Summary
Heterochromatin assembly in budding yeast requires the SIR complex, which contains the NAD-dependent deacetylase Sir2 and the Sir3 and Sir4 proteins. Sir3 binds to nucleosomes containing deacetylated histone H4 lysine 16 (H4K16) and, with Sir4, promotes spreading of Sir2 and deacetylation along the chromatin fiber. Combined action of histone modifying and binding activities is a conserved hallmark of heterochromatin, but the relative contribution of each activity to silencing has remained unclear. Here we reconstitute SIR-chromatin complexes using purified components and show that the SIR complex efficiently deacetylates chromatin templates and promotes the assembly of altered structures that silence Gal4-VP16-activated transcription. Silencing requires all three Sir proteins, even with fully deacetylated chromatin, and involves the specific association of Sir3 with deacetylated H4K16. These results define a minimal set of components that mediate heterochromatic gene silencing and demonstrate distinct contributions for histone deacetylation and nucleosome binding in the silencing mechanism.
Introduction
Assembly of DNA into silent chromatin domains is utilized by all eukaryotic organisms to restrict activity in certain chromosomal loci (Grewal and Moazed, 2003; Richards and Elgin, 2002). This restriction mechanism can stabilize structural regions of the chromosome or prevent gene expression. Silencing is achieved by a set of proteins that spread across the target locus, changing the structure of the chromatin fiber in a manner that can be faithfully inherited through many cell divisions. In most eukaryotes these silenced regions are termed heterochromatin because they have a noticeably different, condensed appearance throughout the cell cycle, compared to actively transcribed parts of the genome called euchromatin. Silent chromatin is also characterized by a distinct pattern of histone hypoacetylation and often methylation on particular lysine residues (Jenuwein and Allis, 2001; Moazed, 2001; Shahbazian and Grunstein, 2007). Silencing is associated with cell differentiation, even in unicellular organisms, where it determines cell type and controls sexual reproduction. In multicellular organisms heterochromatin-like regions are implicated in maintenance of cell identity during development (Ringrose and Paro, 2004).
S. cerevisiae has served as a major model for dissecting steps in assembly of silent chromatin domains. Silent chromatin occurs at three main regions of the S. cerevisiae genome: the mating-type loci, telomeres, and rDNA (reviewed in Rusche et al., 2003; Moazed 2001). Silencing at the mating-type regions and telomeres shares many mechanistic features, while rDNA silencing is achieved by a distinct mechanism. Silencing at the two silent mating-type loci (HMR and HML) has been extensively studied. The ∼3 kilobase regions are flanked by DNA sequences, called silencers, which contain binding sites for factors that, in combination, can recruit the silent information regulator (Sir) proteins. Sir2-4 are important for mating identity, telomeric gene silencing, and telomere clustering in yeast, while the Sir1 protein is only required at the mating type loci. Sir2, Sir3, and Sir4 form a complex, the SIR complex, which is associated continuously across silent regions and Sir2 and Sir4 (Sir2/4) have been purified as a heterodimer (Hecht et al., 1996; Hoppe et al., 2002; Moazed et al., 1997; Moretti et al., 1994; Strahl-Bolsinger et al., 1997).
Silent chromatin at the yeast mating-type loci and telomeres is established in three steps: initiation, nucleation, and spreading (Hoppe et al., 2002; Luo et al., 2002; Rusche et al., 2002). Initiation is characterized by recruitment of the Sir2/Sir4 complex by sequence-specific DNA binding proteins that have bound their target sequence, the silencer element (Figure 1A). Sir3 also becomes associated with the silencer region, primarily through interaction with Sir4. Sir2 is an NAD-dependent histone deacetylase with specificity for H4K16 (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000), while Sir3 and Sir4 bind to deacetylated histone H3 and H4 N termini (Hecht et al., 1995). When Sir2 is recruited to a silencer element it is though to deacetylate adjacent N-terminal histone tails and create a deacetylated nucleosome platform that Sir3 and Sir4 bind. Spreading involves iterative cycles of deacetylation, which are coupled to association of many copies of the SIR complex with nucleosomes across multiple kilobases away from the silencer element in a DNA sequence-independent manner (Figure 1A). In addition to deacetylation, spreading requires self-self interactions within the SIR complex and is dependent on the ability of Sir3 to form a complex with Sir2/4 (Hoppe et al., 2002; Liou et al., 2005; Rudner et al., 2005). Finally, NAD-dependent deacetylation by Sir2 generates O-acetyl-ADP-ribose (OAADPR or AAR), which in vitro studies suggest acts together with the histone H4 N-terminus to promote the association of multiple copies of Sir3 with Sir2/4 (Liou et al., 2005). AAR may contribute to SIR complex spreading in vivo, although a recent study suggests that its synthesis is not required for silencing (Chou et al., 2008).
Figure 1. Reconstitution of acetylated yeast chromatin.
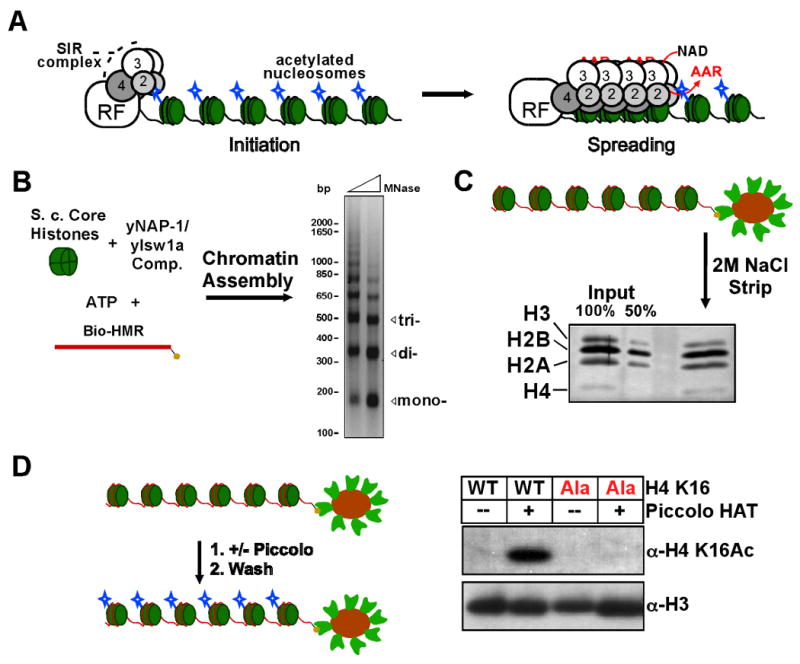
(A) A basic model for SIR complex assembly into silent chromatin. DNA-binding proteins act as recruitment factors (RF) to direct the SIR complex to target chromosomal regions. An iterative cycle of NAD-dependent histone deacetylation and direct chromatin association by the SIR complex leads to spreading of the SIR complex and produces a SIR-coated chromatin fiber that silences transcription. Deacetylation produces the small-molecule O-acetyl-ADP ribose (OAADPR or AAR), which may be incorporated into the SIR complex during assembly on chromatin. (B) Scheme for chromatin assembly using E. coli-expressed yeast histones, purified assembly factors, and a linear biotinylated HMR DNA fragment. Limited micrococcal nuclease (MNase) digestion of the assembled chromatin yields a ladder pattern indicating regularly-spaced nucleosomes. Positions of mono-, di-, and trinucleosomal DNA are indicated. (C) The biotinylated chromatin was immobilized on streptavidin-coated magnetic beads and the assembly factors were washed away. Histones remained stably associated in the proper stoichiometry after conjugation and washing. Input histones were stoichiometric by Coomassie, but each histone stains to a different extent by silver, shown in the panel. (D) The bead-conjugated chromatin is acetylated with the Piccolo HAT complex, which is subsequently washed away. Piccolo treatment produces a strong signal for acetylation of H4K16, which is abolished upon H4K16A mutation.
