Abstract
Purpose:
Most neuroblastomas initially respond to therapy but many relapse with chemoresistant disease. p53 mutations are rare in diagnostic neuroblastomas, but we have previously reported inactivation of the p53/MDM2/p14ARF pathway in 9/17 (53%) neuroblastoma cell lines established at relapse.
Hypothesis:
Inactivation of the p53/MDM2/p14ARF pathway develops during treatment and contributes to neuroblastoma relapse.
Methods:
Eighty-four neuroblastomas were studied from 41 patients with relapsed neuroblastoma including 38 paired neuroblastomas at different stages of therapy. p53 mutations were detected by automated sequencing, p14ARF methylation and deletion by methylation-specific PCR and duplex PCR respectively, and MDM2 amplification by fluorescent in-situ hybridisation.
Results:
Abnormalities in the p53 pathway were identified in 20/41(49%) cases. Downstream defects due to inactivating missense p53 mutations were identified in 6/41 (15%) cases, 5 following chemotherapy and/or at relapse and 1 at diagnosis, post chemotherapy and relapse. The presence of a p53 mutation was independently prognostic for overall survival (hazard ratio 3.4, 95% confidence interval 1.2, 9.9; p = 0.02). Upstream defects were present in 35% cases: MDM2 amplification in 3 cases, all at diagnosis & relapse and p14ARF inactivation in 12/41 (29%) cases: 3 had p14ARF methylation, 2 after chemotherapy, and 9 had homozygous deletions, 8 at diagnosis and relapse.
Conclusions:
These results show that a high proportion of neuroblastomas which relapse have an abnormality in the p53 pathway. The majority have upstream defects suggesting that agents which reactivate wild-type p53 would be beneficial, in contrast to those with downstream defects where p53 independent therapies are indicated.
Keywords: p53, p14ARF, MDM2, neuroblastoma
Introduction
Neuroblastoma is the most common extracranial paediatric solid tumour. It remains one of the most difficult cancers to cure, with less than 40% of patients with high-risk disease (stage 4 over 18 months of age or MYCN amplified disease) becoming long term survivors. Most high risk neuroblastomas do initially respond to cytotoxic therapy, however over half relapse with chemoresistant disease and this often correlates with the intensity of therapy (1).
The p53 gene is inactivated by mutation in over 50% of human malignancies (2). p53 is a key regulator of cell cycle checkpoints and apoptosis, which upon activation by cellular stress, particularly DNA damage, binds DNA in a sequence specific manner to activate the transcription of a large number of downstream genes, including p21 and MDM2, which results in apoptosis, cell-cycle arrest, differentiation and DNA repair (reviewed in (3)). MDM2 functions upstream of p53 as a ubiquitin ligase that targets p53 for proteosome mediated degradation, forming an autoregulatory feedback loop which tightly regulates p53 cellular levels (4). MDM2 amplification has been shown in some tumours and can suppress the activity of p53 by increasing its degradation.
The INK4a/ARF locus on 9p21-22, encodes two intimately linked but distinct tumour suppressor proteins, p16INK4a and p14ARF, which exert active roles in the retinoblastoma protein (RB) and p53 pathways respectively. p14ARF and p16INK4a share common coding sequences for exons 2 and 3, however they have distinct promoter and exon 1 sequences (5). p14ARF is an upstream regulator of p53 which can activate the p53 pathway by directly binding to and antagonizing the E3 ubiquitin ligase activity of MDM2 (6). p14ARF also sequesters MDM2 in the nucleolus, preventing interaction between p53 and MDM2, so releasing p53 from the inhibitory effects of MDM2 (reviewed in (7)). There is also evidence that wild-type p53 down-regulates p14ARF expression, establishing an autoregulatory loop between p53-MDM2-p14ARF which is critical for normal cell cycle progression (8).
In neuroblastoma p53 mutations at diagnosis are rare, occurring in < 2% of cases (reviewed by (9)). We reported an inactivating p53 mutation in a neuroblastoma cell line (BE2c) established at relapse which was more chemoresistant than the paired p53 wild-type cell line (BE1n) established at diagnosis (10). More recently we reported an increased frequency of abnormalities in the p53/MDM2/p14ARF pathway in neuroblastoma cell lines established from tumours following chemotherapy (11). Based on these observations in cell lines we hypothesised that aberrations in the p53/MDM2/p14ARF pathway occur more frequently in neuroblastoma tumours following chemotherapy at relapse and lead to chemoresistance. We performed an analysis of the p53/MDM2/p14ARF pathway in paired neuroblastomas to determine at what stage p53 pathway abnormalities develop.
Methods
Tumours
A mixture of frozen and formalin-fixed, paraffin-embedded tumours were obtained from The Royal Victoria Infirmary Newcastle, other UK Children's Cancer and Leukaemia Group centres (CCLG), Dept. of Paediatrics, Memorial Sloan-Kettering Cancer Center, New York, Münster University and the German national tumour bank, Kiel, Germany, with approvals from the individual Institutional Review Boards. Power calculations based on cell line data informed the study size. Forty one neuroblastoma cases were studied: 34 paired diagnostic and relapsed neuroblastomas (including 4 at diagnosis, post-chemotherapy and relapse), 1 pre and post-chemotherapy, 3 post-chemotherapy and relapse including one at further relapse and 3 relapsed tumours only. Thirty-eight out of 41 patients achieved an initial complete or partial response and 3 progressed during initial therapy. In cases where a p53 pathway abnormality was detected at relapse but not diagnosis, the post-chemotherapy sample was also studied if available. Post-chemotherapy refers to the resected tumour following induction chemotherapy. There were 4 International Neuroblastoma Staging System (12) stage 1 tumours, 7 stage 2, 6 stage 3, 22 stage 4 and 2 stage 4s tumours. Clinical characteristics including age, stage, MYCN status and outcome are shown in Appendix 1. At the time of analysis, 26/41 patients had died, 8 were alive and disease free, and 7 were alive with disease. The median overall and progression free survival was 3.25 and 1.43 years respectively and the median length of follow up of survivors was 8 years. All tumours were reviewed by a consultant histopathologist (K.M. Wood) to ensure > 60% neuroblastoma tumour cell content for analysis as recommended for PCR studies (13).
