Summary
Genetic alterations that promote chromosome missegregation have been proposed to drive tumorigenesis through loss of whole chromosomes containing key tumor suppressor genes. To test this unproven idea, we bred Bub1 mutant mice that inaccurately segregate their chromosomes onto p53+/−, ApcMin/+, Rb+/− or Pten+/− backgrounds. Bub1 insufficiency predisposed p53+/− mice to thymic lymphomas and ApcMin/+ mice to colonic tumors. These tumors consistently lacked the non-mutated tumor suppressor allele, but had gained a copy of the mutant allele. In contrast, Bub1 insufficiency had no impact on tumorigenesis in Rb+/− mice and inhibited prostatic intraepithelial neoplasia formation in Pten+/− mice. Thus, Bub1 insufficiency can drive tumor formation through tumor suppressor gene loss of heterozygosity, but only in restricted genetic and cellular contexts.
Introduction
Aneuploidy, an abnormal number of chromosomes, is a distinctive feature of most human cancers, however its role in neoplastic transformation is somewhat mysterious (Ricke et al., 2008). On the one hand, there is ample evidence that chromosome imbalances reduce cell proliferation and overall organismal fitness (Torres et al., 2008), arguing against a causal role for aneuploidy in malignant cell growth. On the other hand, some of the mouse models for aneuploidy that have been generated are prone to late-life tumors (Iwanaga et al., 2007; Jeganathan et al., 2007; Michel et al., 2001; Sotillo et al., 2007; Weaver et al., 2007), suggesting that aneuploidy has the ability to contribute to tumorigenesis. Although chromosomal instability studies in mice have provided some evidence for a causal link between aneuploidy and cancer, they also raised several key questions (Ricke et al., 2008). For instance, why do chromosomal instability genes that are active in a wide variety of tissues throughout the body promote tumorigenesis only in a few tissues when defective, and why do tumor prone tissues vary between different aneuploidy models? Also, why are some of the models not prone to tumors despite the presence of vast amounts of aneuploid cells, and why do some chromosomal instability gene defects exert tumor suppressive ability in certain genetic contexts? Clearly, the totality of currently available data suggest that the mechanisms by which whole chromosome instability gene defects affect tumor development are highly complex and depend on the particular gene that is defective and the severity of the defect.
One mechanism through which chromosomal instability is thought to promote tumorigenesis is by increasing loss of tumor suppressor genes (Michor et al., 2005). The central idea is that in instances where one copy of a particular tumor suppressor gene has suffered a mutation, the chromosome harboring the second, intact, gene copy is inactivated by a whole chromosome loss during an aberrant mitosis. One study examined this mechanism by crossing mice with only one functional copy of p53 or Rb onto a heterozygous-null genetic background for the mitotic checkpoint gene Bub3. However, no increase in tumorigenesis was observed in this analysis (Kalitsis et al., 2005). Another study found that colon tumor development increased when ApcMin/+ mice had only one copy of the mitotic checkpoint protein BubR1, but whether this increase was associated with accelerated LOH of the Apc+ locus is not known (Rao et al., 2005). The same holds true for a study of p53 and Mad2 double-heterozygous null mice (Chi et al., 2009). These mice showed increased lymphomagenesis, but whether this increase was linked to p53 LOH was not determined. It has also been proposed that chromosomal instability may not drive tumorigenesis by LOH of tumor suppressor genes (Weaver and Cleveland, 2007). This suggestion was based on the observation that aneuploidy resulting from Cenp-E haploinsufficiency delayed tumor formation when both copies of the p19Arf tumor suppressor gene are lacking.
Given the complex relationship between numerical chromosome segregation and tumorigenesis, it will be important to address the question as to whether inaccurate chromosome segregation drives tumorigenesis through tumor-suppressor gene LOH in a comprehensive fashion. The mitotic checkpoint protein Bub1, which is mutated or expressed at low levels in a subset of human cancers (Yuen et al., 2005), has been causally implicated in cancer (Jeganathan et al., 2007; Schliekelman et al., 2009). Mice in which expression of Bub1 is gradually reduced using various combinations of wild-type, hypomorphic and knockout Bub1 alleles, develop spontaneous tumors with reduced latency and increased incidence in a dose-dependent fashion (Jeganathan et al., 2007). We reasoned that using this series of mice in combination with different heterozygous tumor suppressor gene mutant mice, would not only allow for comprehensive analysis of the issue as to whether aneuploidy can promote tumorigenesis through tumor suppressor gene LOH, but also help us understand how the degree of chromosomal instability impacts this mechanism. The tumor suppressor genes that we selected are p53, Rb, Apc and Pten, which are located on mouse chromosomes 11, 14, 18 and 19, respectively.