Investigation of the assembly of the Sir proteins into silent chromatin has suggested that a specialized structure is created during establishment of silencing. Early studies demonstrated that silent chromatin is inaccessible to DNA methyltransferase and restriction endonuclease action (Gottschling, 1992; Loo and Rine, 1994). More recently it has been shown that certain histone mutations produce a situation in vivo where the Sir proteins are associated across a region normally targeted for silencing, yet the region is not silenced, indicating that the association of the SIR complex with chromatin may not be sufficient for gene silencing (Xu et al., 2007; Yang et al., 2008; Yang and Kirchmaier, 2006). In addition, electron micrographs of reactions containing the Sir proteins and affinity-purified chromatin revealed fiber structures that required the presence of all three Sir proteins and were not observed in the absence of NAD or when chromatin with an H4K16 mutation replaced wild-type chromatin (Onishi et al., 2007).
Although genetic studies have identified a requirement for a core set of proteins in gene silencing in budding yeast, the minimal set of proteins that can establish heterochromatin has not yet been defined. Furthermore, while the importance of all three Sir proteins in silencing is appreciated, it is unclear whether the Sir3 and Sir4 proteins perform functions beyond the spreading of Sir2-mediated deacetylation. In this study, we use in vitro assembled chromatin templates to investigate the interactions and silencing activities of the SIR complex. We find that Sir3 interaction with chromatin can be disrupted by histone acetylation or by removal of H4K16. However, Sir2/4 binds to chromatin independently of these changes and can recruit Sir3 to an acetylated chromatin template. Importantly, we demonstrate that the silencing activities of the SIR complex, which are accompanied by a structural alteration in the chromatin template, require NAD-dependent deacetylation even though deacetylation is not required for binding of the full SIR complex to chromatin. Furthermore, mutation of H4K16 disrupts the silencing activities of the SIR complex without affecting its Sir2/4-mediated association with chromatin, demonstrating that specific Sir3-nucleosome interactions are required to achieve silencing.
Results
Reconstitution of acetylated yeast chromatin
In order to analyze the mechanism of silent chromatin assembly using purified components in vitro, a specific chromatin assembly scheme was developed. S. cerevisiae histones were purified from E. coli to avoid post-translational modification (confirmed by mass spectrometry, Figure S1C) as described previously for histone preparations (Figure S1)(Luger et al., 1999). Histone chaperone Nap1 and the nucleosome spacing complex Isw1a were purified and used to assemble a regularly-spaced nucleosome array on a biotinylated PCR fragment bearing the sequence of the HMR locus (Figure 1B). The in vitro assembled chromatin was subsequently conjugated to streptavidin-coated magnetic beads and the assembly factors washed away (Figure 1C and Figure S1E).
The bead-bound chromatin was found to be permissive to acetylation by the catalytic Piccolo subcomplex of the NuA4 histone acetyltransferase (HAT) complex (a kind gift from B. Hnatkovich and S. Tan). Approximately thirteen lysines per histone octamer were acetylated by Piccolo (determined by 3H-acetyl incorporation, data not shown). A robust signal was observed for the acetylated chromatin using an antibody that recognizes acetyl-H4K16 and this signal was lost upon mutation of lysine 16 to alanine (Figure 1D). H4K16 is particularly important for silencing and for the interaction of Sir3 with histone H4 N-terminal peptides and the nucleosome (Liou et al., 2005; Onishi et al., 2007; Rusche et al., 2003; Shahbazian and Grunstein, 2007).
Acetylation or mutation of histone H4 lysine 16 disrupts Sir3 interaction with chromatin
The bead-bound chromatin templates were used in binding assays with the Sir proteins to test the effects of chromatin modification on the interaction. The Sir proteins were overexpressed in yeast and affinity-purified as described (Buchberger et al., 2008; Liou et al., 2005; Onishi et al., 2007)(Figure S2). Sir3 was purified alone and Sir2 and Sir4 were co-expressed and purified as a complex in order to maintain Sir4 integrity during purification.
Sir3 alone was incubated with the bead-conjugated wild-type, unmodified nucleosome array and the two were found to stably interact through washing of the beads (Figure 2A). Sir3 bound to the chromatin template at approximately a 1:2 (Sir3:nucleosome) ratio, though the reaction contained a two-fold molar excess of Sir3. Acetylation of the chromatin caused a dramatic inhibition of the Sir3-chromatin interaction. Consistent with previous studies (Liou 2005, Onishi 2007), we found that the substitution of H4K16 with alanine disrupted the Sir3-chromatin interaction (Figure 2A), indicating a direct role for H4K16 and its acetylation in controlling the association of Sir3 with the chromatin template.
Figure 2. Sir3 binding to chromatin requires unmodified H4K16.
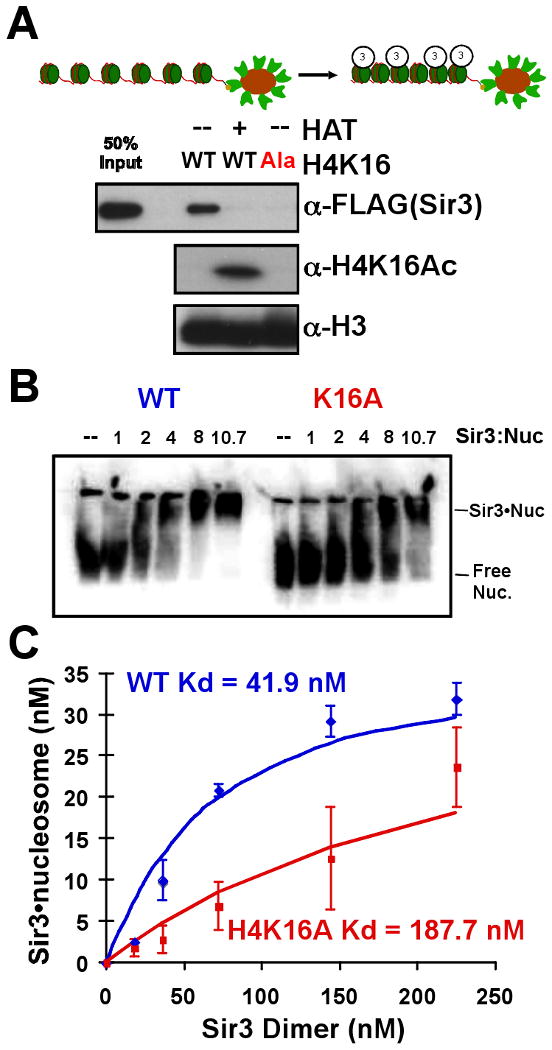
Sir3 was incubated with bead-conjugated chromatin, the beads were washed, and chromatin association was determined by Western blot analysis of the bead-bound fraction. (A) Sir3 alone was incubated with wild-type chromatin (WT) +/- histone acetyltransferase treatment (HAT) or chromatin containing a H4K16 to alanine mutant (H4K16:Ala). (B) Sir3 association with a 32P-labeled mononucleosome was tested by gel shift assay. Nucleosomes with either wild-type (left) or H4K16A (right) histones were pre-incubated with Sir3 (ratio of Sir3 monomer: nucleosome shown) and subsequently run on a native polyacrylamide gel. The gel was dried and visualized by storage phosphor screen. (C) Quantification of bound vs. free nucleosome was used to determine an apparent Kd value for a mononucleosome and a dimer of Sir3.