p53 gene sequencing and p53 mutation-specific PCR
DNA was extracted from frozen and micro-dissected formalin-fixed, paraffin-embedded tumours, and exons 4-9 of the p53 gene amplified by PCR and sequenced using primers and methods described previously (11). Where possible, p53 mutations were also detected using denaturing high performance liquid chromatography (DHPLC) on a DNA fragment analysis system (Transgenomic, San Jose). To test the hypothesis that a small number of p53 mutant cells were present at diagnosis in one case (Case 1), oligonucleotide primer pairs specific for the codon 270 mutation present and the wild-type allele were designed; sense wild-type 5′CTACTGGGACGGAACAGCTTT 3′, sense mutant 5′CTACTGGGACGGAACAGCTTA3′, antisense 5′GCATAACTGCACCCTTGGT 3′. PCR conditions used were 95°C-10 mins, and then 14 cycles each consisting of 94°C-20 seconds, 67°C annealing temperature which decreased by 0.5°C with each cycle ending at 60°C, and finally 1 minute extension at 72°C. When the annealing temperature was at 60°C a further 26 cycles, as above were performed.
Fluorescence in situ hybridisation (FISH)
FISH to detect MDM2 amplification was performed on tumour imprints and paraffin sections using the MDM2 amplified NGP neuroblastoma cell line as a positive control (11). For tumour imprints a directly labelled spectrum orange-labelled MDM2 probe was used in combination with a spectrum green-labelled chromosome 12 centromeric probe (Abbott Molecular, IL) and hybridisations performed as described previously (11). For paraffin sections FISH was performed using previously described methods (14), with probes for MDM2 (12q15) and chromosome 12 centromere, generated from plasmid DNA (University of Bari, Italy) using the Nucleobond BAC100 Kit (ABgene, Epsom, UK). Amplification at this locus was defined as the presence of > 4 fold increase in copy number of the MDM2 signal relative to the chromosome 12 centromeric signal.
Single nucleotide polymorphism (SNP) array analysis
The presence of MDM2 amplification in the absence of MYCN amplification was confirmed using Affymetrix 10K and 50K SNP array analysis (Affymetrix UK Ltd, High Wycombe, UK) on diagnostic and relapsed tumour DNA from case 4 using methods described previously (15).
p14ARF gene promoter methylation, homozygous deletion and quantitative RT-PCR
The DNA methylation status of the p14ARF gene was determined by methylation specific PCR (MSP), and homozygous deletion of the p14ARF locus by duplex PCR using primers for exon 1β and the Na channel control gene and methods described previously (11). An alternative primer pair specific for exon 1β of p14ARF was also used in 6 constitutional DNA samples; sense 5′CTGCTCACCTCTGGTGCCAA 3′ and anti-sense 5′CGAGGGCCTTTCCTACCTGGT 3′. The relationship between p14ARF gene promoter methylation and gene silencing was investigated using quantitative RT-PCR, where frozen tumour was available using methods described previously (11). The p14ARF homozygously deleted cell lines LAN-6 and SHEP and the non-deleted SKNRA cell line were used as controls (11).
Immunohistochemistry
In case 1 immunohistochemistry was performed on formalin-fixed, paraffin-embedded sections using methods and antibodies described previously (16).
Statistical Analysis
Fisher's exact test was used to compare proportions between study groups and the log-rank test was used for survival analysis with Kaplan-Meier survival curves using Stata version 10 (StataCorp. 2007, TX) and Prism version 4, (Graphpad Software Inc, CA). Multivariate analysis was performed using a Cox proportional hazards model (Stata).
Results
A p53 pathway abnormality was detected in 20/41 (49%) cases, 13 cases at presentation and 8 cases post-chemotherapy and/or at relapse (Table 1). In one case (case 9) a p53 mutation and p14ARF methylation were detected (Table 1). p53 mutations were detected following chemotherapy in all but one case, whereas MDM2 amplification and p14ARF inactivation were present at both diagnosis and relapse in the majority of cases (Table 1).
Table 1.
Neuroblastoma cases with abnormalities of the p53/MDM2/p14ARF pathway. Key, Amp = Amplified, DOD = Died of disease, ADF = Alive disease free, AWD = Alive with disease, Post = Post induction chemotherapy, WT= wild-type, Meth = Methylated, Del = Homozygous deletion.