Results
Bub1 insufficiency causes thymic lymphomagenesis in p53+/− mice
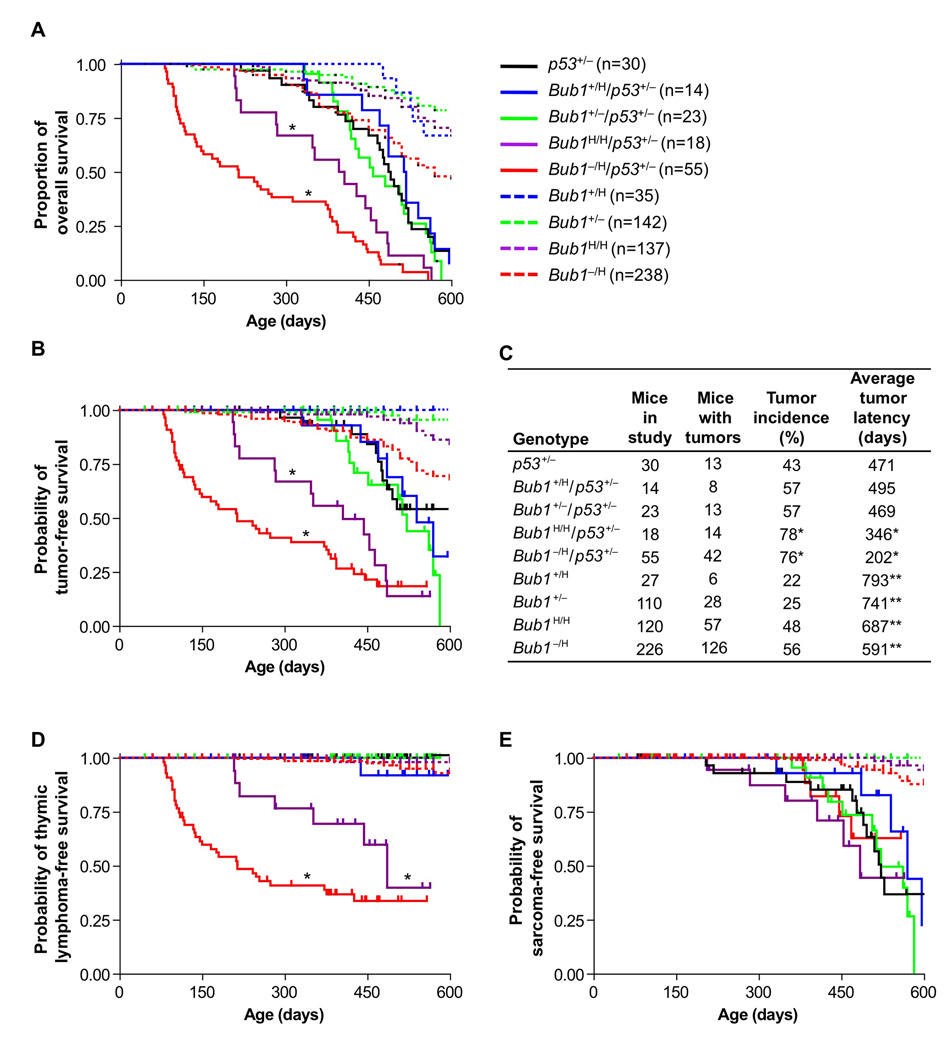
Mice with two inactivated copies of p53 typically develop lethal thymic lymphomas within the first 9 months of life (Jacks et al., 1994). p53 heterozygous null mice are also prone to tumors, primarily sarcomas. However, these tumors rarely surface within the first year of life. The majority of tumors in p53+/− mice display loss of heterozygosity of the remaining wild-type allele, suggesting that a critical barrier to neoplastic growth is established by retention of this wild-type allele. To determine whether chromosomal instability can decrease the tumor latency in p53+/− mice by accelerating loss of the wild-type p53 allele, we used a mouse series with graded reduction in Bub1 expression. We have previously shown that Bub1 expression inversely correlates with aneuploidy in mouse embryonic fibroblasts (MEFs) and mouse splenocytes (Jeganathan et al., 2007). Furthermore, chromosome counts on various kinds of cultured primary cells from Bub1 mutant mice, including vascular smooth muscle cells, lung fibroblasts, mammary gland epithelial cells and prostate cells, suggests that increased aneuploidy is likely to be a common consequence of Bub1 insufficiency (Figure S1). Through two rounds of breeding, we established cohorts of p53+/−, Bub1+/H/p53+/− (~75% of normal Bub1 levels), Bub1+/−/p53+/− (~50% Bub1), Bub1H/H/p53+/− (~30% Bub1) and Bub1−/H/p53+/− (~20% Bub1) mice, and monitored these mice for development of overt tumors or signs of ill health for up to 600 days. Bub1+/H/p53+/− and Bub1+/−/p53+/− mice showed similar survival as p53+/− mice. However, survival of Bub1H/H/p53+/− and Bub1−/H/p53+/− mice was markedly decreased, with the latter mice showing the most dramatic reduction (Figure 1A). Tumor-free survival curves revealed that decreased survival was due to tumor development (Figure 1B). The tumor incidence in Bub1H/H/p53+/− and Bub1−/H/p53+/− mice was significantly increased, while the average tumor latency was considerably decreased (Figure 1C). In contrast, no such changes were observed in Bub1+/−/p53+/− and Bub1+/H/p53+/− mice. Strikingly, Bub1H/H/p53+/− and Bub1−/H/p53+/− mice were almost exclusively predisposed to the development of thymic lymphoma, a tumor type that was not observed in p53+/− mice and only at low rates in Bub1H/H and Bub1−/H single mutant mice (Figure 1D). Thymic lymphomas arose significantly earlier in Bub1−/H/p53+/− mice than in Bub1H/H/p53+/− mice, with a mean time to tumor onset of 163 and 314 days, respectively (p = 0.0004). The most common tumors observed in p53+/− animals are sarcomas (Jacks et al., 1994), but the incidence and latency with which these tumors formed were unaffected by Bub1 protein levels (Figure 1E). These results indicate that the level of Bub1 needs to drop below a critical threshold in order to progressively promote tumorigenesis in p53 heterozygous mice, and that thymic T cells are particularly sensitive to this decline in Bub1.
Figure 1.
(A) Kaplan-Meier overall survival curves of p53+/−, Bub1+/H/p53+/−, Bub1+/−/p53+/−, Bub1H/H/p53+/−, Bub1−/H/p53+/−, Bub1+/H, Bub1+/−, Bub1H/H and Bub1−/H mice. *p < 0.05 compared to p53+/− mice (Log rank test). Mice in these cohorts were monitored for 600 days. Animals represented in the curves are mice that died or that had to be sacrificed due to ill health or overt tumors. Cohorts of Bub1+/−, Bub1H/H and Bub1−/H mice are as described (Jeganathan et al., 2007).
(B) Kaplan-Meier tumor-free survival curves of mice presented in (A). Animals that died without tumors were censored from the data. We note that the percentage of p53+/− animals dying without tumors was similar to that previously reported for this strain (Jacks et al., 1994). *p < 0.05 compared to p53+/− mice (Log rank test). Legend is as in (A).
(C) Tumor incidence and average tumor latency of mice shown in (A). *p < 0.05 versus p53+/− mice (Fisher’s exact test for incidence; Log rank test for latency). **These cohorts were monitored for 1000 days, instead of 600 days.
(D) Kaplan-Meier curves showing the thymic lymphoma-free survival of mice presented in (A). *p < 0.05 compared to p53+/− mice (Log rank test). Legend is as in (A).
(E) Same as (D) with sarcoma incidence plotted. Legend is as in (A).
We note that for groups heterozygous for p53, after 500 days no individual group had more than 10 animals at risk for developing neoplasia. The large steps that appear during late age are because individual tumors that occurred in these small sample size groups reflect larger proportional changes. Furthermore, note that all Bub1H/H/p53+/− and Bub1−/H/p53+/− mice in the study died before 600 days. Therefore, the curves for these animal groups in (B), (D) and (E) do not extend to 600 days. Tick marks indicate censored animals in curves plotted in (B), (D) and (E).