The above experiment, together with the demonstrated specific affinity of Sir3 for H4 peptides that contain a deacetylated lysine 16 (Liou et al., 2005), eliminate the possibility that H4K16 affects Sir3 binding indirectly through effects on other histone modifications. Nonetheless, it remains possible that altering H4K16 also changes the conformation of the chromatin such that Sir3 cannot bind, because acetylation and mutation of H4K16 is known to affect chromatin structure (Dorigo et al., 2003; Shogren-Knaak et al., 2006). To address this possibility we tested the interaction between Sir3 and a mononucleosome, avoiding the effects of higher-order chromatin structure. Mononucleosomes were reconstituted by salt dialysis onto a DNA fragment bearing the 601 nucleosome positioning sequence (Li and Widom, 2004). A gel shift assay was used to compare the ability of Sir3 to complex with wild-type and mutant nucleosomes. When a mononucleosome reconstituted with H4K16A was used in the assay, Sir3 associated with the nucleosome with 4.5-fold less affinity, indicating a direct role for H4K16 in Sir3 binding to the nucleosome (Figure 2B and C).
Sir2/4 bridges Sir3 to chromatin
Next, we tested the ability of the Sir2/4 complex to interact stably with chromatin. Sir2/4 bound efficiently to the wild-type chromatin (Figure 3A). Surprisingly, modification by acetylation or H4K16 mutation had no observable effect on the interaction of Sir2/4 with the nucleosome array, suggesting that factors other than H4K16 acetylation regulate Sir2/Sir4 chromatin interactions. Moreover, when Sir3 and Sir2/4 were pre-assembled into the SIR complex (Sir2/3/4) all three proteins stably associated with chromatin, irrespective of acetylation or H4K16A mutation (Figure 3B). Sir2/4 promoted approximately twice as much Sir3 association with wild-type chromatin as Sir3 alone, leading to a ratio of ∼2:1:2 (Sir3:Sir2/4:nucleosome). We presumed that the association of Sir3 with modified chromatin was mediated through the interaction between Sir3 and Sir4, which involves a coiled-coil region on Sir4 and the extreme C-terminus of Sir3 (Chang et al., 2003; Moazed et al., 1997; Rudner et al., 2005). To test this, Sir2/4 complex containing an I1311N mutation in Sir4 was purified. This mutation was previously shown to disrupt Sir3-Sir4 co-immunoprecipitation and Sir3 recruitment to silent loci (Rudner et al., 2005). The mutant Sir2/4 complex was not able to recruit Sir3 to the H4K16A chromatin template (Figure 3C), confirming that Sir4 serves to bridge Sir3 to modified chromatin. This is likely to reflect different modes of binding to acetylated versus unmodified chromatin during the initial recruitment and stable binding steps in silent chromatin assembly, respectively.
Figure 3. Sir2/4 activities on chromatin.
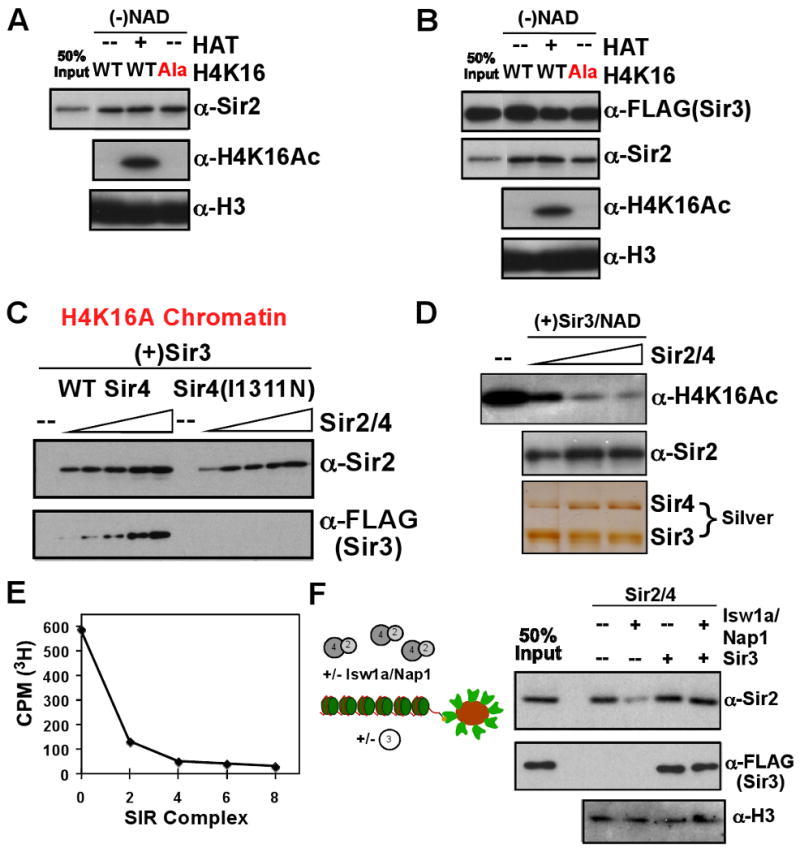
Sir2/4 (A) and the SIR complex (B) were used in chromatin association assays similar to those in Figure 2A, in the absence of NAD. (C) Titration of either wild-type or Sir4I1311N Sir2/4 subcomplex into a chromatin pull-down reaction containing a constant amount of Sir3 and H4K16A chromatin. (D) Sir2/4 was titrated into a reaction containing HAT-treated chromatin, Sir3, and NAD. The presence of Sir3 and Sir4 was determined by silver stain. (E) Bead-bound chromatin was acetylated in the presence of 3H-acetyl-CoA and subsequently treated with the SIR complex in the presence of NAD. Beads were washed and counted for 3H by liquid scintillation. (F) Sir2/4 binding to chromatin was assayed in the presence of the chromatin assembly components Isw1a and Nap1. The effect of addition of Sir3 was further tested.
Sir2 can fully deacetylate a chromatin template
In previous studies, yeast Sir2 deacetylation activity has not been observed on nucleosomal substrates, compared with acetylated histone peptides and free histones (Parsons et al., 2003; Tanny et al., 2004). We tested the ability of Sir2, in the context of the SIR complex, to deacetylate the chromatin assembled from recombinant histones and acetylated in vitro. When titrated into a reaction in the presence of NAD, Sir2/4 caused a near-complete loss of acetylation on H4K16 (Figure 3D). Although Sir2 has been shown previously to have some specificity for H4K16 on peptide substrates, we observed deacetylation of many lysine residues when using nucleosome arrays (data not shown). Monitoring overall deacetylation of 3H-acetyl-chromatin, Sir2 removed virtually all acetyl groups from the chromatin substrate, independently of Sir3 (Figures 3E,S3). Thus, consistent with the general state of deacetylation of silent chromatin in vivo (Suka et al., 2001), these results demonstrate that Sir2 can efficiently deacetylate nucleosomes in the absence of other co-factors.
Isw1a/Nap1 disrupts the association of Sir2/Sir4 with chromatin
The above results demonstrate that Sir2/4 association with chromatin is not sensitive to H4K16 mutation or acetylation. What then modulates Sir2/4 interaction with chromatin to prevent nonspecific silent chromatin assembly? In the course of this study we observed that the components of the chromatin assembly reaction, which have also been shown to have chromatin remodeling activity (Vary et al., 2003), could disrupt Sir2/4 association with chromatin when present during the chromatin binding reaction (Figure 3F). This effect was mainly through Nap1, though the presence of the Isw1a complex and ATP was necessary for maximal disruption of Sir2/4 chromatin association (data not shown). This observation suggests that the association of Sir2/Sir4 with chromatin is readily reversible in the presence of nucleosome-binding or remodeling complexes. In contrast, binding of Sir3 to the chromatin template prevented the disruption of the Sir2/4-chromatin interaction by the assembly factors (Figure 3F), suggesting that Sir3 can stably bind to unmodified chromatin in the presence of competing assembly components and stabilizes the association of Sir2/4 with chromatin.
