Abstract
In recent years, the importance of horizontal gene transfer (HGT) in bacterial evolution has been elevated to such a degree that many bacteriologists now question the very existence of bacterial species. If gene transfer is as rampant as comparative genomic studies have suggested, how could bacterial species survive such genomic fluidity? And yet, most bacteriologists recognize, and name, as species, clusters of bacterial isolates that share complex phenotypic properties. The Core Genome Hypothesis (CGH) has been proposed to explain this apparent paradox of fluid bacterial genomes associated with stable phenotypic clusters. It posits that there is a core of genes responsible for maintaining the species-specific phenotypic clusters observed throughout bacterial diversity and argues that, even in the face of substantial genomic fluidity, bacterial species can be rationally identified and named.
Keywords: Bacterial species concept, core genome hypothesis, population genomics
1. Introduction
The impact of molecular systematics on bacterial classification has been profound. Indeed, phylogenies based on highly conserved molecules, such as ribosomal RNA, have fundamentally changed our view of biological diversity (1, 2). These molecular phylogenies have confirmed the existence of three primary divisions of life (Archaea, Bacteria, and Eukarya), rather than the five that had emerged from phenotype studies (Animalia, Plantae, Fungi, Protista, and Monera), and reveal that microbes comprise, by far, the greatest amount of biological diversity (1, 3–5). Further, as additional highly conserved genes are examined, such as elongation factor-1α, actin, α-tubulin, and β-tubulin, we gain confidence that molecular-based phylogenies can provide a fairly robust description of the major evolutionary lineages (6, 7).
Just as molecules appear to have solved some of the outstanding phylogenetic questions, their application has generated an entirely new and unexpected controversy. They have revealed that horizontal gene transfer (HGT) may play an important and unexpectedly large role in evolution (Fig. 21.1) (8–11). Recent observations of putative gene transfer events between some of the deepest branches in the 16S ribosomal RNA-based tree of life have raised the question of whether we should employ networks, rather than dichotomously branching trees, to represent the relationships of evolutionary lineages over time (12). In fact, the importance of HGT in bacterial evolution has been elevated to such a degree that numerous bacteriologists now question the very existence of bacterial species (13–15). If gene transfer is as rampant as some propose, how could bacterial species survive such genomic fluidity?
Fig. 21.1. The impact of gene transfer on molecular phylogenies.
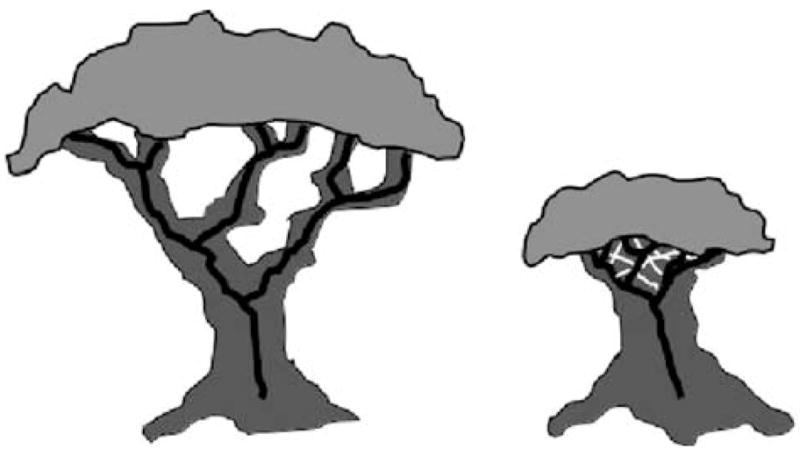
The tree on the left represents a dichotomously branching set of lineages. Over time the lineages diverge and result in extensive biological diversity. The tree on the right shows the impact of extensive HGT, which serves to limit divergence and the resulting diversity. Such lineages are homogenized by HGT and appear to have a shortened evolutionary history.
Traditional bacterial species designations were based upon extensive phenotypic characterization of a large number of isolates. Although current methods now require the use of 16S rRNA sequence comparisons to identify the closest relatives of a proposed species, phenotype still remains the primary criterion by which species are identified (16). The emerging phylo-phenetic bacterial species concept posits that a bacterial species is “a monophyletic and genomically coherent cluster of individual organisms that show a high degree of overall similarity with respect to many independent characteristics, and is diagnosable by a discriminative phenotypic property.” (16).