Low Bub1 promotes p53 LOH through whole chromosome missegregation
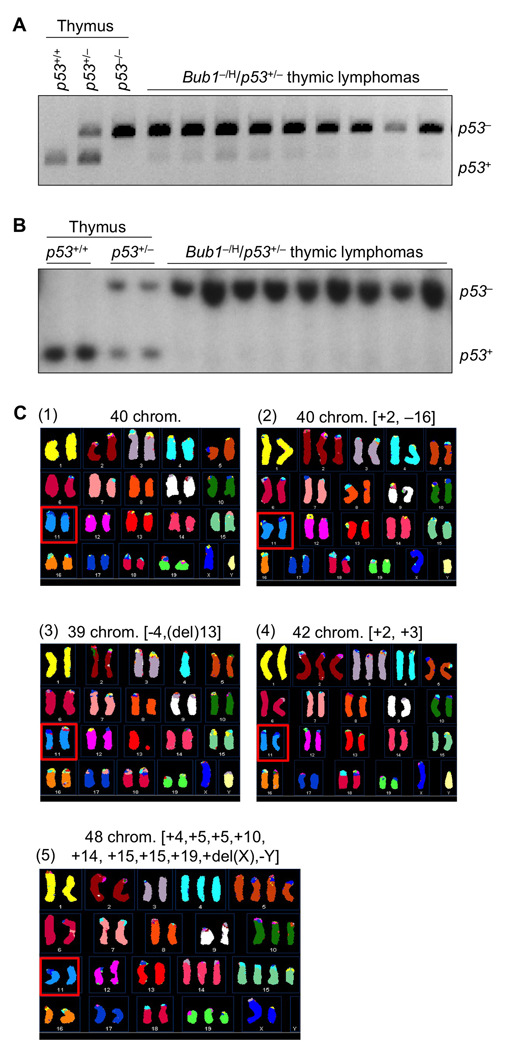
The tumor profile that we observed in Bub1−/H/p53+/− mice is similar to that of p53−/− mice (Jacks et al., 1994). A potential mechanism underlying this similarity might be that the high rate of chromosomal missegregation resulting from low Bub1 expression dramatically increases the frequency of loss of the chromosome containing the remaining wild-type p53 allele. To test this hypothesis, nine thymic lymphomas from Bub1−/H/p53+/− mice were screened for p53 status using a previously established PCR method (Jacks et al., 1994). In all instances, we found that the PCR fragment representing wild-type p53 was either highly underrepresented or completely absent (Figure 2A). Southern blot analysis of the same set of tumors confirmed the absence of wild-type p53 (Figure 2B), indicating that Bub1−/H/p53+/− thymocytes that develop into lethal tumors consistently lose wild-type p53. Thymic lymphomas from Bub1H/H/p53+/− animals also lacked wild-type p53 (data not shown), lending further support to the notion that p53 LOH is a requisite for tumorigenesis to occur in the thymus.
Figure 2. p53Bub1−/H/p53+/− and Bub1H/H/p53+/− mice.
(A) p53 status of lymphomas from Bub1−/H/p53+/− mice analyzed by PCR. Genomic DNA samples prepared from thymus of 6-week-old p53+/+, p53+/− and p53−/− mice were used as PCR controls.
(B) Analysis of the p53 status of lymphomas shown in (A) using Southern blotting as described (Jacks et al., 1994). Genomic DNA samples prepared from thymus of 6-week-old p53+/+ and p53+/− mice were used as controls.
(C) Spectral karyotype analysis of lymphoma cells harvested from a representative Bub1−/H/p53+/− mouse. Shown are examples of cells (from the same tumor) that: (1) are karyotypically normal; (2) have 40 chromosomes but are karyotypically abnormal; (3 and 4) have minor numerical abnormalities (upper right and lower left); or, (5) exhibit combined numerical and structural chromosome changes (lower right). About 10–11% of tumor cells of three independent lymphomas had structural chromosome changes, which is to be expected for cells that lack p53, a key regulator of genome maitenance. Note that all cells have two copies of chromosome 11 (see red squares).
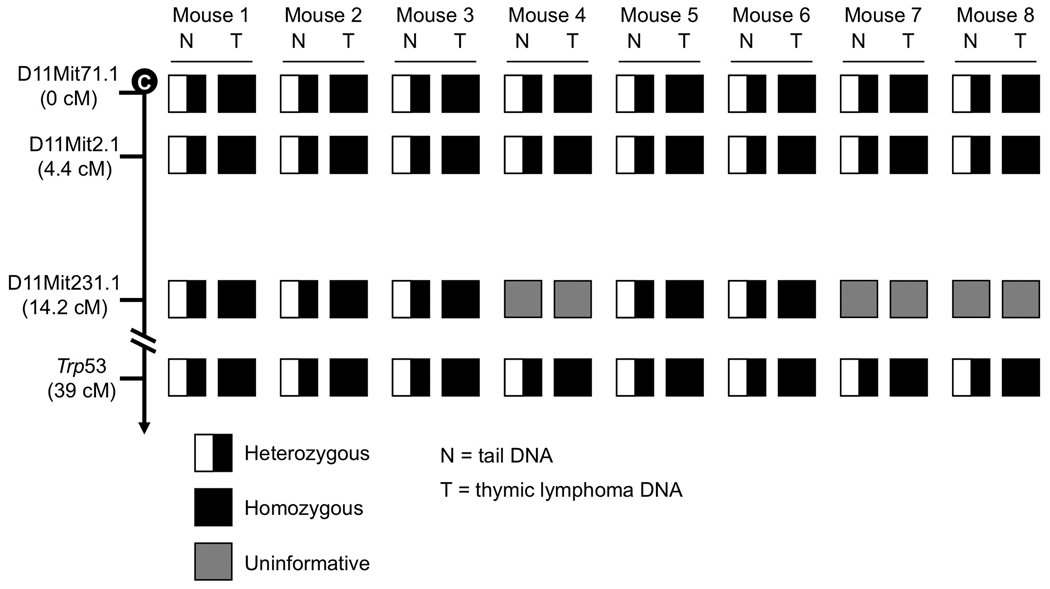
Given that Bub1 insufficiency causes chromosome missegregation (Jeganathan et al., 2007), the simplest explanation for the observed p53 LOH would be that the copy of chromosome 11 containing the wild-type p53 allele gets lost during an aberrant cell division. To test for this possibility, we performed spectral karyotype (SKY) analysis on three independent lymphomas from Bub1−/H/p53+/− mice. We found that all of the spreads examined had two chromosome 11 copies, instead of one (Figure 2C, panels 1–5). This, combined with our Southern data suggested that instead of simply losing chromosome 11 bearing wild-type p53, lymphoma cells had gained an extra copy of chromosome 11 harboring the knockout allele. This might have occurred in one step through non-disjunction of the mutant chromosome pair and coincidental loss of the chromosome carrying wild-type p53, or in two separate cell divisions: loss of chromosome 11 carrying wild-type p53 follow by a gain of chromosome 11 containing mutant p53, or vice versa. However, two alternative mechanisms that could explain the presence of two chromosome 11 copies containing the knockout allele are mitotic recombination and gene conversion. To determine whether these mechanisms might be involved, we performed simple sequence length polymorphism (SSLP) analysis on a series of thymic lymphomas from Bub1−/H/p53+/− mice. We selected six SSLP markers along the 0–36.1 cM region of chromosome 11 (Figure 3). Three of these markers were informative on normal tail DNA from eight lymphoma-positive Bub1−/H/p53+/− mice. Corresponding lymphoma DNA samples from these mice were then subjected to the same analysis. In all of these tumors, we observed LOH at all informative SSLP markers (Figure 3), indicating that whole chromosome missegregation rather than mitotic recombination was responsible for p53 LOH. Thus, mitotic recombination and gene conversion, two alternative mechanisms by which cells could become homozygous for an inactivate tumor suppressor gene, are unlikely to drive p53 LOH in our Bub1−/H/p53+/− model.