| Case No | Age (years) |
Stage | MYCN | Current Status |
Pre/Post /Rel |
p53 | p14 ARF | MDM2 |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.6 | 4 Amp | DOD | Pre | WT | Normal | Non-Amp | |
| Post | Phe270Leu | Normal | Non-Amp | |||||
| Relapse | Phe270Leu | Normal | Non-Amp | |||||
|
| ||||||||
| 2 | 2 | 4 Non-Amp | DOD | Pre | WT | Meth | No result | |
| Post | WT | Meth | ||||||
| Relapse | WT | Meth | ||||||
|
| ||||||||
| 3 | 2.5 | 3 Amp | DOD | Pre | WT | Normal | No result | |
| Post | WT | Meth | ||||||
| Relapse | WT | Meth | ||||||
|
| ||||||||
| 4 | 3.6 | 3 Non-Amp | ADF | Pre | WT | Normal | Amp | |
| Relapse | WT | Normal | Amp | |||||
|
| ||||||||
| 5 | 8.95 | 4 Non-Amp | AWD | Pre | WT | Del | No result | |
| Relapse | WT | Del | ||||||
|
| ||||||||
| 6 | 2.35 | 4 Non-Amp | DOD | Pre | WT | Del | No result | |
| Relapse | WT | Del | ||||||
|
| ||||||||
| 7 | 10.9 | 4 Non-Amp | AWD | Pre | WT | Del | No result | |
| Relapse | WT | Del | ||||||
|
| ||||||||
| 8 | 2.65 | 2B | Amp | DOD | Pre | WT | Del | Non-Amp |
| Relapse | WT | Del | Non-Amp | |||||
|
| ||||||||
| 9 | 0.6 | 4 Non-Amp | DOD | Pre | WT | Normal | Non-Amp | |
| Progression | Val157Gly | Meth | Non-Amp | |||||
|
| ||||||||
| 10 | 23.56 | 4 Non-Amp | AWD | Pre | WT | Del | No result | |
| Relapse | WT | Del | ||||||
|
| ||||||||
| 11 | 2.26 | 4 Non-Amp | DOD | Pre | WT | Normal | No result | |
| Relapse | Asp259Tyr | Normal | ||||||
|
| ||||||||
| 12 | 5.8 | 4 Non-Amp | DOD | Pre | WT | Normal | No result | |
| Post | Asp259Tyr | Normal | ||||||
|
| ||||||||
| 13 | 6.39 | 1 Non-Amp | DOD | Pre | WT | Normal | Amp | |
| Relapse | WT | Normal | Amp | |||||
|
| ||||||||
| 14 | 15.33 | 2B | Non-Amp | DOD | Pre | Val203Met | Normal | Non-Amp |
| Post | Val203Met | Normal | Non-Amp | |||||
| Relapse | Val203Met | Normal | Non-Amp | |||||
|
| ||||||||
| 15 | 1.5 | 1 Amp | ADF | Pre | WT | Normal | Non-Amp | |
| Relapse | WT | Del | Non-Amp | |||||
|
| ||||||||
| 16 | 16.3 | 2B | Non-Amp | ADF | Post | Cys238Tyr | Normal | No result |
| Relapse | Cys238Tyr | Normal | ||||||
| Further relapse | Cys238Tyr | Normal | ||||||
|
| ||||||||
| 17 | 1.6 | 4 Amp | DOD | Pre | WT | Del | No result | |
| Relapse | WT | Del | ||||||
|
| ||||||||
| 18 | 5.4 | 4 Non-Amp | DOD | Post | WT | Del | Non-Amp | |
| Relapse | WT | Del | Non-Amp | |||||
|
| ||||||||
| 19 | 0.54 | 4 Amp | ADF | Pre | WT | Normal | Amp | |
| Relapse | WT | Normal | Amp | |||||
|
| ||||||||
| 20 | 4.33 | 4 Non-Amp | DOD | Pre | WT | Del | No result | |
| Relapse | WT | Del | ||||||
p53 mutations
Inactivating p53 mutations were detected in 6/41(15%) cases (Table 1). Four were stage 4 tumours (1 MYCN amplified and 3 non-MYCN amplified) and two were stage 2B tumours (both non-MYCN amplified). All mutations were missense mutations, 5 transversions and 1 transition. Five out of 6 patients with p53 mutations died from disease (p=0.07, log rank, progression free survival) (data not shown). There was no relationship between p53 mutation and tumour stage or MYCN amplification (Table 2), or patient age < or ≥ 18 months (data not shown).
Table 2.
Relationship between p53 pathway abnormality, stage and MYCN amplification. Key, Amp = Amplified, Meth = Methylated, Del = Homozygous deletion, Low = Stage 1, 2, 3, 4s, High = Stage 4, Codon 72 = Codon 72 polymorphism. Fisher's exact test was used for p14ARF, p53 and MDM2 analysis and χ2 for codon 72 polymorphism.
| p53 pathway abnormality |
Stage | MYCN Amp | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Low | High | P | + | − | p | ||
| p14ARF meth/ del | + | 2 | 10 | 0.02 | 3 | 9 | 1.0 |
| − | 17 | 12 | 8 | 21 | |||
|
| |||||||
| p53 mutation | + | 2 | 4 | 0.7 | 1 | 5 | 1.0 |
| − | 17 | 18 | 10 | 25 | |||
|
| |||||||
| MDM2 amp | + | 2 | 1 | 1.00 | 1 | 2 | 1.0 |
| − | 12 | 8 | 6 | 14 | |||
|
| |||||||
| Codon 72 Arg/Pro | 5 | 4 | 0.007 | ||||
| Arg/Arg | 4 | 26 | |||||
| Pro/Pro | 1 | 0 | |||||
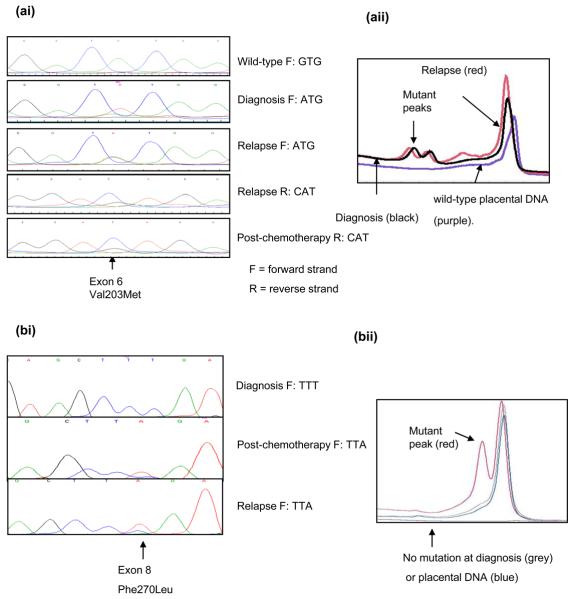
In case 14, a p53 mutation was detected at diagnosis, post-chemotherapy and relapse (Figure 1a), whereas in the remaining cases p53 mutations were detected following induction chemotherapy and/or relapse, and not at diagnosis (Table 1 and Figure 1b). However, diagnostic tumour was unavailable from case 16. In 3 cases mutations were also detected by DHPLC (Figure 1a&b and data not shown). Immunocytochemistry on case 1 showed nuclear p53 expression in all 3 samples with a reduction post-chemotherapy and an increase at relapse. p21 expression was detectable in diagnostic, but not relapsed tumour consistent with non-functional p53 (Figure 1c).
Figure 1. p53 status and function in neuroblastoma at different stages of treatment.
(a) Case 14, (ai) codon 203 mutation (aii) DHPLC chromatogram. (b-d) Case 1 (bi) codon 270 mutation present post-chemotherapy and relapse but not at diagnosis. (bii) DHPLC chromatogram (c) p53 and p21 immunohistochemistry showing lack of p21 expression at relapse (scale bar = 50μm) (d) Codon 270 mutation-specific PCR.
