521(96)90280-x. [DOI] [PubMed] [Google Scholar]
- 176.Jencks WP. Proc Natl Acad Sci. 1981;78:4046. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Nature. 2000;403:669. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 178.Zhang Z, Merritt EA, Ahn M, Roach C, Hu Z, Verlinde CLMJ, Hol WGJ, Fan E. J Am Chem Soc. 2002;124:12991. doi: 10.1021/ja027584k. [DOI] [PubMed] [Google Scholar]
- 179.Fan E, Zhang Z, Minke WE, Hou Z, Verlinde CLMJ, Hol WGJ. J Am Chem Soc. 2000;122:2663. [Google Scholar]
- 180.Banerjee AL, Eiler D, Roy BC, Jia X, Haldar MK, Mallik S, Srivastava DK. Biochemistry. 2005;44:3211. doi: 10.1021/bi047737b. [DOI] [PubMed] [Google Scholar]
- 181.Howorka S, Nam J, Bayley H, Kahne D. Angew Chem Int Ed. 2004;43:842. doi: 10.1002/anie.200352614. [DOI] [PubMed] [Google Scholar]
- 182.Mammen M, Dahmann G, Whitesides GM. J Med Chem. 1995;38:4179. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 183.Sheetz MP, Felsenfeld DP, Galbraith CG. Trends Cell Biol. 1998;8:51. doi: 10.1016/s0962-8924(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 184.Schwartz MA. Trends Cell Biol. 2001;11:466. doi: 10.1016/s0962-8924(01)02152-3. [DOI] [PubMed] [Google Scholar]
- 185.Schoenwaelder SM, Burridge K. Curr Opin Cell Biol. 1999;11:274. doi: 10.1016/s0955-0674(99)80037-4. [DOI] [PubMed] [Google Scholar]
- 186.Bouvard D, Brakebusch C, Gustafsson E, Aszódi A, Bengtsson T, Berna A, Fässler R. Circ Res. 2001;89:211. doi: 10.1161/hh1501.094874. [DOI] [PubMed] [Google Scholar]
- 187.Hynes RO. Cell. 2002;110:673. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 188.Li RH, Mitra N, Gratkowski H, Vilaire G, Litvinov R, Nagasami C, Weisel JW, Lear JD, DeGrado WF, Bennett JS. Science. 2003;300:795. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 189.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 190.Graef IA, Chen F, Kuo A, Crabtree GR. Cell. 2001;105:863. doi: 10.1016/s0092-8674(01)00396-8. [DOI] [PubMed] [Google Scholar]
- 191.Yap AS, Brieher WM, Pruschy M, Gumbiner BM. Curr Biol. 1997;7:308. doi: 10.1016/s0960-9822(06)00154-0. [DOI] [PubMed] [Google Scholar]
- 192.Muthuswamy SK, Gilman M, Brugge JS. Mol Cell Biol. 1999;19:6845. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Amara JF, Clackson T, Rivera VM, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage NL, Holt DA, Gilman M. Proc Natl Acad Sci USA. 1997;94:10618. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clemons PA, Gladstone BG, Seth A, Chao ED, Foley MA, Schrieber SL. Chem Biol. 2002;9:49. doi: 10.1016/s1074-5521(02)00085-6. [DOI] [PubMed] [Google Scholar]
- 195.Hato T, Pampori N, Shattil SJ. J Cell Biol. 1998;141:1685. doi: 10.1083/jcb.141.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Boudreau NJ, Jones PL. Biochem J. 1999;339:481. [PMC free article] [PubMed] [Google Scholar]
- 197.Welsh ER, Tirrell DA. Biomacromolecules. 2000;1:23. doi: 10.1021/bm0002914. [DOI] [PubMed] [Google Scholar]