Anti-silencing chromatin prevents Sir3 association
Certain chromatin marks are thought to prevent silencing proteins from binding to regions of the genome that should not be silenced. This “anti-silencing” has the added effect of directing the Sir proteins to the appropriate place in the genome, avoiding dilution of the limited pool of Sir proteins, which can disrupt silencing. The most important of the anti-silencing marks are the methylation of histone H3 lysine 79 (H3K79) (Ng et al., 2003; Ng et al., 2002; van Leeuwen et al., 2002) and the replacement of histone H2A with the histone variant Htz1 in the nucleosome (Meneghini et al., 2003; Zhang et al., 2004). We tested whether these anti-silencing chromatin modifications play a direct role in regulating the interactions of Sir proteins with chromatin.
We observed a disruption of Sir3 association with chromatin containing H3K79 mutated to alanine, similar to the disruption observed with chromatin containing the H4K16A mutation (Figure 4A). We found that Htz1-containing chromatin was also impaired for association with Sir3 (Figure 4A). Surprisingly, little difference was observed in the affinity of Sir3 for wild-type and H3K79A mononucleosomes (Figure 4B). The tight interaction between H4K16 may be a dominant influence by gel-shift, compared to using a nucleosome array.
Figure 4. Anti-silencing chromatin is refractory to Sir3 binding.
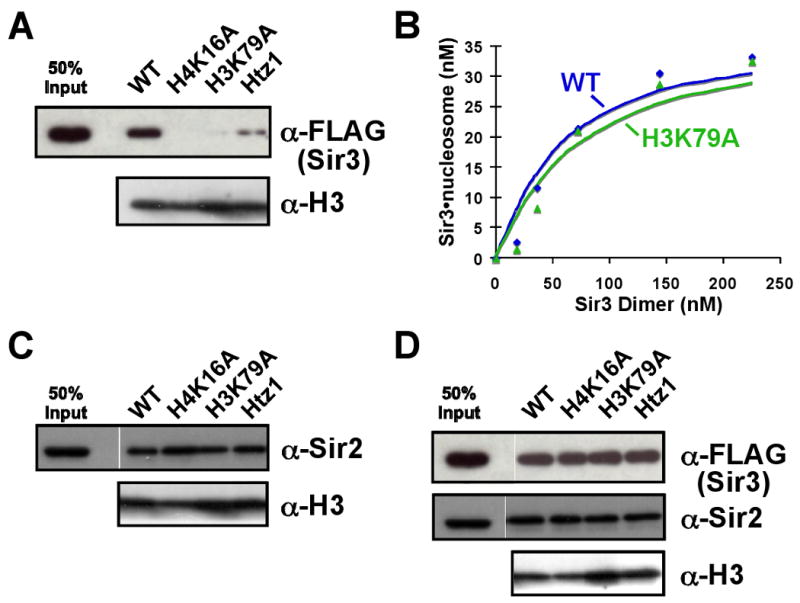
Magnetic bead pull-down experiments were performed similarly to those in Figure 2. Sir3 alone (A), Sir2/4 (C) or the SIR complex (D) were incubated with wild-type chromatin, or chromatin containing either H4K16A, H3K79A, or the histone variant Htz1 replacing histone H2A. (B) Quantification of a mononucleosome gel-shift assay with either a wild-type or H3K79A nucleosome and Sir3.
Sir2/4 alone was able to bind to H3K79A and Htz1 chromatin arrays with a similar affinity as to wild-type chromatin (Figure 4C). Moreover, similarly to the H4K16A chromatin, the presence of Sir2/4 in the chromatin pull-down assay promoted Sir3 association with these modified chromatin templates (Figure 4D). Thus, anti-silencing chromatin modifications function by directly disrupting the association of Sir3, but not Sir2/4, with chromatin.
In vitro-assembled silent chromatin prevents restriction enzyme access
We next wished to test whether the SIR complex–chromatin assembly displays characteristics of in vivo silent chromatin. A previous report demonstrated that the HMR region of silent chromatin is inaccessible to digestion by several restriction enzymes in isolated nuclei (Loo and Rine, 1994). We tested whether the chromatinized HMR template used this study (Figure 5A) was similarly affected by association of the SIR complex. Deacetylation-coupled SIR-chromatin assembly was treated with the BglII restriction enzyme (scheme in Figure 5B, left). A clear inhibition of BglII activity was observed in the presence of the SIR complex (Figure 5C, left panel). Using H4K16A chromatin, we observed ∼8-fold reduction in the ability of the SIR complex to inhibit BglII access (Figure 5C, right panel, and Figure 5D). The SIR complex produced similar effects in a kinetic analysis of BglII cleavage (Table S1). These results demonstrate that the in vitro-assembled SIR-chromatin complex displays accessibility properties that are similar to those described for silent chromatin domains in vivo and suggest that there is a difference in the structure of the SIR-chromatin assembly when Sir3 is unable to bind directly to chromatin.
Figure 5. NAD-dependent deacetylation by the SIR complex alters the structure of chromatin.
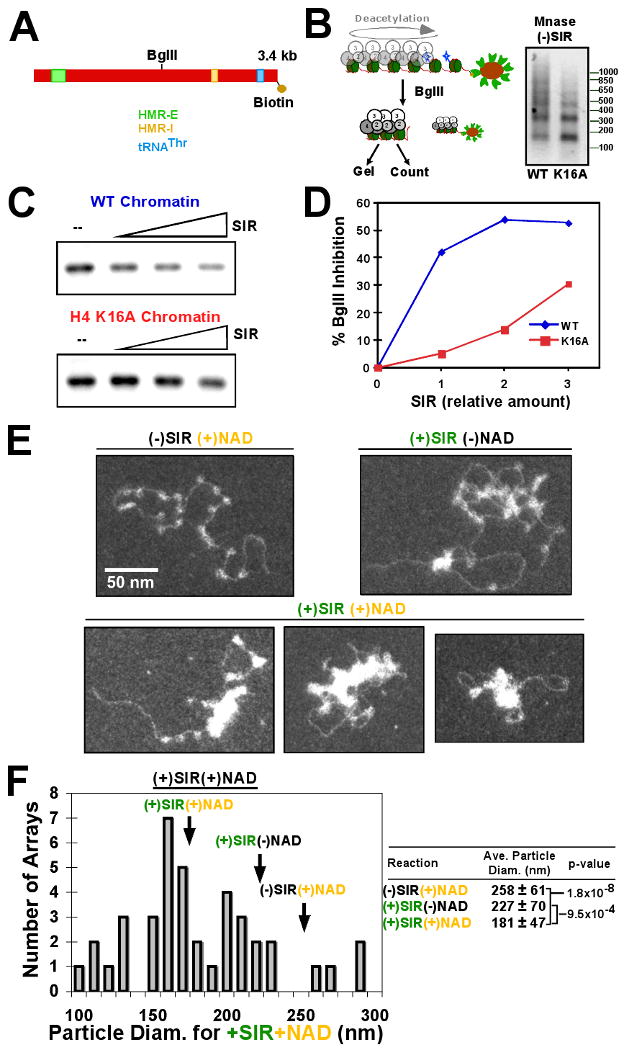
(A) Diagram of the HMR-containing DNA template and scheme (B) for this assay. (right) The fragment released from the beads by BglII cleavage was digested by micrococcal nuclease. (C) Acetylated wild-type or H4K16A chromatin was analyzed as in the scheme. Reactions were quenched with EDTA and either run on an agarose gel and imaged by storage phosphor screen (C) or analyzed by liquid scintillation counting (D). (E) Desthiobiotinylated-chromatin was acetylated on magnetic beads and eluted with free biotin. Acetylated chromatin was incubated with NAD only, SIR complex without NAD, or SIR complex with NAD and subsequently imaged by electron microscopy. (F) Histogram of particle diameters from (+)SIR(+)NAD sample. Arrows indicate average particle diameter of indicated samples. (right) Table containing average particle diameters from EM experiments with standard deviations and p-values from two-tailed T-test analysis as indicated.