Numerous studies have revealed clusters of bacterial isolates that share complex phenotypes and these clusters are often designated as species (17–21). In fact, Cohan uses the mere existence of these clusters as prima facie evidence of the existence of bacterial species. He notes “Bacterial species exist … bacterial diversity is organized into discrete phenotypic and genetic clusters, which are separated by large phenotypic and genetic gaps, and these clusters are recognized as species.” (22).
2. Molecular-Based Species Distinctions
More recent investigations into microbial species distinctions have sought to incorporate estimates of molecular diversity into the process of species identification. The assumption is that this molecular diversity will fall into discrete clusters that correspond with observed phenotype-based species clusters. Can sequence variability be employed to inform the division of a genus into species, to distinguish among similar species, or to address whether bacterial species even exist (23–28)?
Early attempts to use molecular data to delineate bacterial species began with the introduction of DNA–DNA hybridization, in which bacterial species were defined as those isolates sharing at least 70% hybridization under standardized conditions (29). Given the enormous range of genetic variation detected in different clearly recognized species, it became clear that a variability cutoff, such as is imposed with hybridization methods, was not appropriate. Levels of variability will vary over the lifetime of a species and will reflect aspects of its life history, particularly the process by which it adapts to its range of habitats.
The use of DNA–DNA hybridization has largely been replaced by the use of 16S rRNA sequences to determine the closest relatives of an isolate, combined with extensive phenotypic data. A disturbingly large number of publications now report species diversity based solely upon the quantity of novel 16S rRNA sequences detected (30–32). This approach has no valid systematic basis and should be avoided at all cost (16, 33). Variability in 16S rRNA sequences can provide a valid estimate of molecular diversity, but that estimate cannot necessarily be equated with species diversity.
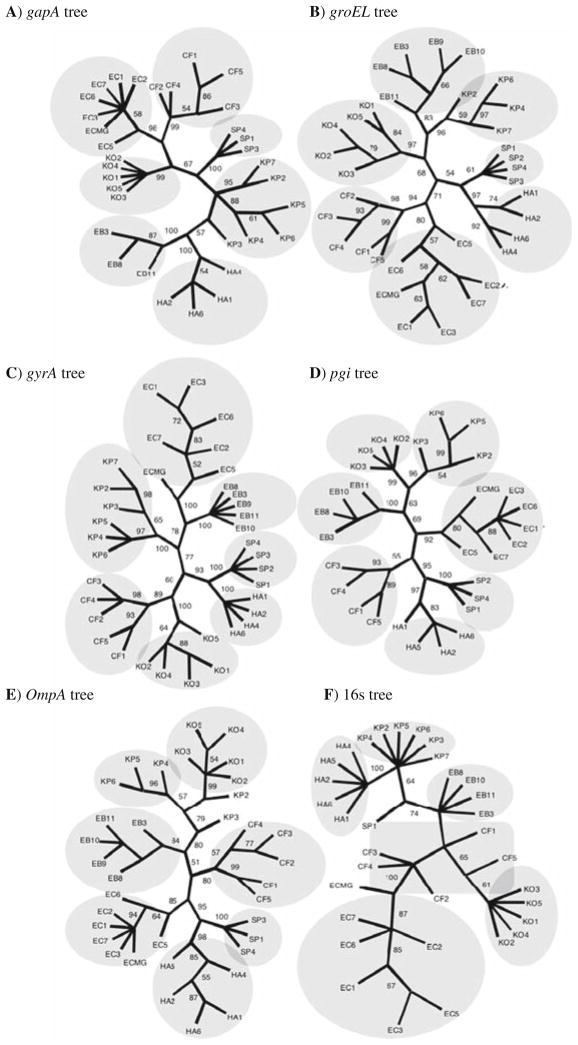
One of the first gene-based investigations into the microbial species concept was conducted in 2003 by Wertz et al., who sequenced six housekeeping genes from a sample of environmental bacteria representing seven species of Enterobacteriaceae (34, 35). Molecular phylogenies for each of the genes were inferred (Fig. 21.2) and the branching patterns of the resulting trees compared. In almost every case, isolates from a species formed a well-supported monophyletic group, which corresponded precisely with the clusters identified by phenotypic data, and upon which species distinctions were initially delineated (16,36). A molecular-based enteric species phylogeny was inferred from the composite data by concatenating the sequences of the six genes (34). The concatenated sequence contained enough phylogenetic signal to resolve all of the interspecies nodes and thus provided a robust estimate of the enteric phylogeny, which corresponded with the existing phenotype-based phylogeny. The authors concluded that, at least for these enteric bacteria, bacterial species clearly do exist and identical species designations emerge from both phenotypic and genotypic data.