Mutation specific PCR for the Phe270Leu, TTT → TTA mutation in case 1 (Figure 1d), showed that using wild-type p53 primers it was possible to generate a PCR product from tumour DNA at diagnosis and relapse. However, using mutation-specific primers, only relapsed tumour DNA could be amplified. The minimum concentration of mutant p53 DNA required to generate a PCR product using the mutation-specific primers was 0.05% suggesting that the mutation, if present at diagnosis, occurred in less than 1 in 2000 cells (Figure 1d).
There was an association between the Arg/Arg (CGC) p53 codon 72 polymorphism and non-MYCN amplified tumours (p = 0.007, Table 2).
MDM2 amplification
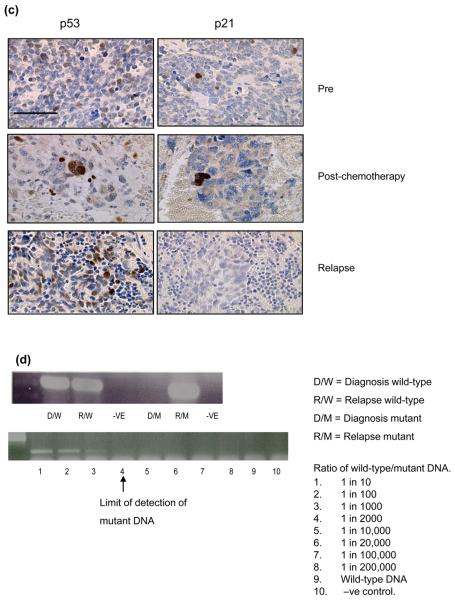
Of the remaining 85% of patients that had wild-type p53 at relapse 3/23 cases showed amplification of MDM2, at both diagnosis and relapse (Figure 2). One case was stage 4 (MYCN amplified) and the others were stage 1 & 3 (non-MYCN amplified) (Table 1). There was no relationship between MDM2 amplification and tumour stage (Table 2) or survival (data not shown). The presence of MDM2 amplification at 12q15 in the absence of MYCN amplification at 2p24 in case 4 was confirmed by SNP arrays (Figure 2b).
Figure 2. MDM2 amplification in neuroblastoma.
(a & b) Case 4 (ai) MDM2 amplification by FISH on a relapsed tumour imprint (×100), (aii) Non-MDM2 amplified tumour. (b) 50K SNP arrays log2ratio from relapsed tumour DNA showing (bi) MDM2 amplification (12q15) (bii) no MYCN amplification (2p24). (c) Case 19, MDM2 amplification by FISH on paraffin sections from (ci) diagnostic tumour and (cii) relapse.
p14ARF methylation/deletion
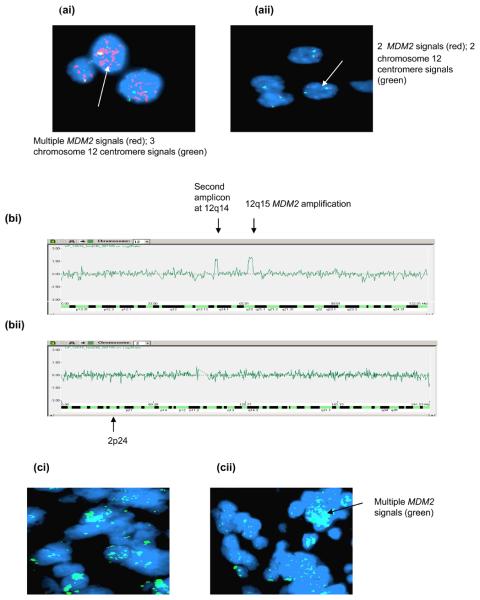
p14ARF inactivation was observed in 12/41 (29%) of tumours. Three cases were partially methylated for p14ARF (Figure 3a). In 2/3 cases methylation was observed following cytotoxic treatment but not at diagnosis (Table 1). Decreased p14ARF mRNA expression was shown in a methylated sample obtained at progression compared with the unmethylated diagnostic sample (Figure 3b). Homozygous p14ARF deletion was detected in 9 cases, 8 at diagnosis and relapse and 1 at relapse alone (Table 1 and Figure 3ci). Constitutional DNA from cases with p14ARF homozygous deletion at diagnosis and relapse was obtained in 6/8 cases, and in all cases, p14ARF was not deleted (Figure 3cii). Nine patients with p14ARF methylation or deletion have died from disease and p14ARF inactivation was found to be associated with stage 4 disease (p = 0.02, Table 2), but not survival.
Figure 3. p14ARF abnormalities in neuroblastoma.
(a & b) Case 9 (a) MSP showing unmethylated p14ARF (diagnosis) and partial methylation (progression) (b) p14ARF mRNA expression showing decreased expression at progression. (c) Duplex PCR showing (ci) homozygous deletion at diagnosis and relapse (case 20) and other relapsed tumours, (cii) the presence of p14ARF in constitutional DNA from cases homozygously deleted for p14ARF.
Forward stepwise Cox regression analysis considering age < or ≥ 18 months, stage group (low- 1, 2,3 or 4s or high- 4), MYCN amplification, p53 mutation, p14ARF abnormality and codon 72 status, showed that a p53 mutation, stage 4 disease and MYCN amplification were all independently prognostic for overall survival. The hazard ratios (95% confidence intervals) and p values were 3.4 (1.2, 9.9), p = 0.02 for a p53 mutation, 2.7 (1.1, 6.2), p = 0.02 for stage 4 disease and 4.4 (1.8, 10.9), p = 0.001 for MYCN amplification. MYCN amplification was the only independent prognostic indicator for progression free survival (data not shown).
Discussion
We investigated the frequency of aberrations of the p53/MDM2/p14ARF pathway in paired neuroblastoma tumours. To our knowledge, there have been no previous studies of the p53 pathway in paired neuroblastomas, giving an important insight into the stage at which p53 pathway abnormalities develop. Neuroblastomas are rarely biopsied at relapse so to obtain a sufficient number of samples it was necessary to include tumours from different risk groups obtained from international collaborations.