Figure 3. p53 LOH in lymphomas of Bub1−/H/p53+/− mice occurred by loss of chromosome 11 harboring the p53+ locus.
Haplotypes of lymphomas and matched tail (normal) tissue from Bub1−/H/p53+/− mice.
To determine the degree of aneuploidy in lymphomas of Bub1−/H/p53+/− mice, we performed chromosome counts on metaphase spreads of 6 independent thymic tumors. On average 56% of tumor cells were aneuploid, with most cells losing or gaining one or several chromosomes (near diploid) and some cells gaining more than 10 chromosomes (Table 1). SKY data confirmed that a substantial proportion of tumor cells had 40 chromosomes (Figure 2C, panels 1 and 2). Some of these cells had completely normal karyotypes (Figure 2C, panel 1), but according to our LOH analysis, these cells had to have two copies of the p53 knockout allele (Figure 2A and 2B). Other cells with 40 chromosomes had non-modal numbers of specific chromosomes (Figure 2C, panel 2), indicating that our chromosome counts on metaphase spreads underrated the incidence of aneuploidy. Thymic lymphomas from Bub1−/H/p53+/− had remarkably similar rates of aneuploidy as thymic lymphomas from p53−/− (Table 1), indicating that cells with low levels of Bub1 do not necessarily produce cancer cells with more profound aneuploidy. Chromosome counts on metaphase spreads from healthy 5-month-old p53+/− and Bub1−/H/p53+/− mice, however, demonstrated that Bub1 insufficiency strongly accelerates chromosome missegregation in non-transformed p53+/− cells (Table 1). We note that even though the percentage of aneuploid splenocytes was relatively high in Bub1−/H/p53+/− mice, it was considerably lower than that of Bub1−/H mice (19% versus 39%; Jeganathan et al., 2007). The reason for this is currently unclear. Collectively, the above data suggest that loss of chromosome 11 containing wild-type p53 followed by (or coinciding with) gain of chromosome 11 containing the knockout p53 allele is a key mechanism by which Bub1 insufficiency drives thymic lymphomagenesis in p53+/− mice.
Table 1.
Bub1 insufficiency promotes chromosome loss in p53 heterozygous splenocytes
| Mouse genotype |
Tumor/tissue | Mitotic figures inspected |
Percent aneuploid figures |
Karyotypes with indicated chromosome number | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 46 | >50 | ||||
| Bub1−/H/p53+/− | Thymic lymphoma 1 | 55 | 45 | 1 | 1 | 2 | 2 | 1 | 3 | 1 | 30 | 7 | 7 | ||||||
| Thymic lymphoma 2 | 50 | 54 | 1 | 2 | 6 | 9 | 23 | 2 | 1 | 6 | |||||||||
| Thymic lymphoma 3 | 52 | 65 | 1 | 1 | 2 | 2 | 1 | 7 | 4 | 18 | 6 | 2 | 1 | 7 | |||||
| Thymic lymphoma 4 | 98 | 51 | 2 | 2 | 1 | 2 | 3 | 4 | 13 | 48 | 20 | 3 | |||||||
| Thymic lymphoma 5 | 49 | 55 | 1 | 3 | 2 | 1 | 4 | 6 | 22 | 6 | 1 | 1 | 3 | ||||||
| Thymic lymphoma 6 | 54 | 63 | 2 | 1 | 1 | 3 | 6 | 12 | 20 | 3 | 3 | 1 | 2 | ||||||
| p53−/− | Thymic lymphoma 1 | 51 | 55 | 1 | 3 | 4 | 8 | 23 | 4 | 3 | 5 | ||||||||
| Thymic lymphoma 2 | 52 | 63 | 2 | 4 | 4 | 4 | 6 | 19 | 2 | 3 | 1 | 4 | |||||||
| Thymic lymphoma 3 | 55 | 62 | 1 | 1 | 3 | 1 | 7 | 10 | 21 | 1 | 1 | 1 | 8 | ||||||
| p53+/+ | Spleen 1 (5 mo) | 50 | 0 | 50 | |||||||||||||||
| Spleen 2 (5 mo) | 50 | 0 | 50 | ||||||||||||||||
| Spleen 3 (5 mo) | 50 | 0 | 50 | ||||||||||||||||
| p53+/− | Spleen 1 (5 mo) | 49 | 6 | 1 | 46 | 2 | |||||||||||||
| Spleen 2 (5 mo) | 38 | 11 | 3 | 34 | 1 | ||||||||||||||
| Spleen 3 (5 mo) | 19 | 6 | 1 | 18 | |||||||||||||||
| Bub1−/H/p53+/− | Spleen 1 (5 mo) | 18 | 17 | 1 | 1 | 1 | 15 | ||||||||||||
| Spleen 2 (5 mo) | 47 | 23 | 1 | 2 | 1 | 1 | 5 | 36 | 1 | ||||||||||
| Spleen 3 (5 mo) | 67 | 18 | 1 | 3 | 3 | 4 | 55 | 1 | |||||||||||
| p53+/+ | Testis 1 (5 mo) | 50 | 0 | 50 | |||||||||||||||
| Testis 2 (5 mo) | 50 | 0 | 50 | ||||||||||||||||
| Bub1−/H | Testis 1 (5 mo) | 50 | 0 | 50 | |||||||||||||||
| Testis 2 (5 mo) | 50 | 0 | 50 | ||||||||||||||||
Although p53 LOH is not a prerequisite for sarcomagenesis in p53+/− mice, the p53 wild-type allele is either absent or under-represented in most sarcomas that develop in these mice (Jacks et al., 1994). The number of chromosome 11 copies in sarcomas with p53 LOH, however, is unknown. To test for this, we first screened six sarcomas from p53+/− mice for p53 LOH by Southern blot analysis and identified four sarcomas with little or no wild-type p53 (Figure S2A). Like lymphomas from Bub1−/H/p53+/− mice with two chromosome 11 copies harboring the p53 knockout allele, the p53 knockout allele was highly over-represented in these sarcomas (Figure S2A), indicating that the knockout allele was duplicated. To examine this further, we performed interphase FISH for chromosome 11 on paraffin sections from these tumors. The percentage of cells with two chromosome 11 signals was 80% or more for each tumor, which was the same as for non-tumor tissue (Figure S2B and S2C). Collectively, these data demonstrate that sarcomas that lose the wild-type copy of p53 retain two copies of chromosome 11, suggesting that there is selective pressure against cells losing a copy of this chromosome.