A distinct structural transition is induced upon deacetylation-coupled association of the SIR complex
To further investigate the structural changes of chromatin upon SIR complex association, images of single chromatin particles were analyzed using electron microscopy (EM). Chromatin was assembled using a DNA template that was modified on one 5′ end with desthiobiotin, a modified form of biotin that can be eluted from streptavidin with free biotin (Hirsch et al., 2002). Bead-bound chromatin was acetylated and eluted from the resin with free biotin (see scheme in Figure S4A). Eluted chromatin samples were incubated with or without the SIR complex in the absence or presence of NAD and subsequently prepared for analysis by EM. Samples of chromatin alone displayed the expected “beads-on-a-string” appearance, corresponding to a spaced nucleosome array (Figure 5E). Upon addition of the SIR complex in the presence of NAD a significant structural change was observed. Chromatin particles in these samples no longer displayed individual nucleosomes, instead regions of higher density were observed with loops of free DNA emanating out (Figure 5E, lower panels). More than one class of chromatin particle was present in the samples containing NAD, including both highly compact particles and those that were more elongated. Some particles contained both compact clusters as well as a region with discernable free nucleosomes. In samples containing the SIR complex we also observed certain chromatin particles with considerable nucleosome-free regions, perhaps suggesting nucleosome movement (Figure S4B). In contrast to the samples with SIR complex and NAD, images of samples containing the SIR complex, but lacking NAD, contained a higher proportion of more open chromatin with more discontinuous regions of high density (Figure 5E, upper right panel).
Quantification of nucleosome array diameter (see Francis et al., 2004) demonstrated a highly significant shift to a smaller average diameter for samples containing SIR complex and NAD, compared to omitting either (Figure 5F). These results demonstrate that the SIR complex can alter the structure of chromatin and suggests that NAD-dependent deacetylation, which allows direct Sir3 association with chromatin, contributes to this transition.
In vitro-assembled silent chromatin represses RNA Polymerase III transcription
The above results indicated that the SIR-chromatin complexes assembled in this system share basic properties of yeast heterochromatin; therefore, we wished to test whether this minimal assembly could repress transcription. A whole-cell yeast extract was prepared as previously described (Schultz, 1999) that was shown to have transcription activity. When the HMR fragment used in this study was incubated with the extract and radiolabeled NTPs, a band was observed at ∼85 nt in addition to a smaller product that was enriched upon longer incubations (Figure 6A). The length of this product was consistent with that of the tRNAThr gene included in this HMR fragment and the change in size is likely due to processing to the 72 nt mature form. Indeed, if a similar DNA that lacked the tRNA gene was used as a template in the reaction, no product of this size was observed (Figure 6A). Previous reports have indicated that RNA polymerase III (Pol III) genes can be silenced by neighboring SIR-dependent silent chromatin (Huang et al., 1997; Schnell and Rine, 1986), similar to RNA polymerase II genes.
Figure 6. The SIR complex inhibits RNA Polymerase III transcription on chromatin.
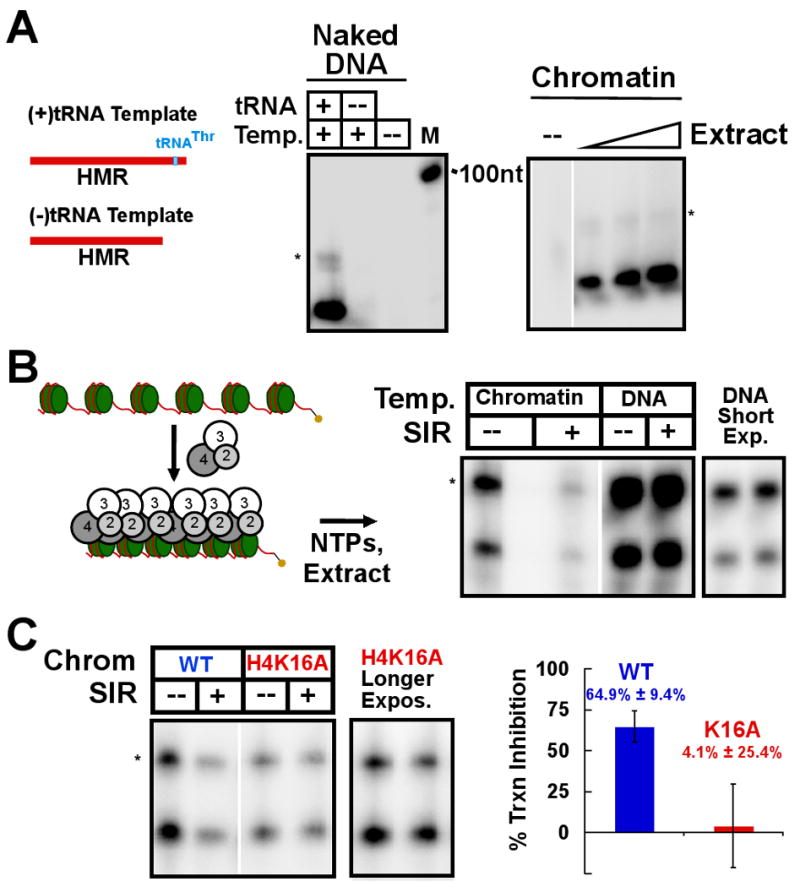
Transcription assays were performed using a whole-cell yeast extract enriched for transcription activity and RNA products were followed by α-32P-labeled UTP incorporation. Products were purified and separated on a 5 % acrylamide-urea-TBE gel and visualized on a storage phosphor screen. (A) The naked DNA template used in this study, a fragment of the HMR region including the flanking tRNA gene, was used in the transcription assay (left). Control reactions without template or with the DNA template truncated to remove the tRNA gene were also performed. (right) The chromatinized DNA was used as a template for extract-dependent transcription. (B) The SIR complex was pre-incubated with the chromatin template prior to initiation of transcription (scheme at left). Transcriptional repression of the SIR complex on the chromatin template was compared to an equal amount of the naked DNA template. The asterisk (*) indicates the unprocessed tRNA transcript. (C) Wild-type and H4K16A chromatin were compared in the ability to allow SIR-dependent repression of transcription. (right) Quantification by storage phosphor screen of three such experiments with standard deviations.
In vitro-assembled chromatin was also a suitable template for the transcription reaction from the extract (Figure 6A). Reconstituted chromatin was incubated with the SIR complex, followed by the addition of NTPs, an ATP regeneration system, and transcription extract. A clear repression of the transcription activity was observed upon pre-incubation of the SIR complex with the chromatin template (Figure 6B). This repression did not occur when either Sir2/4 or Sir3 alone were used in the assay (Figure S5A), but a synergistic effect was observed as Sir2/4 was titrated into a reaction containing a constant amount of Sir3 (Figure S5B). Thus, the Sir2/4 complex contributes to transcriptional gene silencing beyond histone deacetylation. The SIR complex did not inhibit transcription from a naked DNA template, suggesting that specific SIR-chromatin contacts were required for the repression of transcription (Figure 6B).
To more definitively determine whether specific SIR-chromatin complex formation mediated transcriptional silencing in this system, we tested whether the SIR complex was able to inhibit transcription from a chromatin template bearing the H4K16 to alanine mutation. The H4K16A chromatin template did not produce as much product as wild-type chromatin, but transcription was approximately 20-fold above background, allowing a reasonable measurement. Transcription from wild-type chromatin was repressed ∼65% by association with the SIR complex. In contrast, transcriptional repression with H4K16A chromatin was only ∼4% (Figure 6C). These results suggest that the SIR complex can block the RNA polymerase III transcription machinery from acting on the chromatin and removal of a critical contact such as H4K16 prevents the action of the SIR complex.