Fig. 21.2. Molecular phylogenies of six housekeeping genes for six species of enteric bacteria.
Each molecular phylogeny was inferred from the DNA sequences of one housekeeping gene using maximum likelihood. Cladograms were based on maximum likelihood for six putative core genes. (a) gapA, (b) groEL, (c) gyrA, (d) pgi, (e) ompA, and (f) 16S rRNA. Bootstrap values less than 50% (of 500 replicates) were omitted. Confidence in the branching patterns is indicated along the branches. Shading denotes a cluster of lineages that correspond to a named species. Taxa abbreviations are CF: Citrobacter freundii; EB: Enterobacter cloacae; EC: Escherichia coli; HA: Hafnia alvei; KO: Klebsiella oxytoca; KP: Klebsiella pneumoniae, and SP: Serratia plymuthica. ECMG refers to E. coli strain MG1655; see (34).
The use of multi-locus sequence typing (MLST), in which portions of, generally, seven housekeeping genes are sequenced, has become the norm for characterizing genetic diversity within a bacterial species (37, 38). This method permits the analysis of large numbers of related bacterial isolates, which is essential to the determination of species designations (24, 38). Such studies have confirmed that species designations based upon phenotypic criteria have a corresponding underlying MLST-based genotypic clustering (23, 26, 39, 40).
3. Population Genomics Informs Species Designations
The availability of whole genome sequences for multiple isolates of E. coli provided our first glimpse into the dynamic nature of a species genome. Glasner and Perna (41) and Mau et al. (42) compared six complete genomes of E. coli (including Shigella flexneri) and revealed a highly conserved genomic backbone of more than 3,000 genes, each with an average of greater than 98% sequence similarity among the isolates. Further, Mau et al. (42) detected a high level of homologous recombination among these shared genes, confirming earlier studies by Roger Milkman (43, 44). More surprising was the observation that this conserved backbone was punctuated with hundreds of “sequence islands” specific to one strain or another.
Edwards et al. (45) provided a similar comparison of two complete and three draft genome sequences of Salmonella enterica. As in E. coli, a backbone of highly conserved genes was identified, each with an average of greater than 99% sequence similarity and a similar pattern of unique sequence islands specific to one strain or another. This pattern of shared and unique sequences appears to be common among many bacterial species (46–48).
Studies with subtractive hybridization and comparative genome hybridization revealed that for Helicobacter pylori, E. coli, and Staphylococcus aureus, strains within a species share roughly 75–85% of their genome. A comparison of eight genomes of group B Streptococci revealed a core of 1,806 genes present in every genome and 907 genes absent in one or more genomes. A similar comparison between five genomes of Streptococcus pyogenes revealed a comparable level of genomic diversity and predicted that each new genome added approximately 27 strain-specific genes to the species total pan-genome. In contrast, eight genomes of Bacillus anthracis revealed very few strain-specific genes. In fact, after the addition of four genomes to the comparison, no new unique genes were identified. The general pattern that emerges is that members of a bacterial species share some large fraction of their genomes, but often carry unique, strain-specific sequences. The fraction of the genome that is shared versus unique varies greatly from one bacterial species to the next.