Bub1 hypomorphism selectively drives colon tumorigenesis in ApcMin/+ mice
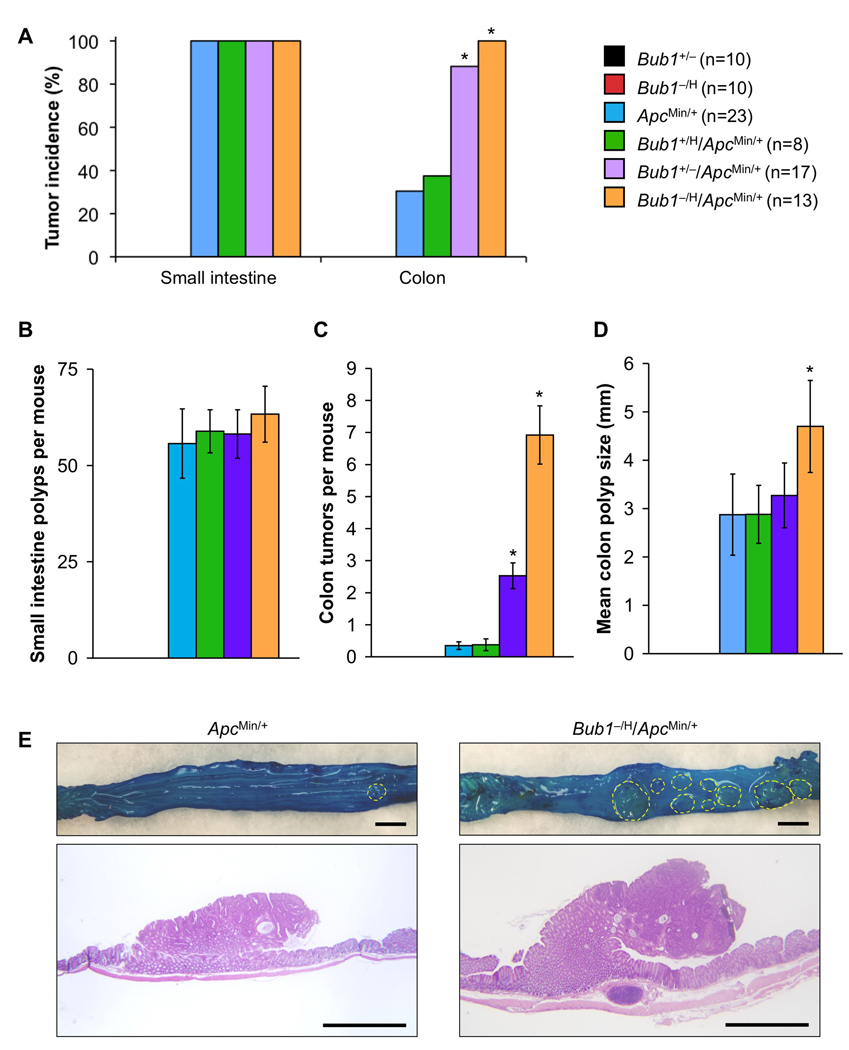
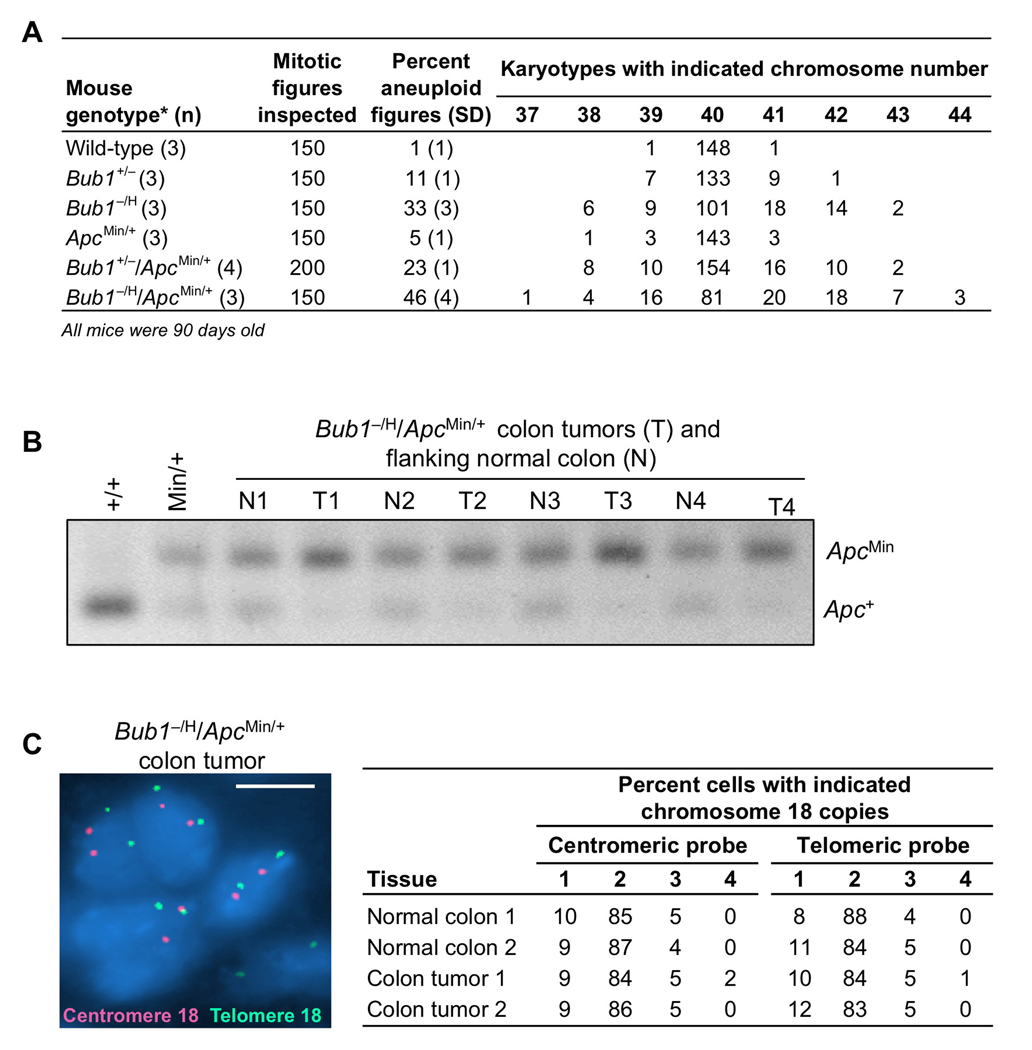
We wondered if Bub1 insufficiency would also promote tumorigenesis in other heterozygous tumor-suppressor models that rely on LOH, such as the ApcMin/+ mouse model for intestinal cancer (Luongo et al., 1994). This model is particularly attractive for further study, because Bub1 is either mutated or expressed at low levels in a subset of human colon carcinomas (Lengauer et al., 1997; Shichiri et al., 2002). Because tumor penetrance in ApcMin/+ mice is dependent on the genetic background, we backcrossed Bub1−/H mice to C57BL/6 mice for ten generations before breeding them to ApcMin/+ C57BL/6 mice. Cohorts of Bub1+/H/ApcMin/+, Bub1+/−/ApcMin/+, Bub1−/H/ApcMin/+, Bub1+/−, Bub1−/H and ApcMin/+ mice were established, and, at 90 days, mice were sacrificed and screened for intestinal tract polyps. Consistent with the literature, ApcMin/+ mice developed small intestinal tumors with 100% incidence (Figure 4A). On average, these mice had ~55 small intestinal tumors (Figure 4B). We found that the number of small intestinal tumors did not change with decreasing amounts of Bub1 (Figure 4A and B), demonstrating that low amounts of Bub1 are not capable of promoting small intestinal tumorigenesis. In striking contrast, however, tumor formation in the large intestine strongly increased with decreasing Bub1 levels. Whereas colon tumors were observed in 30% of ApcMin/+ mice, 88% of Bub1+/−/ApcMin/+ mice and 100% of Bub1−/H/ApcMin/+ mice had such tumors (Figure 4A). In addition, Bub1+/−/ApcMin/+ and Bub1−/H/ApcMin/+ mice had on average 8- and 23-fold more tumors than ApcMin/+ mice, respectively (Figure 4C and E). Typically, colon tumors of Bub1−/H/ApcMin/+ were largest in size (Figure 4D and 4E). Interestingly, colon tumor incidence and burden were not affected in Bub1+/H/ApcMin/+ mice (Figures 4A–D), indicating that accelerated colonic tumorigenesis requires a certain threshold level of Bub1 reduction.
Figure 4. Colon tumor formation dramatically increases in ApcMin/+ mice when Bub1 levels decline.
(A) Small intestine and colon tumor incidences in mutant mice of the indicated genotypes. Bub1 mutant strains not bred onto an ApcMin/+ background did not develop any intestinal tumors. *p < 0.05 compared to ApcMin/+ mice (Fisher’s exact test).
(B) Small intestine tumor multiplicity. Color codes are as indicated in (A). Data are mean ± SEM.
(C) Colon tumor burden. Color codes are as indicated in (A). Data are mean ± SEM. *p < 0.05 compared to ApcMin/+ mice (unpaired t test).
(D) Mean size of colon tumors. Data are mean ± SEM. *p < 0.05 compared to ApcMin/+ mice (unpaired t test).
(E) Colonic tumors in ApcMin/+ and Bub1−/H/ApcMin/+ mice (Top). Colons were stained with methylene blue. Hatched yellow lines surround individual colon tumors. (Bottom) Hematoxylin-eosin stained sections of colon tumors from ApcMin/+ and Bub1−/H/ApcMin/+ mice. Bars represent 2 mm.