NAD-dependent Silencing of Activator-dependent RNA Polymerase II Transcription
The a1 and a2 genes included on the HMR-containing chromatin template that we used in this study were not transcribed in our in vitro reactions (Figure 6). We therefore used a well-studied template with a CYC1 promoter and activator sites upstream of a G-less cassette (Figure 7A) that yields three RNase T1-resistant transcripts (for details see Sawadogo and Roeder, 1985; Woontner et al., 1991). Gal4-VP16 activator-dependent transcription from this template is known to be dependent primarily upon the RNA Polymerase II machinery (Keogh et al., 2002).
Figure 7. The SIR complex represses activator-dependent RNA Polymerase II transcription.
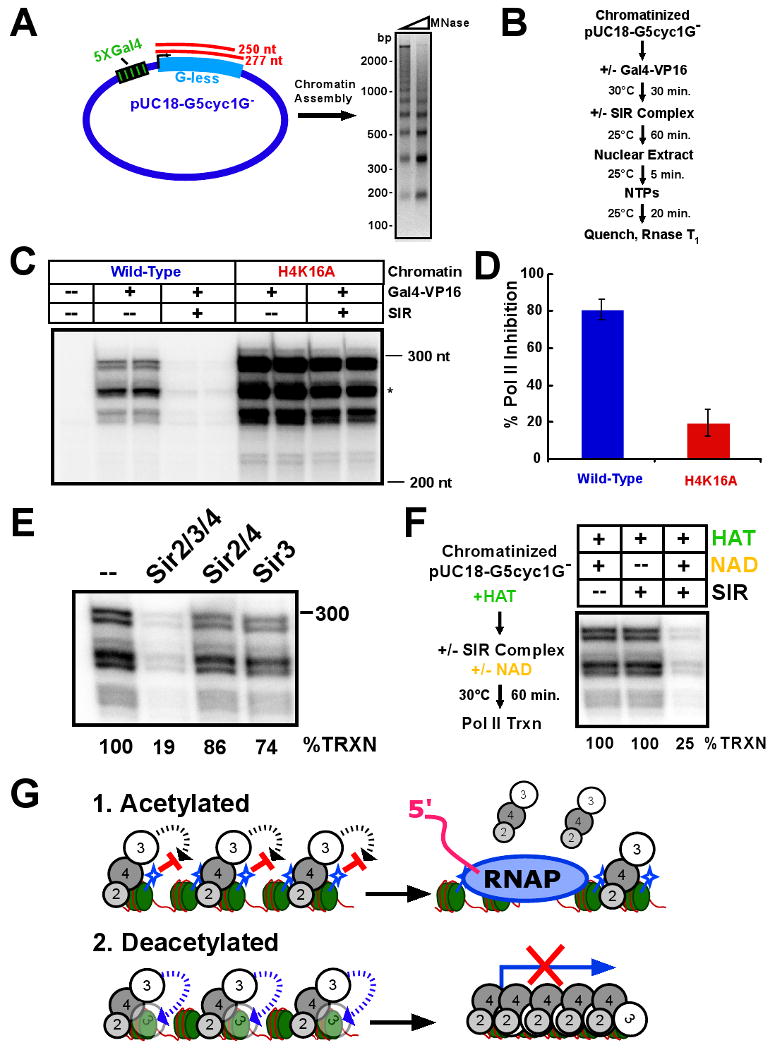
(A) Diagram of a plasmid template bearing an array of Gal4 binding sites upstream of a CYC1 promoter-driven G-less cassette with two expected transcripts, 250 and 277 nt in length (left). The plasmid was assembled into chromatin and analyzed as in Figure 1B for chromatinization of the DNA by limited micrococcal nuclease digestion. (B) Scheme of the activator-dependent transcription experiment. (C) Chromatinized G-less cassette plasmid, assembled with either wild-type or H4K16A histone octamer, was incubated with or without the SIR complex before initiation of transcription. RNase T1-treated products from duplicate reactions were resolved by denaturing PAGE. Migration positions of 300 and 200 nt RNA markers is shown. The asterisk marks the larger of the two expected transcripts. The topmost doublet band was most likely a read-through transcription product from the G-less region. (D) Quantification of transcription inhibition by the SIR complex on either wild-type or H4K16A templates by storage phosphor screen analysis. Shown are the averages of three experiments with standard deviations. (E) Individual components of the SIR complex were tested for Pol II repression as in (C). (F) Chromatinized G-less cassette plasmid was acetylated prior to SIR complex incubation performed in the absence or presence of NAD (scheme at left). Transcription assays were performed and products analyzed as above (right). (G) Model for transcriptional silencing by Sir3 lockdown onto chromatin, mediated by Sir4 and deacetylated H4K16. 1. Acetylated chromatin allows Sir2/4 binding, but prevents direct association of Sir3 with nucleosomes. This loosely associated SIR complex is permissive to transcription. 2. NAD-dependent deacetylation generates a high-affinity substrate for Sir3 lockdown onto chromatin and promotes an altered SIR-chromatin complex that is repressive to transcription.
When the chromatinized, Gal4-VP16-bound template was pre-incubated with the SIR complex (scheme in Figure 7B), approximately 5-fold repression of transcription was observed (Figure 7C and 7D). The template assembled into chromatin with the H4K16A mutation yielded significantly more transcript compared to wild-type chromatin; however, addition of the SIR complex produced only a 1.25-fold decrease in transcription. Neither the Sir2/4 subcomplex nor Sir3 alone could repress transcription comparable to the full SIR complex (1.16- and 1.35-fold repression, respectively)(Figure 7E). Finally, we tested the ability of the SIR complex to silence Pol II transcription of an acetylated chromatin template. We found that only in the presence of NAD could the SIR complex repress transcription (Figure 7F). The effect was nearly equal to that observed on unmodified chromatin, even in the presence of HAT activity. These results demonstrate that the specific association of the SIR complex with deacetylated chromatin represses transcription and define a minimal system, containing chromatin templates assembled with the four core histones and the purified Sir2, Sir3, and Sir4 proteins, for heterochromatin-mediated gene silencing.
Discussion
Our study set out to address whether the SIR complex itself is sufficient to assemble a repressive state of chromatin that recapitulates the salient properties of silent chromatin, as established by extensive previous studies using in vivo approaches. Taken together, our results define a minimal system that displays key features of silent chromatin. In this in vitro system, the ability of the Sir3 protein to associate with a chromatin template is regulated by the acetylation state of H4K16 and is inhibited by well-established anti-silencing regulators. We further demonstrate that the Sir2/4 sub-complex recruits Sir3 to a chromatin template, even when Sir3 cannot bind to the template directly. Additionally, the Sir2/4 subcomplex can efficiently deacetylate nucleosomal histones in an NAD-dependent manner in the absence of any added co-factors. This activity promotes a structural transition of the chromatin leading to a dense particle with a continuous cluster of nucleosomes. Finally, we demonstrate that the SIR-chromatin assembly reduces accessibility to restriction enzyme digestion and silences transcription. The silencing activities of the SIR complex require a specific mode of association between the full SIR complex and its target chromatin that is regulated by H4K16. Furthermore, the requirement for all three Sir proteins establishes a clear role for nucleosome binding as well as histone deacetylation in silencing.
Sir3 interactions with chromatin dictate specificity of silencing
Our experiments suggest that Sir3 dictates the specificity of direct and functional interactions between the SIR complex and chromatin. We observed that Sir3 association with chromatin was disrupted by acetylation or mutation of critical histone residues that have previously been implicated in assembly of silent chromatin (Figures 2 and 4). The requirement for H4K16 to remain unacetylated for Sir3 to associate with nucleosomes was clear from the nucleosome array template experiments and was further supported by loss of Sir3 binding to the nucleosome array and mononucleosome when H4K16 was substituted with alanine, mimicking the acetylated state. In principle, multiple factors may contribute to the effect of H4K16 mutation on Sir3 interaction with the nucleosome array, but the mononucleosome-binding results presented here and previous binding experiments with histone peptides demonstrate a direct role for H4K16 in the association of Sir3 with the nucleosome (Liou et al., 2005) and (Figure 2B,C).