4. The Core Genome Hypothesis and the Bacterial Species Concept
Lan and Reeves were the first to recognize the potential link between the observation of shared versus unique sequences in bacterial genomes and its implication for discriminating bacterial species (49). They proposed the Core Genome Hypothesis (CGH), which distinguishes between that fraction of the genome (the core) shared by all members of a species and the fraction found in only a subset of the population (the auxiliary). Core genes encode essential metabolic housekeeping and informational processing functions (50). They are ubiquitous in a species and define the species-specific characteristics. Auxiliary genes may or may not be present in a strain and are generally genes that encode supplementary biochemical pathways, are associated with phage or other mobile elements, or encode products that serve to interact with the external environment. Thus, auxiliary genes serve in the adaptation of strains to local competitive or environmental pressures (14). They are likely to encode antibiotic resistance, novel metabolic functions, toxin production, and the like (51–53).
The CGH has dramatically influenced how bacteriologists think about the nature of bacterial species. Prior to the CGH, the strongest argument against the recognition of bacterial “species” was the simple observation of HGT between bacterial lineages. The fact that bacterial species gene pools may not be tightly closed was enough reason for many microbiologists to conclude bacterial species could not survive such exchange. This contradicts the clearly demonstrated fact that bacteria exist in phenotypic clusters, which many microbiologists recognize as species. Even more compelling, it is becoming clear that these well-defined phenotypic clusters correspond to underlying genotype clusters (26, 39, 48, 54).
Some have argued that it is futile to expect a bacterial species to ever be characterized fully at the genome level, particularly since as more genome sequences are obtained, the pan-genomes (i.e., the sum of all genes identified within a species) of numerous species continues to grow (55, 56). In fact, some predict that hundreds of thousands of genome sequences are required to fully define certain bacterial species (55). Others suggest that the wide range of intra-species variation observed for bacterial species reflects the lack of a universal and meaningful species definition (50).
Many ecological and evolutionary factors will impact how many unique genes a species pan-genome may encode and how much genetic variation it harbors. There is no “one size fits all” concept that can, or should, be applied. In fact, no existing species definition requires that either the pan-genome or the level of genetic variation be known in order to delineate members of a species.
One of the more commonly accepted species concepts is the Biological Species Concept (BSC) proposed by Ernst Mayr (57). Mayr proposed that a biological species is comprised of groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups (57). Although Mayr developed this definition specifically for eukaryotes, it can be modified to apply to bacteria. However, it is important to note that at this juncture, the BSC should not be taken to imply any particular process of speciation, merely that the observation of more gene “sharing” (via recombination and/or LGT) is observed within versus between putative bacterial species. The Core Genome Hypothesis provides a perfect backdrop from which to articulate this bacterial-based modification of the BSC. According to the CGH, a bacterial species is comprised of groups of strains that frequently exchange, or could exchange, core genes, but which are relatively restricted from such exchange with other groups.
The CGH predicts that a subset of genes, the core, is present in all, or nearly all, individuals within a species. These are the genes that provide the defining characteristics of a species and are assumed to experience primarily purifying selection, to remove deleterious mutations, and to maintain existing functions. As a species evolves, its core genome will evolve as a complex of co-evolved functions. When transferred between species, such genes will most likely experience a selective disadvantage, as this will disrupt co-evolved functions. Such transfer will rarely survive. Thus, core genes will diverge as the species diverge (Fig. 21.1).
In contrast, auxiliary genes will be found in only a subset of individuals within a species. The CGH predicts that these genes experience intermittent positive selection, when their function enhances survival in a varied and ever-changing environment. When such genes are exchanged between species, their functions will often provide a selective advantage to the recipient. Frequent successful transfers between species will serve to limit the divergence of auxiliary genes, relative to the core (Fig. 21.1).
The most specific prediction that emerges from the CGH concerns the rate at which core and auxiliary genes accumulate variability. Core genes will, on average, display a neutral rate of evolution, while auxiliary genes will experience a variety of selective pressures, including diversifying selection (acting to increase levels of variation), directional selection (acting to decrease levels of variation), balancing selection (acting to maintain particular alleles in the species), and purifying selection (acting to weed out deleterious mutations). Thus, the average rate of evolution for auxiliary genes could be just about anything, and the variance around this rate should be extreme. These expectations, based upon the neutral theory (58), are quite useful for testing predictions from the CGH (59). However, such tests require population-based samples of multiple genomes per species and, unfortunately, most existing species-based genome samples are chosen to represent the diversity of clinical isolates of human pathogens and thus will often underestimate standing levels of genome diversity. The appropriate data are in the pipeline and should soon be available to permit the sort of population genomics required to address this complex and fascinating matter.