Karyotype analysis showed that splenocytes from ApcMin/+ mice had moderately increased aneuploidy over wild-type splenocytes (Figure 5A), which is in accordance with earlier studies reporting elevated aneuploidy rates in cultured ApcMin/+ MEFs and ES cells (Fodde et al., 2001; Kaplan et al., 2001; Rao et al., 2005). However, relative to ApcMin/+ splenocytes, the percentages of aneuploidy in Bub1+/−/ApcMin/+ and Bub1−/H/ApcMin/+ splenocytes were dramatically increased (18% and 41%, respectively; see Figure 5A). Interestingly, aneuploidy rates of Bub1−/H/ApcMin/+ compound mutant mice were significantly higher than the rates of Bub1−/H and ApcMin/+ single mutants combined (46% versus 38%), indicating that Bub1 and Apc synergize in chromosome missegregation. The exact same phenomenon was observed in Bub1+/−/ApcMin/+ mice (Figure 5A). The above data suggest that Bub1 insufficiency accelerates the rate at which whole chromosomes are lost or gained in ApcMin/+ mice, thereby perhaps accelerating Apc LOH and driving colonic tumorigenesis. To test for LOH at the Apc locus, we collected colon tumors (T) of several Bub1−/H/ApcMin/+ mice and analyzed these tumors for the presence of the Apc+ allele using a previously established PCR-based assay. Normal tissue flanking each of the tumors (N) was used as a control. We found that the remaining Apc wild-type allele was lost in 100% of tumors examined (Figure 5B). To investigate whether Apc LOH was due to loss of the chromosome 18 copy containing the wild-type Apc allele, we performed interphase FISH on colon tumor sections of Bub1−/H/ApcMin/+ mice using centromeric and telomeric chromosome 18 probes. The vast majority of colon tumor cells had 2 copies of chromosome 18 (Figure 5C), suggesting that colon tumors, similar to thymic lymphomas of Bub1−/H/p53+/− mice, have a duplication of the mutant allele. Given that Bub1 is a whole chromosome instability gene, it is reasonable to assume that this duplication occurred through a gain of whole chromosome 18 containing the ApcMin allele. The mice were on a pure C57BL/6 genetic background, precluding the use of SSLP analysis to fully prove this. The only alternative mechanism through which the ApcMin allele might have been duplicated is mitotic recombination. However, this mechanism is unlikely because sister chromatid exchange (SCE) analysis on Bub1−/H and wild-type MEFs revealed that Bub1 insufficiency has no impact on the rate of mitotic recombination (Figures S3A and S3B). Interphase FISH analysis for chromosomes 4 and 7 on cells from collagenase-treated Bub1−/H/ApcMin/+ colon tumors provided evidence for significant chromosome reshuffling (Figure S3C–E), further supporting that Bub1 insufficiency drives colon tumor formation in ApcMin/+ mice by inducing loss of chromosome 18 containing the APC+ allele and gain of chromosome 18 bearing the ApcMin allele.