Downstream inactivation of the p53 pathway via inactivating p53 mutations within the p53 DNA binding domain were observed in 6 cases (15%). The frequency of reported p53 mutations in neuroblastomas is low ~ 2% (reviewed in (9)). Out of around 340 tumours sequenced for p53, inactivating mutations were detected in 7 cases with 4/7 tumours from patients with progressive or relapsed disease (17-20) (Table 3). In 2 of the 4 previously reported cases the corresponding diagnostic tumour was p53 wild-type (18, 19), in another case the p53 mutation was reported in a bone marrow metastasis but not the primary tumour (18) and in a further case, the mutation was detected in a stage 4 tumour after 2 cycles of chemotherapy (17). In the current study 5/6 cases had p53 mutations detected after chemotherapy and/or at relapse, but not at diagnosis. The case in which a p53 mutation was detected at both diagnosis and relapse was 15 years old at diagnosis, which is unusual for a patient with neuroblastoma as < 1% are diagnosed in children > 10 years of age. The presence of a p53 mutation at diagnosis may reflect environmental exposure to p53 mutagens. Another neuroblastoma with a p53 mutation detected post-chemotherapy and relapse was from a 16 year old patient (Table 1, case 16). Diagnostic tumour was unavailable, but it is tempting to speculate that the mutation may also have been present at diagnosis for the same reason. If so this suggests that adolescents with neuroblastoma may have a higher frequency of p53 mutations at diagnosis.
Table 3.
Comparison of inactivating p53 mutations in neuroblastomas reported in this study with those previously reported including details of cytotoxic therapy received prior to the detection of the p53 mutation where available.
| Authors | Stage MYCN |
p53 mutation Pre/Post/Rel |
Exon | Codon | Nucleotide Change |
Amino Acid change |
Therapy |
|---|---|---|---|---|---|---|---|
| 18 | 3→ 4 Non-Amp |
Relapse | 5 | 135 | GAG → GTG | Cys to Tyr | Not stated |
| 4 Non-Amp |
Pre | 6 | 204 | TGC → TAC | Gly to Val | ||
| 17 | 4 Non-Amp |
Post | 7 | 259 | GAC → TAC | Asp to Tyr | 2 courses of CTX |
| 19 | 4 Amp |
Post* | 8 | 277 | TGT → TTT | Cys to Phe | JM-8, VP- 16,Cyclo |
| 20 | A Non-Amp |
Pre | 8 | 273 | CGT → CTT | Arg to Leu | Present at diagnosis |
| B Non-Amp |
Pre | 8 | 283 | CGC → TGC | Arg to Cys | Present at diagnosis |
|
| Ds Non-Amp |
Pre | 8 | 283 | CGC → TGC | Arg to Cys | Present at diagnosis |
|
| Case 1 | 4 Amp |
Post & Relapse | 8 | 270 | TTT → TTA | Phe to Leu | ENSG5- COJEC, S, |
| Case 9 | 4 Non-Amp |
Progression* | 5 | 157 | GTC→ GGC | Val to Gly | OPEC/OJEC, CADO |
| Case 11 | 4 Non-Amp |
Relapse* | 7 | 259 | GAC → TAC | Asp to Tyr | N7, 3F8. S, RT |
| Case 12 | 4 Non-Amp |
Post* | 7 | 259 | GAC → TAC | Asp to Tyr | N7 |
| Case 14 | 2B Non-Amp |
Pre, Post & Rel | 6 | 203 | GTG → ATG | Val to Met | Present at diagnosis |
| Case 16 | 2B Non-Amp |
Post, Relapse x 2 | 7 | 238 | TGT →TAT | Cys to Tyr | NB-90 |
Paired sample was wild-type at diagnosis.
CTX = chemotherapy, JM-8 = carboplatin, VP-16 = etoposide, cyclo = cyclophosphamide, S = surgery, ENSG5-COJEC (cisplatin, carboplatin, etoposide, vincristine, cyclophosphamide) (47), OPEC/OJEC = cisplatin, carboplatin, etoposide, vincristine, cyclophosphamide (48), CADO = cyclophosphamide, doxorubicin, vincristine, N7= vincristine, cyclophosphamide, doxorubicin, cisplatin, etoposide (49), 3F8= Anti-GD2 monoclonal antibody, RT = Radiotherapy, NB-90 = cisplatin, etoposide, vindesine, ifosfamide, vincristine, DTIC, doxorubicin + High dose melphalan and autologous bone marrow transplant /or 1 year chemotherapy (50).
The mutation detected in codon 259 GAC →TAC was previously reported in a neuroblastoma tumour following cytotoxic treatment (17) (Table 3). The Universal Mutation Database (UMD) shows 31 reports of this mutation (www.umd.be:2072), so although not classed as a hotspot, it is a frequent mutation. The amino acid substitution from Asp to Tyr results in a change in the charge of the p53 protein, suggesting this missense mutation is likely to have contributed to tumour progression (17). The mutation detected at codon 238 is also a frequent mutation; UMD shows 98 reports of this mutation. Cysteine residues are critical to both protein structure and function (21). Removal of the zinc ion at Cys-238 results in destabilisation of the p53 protein leading to a loss of sequence specific DNA binding (22).
Loss of p53 function due to the codon 270 mutation in case 1, was confirmed by the lack of p21 expression in the relapsed tumour (Figure 1c). Detection of this mutation after less than 3 months of cisplatin-based chemotherapy suggests that a small number of mutant p53 cells may have been present at diagnosis and selected for by p53 dependent cytotoxic drugs leading to drug resistance. However, mutation-specific PCR showed that only the relapsed tumour DNA could be amplified, and that if the mutation was present at diagnosis, it was in < 1 in 2000 cells (Figure 1d). It is also possible that the DNA damaging effect of the chemotherapy itself caused the mutation, which was subsequently selected for, and persisted at relapse. Clonal expansion of mutant p53 cells has been reported in malignant gliomas (23, 24) and in the malignant progression of leukaemias and sarcomas (25, 26). In ovarian carcinoma cells, selection of the same codon 270 T → A mutation led to cisplatin resistance (27).