Although we are on the verge of obtaining the type and amount of genotypic data required to examine bacterial species definitions, it is important to note that there is little, if any, substantive data to support the conclusion that bacterial species do not exist. Hence, the real argument remaining is not do they exist, but rather how can they exist in the face of potentially high levels of HGT. Our job is to develop an understanding of bacterial evolution rich enough to explain this apparent paradox. The CGH provides a set of testable hypotheses from which to launch this exploration.
5. Conclusions
The community of bacteriologists has failed to establish a uniformly accepted definition of bacterial species. In part, this is due to the extraordinary levels of bacterial diversity and its complexity in terms of culturability, levels of observed HGT, importance in human health, and a variety of other scientific and social factors. However, we are poised to witness a synthesis of the general acknowledgment that bacteria are found in clusters of complex phenotypes (often designated as species) and the underlying genetic basis for these clusters. The Core Genome Hypothesis has, so far, provided the most compelling explanation for the apparent paradox observed for bacteria, in which the observation of stable phenotypic clusters apparently contradicts the existence of dynamic, fluid genomes. The CGH recognizes a species core genome, responsible for maintaining a species identity, and an auxiliary genome, responsible for gene transfer and rapid adaptation of strains to an ever-changing environment. The CGH argues that, even in the presence of substantial genomic fluidity, bacterial species can be rationally identified and named.
Acknowledgments
The authors acknowledge the valuable input provided by Carla Goldstone, Chris Vriezen, and Emma White. This article was written with support from NIH R01 GM068657-01A2 and R01 A1064588-01A2 grants to MAR.
References
- 1.Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, Zablen LB, Blakemore R, Gupta R, Bonen L, Lewis BJ, Stahl DA, Luehrsen KR, Chen KN, Woese CR. The phylogeny of prokaryotes. Science. 1980;209:457–63. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 2.Olsen GJ, Woese CJ. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–23. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 3.Pace NR, Stahl DA, Lane DJ, Olsen GJ. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 4.Woese CR. Bacterial evolution. Microbiological Reviews. 1987;51:221–71. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese C, Kandler O, Wheelis M. Towards a natural system of organisms – proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig W, Neumaier J, Klugbauer N, Brockmann E, Roller C, Jilg S, Reetz K, Schachtner I, Ludvigsen A, Bachleitner M, Fischer U, Schleifer KH. Phylogenetic relationships of Bacteria based on comparative sequence analysis of elongation factor Tu and ATP-synthase beta-subunit genes. Antonie Van Leeuwenhoek. 1993;64:285–305. doi: 10.1007/BF00873088. [DOI] [PubMed] [Google Scholar]
- 7.Brown J, Douady C, Italia M, Marshall W, Stanhope M. Universal trees based on large combined protein sequence data sets. Nat Genet. 2001;28:281–5. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- 8.Kidwell M. Lateral transfer in natural populations of eukaryotes. Ann Rev Genetics. 1993;27:235–56. doi: 10.1146/annurev.ge.27.120193.001315. [DOI] [PubMed] [Google Scholar]
- 9.Nelson K, Selander RK. Inter-generic transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci U S A. 1994;91:10227–31. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nesbo CL, L’Haridon S, Stetter KO, Doolittle WF. Phylogenetic analyses of two “Archaeal” genes in thermotoga maritima reveal multiple transfers between Archaea and Bacteria. Mol Biol Evol. 2001;18:362–75. doi: 10.1093/oxfordjournals.molbev.a003812. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–8. [PubMed] [Google Scholar]
- 13.Doolittle WF, Papke RT. Genomics and the bacterial species problem. Genome Biology. 2006;7:116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohan F. What are bacterial species? Annu Rev Microbiol. 2002;56:457–87. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 15.Sapp J. The Bacteriums Place in Nature. Oxford University Press; New York: 2005. Microbial phylogeny and evolution: Concepts and controversies. [Google Scholar]