Figure 5. Apc LOH in colon tumors from Bub1−/H/ApcMin/+ mice.
(A) Chromosome counts on splenocytes of 3-month-old wild-type, Bub1+/−, Bub1−/H, ApcMin/+, Bub1+/−/ApcMin/+ and Bub1−/H/ApcMin/+ mice.
(B) PCR-based screening for Apc LOH in colon tumors from Bub1−/H/ApcMin/+. Tail DNA samples from wild-type and ApcMin/+ mice were used as PCR controls. Note that colon tumors from Bub1−/H/ApcMin/+ mice consistently show preferential amplification of the ApcMin allele, indicating loss of the Apc+ allele.
(C) Interphase FISH image of a 5 µm colon tumor section of a Bub1−/H/ApcMin/+ mouse hybridized to both centromeric (red) and telomeric (green) chromosome probes. Quantification of the chromosome 18 copies in colon cancer cells and flanking normal colon epithelial cells. FISH signals of one hundred cells were counted per section. Scale bar is 5 µm.
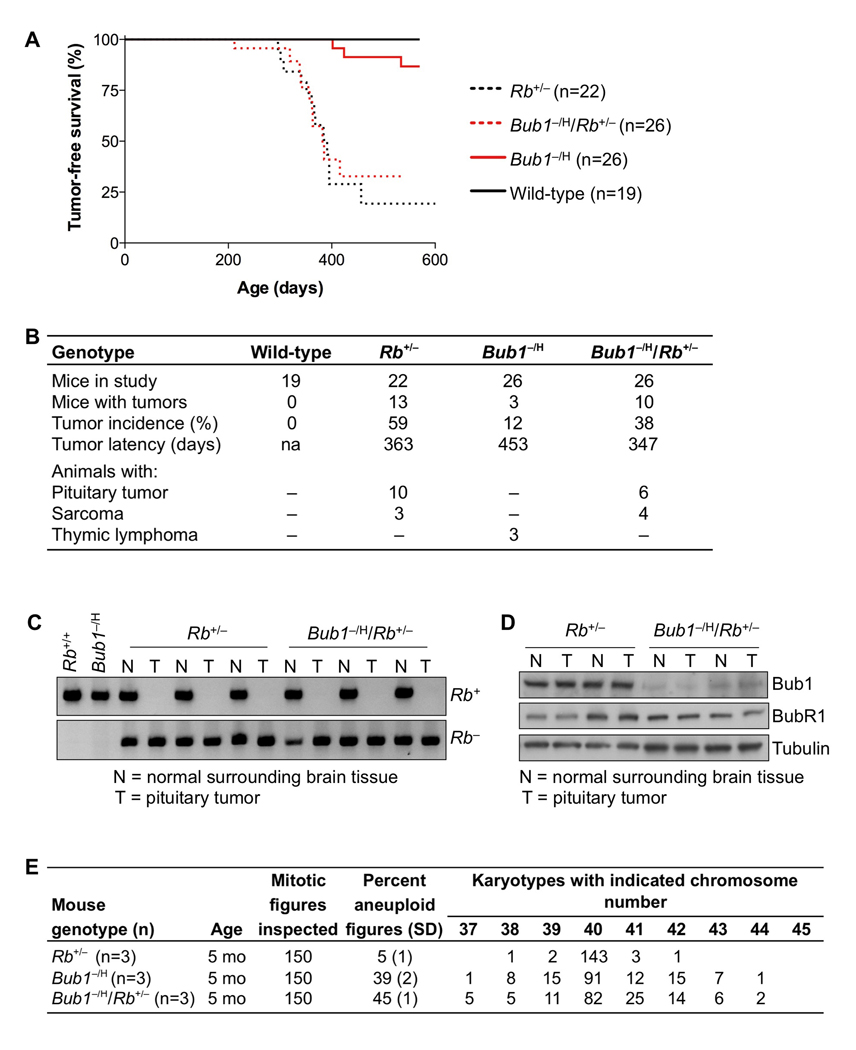
Bub1 insufficiency has no effect on tumorigenesis in Rb+/− mice
Mice in which one allele of Rb is disrupted develop pituitary tumors at nearly 100% incidence (Jacks et al., 1992; Lee et al., 1992). These tumors invariably lose expression of Rb protein due to loss of the remaining wild-type Rb allele (Hu et al., 1994). To determine the impact of whole chromosome instability resulting from low Bub1 on tumor development in this animal model, cohorts of Rb+/−, Bub1−/H/Rb+/−, Bub1−/H and wild-type mice were generated and monitored. In contrast to our observations in p53+/− and ApcMin/+ mice, low Bub1 levels had absolutely no impact on tumor development in Rb+/− mice. As shown in Figure 6A, the tumor-free survival curves of Rb+/− and Bub1−/H/Rb+/− mice were almost completely overlapping. Consistent with earlier studies (Jacks et al., 1992; Lee et al., 1992), Rb+/− mice developed primarily pituitary tumors and an occasional sarcoma (Figure 6B). Although the incidence of pituitary tumors in Bub1−/H/Rb+/− mice was somewhat lower than in Rb+/− mice (Figure 6B), this difference did not reach statistical significance.
Figure 6. Tumorigenesis in Rb+/− mice is not impacted by Bub1 insufficiency.
(A) Kaplan-Meier tumor-free survival curves of Rb+/−, Bub1−/H/Rb+/−, Bub1−/H and wild-type mice. Curves of Rb+/− and Bub1−/H/Rb+/− mice are not significantly different from each other (Log rank test).
(B) Tumor spectrum of mice represented in (A). There were no statistically significant changes in tumor spectrum between any of the mouse cohorts (Fisher’s exact test).
(C) PCR-based screening for Rb LOH in pituitary tumors from Rb+/− and Bub1−/H/Rb+/− mice. As controls we used DNA samples from flanking normal brain tissue and brain DNA samples of wild-type and Bub1−/H mice.
(D) Western blot analysis of normal brain (N) and pituitary tumors (T) of Rb+/− and Bub1−/H/Rb+/− animals. Note that Bub1, but not the Bub1-related protein BubR1, was expressed at reduced levels in both normal and tumor tissues of Bub1−/H/Rb+/− mice. Tubulin was used as a loading control.
(E) Chromosome counts on splenocytes of 5-month-old Rb+/−, Bub1−/H/Rb+/− and Bub1−/H mice.