- 198.Stupack DG, Li E, Silletti SA, Kehler JA, Geahlen RL, Hahn K, Nemerow GR, Cheresh DA. J Cell Biol. 1999;144:777. doi: 10.1083/jcb.144.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.George SR, O'Dowd BF, Lee SP. Nature Reviews Drug Discovery. 2002;1:808. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 200.Mellado M, Rodríguez-Frade J, Mañes S, Martínez-A C. Annu Rev Immunol. 2001;19:397. doi: 10.1146/annurev.immunol.19.1.397. [DOI] [PubMed] [Google Scholar]
- 201.Kramer RH, Karpen JW. Nature. 1998;395:710. doi: 10.1038/27227. [DOI] [PubMed] [Google Scholar]
- 202.Glick GD, Toogood PL, Wiley DC, Skehel JJ, Knowles JR. J Biol Chem. 1991;266:23660. [PubMed] [Google Scholar]
- 203.England BP, Balasubramanian P, U I, Bethell S, Chen MJ, Schatz PJ, Yin Q, Chen YF, Whitehorn EA, Tsavaler A, Martens CL, Barrett RW, McKinnon M. Proc Natl Acad Sci USA. 2000;97:6862. doi: 10.1073/pnas.110053997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.He Y, Karpen JW. Biochemistry. 2001;40:286. doi: 10.1021/bi002014n. [DOI] [PubMed] [Google Scholar]
- 205.Paar JM, Harris NT, Holowka D, Baird B. J Immunol. 2003;169:856. doi: 10.4049/jimmunol.169.2.856. [DOI] [PubMed] [Google Scholar]
- 206.Chan D, Gera L, Stewart J, Helfrich B, Verella-Garcia M, Johnson G, Baron A, Yang J, Bunn JP. Proc Natl Acad Sci USA. 2002;99:4608. doi: 10.1073/pnas.072077299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bhushan RG, Sharma SK, Xie Z, Daniels DJ, Portoghese PS. J Med Chem. 2004;47:2969. doi: 10.1021/jm0342358. [DOI] [PubMed] [Google Scholar]
- 208.Miyazaki M, Kodama H, Fujita I, Hamasaki Y, Miyazaki S, Kondo M. J Biochem. 1995;117:489. doi: 10.1093/oxfordjournals.jbchem.a124734. [DOI] [PubMed] [Google Scholar]
- 209.Yano K, Kimura S, Imanishi Y. Eur J Pharm Sci. 1998;7:41. doi: 10.1016/s0928-0987(98)00002-5. [DOI] [PubMed] [Google Scholar]
- 210.He L, Fong J, von Zastrow M, Whistler JL. Cell. 2002;108:271. doi: 10.1016/s0092-8674(02)00613-x. [DOI] [PubMed] [Google Scholar]
- 211.Dustin ML. J Clin Invest. 2002;109:155. doi: 10.1172/JCI14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Schamel WWA, Arechaga I, Risueno RM, van Santen HM, Cabezas P, Risco C, Valpuesta JM, Alarcon B. Journal of Experimental Medicine. 2005;202:493. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Boniface JJ, Rabinowitz JD, Wülfing C, Hampl J, Reich Z, Altman JD, Kantor RM, Beeson C, McConnell HM, Davis MM. Immunity. 1998;9:459. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 214.Cochran JR, Aivazian D, Cameron TO, Stern LJ. Trends Biochem Sci. 2001;26:304. doi: 10.1016/s0968-0004(01)01815-1. [DOI] [PubMed] [Google Scholar]
- 215.Cochran JR, Stern LJ. Chem Biol. 2000;7:683. doi: 10.1016/s1074-5521(00)00019-3. [DOI] [PubMed] [Google Scholar]
- 216.Miyamoto S, Akiyama SK, Yamada KM. Science. 1995;267:883. doi: 10.1126/science.7846531. [DOI] [PubMed] [Google Scholar]
- 217.Hubbell JA. Curr Opin Biotechnol. 1999;10:123. doi: 10.1016/s0958-1669(99)80021-4. [DOI] [PubMed] [Google Scholar]
- 218.Lowin-Kropf B, Shapiro VS, Weiss A. J Cell Biol. 1998;140:861. doi: 10.1083/jcb.140.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Monks CRF, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Nature. 1998;395:82. [Google Scholar]