- 16.Rossello-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 17.Shute LA, Gutteridge CS, Norris JR, Berkeley RC. Curie-point pyrolysis mass spectrometry applied to characterization and identification of selected Bacillus species. J Gen Microbiol. 1984;130:343–55. doi: 10.1099/00221287-130-2-343. [DOI] [PubMed] [Google Scholar]
- 18.Sneath P, Stevens M. A numerical taxonomic study of Actinobacillus, Pasteurella, and Yersinia. J Gen Microbiol. 1985;131:2711–38. doi: 10.1099/00221287-131-10-2711. [DOI] [PubMed] [Google Scholar]
- 19.Barrett S, Sneath P. A numerical phenotypic taxonomic study of the genus Neisseria. Microbiology. 1994;140:2867–91. doi: 10.1099/00221287-140-10-2867. [DOI] [PubMed] [Google Scholar]
- 20.Mauchline W, Keevil C. Development of the BIOLOG substrate utilization system for identification of Legionella spp. Appl Environ Microbiol. 1991;57:3345–9. doi: 10.1128/aem.57.11.3345-3349.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner C, Maquelin K, Pina P, Thi NN, Choo-Smith L, Sockalingum G, Sandt C, Ami D, Orsini F, Doglia S, Allouch P, Mainfait M, Puppels G, Naumann D. Classification and identification of enterococci: A comparative phenotypic, genotypic, and vibrational spectroscopic study. J Clin Microbiol. 2001;39:1763–70. doi: 10.1128/JCM.39.5.1763-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohan F. Sexual isolation and speciation in bacteria. Genetica. 2002;116:359–70. [PubMed] [Google Scholar]
- 23.Godoy AP, Ribeiro ML, Benvengo YH, Vitiello L, Miranda MdeC, Mendonca S, Pedrazzoli J., Jr Analysis of antimicrobial susceptibility and virulence factors in Helicobacter pylori clinical isolates. BMC Gastroenterol. 2003;3:20–26. doi: 10.1186/1471-230X-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanage WP, Fraser C, Spratt BG. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priest F, Barker M, Baillie L, Holmes E, Maiden M. Population structure and evolution of the Bacillus cereus group. J Bacteriol. 2004;186:7959–70. doi: 10.1128/JB.186.23.7959-7970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson J, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt D, Benoit J, Sarma-Rupavtarm R, Distel D, Polz M. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–3. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- 27.Baldwin A, Mahenthiralingam E, Thickett K, Honeybourne D, Maiden M, Govan J, Speert D, Lipuma J, Vandamme P, Dowson C. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol. 2005;43:4665–73. doi: 10.1128/JCM.43.9.4665-4673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanage W, Kaijalainen T, Syrjanen R, Auranen K, Leinonen M, Makela P, Spratt B. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect Immun. 2005;73:431–5. doi: 10.1128/IAI.73.1.431-435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P, Maiden MC, Nesme X, Rosselló-Mora R, Swings J, Trüper HG, Vauterin L, Ward AC, Whitman WB. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–7. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 30.Lindh J, Terenius O, Faye I. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol. 2005;71:7217–23. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drancourt M, Berger P, Raoult D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004;42:2197–202. doi: 10.1128/JCM.42.5.2197-2202.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarridge JR. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–62. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton R, Sutton G, Hinkle PJ, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–9. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 34.Wertz JE, Valletta-Goldstone CM, Gordon DM, Riley MA. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J Evol Biol. 2003;16:1236–48. doi: 10.1046/j.1420-9101.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- 35.Gordon DM, Fitzgibbon F. The distribution of enteric bacteria from Australian mammals: Host and geographical effects. Microbiology. 1999;145:2663–71. doi: 10.1099/00221287-145-10-2663. [DOI] [PubMed] [Google Scholar]
- 36.Holt JG. Bergey’s Manual of Determinative Bacteriology. Williams and Wilkins; Baltimore, MD: 1994. [Google Scholar]
- 37.Maiden M. Horizontal genetic exchange, evolution and spread of antibiotic resistance in bacteria. Clin Infect Dis. 1998;27:S12–20. doi: 10.1086/514917. [DOI] [PubMed] [Google Scholar]