Previous studies have demonstrated that H4K16 is also important for higher-order chromatin folding and must remain accessible for optimal chromatin dynamics, such as compaction of a nucleosome array (Dorigo et al., 2003; Shogren-Knaak et al., 2006). The effect of acetylation or mutation of this residue on these higher-order dynamics may also contribute to disruption of a stable association between Sir3 and the nucleosome array, perhaps preventing Sir3-mediated nucleosome packing. The exact nature of Sir3-nucleosome interaction will require future structural studies. Nonetheless, our results provide unequivocal support for the idea that post-translational modifications, involving the acetylation of a single lysine side chain, can dramatically reduce the association of an effector protein with chromatin templates.
We also demonstrate in this study the effect of anti-silencing marks on Sir3 interaction with chromatin (Figure 4A). Both H3K79 methylation and Htz1 incorporation into chromatin are thought to be involved in preventing Sir3 from binding to regions of the genome that are not targeted for silencing. Surprisingly, the H3K79A mutation did not have a strong effect on Sir3 binding to a mononucleosome compared to the nucleosome array (Figure 4B). This may be due to the mechanistic differences of Sir3 binding to a single nucleosome versus an array of nucleosomes and may be related to previous observations of non-specific associations between the SIR complex and DNA or chromatin (Georgel et al., 2001; Martino et al., 2009). Our study also provides direct evidence that incorporation of Htz1 by replacement of histone H2A generates an altered nucleosome that is no longer conducive to Sir3 association (Figure 4A). Structural studies have demonstrated that the H2A.Z variant, for which Htz1 is the S. cerevisiae member, alters the nucleosome surface charge in a way that would likely promote the tighter association of the H4 tail of one nucleosome with its neighbor (Fan et al., 2002; Suto et al., 2000). Perhaps the Htz1-conataining chromatin does not present the H4 tail for Sir3 binding, thus preventing a strong association.
Deacetylation of chromatin by Sir2
Previous studies on Sir2 and other members of the Sirtuin family suggested that these proteins were unable to deacetylate histones in a nucleosomal substrate (Parsons et al., 2003; Tanny et al., 2004). It was unclear whether the native histones used in such experiments were inhibitory to Sir2 activity or whether another factor, such as a chromatin remodeling complex, was necessary for deacetylation. We provide evidence for the ability of Sir2, within the Sir2/4 complex, to efficiently and directly deacetylate nucleosomal arrays, in the presence or absence of Sir3 (Figure 3D-E, and S3). In contrast to previous studies, which used chromatin prepared using acid-extracted Drosophila or mammalian histones, the chromatin arrays in our experiments were prepared with yeast histones produced in E. coli that lack post-translational modifications, which we believe may explain why they can be readily deacetylated. Future studies will be necessary to determine which modification or modifications interferes with Sir2 activity.
Sir2/4 can bridge Sir3 to chromatin—a glimpse at silent chromatin initiation
Models for silent chromatin initiation include steps that precede SIR complex association with chromatin where the silencer-binding proteins recruit the SIR complex to a target locus. Although our study does not use any sequence-specific DNA binding proteins, we observed that Sir2/4 can bypass the requirement for these factors and bind to acetylated chromatin in the absence of NAD and facilitate Sir3 association with chromatin. In this case Sir3 associates with the chromatin indirectly, through interaction with the coiled-coil domain of Sir4 (Figure 3C). This mode of interaction of the SIR complex with chromatin was observed in in vivo studies using a Sir2 activity mutant that was unable to create a deacetylated histone tail platform for Sir3 to bind (Hoppe et al., 2002; Rudner et al., 2005; Rusche et al., 2002). The fact that Sir3 is only recruited to acetylated chromatin via Sir4 and that Sir2 can deacetylate the template, facilitating direct binding of Sir3 to chromatin, implies that our system recapitulates the iterative mechanism of SIR complex assembly along the chromatin fiber.
What regulates the interaction between Sir2/4 and chromatin? Certainly DNA-binding proteins such as Rap1, Abf1, and the ORC complex will recruit Sir2/4 to target loci, leading to enrichment at these regions. The dosage of Sir4 is critical for the proper establishment of silencing (Sussel et al., 1993), as over-expression of Sir4 leads to a loss of silencing at telomeres and the mating type loci. Our observation that the Sir2/4 complex could associate with chromatin non-specifically may provide an explanation for the dominant negative effect of Sir4 over-expression, as this may lead to random recruitment of limiting amounts of Sir3 away from target loci. In addition, we observed that the chromatin assembly components could disrupt Sir2/4 interaction with chromatin and Sir3 prevented this disruption, allowing the SIR complex to bind chromatin (Figure 3F). Such displacement may provide another layer of control that would prevent non-specific nucleation of silent chromatin.
Structural changes upon SIR complex association with chromatin
The EM analysis of SIR-chromatin complexes described in this work highlights the general features of the effect of the SIR complex on chromatin. It is clear that the SIR complex targets nucleosomes while intervening DNA is looped out of the core. This feature is consistent with analysis of heterochromatin in purified yeast nuclei demonstrating that linker DNA within silent chromatin is accessible to micrococcal nuclease and DNase I (Nasmyth, 1982; Ravindra et al., 1999; Weiss and Simpson, 1998). We observed a subset of particles in the samples containing SIR and NAD that were rod-like in appearance, seeming to project density along an axis. It is possible that these structures are of particular significance as a reflection of directional spreading of the SIR complex along the chromatin fiber. Previous EM studies using purified Sir proteins and native chromatin fragments reported similar fiber-like SIR-nucleosome structures (Onishi et al., 2007). Under conditions of higher SIR:nucleosome ratios using the templates described in this report, highly-uniform fiber-like structures were also observed (unpublished results, A.J., C.W., and D.M.), though the degree of specificity of these structures is not yet evident, requiring future investigations.
Requirements for the transition to silent chromatin
Our results show that chromatin-mediated repression of transcription requires the full SIR complex. Although it is clear that binding of the Sir3 protein to chromatin can inhibit histone methylation by Dot1 (Altaf et al., 2007; Fingerman et al., 2007) and formation of recombination intermediates by Rad51 (M. Sinha and C. Peterson, submitted), transcriptional silencing in vitro evidently requires the association of all three Sir proteins with chromatin in conjunction. We observed that the association of the full SIR complex with an acetylated chromatin template, or the association of Sir3 with deacetylated chromatin in the absence of Sir2/4, could not efficiently repress transcription, indicating that the direct or indirect association of Sir3 with chromatin is not sufficient to repress transcription without proper coordination by Sir4 and direct binding of Sir3 to deacetylated H4K16 (Figure 7G). Together with previous chromatin immunoprecipitation experiments that uncouple SIR complex binding from silencing in vivo (Xu et al., 2007; Yang et al., 2008), our results suggest that transcriptional gene silencing requires the formation of structures in which the silencing complex is locked down on its chromatin template via interactions with specific histone residues.
Experimental Procedures
Proteins and DNA Templates
See Supplemental Data for details.
Chromatin Assembly
Chromatin assembly reactions were performed as described previously (Vary et al., 2004) with modifications. Histone octamer, Nap1, Isw1a, and DNA were incubated at 30°C for 5 hr. Chromatin was subsequently conjugated to magnetic beads for 1hr at room temperature. To generate an acetylated nucleosome template, bead-conjugated chromatin was incubated with an equal amount of Piccolo HAT complex v7 (a gift from B. Hnatkovich and S. Tan) for 1 hr at 30°C.