- 220.Stock J, Levit M. Curr Biol. 2000;10:R11. doi: 10.1016/s0960-9822(99)00248-1. [DOI] [PubMed] [Google Scholar]
- 221.Hazelbauer GL, Berg HC, Matsumura P. Cell. 1993;73:15. doi: 10.1016/0092-8674(93)90156-k. [DOI] [PubMed] [Google Scholar]
- 222.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. Annu Rev Cell Dev Biol. 1997;13:457. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jasuja R, Yu-Lin, Trentham DR, Khan S. Proc Natl Acad Sci USA. 1999;96:11346. doi: 10.1073/pnas.96.20.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Stock AM. Proc Natl Acad Sci USA. 1999;96:10945. doi: 10.1073/pnas.96.20.10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Armitage JP, Dorman CJ, Hellingwerf K, Schmitt R, Summers D, Holland B. Mol Microbiol. 2003;47:583. doi: 10.1046/j.1365-2958.2003.03324.x. [DOI] [PubMed] [Google Scholar]
- 226.Feng X, Baumgartner JW, Hazelbauer GL. J Bacteriol. 1997;179:6714. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mello BA, Tu Y. Proc Natl Acad Sci USA. 2003;100:8223. doi: 10.1073/pnas.1330839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gestwicki JE, Lamanna AC, Harshey RM, McCarter LL, Kiessling LL, Adler J. J Bacteriol. 2000;182:6499. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kim KK, Yokota H, Kim SH. Nature. 1999;400:787. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 230.Gestwicki JE, Cairo CW, Mann DA, Owen RM, Kiessling LL. Anal Biochem. 2002;305:149. doi: 10.1006/abio.2002.5652. [DOI] [PubMed] [Google Scholar]
- 231.Sourjik V. Trends Micobiol. 2004;12:569. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 232.Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, Zhan H, Osslund TD, Chirino AJ, Zhang J, Finer-Moore J, Elliott S, Sitney K, Katz BA, Matthews DJ, Wendoloski JJ, Egrie J, Stroud RM. Nature. 1998;395:511. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 233.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Current Opinion in Structural Biology. 2004;14:690. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 234.Beyermann M, Rothemund S, Heinrich N, Fecher K, Furkert J, Dathe M, Winter R, Krause E, Bienert M. J Biol Chem. 2000;275:5702. doi: 10.1074/jbc.275.8.5702. [DOI] [PubMed] [Google Scholar]
- 235.Qin D, Sullivan R, Berkowitz WF, Bittman R, Rotenberg SA. J Med Chem. 2000;43:1413. doi: 10.1021/jm990340z. [DOI] [PubMed] [Google Scholar]
- 236.Park SH, Zarrinpar A, Lim WA. Science. 2003;299:1061. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 237.Hasler P, Zouali M. FASEB J. 2001;15:2085. doi: 10.1096/fj.00-0860rev. [DOI] [PubMed] [Google Scholar]
- 238.Dintzis HM, Dintzis RZ, Vogelstein B. Proc Natl Acad Sci USA. 1976;73:3671. doi: 10.1073/pnas.73.10.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dintzis RZ, Vogelstein B, Dintzis HM. Proc Natl Acad Sci USA. 1982;79:884. doi: 10.1073/pnas.79.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sulzer B, Perelson AS. Mol Immunol. 1997;34:63. doi: 10.1016/s0161-5890(96)00096-x. [DOI] [PubMed] [Google Scholar]
- 241.Mongini PKA, Blessinger CA, Highlet PF, Inman JK. J Immunol. 1992;148:3892. [PubMed] [Google Scholar]