- 38.Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, Peacock SJ, Smith JM, Murphy M, Spratt BG, Moore CE, Day NP. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitaker R, Grogan D, Taylor J. Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol Biol Evol. 2005;22:2354–61. doi: 10.1093/molbev/msi233. [DOI] [PubMed] [Google Scholar]
- 40.Woodward M, Sojka M, Sprigings K, Humphrey T. The role of SEF14 and SEF17 fimbriae in the adherence of Salmonella enterica serotype Enteritidis to inanimate surfaces. J Med Microbiol. 2000;49:481–7. doi: 10.1099/0022-1317-49-5-481. [DOI] [PubMed] [Google Scholar]
- 41.Glasner J, Perna N. Comparative genomics of E. coli. Microbiol Today. 2004:31. [Google Scholar]
- 42.Mau B, Glasner J, Darling A, Perna N. Genome-wide detection and analysis of homologous recombination among sequenced strains of Escherichia coli. Genome Biology. 2006;7:R44. doi: 10.1186/gb-2006-7-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milkman R, Raleigh E, McKanea M, Crydermanc D, Bilodeau P, Mcweeny K. Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics. 1999;153:539–54. doi: 10.1093/genetics/153.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milkman R, Jaeger E, Mcbride R. Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics. 2003;163:475–83. doi: 10.1093/genetics/163.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards SV, Fertil B, Giron A, Deschavanne PJ. A genomic schism in birds revealed by phylogenetic analysis of DNA strings. Systematic Biology. 2002;51:599–613. doi: 10.1080/10635150290102285. [DOI] [PubMed] [Google Scholar]
- 46.Juhas M, Power PM, Harding RM, Ferguson DJ, Dimopoulou ID, Elamin AR, Mohd-Zain Z, Hood DW, Adegbola R, Erwin A, Smith A, Munson RS, Jr, Harrison A, Mansfield L, Bentley S, Crook DW. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007;8:R237. doi: 10.1186/gb-2007-8-11-r237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coleman M, Sullivan M, Martiny A, Steglich C, Barry K, Delong E, Chisholm S. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–9. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 48.Waterfield N, Daborn P, Dowling A, Yang G, Hares M, Ffrench-Constant R. The insecticidal toxin makes caterpillars floppy 2 (Mcf2) shows similarity to HrmA, an avirulence protein from a plant pathogen. FEMS Microbiol Lett. 2003;229:265–70. doi: 10.1016/S0378-1097(03)00846-2. [DOI] [PubMed] [Google Scholar]
- 49.Lan R, Reeves PR. Gene transfer is a major factor in bacterial evolution. Mol Biol Evol. 1996;13:47–55. doi: 10.1093/oxfordjournals.molbev.a025569. [DOI] [PubMed] [Google Scholar]
- 50.Feil E. Small change: Keeping pace with microevolution. Nat Rev Microbiol. 2004;2:483–95. doi: 10.1038/nrmicro904. [DOI] [PubMed] [Google Scholar]
- 51.Dobrindt U, Reidl J. Pathogenicity islands and phage conversion: Evolutionary aspects of bacterial pathogenesis. Int J Med Microbiol. 2000;290:519–27. doi: 10.1016/S1438-4221(00)80017-X. [DOI] [PubMed] [Google Scholar]
- 52.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–43. doi: 10.1016/s0966-842x(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 53.White PA, Mciver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:2658–61. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godoy A, Ribeiro M, Benvengo Y, Vitiello L, Miranda CM, Mendonca S, Pedrazzoli JJ. Analysis of antimicrobial susceptibility and virulence factors in Helicobacter pylori clinical isolates. BMC Gastroenterol. 2003;3:20. doi: 10.1186/1471-230X-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–94. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Tettelin H, Masignani V, Ciesiewicz M, Donati C, Medini D, Ward N, Angiuli S, Crabtree J, Jones A, Durkin A. Genome analysis of multiple pathogenic isolates of Streptoccocus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci USA. 2005;102:13950–5. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayr E. Systematics and the Origin of Species. Columbia University Press; New York: 1942. [Google Scholar]
- 58.Kimura M. Genetic variability maintained in a finite population due to mutation production of neutral and nearly neutral isoalleles. Genetic Res Camb. 1968;11:247–69. doi: 10.1017/s0016672300011459. [DOI] [PubMed] [Google Scholar]
- 59.Fay J, Wyckoff G, Wu C. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature. 2002;415:1024–6. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]