It was not possible to determine whether the mutations in this study were heterozygous or homozygous, as although the tumour cell content was always confirmed to be > 60%, some contaminating normal tissue might have been present. Data from in-vitro studies suggests that many missense mutations, as well as disrupting the ability of the p53 protein to bind DNA and activate transcription, can inhibit the function of the wild-type protein in a dominant negative manner, which could result in functional inactivation of cellular p53 (28, 29).
In this study 5/6 patients with p53 mutations died from disease and the presence of a p53 mutation was independently prognostic for overall survival. The observation that p53 mutations are rare at diagnosis in neuroblastoma may explain the initial good response to chemotherapy in the majority of cases, but the development of p53 mutations or loss of p53 function during therapy may contribute to relapse. In neuroblastoma cell lines derived from tumours at relapse, the presence of p53 mutations or loss of function is associated with drug resistance (1, 10, 11, 30).
An association was observed between the p53 codon 72 Arg/Arg polymorphism and non-MYCN amplified tumours (Table 2). This polymorphism has been reported to be more susceptible to degradation by the human papillomavirus HPV 18 E6 protein (31), but is more efficient at inducing apoptosis than the Pro variant, partly due to a greater localisation to the mitochondria (32). This could have implications for chemosensitivity in non-MYCN amplified tumours since many cytotoxic agents induce tumour cells to undergo p53 mediated apoptosis.
In contrast to the observation of downstream inactivation of p53 via mutation predominantly at relapse, upstream inactivation via MDM2 amplification was observed at both diagnosis and relapse in all 3 cases and in 2 cases was detected in the absence of MYCN amplification. 12q amplification in the absence of MYCN amplification has previously been reported in 3/95 neuroblastomas by FISH in one series (33) and in 2/90 neuroblastomas by array comparative genomic hybridisation in another (34). However, in cell lines, MDM2 amplification has so far only been reported in the presence of MYCN amplification (reviewed in (9)). The relatively high proportion of MDM2 amplification detected by FISH in the current study probably reflects the fact that this is a selected sample of cases that went on to relapse.
The presence of MDM2 amplification at both diagnosis and relapse in all cases suggests that upstream suppression of p53 via MDM2 may be important in neuroblastoma pathogenesis. Epidemiological studies have shown that the presence of a T > G polymorphism in the MDM2 promoter (SNP 309) increases the affinity between the MDM2 promoter and the transcriptional activator Sp1, resulting in high MDM2 expression and subsequent attenuation of p53. In a study of 239 neuroblastoma patients, homozygous individuals (G/G) and those with heterozygous SNP (T/G) had an increased risk of neuroblastoma development compared with controls. Furthermore patients who were homozygous/heterozygous SNP309 variant carriers presented at a more advanced stage as well as having a shorter 5-year overall survival than patients with the wild-type allele (T/T) (35) Another study showed that MDM2 SNP309 had an adverse effect on stage 4 neuroblastoma disease progression and survival. Homozygous GG patients had a worse overall survival post relapse than patients who were TT homozygotes particularly in the presence of MYCN amplfication (36).
Comparable with MDM2 amplification, upstream inactivation of the p53 pathway via p14ARF abnormalities were also detected at diagnosis and relapse in 9/12 (75%). Methylation of the p14ARF gene was observed in 7% tumours, in contrast to a previous study which reported p14ARF methylation in 14% of diagnostic neuroblastomas (37). Inactivation of p14ARF by epigenetic mechanisms can interfere with the p53 pathway, as this increases MDM2 levels, which in turn inactivates p53 (38). Homozygous deletion of p14ARF was observed in a higher proportion of cases than previous reports (20, 39-42). It is also possible that small deletions in the 9p21 locus, specifically at the exon 1β locus exist, which are being detected by duplex PCR but not other methods.
The presence of p14ARF silencing predominantly at diagnosis and relapse further suggests that upstream modulation of p53 activity may be involved in neuroblastoma pathogenesis. This data shows concordance with the recent findings of Chen et al who showed a lack of ARF expression in MYCN transgenic tumours with MDM2 haploinsufficiency, suggesting that these tumours have a selective pressure to silence the p19ARF locus, and that low p19ARF expression is important in the development and progression of p53 wild-type neuroblastoma (43).
In conclusion, 15% of cases in this series had a p53 downstream pathway defect in the form of a p53 mutation whereas 35% of patients had upstream defects from p14ARF and MDM2 alterations. From this data it would appear that attenuation of the p53/MDM2/p14ARF axis, particularly increased MDM2 activity from p14ARF inactivation or MDM2 amplification is a critical mediator of p53 inactivation in neuroblastoma tumours. These results have important clinical implications as reactivation of p53 using p53/MDM2 inhibitors offers a novel therapeutic strategy in neuroblastoma alone or in combination with other therapies including non-genotoxic therapies. The p53/MDM2 inhibitor Nutlin 3a has been found to stabilise p53, induce expression of p53 target genes in wild-type p53 neuroblastoma cell lines (44), sensitise cells to conventional chemotherapeutic agents (45), and very recently Nutlin 3a has been found to be active in wild-type p53 chemoresistant cell lines and xenografts (46). From the current study p53/MDM2 inhibitors might be expected to benefit 85% of patients with recurrent neuroblastoma.
However, for neuroblastomas with downstream defects of the p53 pathway notably inactivating p53 mutations, which were associated with inferior survival in the current study, reactivating p53 would be ineffective. In these cases it would be important to include therapies that act independently of the p53 pathway e.g. temozolamide, taxol and arsenic tri-oxide to try to prevent relapse and the development of chemoresistance.
Statement of translational relevance.
Neuroblastoma remains one of the most difficult childhood cancers to cure. Over half of children with high-risk neuroblastoma relapse with chemoresistant disease and are incurable. We investigated paired neuroblastomas at different stages of therapy for abnormalities in the p53/MDM2/p14ARF pathway. Almost half of cases 20/41 (49%) had an abnormality in the p53 pathway detected at relapse, which was also present at diagnosis in 13 cases. Downstream defects due to inactivating p53 mutations were observed in 6/41 (15%) cases and were independently prognostic for overall survival (p = 0.02). Upstream defects were present in 15/41(35%) cases due to MDM2 amplification and p14 abnormalities. These results indicate that p53 pathway abnormalities are common in relapsed neuroblastoma. For upstream defects therapies to reactivate wild-type p53 such as p53/MDM2 inhibitors may be beneficial whereas for downstream defects agents which act independently of p53 should be considered upfront to try to prevent relapse and chemoresistance.