- 242.Ragupathi G, Howard L, Capello S, Koganty RR, Qiu D, Longnecker BM, Reddish MA, Lloyd KO, Livingston PO. Cancer Immunol Immunother. 1999;48:1. doi: 10.1007/s002620050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Dintzis RZ, Okajima M, Middleton MH, Greene G, Dintzis HM. J Immunol. 1989;143:1239. [PubMed] [Google Scholar]
- 244.McEver RP, Moore KL, Cummings RD. J Biol Chem. 1995;270:11025. doi: 10.1074/jbc.270.19.11025. [DOI] [PubMed] [Google Scholar]
- 245.Hasslen SR, von Andrian UH, Butcher EC, Nelson RD, Erlandsen SL. Histochem J. 1995;27:547. [PubMed] [Google Scholar]
- 246.Hasslen SR, Burns AR, Simon SI, Smith W, Starr K, Barclay AN, Michie SA, Nelson RD, Erlandsen SL. J Histochem Cytochem. 1996;44:1115. doi: 10.1177/44.10.8813076. [DOI] [PubMed] [Google Scholar]
- 247.Rosen SD. Annual Review of Immunology. 2004;22:129. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 248.Simanek EE, McGarvey GJ, Jablonowski JA, Wong CH. Chem Rev. 1998;98:833. doi: 10.1021/cr940226i. [DOI] [PubMed] [Google Scholar]
- 249.Thoma G, Duthaler RO, Magnani JL, Patton JT. J Am Chem Soc. 2001;123:10113. doi: 10.1021/ja0164430. [DOI] [PubMed] [Google Scholar]
- 250.Thoma G, Patton JT, Magnani JL, Ernst B, Ohrlein R, Duthaler RO. Journal of the American Chemical Society. 1999;121:5919. doi: 10.1021/ja0164430. [DOI] [PubMed] [Google Scholar]
- 251.Manning DD, Hu X, Beck P, Kiessling LL. J Am Chem Soc. 1997;119:3161. [Google Scholar]
- 252.Sanders WJ, Gordon EJ, Dwir O, Beck PJ, Alon R, Kiessling LL. J Biol Chem. 1999;274:5271. doi: 10.1074/jbc.274.9.5271. [DOI] [PubMed] [Google Scholar]
- 253.Stahn R, Schafer H, Kernchen F, Schreiber J. Glycobiology. 1998;8:311. doi: 10.1093/glycob/8.4.311. [DOI] [PubMed] [Google Scholar]
- 254.Owen RM, Gestwicki JE, Young T, Kiessling LL. Org Lett. 2002;4:2295. doi: 10.1021/ol0259239. [DOI] [PubMed] [Google Scholar]
- 255.Gordon EJ, Gestwicki JE, Strong LE, Kiessling LL. Chem Biol. 2000;7:9. doi: 10.1016/s1074-5521(00)00060-0. [DOI] [PubMed] [Google Scholar]
- 256.Gordon EJ, Strong LE, Kiessling LL. Bioorg Med Chem Lett. 1998;6:1293. doi: 10.1016/s0968-0896(98)00122-9. [DOI] [PubMed] [Google Scholar]
- 257.Mowery P, Yang ZQ, Gordon EJ, Dwir O, Spencer AG, Alon R, Kiessling LL. Chemistry & Biology. 2004;11:725. doi: 10.1016/j.chembiol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 258.Gordon EJ, Strong LE, Kiessling LL. Bioorg Med Chem. 1998;6:1293. doi: 10.1016/s0968-0896(98)00122-9. [DOI] [PubMed] [Google Scholar]
- 259.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Nature Biotechnol. 2001;19:958. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 260.Bracci L, Lozzi L, Pini A, Lelli B, Falciani C, Niccolai N, Bernini A, Spreafico A, Soldani P, Neri P. Biochemistry. 2002;41:10194. doi: 10.1021/bi0256025. [DOI] [PubMed] [Google Scholar]
- 261.Byrne GW, Schwarz A, Fesi JR, Birch P, Nepomich A, Bakaj I, Velardo MA, Jiang C, Manzi A, Dintzis H, Diamond LE, Logan JS. Bioconj Chem. 2002;13:571. doi: 10.1021/bc015565e. [DOI] [PubMed] [Google Scholar]