Chromatin Association Assays
Sir protein association with magnetic bead-conjugated chromatin was assayed by co-precipitation by magnetic concentration. Sir3 (1.05 pmol) and the Sir2/Sir4 (Sir2/4) subcomplex (340 fmol) were preincubated on ice in experiments where the SIR complex was used. Sir proteins were mixed with bead-conjugated chromatin (60 ng DNA, ∼ 480 fmol nucleosomes, assuming 18 nucleosomes per ∼3.4 kb DNA fragment) in 20 μL. Reactions were incubated at room temperature with rotation for 10 min followed by magnetic concentration and one wash in 200 μL reaction buffer. Proteins were stripped from beads in SDS, separated by SDS-PAGE, and subjected to standard Western analysis as described previously (Onishi et al., 2007).
Deacetylation reactions were performed as above with acetylated chromatin in the presence of 0.2 mM NAD. Reactions were incubated for 1 hr. at 30°C with rotation then at 4°C for 1 hr.
Mononucleosome Binding Assays
Mononucleosome binding assays were performed by incubating reconstituted mononucleosome (36 nM) with increasing amounts of Sir3 protein. The binding reaction was incubated at 30°C for 1 hour and resolved by native acrylamide gel. The amount of Sir3-nucleosome complex was determined by the difference in free nucleosome for each sample. The data were fit to determine an apparent Kd using a simple model (A + B → AB, where A refers to the mononucleosome, B refers to a Sir3 dimer) analyzed by the Kaleidograph software (Synergy Software).
Restriction Enzyme Protection
Chromatin was assembled with the biotinylated HMR PCR product produced in a PCR reaction with α-32P-labeled dCTP. The chromatin was conjugated to magnetic beads and acetylated as above. Bead-conjugated acetylated wild-type or H4K16A mutant chromatin (100 ng DNA, ∼800 fmol nucleosomes) was incubated with an increasing amount of the SIR complex (Sir3 and Sir2/4 pre-incubated at a ratio of 3.3:1 and titrated as 170, 340, and 510 fmol Sir2/4) in 50 μL at room temperature for 45 min with rotation. Subsequently, reactions were initiated with 250 U BglII restriction enzyme (NEB) and incubated at 31°C with rotation for 20 hr. A 25 μL aliquot was removed and quenched with 3 μL 0.5 M EDTA, the beads separated, the sample split and either counted by liquid scintillation or deproteinated and separated by agarose gel and visualized by storage phosphor screen.
Electron Microscopy
Desthiobiotin-labeled, bead-bound chromatin was assembled, acetylated and eluted (see Supplemental Data for details). Chromatin samples were incubated for 1 hr with or without SIR complex and NAD and subsequently fixed in glutaraldehyde. Preparation of samples for electron microscopy was essentially as described (Nikitina et al., 2007). See Supplemental Data for details.
RNA Polymerase III Transcription Assays
A whole-cell yeast extract was prepared as described previously (Schultz, 1999) using the protease-deficient strain SF10 (Hoppe et al., 2002). Transcription assays were performed in a final volume of 25 μL. Either 90 ng of naked biotinylated-HMR PCR product or a 90 ng (DNA) aliquot of a chromatin assembly reaction (∼720 fmol nucleosomes) as described above was used as template and pre-incubated with the SIR complex (950 fmol Sir2/4 and 3.5 pmol Sir3), when present, for 1 hr at room temperature. The NTPs were added on ice, and the reaction was initiated with 2μL (80 μg) of whole cell extract. Reactions were quenched after 10 min for reactions in panels 5B-C or 1 hr. for panel 5A. Quenched reactions were purified and separated on urea-polyacrylamide gel, dried, and analyzed by storage phosphor screen as above.
RNA Polymerase II Transcription Assays
A nuclear extract was prepared essentially as described previously (Ponticelli and Struhl, 1990; Ranish et al., 1999)(see Supplemental Data). Chromatin assembly reactions were performed with plasmid pUC18-G5cyc1G- and the template was incubated with HAT complex or Gal4-VP16, as indicated. SIR complex (950 fmol Sir2/4 and 3.5 pmol Sir3) was pre-incubated with the chromatin template (100 ng DNA, 800 fmol nucleosomes) for 1 hr at room temperature and then placed on ice. A 10× mixture of magnesium acetate, RNase inhibitor, phosphocreatine, and creatine kinase was added. This mixture was pre-incubated at room temperature with 320 μg nuclear extract, NTPs were added and the reaction quenched after 20 min (30 μL final volume). Reactions were processed and analyzed as described in Supplemental Data.
Supplementary Material
Acknowledgments
We thank Tim Richmond for histone plasmids, Brian Hnatkovich and Song Tan for purified Piccolo acetyltransferase complex, Eun-Jung Cho for construction of plasmid pUC18-G5cyc1G-, Toshio Tsukiyama for advice on chromatin assembly, Cecilia Centrella for technical assistance, and Megumi Onishi and Karim Mekhail for comments on the manuscript. This work was supported by a Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship through the National Institute of General Medical Sciences, F32 GM078799 (A.J.), a National Defense Science and Engineering Graduate Fellowship from the Department of Defense (T.W.S), and grants from the National Institutes of Health (GM061641 to D.M. and GM46498 to S.B.). D.M. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger JR, Onishi M, Li G, Seebacher J, Rudner AD, Gygi SP, Moazed D. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28:6903–6918. doi: 10.1128/MCB.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JF, Hall BE, Tanny JC, Moazed D, Filman D, Ellenberger T. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure. 2003;11:637–649. doi: 10.1016/s0969-2126(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Chou CC, Li YC, Gartenberg MR. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell. 2008;31:650–659. doi: 10.1016/j.molcel.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 2007;21:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Georgel PT, Palacios DeBeer MA, Pietz G, Fox CA, Hansen JC. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc Natl Acad Sci U S A. 2001;98:8584–8589. doi: 10.1073/pnas.151258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci U S A. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal Biochem. 2002;308:343–357. doi: 10.1016/s0003-2697(02)00201-4. [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kahana A, Gottschling DE, Prakash L, Liebman SW. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6693–6699. doi: 10.1128/mcb.17.11.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Cho EJ, Podolny V, Buratowski S. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol Cell Biol. 2002;22:1288–1297. doi: 10.1128/mcb.22.5.1288-1297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci U S A. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Nasmyth KA. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982;30:567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci U S A. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Parsons XH, Garcia SN, Pillus L, Kadonaga JT. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc Natl Acad Sci U S A. 2003;100:1609–1614. doi: 10.1073/pnas.0434064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Struhl K. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol Cell Biol. 1990;10:2832–2839. doi: 10.1128/mcb.10.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra A, Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol Cell Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Sawadogo M, Roeder RG. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci U S A. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC. Chromatin assembly in yeast cell-free extracts. Methods. 1999;17:161–172. doi: 10.1006/meth.1998.0727. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Sussel L, Vannier D, Shore D. Epigenetic switching of transcriptional states: cis-and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3919–3928. doi: 10.1128/mcb.13.7.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol Cell Biol. 2004;24:6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Vary JC, Jr, Fazzio TG, Tsukiyama T. Assembly of yeast chromatin using ISWI complexes. Methods Enzymol. 2004;375:88–102. doi: 10.1016/s0076-6879(03)75006-x. [DOI] [PubMed] [Google Scholar]
- Vary JC, Jr, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woontner M, Wade PA, Bonner J, Jaehning JA. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Britton J, Kirchmaier AL. Insights into the impact of histone acetylation and methylation on Sir protein recruitment, spreading, and silencing in Saccharomyces cerevisiae. J Mol Biol. 2008;381:826–844. doi: 10.1016/j.jmb.2008.06.059. [DOI] [PubMed] [Google Scholar]
- Yang B, Kirchmaier AL. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:5287–5297. doi: 10.1091/mbc.E06-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