Supplementary Material
(on line only) Clinical information and p53 pathway status of all cases studied.
Acknowledgements
We thank Lorna More, Gavin Whyman and Caroline Ellershaw for clinical information on UK cases, and Josef Vormoor and Barbara Hero for clinical information on German cases. We thank Holger Christiansen for constitutional DNA and Helen Beris (Abbott Molecular) for the MDM2 probe.
This work was supported by the U.K Department of Health (Clinician Scientist Fellowship to DAT), The North of England Children's Cancer Research Fund (DAT), The Wellcome Trust (LE) and a donation from a parents' group.
References
- 1.Keshelava N, Zuo JJ, Chen P, et al. Loss of p53 function confers high-level multidrug resistance in neuroblastoma cell lines. Cancer Res. 2001;61:6185–93. [PubMed] [Google Scholar]
- 2.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 3.Michalak E, Villunger A, Erlacher M, Strasser A. Death squads enlisted by the tumour suppressor p53. Biochem Biophys Res Commun. 2005;331:786–98. doi: 10.1016/j.bbrc.2005.03.183. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. The EMBO journal. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stone S, Jiang P, Dayananth P, et al. Complex structure and regulation of the P16 (MTS1) locus. Cancer Res. 1995;55:2988–94. [PubMed] [Google Scholar]
- 6.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. The EMBO journal. 1999;18:22–7. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher SJ, Kefford RF, Rizos H. The ARF tumour suppressor. The international journal of biochemistry & cell biology. 2006;38:1637–41. doi: 10.1016/j.biocel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Molecular and cellular biology. 1998;18:6457–73. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tweddle DA, Pearson AD, Haber M, et al. The p53 pathway and its inactivation in neuroblastoma. Cancer letters. 2003;197:93–8. doi: 10.1016/s0304-3835(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 10.Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61:8–13. [PubMed] [Google Scholar]
- 11.Carr J, Bell E, Pearson AD, et al. Increased frequency of aberrations in the p53/MDM2/p14(ARF) pathway in neuroblastoma cell lines established at relapse. Cancer Res. 2006;66:2138–45. doi: 10.1158/0008-5472.CAN-05-2623. [DOI] [PubMed] [Google Scholar]
- 12.Brodeur GM, Pritchard J, Berthold F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–77. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 13.Ambros IM, Benard J, Boavida M, et al. Quality assessment of genetic markers used for therapy stratification. J Clin Oncol. 2003;21:2077–84. doi: 10.1200/JCO.2003.03.025. [DOI] [PubMed] [Google Scholar]
- 14.Lamont JM, McManamy CS, Pearson AD, Clifford SC, Ellison DW. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004;10:5482–93. doi: 10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
- 15.Carr J, Bown NP, Case MC, Hall AG, Lunec J, Tweddle DA. High-resolution analysis of allelic imbalance in neuroblastoma cell lines by single nucleotide polymorphism arrays. Cancer genetics and cytogenetics. 2007;172:127–38. doi: 10.1016/j.cancergencyto.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Malcolm AJ, Wood KM, et al. Cell cycle. Vol. 6. Georgetown, Tex: 2007. p53 is nuclear and functional in both undifferentiated and differentiated neuroblastoma; pp. 2685–96. [DOI] [PubMed] [Google Scholar]
- 17.Hosoi G, Hara J, Okamura T, et al. Low frequency of the p53 gene mutations in neuroblastoma. Cancer. 1994;73:3087–93. doi: 10.1002/1097-0142(19940615)73:12<3087::aid-cncr2820731230>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Imamura J, Bartram CR, Berthold F, Harms D, Nakamura H, Koeffler HP. Mutation of the p53 gene in neuroblastoma and its relationship with N-myc amplification. Cancer Res. 1993;53:4053–8. [PubMed] [Google Scholar]
- 19.Manhani R, Cristofani LM, Odone Filho V, Bendit I. Concomitant p53 mutation and MYCN amplification in neuroblastoma. Medical and pediatric oncology. 1997;29:206–7. doi: 10.1002/(sici)1096-911x(199709)29:3<206::aid-mpo7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 20.Omura-Minamisawa M, Diccianni MB, Chang RC, et al. p16/p14(ARF) cell cycle regulatory pathways in primary neuroblastoma: p16 expression is associated with advanced stage disease. Clin Cancer Res. 2001;7:3481–90. [PubMed] [Google Scholar]
- 21.Rainwater R, Parks D, Anderson ME, Tegtmeyer P, Mann K. Role of cysteine residues in regulation of p53 function. Molecular and cellular biology. 1995;15:3892–903. doi: 10.1128/mcb.15.7.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 23.Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, Vogelstein B. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–7. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 24.Ishii N, Tada M, Hamou MF, et al. Cells with TP53 mutations in low grade astrocytic tumors evolve clonally to malignancy and are an unfavorable prognostic factor. Oncogene. 1999;18:5870–8. doi: 10.1038/sj.onc.1203241. [DOI] [PubMed] [Google Scholar]
- 25.Pollock RE, Lang A, Luo J, El-Naggar AK, Yu D. Soft tissue sarcoma metastasis from clonal expansion of p53 mutated tumor cells. Oncogene. 1996;12:2035–9. [PubMed] [Google Scholar]
- 26.Wada H, Asada M, Nakazawa S, et al. Clonal expansion of p53 mutant cells in leukemia progression in vitro. Leukemia. 1994;8:53–9. [PubMed] [Google Scholar]
- 27.Righetti SC, Perego P, Corna E, Pierotti MA, Zunino F. Emergence of p53 mutant cisplatin-resistant ovarian carcinoma cells following drug exposure: spontaneously mutant selection. Cell Growth Differ. 1999;10:473–8. [PubMed] [Google Scholar]
- 28.de Vries A, Flores ER, Miranda B, et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2948–53. doi: 10.1073/pnas.052713099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wijnhoven SW, Speksnijder EN, Liu X, et al. Dominant-negative but not gain-of-function effects of a p53.R270H mutation in mouse epithelium tissue after DNA damage. Cancer Res. 2007;67:4648–56. doi: 10.1158/0008-5472.CAN-06-4681. [DOI] [PubMed] [Google Scholar]
- 30.Xue C, Haber M, Flemming C, et al. p53 determines multidrug sensitivity of childhood neuroblastoma. Cancer Res. 2007;67:10351–60. doi: 10.1158/0008-5472.CAN-06-4345. [DOI] [PubMed] [Google Scholar]
- 31.Storey A, Thomas M, Kalita A, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229–34. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 32.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nature genetics. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 33.Su WT, Alaminos M, Mora J, Cheung NK, La Quaglia MP, Gerald WL. Positional gene expression analysis identifies 12q overexpression and amplification in a subset of neuroblastomas. Cancer genetics and cytogenetics. 2004;154:131–7. doi: 10.1016/j.cancergencyto.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Spitz R, Oberthuer A, Zapatka M, et al. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer. 2006;45:1130–42. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- 35.Cattelani S, Defferrari R, Marsilio S, et al. Impact of a single nucleotide polymorphism in the MDM2 gene on neuroblastoma development and aggressiveness: results of a pilot study on 239 patients. Clin Cancer Res. 2008;14:3248–53. doi: 10.1158/1078-0432.CCR-07-4725. [DOI] [PubMed] [Google Scholar]
- 36.Perfumo C, Parodi S, Mazzocco K, et al. MDM2 SNP309 genotype influences survival of metastatic but not of localized neuroblastoma. Pediatr Blood Cancer. 2009;53:576–83. doi: 10.1002/pbc.22132. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Gomez P, Bello MJ, Lomas J, et al. Aberrant methylation of multiple genes in neuroblastic tumours. relationship with MYCN amplification and allelic status at 1p. Eur J Cancer. 2003;39:1478–85. doi: 10.1016/s0959-8049(03)00312-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 39.Bassi CL, Martelli L, Cipolotti R, Scrideli CA, Defavery R, Tone LG, et al. Lack of evidence for mutations or deletions in the CDKN2A/p16 and CDKN2B/p15 genes of Brazilian neuroblastoma patients. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas / Sociedade Brasileira de Biofisica. 2004;37:1683–7. doi: 10.1590/s0100-879x2004001100014. [DOI] [PubMed] [Google Scholar]
- 40.Castresana JS, Gomez L, Garcia-Miguel P, Queizan A, Pestana A. Mutational analysis of the p16 gene in human neuroblastomas. Molecular carcinogenesis. 1997;18:129–33. doi: 10.1002/(sici)1098-2744(199703)18:3<129::aid-mc1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 41.Diccianni MB, Chau LS, Batova A, Vu TQ, Yu AL. The p16 and p18 tumor suppressor genes in neuroblastoma: implications for drug resistance. Cancer letters. 1996;104:183–92. doi: 10.1016/0304-3835(96)04250-4. [DOI] [PubMed] [Google Scholar]
- 42.Thompson PM, Maris JM, Hogarty MD, et al. Homozygous deletion of CDKN2A (p16INK4a/p14ARF) but not within 1p36 or at other tumor suppressor loci in neuroblastoma. Cancer Res. 2001;61:679–86. [PubMed] [Google Scholar]
- 43.Chen Z, Lin Y, Barbieri E, et al. Mdm2 deficiency suppresses MYCN-Driven neuroblastoma tumorigenesis in vivo. Neoplasia. 2009;11:753–62. doi: 10.1593/neo.09466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Maerken T, Speleman F, Vermeulen J, et al. Small-Molecule MDM2 Antagonists as a New Therapy Concept for Neuroblastoma. Cancer Res. 2006;66:9646–55. doi: 10.1158/0008-5472.CAN-06-0792. [DOI] [PubMed] [Google Scholar]
- 45.Barbieri E, Mehta P, Chen Z, et al. MDM2 inhibition sensitizes neuroblastoma to chemotherapy-induced apoptotic cell death. Mol Cancer Ther. 2006;5:2358–65. doi: 10.1158/1535-7163.MCT-06-0305. [DOI] [PubMed] [Google Scholar]
- 46.Van Maerken T, Ferdinande L, Taildeman J, et al. Antitumor Activity of the Selective MDM2 Antagonist Nutlin-3 Against Chemoresistant Neuroblastoma With Wild-Type p53. J Natl Cancer Inst. 2009;101:1562–74. doi: 10.1093/jnci/djp355. [DOI] [PubMed] [Google Scholar]
- 47.Pearson AD, Pinkerton CR, Lewis IJ, Imeson J, Ellershaw C, Machin D. High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: a randomised trial. The lancet oncology. 2008;9:247–56. doi: 10.1016/S1470-2045(08)70069-X. [DOI] [PubMed] [Google Scholar]
- 48.Tweddle DA, Pinkerton CR, Lewis IJ, Ellershaw C, Cole M, Pearson AD. OPEC/OJEC for stage 4 neuroblastoma in children over 1 year of age. Medical and pediatric oncology. 2001;36:239–42. doi: 10.1002/1096-911X(20010101)36:1<239::AID-MPO1058>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 49.Cheung NK, Kushner BH, LaQuaglia M, et al. N7: a novel multi-modality therapy of high risk neuroblastoma (NB) in children diagnosed over 1 year of age. Medical and pediatric oncology. 2001;36:227–30. doi: 10.1002/1096-911X(20010101)36:1<227::AID-MPO1055>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 50.Berthold F, Hero B, Kremens B, et al. Long-term results and risk profiles of patients in five consecutive trials (1979-1997) with stage 4 neuroblastoma over 1 year of age. Cancer letters. 2003;197:11–7. doi: 10.1016/s0304-3835(03)00076-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(on line only) Clinical information and p53 pathway status of all cases studied.