Pituitary tumors of Rb+/− and Bub1−/H/Rb+/− mice consistently lacked the wild-type allele of Rb (Figure 6C), indicating that, both with and without Bub1 insufficiency, Rb LOH is a requisite for tumor formation. Reduced levels of Rb leads to upregulation of E2F target genes, including some mitotic regulators (Bracken et al., 2004). A trivial explanation for the lack of accelerated tumorigenesis might therefore be that Rb loss counteracts Bub1 insufficiency. However, western blotting confirmed that both normal tissue and pituitary tumors of Bub1−/H/Rb+/− mice were hypomorphic for Bub1 (Figure 6D). Consistent with this, we observed much higher percentages of aneuploid splenocytes in Bub1−/H/Rb+/− muice than in Rb+/− mice (Figure 6E). We were unable to detect significant aneuploidy in pituitary tumors by interphase FISH on paraffin sections (Figure S4A). However, it is important to note that we find that interphase FISH on paraffin sections is not sensitive enough to detect near diploid aneuploidies such as those induced by Bub1 insufficiency (Figure S4B–D). Therefore, it cannot be concluded that these tumors are not aneuploid.
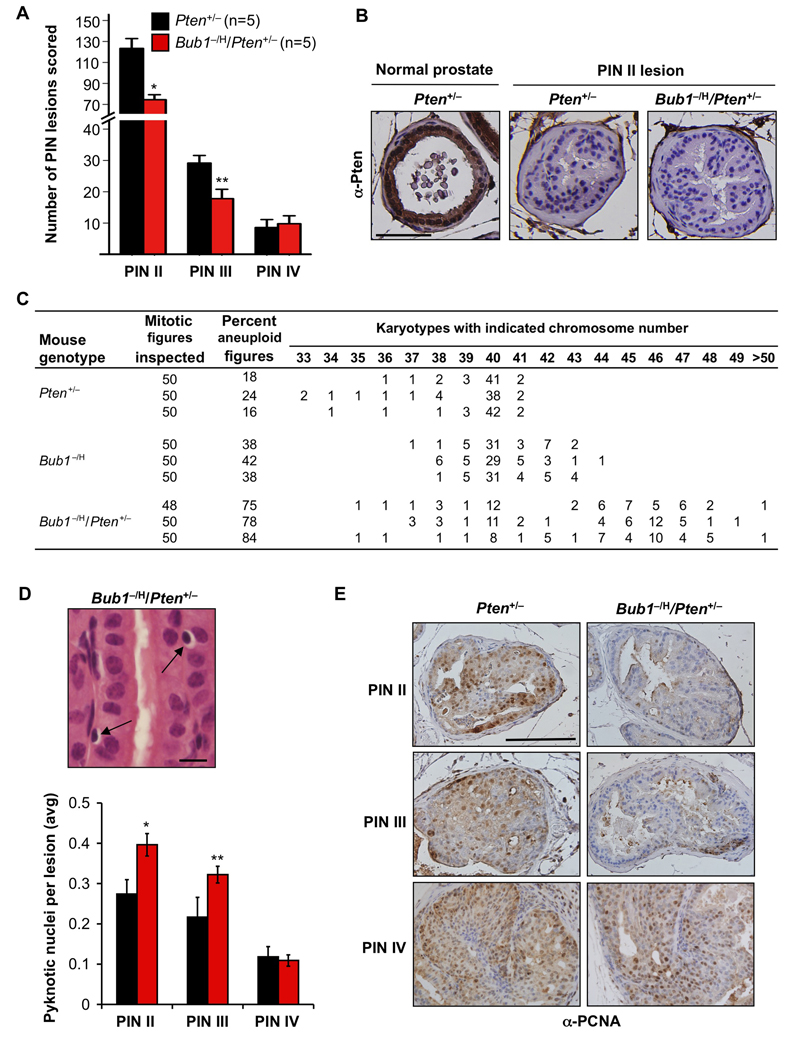
Bub1 loss suppresses prostatic intraepithelial neoplasia in Pten+/− mice
Pten LOH is a common event in human prostate cancer (Di Cristofano and Pandolfi, 2000). This, combined with the observation that chromosomal instability increases with prostate cancer grade in humans (Hawkins et al., 1995), prompted us to test whether the Bub1−/H genotype might drive prostate cancer development in Pten+/− mice (Di Cristofano et al., 2001; Podsypanina et al., 1999). To this end, small cohorts of Pten+/− and Bub1− /H/Pten+/− males were generated and sacrificed at six months of age. Whole prostates were dissected and screened for overt prostate tumors using a dissection microscope; however, no such tumors were detected irrespective of genotype. Following fixation, prostates were serially sectioned and screened for PIN II, III and IV lesions (Park et al., 2002). Strikingly, Bub1−/H/Pten+/− mice had about 40% less PIN II and III lesions than Pten+/− mice (Figure 7A). On the other hand, the numbers of PIN IV lesions were very similar between the two genotypes, although it should be noted that the number of mice in the study is insufficient to detect potential differences. No PIN II, III or IV lesions were observed in 6-month-old Bub1−/H males (data not shown). Immunohistochemisty on paraffin sections of Pten+/− and Bub1−/H/Pten+/− prostates revealed that Pten is either undetectable or barely detectable in PIN II lesions, irrespective of Bub1 level of expression (Figure 7B).
Figure 7. Low Bub1 decreases high-grade PIN lesions in Pten+/− males.
(A) At 6 months, Pten+/− males have significantly more PIN II and III lesions than age-matched Bub1−/H/Pten+/− mice. *p = 0.0017; **p = 0.0186 (unpaired t tests comparing similar grade PIN lesions). Data represented are mean ± SD. PIN IV multiplicity was similar for Pten+/− and Bub1−/H/Pten+/− males.
(B) PIN II lesions of 6-month-old Pten+/− and Bub1−/H/Pten+/− males analyzed for Pten by immunohistochemistry. Pten was either undetectable (shown) or barely detectable (not shown) in PIN II lesions of both mouse strains. Normal prostate epithelium of a Pten+/− male was used as a positive control for Pten staining. Scale bar is 100 µm.
(C) Pten+/− and Bub1−/H genotypes strongly synergize in causing chromosome missegregation in splenocytes. Chromosome counts were performed on splenocytes of 5-month-old mice of the indicated genotypes.
(D) Analysis of dead cells in PIN lesions of Pten+/− and Bub1−/H/Pten+/− animals (at 6 months). (Top) High resolution image of a hematoxylin and eosin stained section of a PIN II lesion from a Bub1−/H/Pten+/− male showing cells with highly condensed pyknotic nuclei with halo like appearance. (Bottom) Quantification of pyknotic nuclei in various PIN lesions of Pten+/− and Bub1−/H/Pten+/− animals. N = 270 PIN II, 100 PIN III and 12 PIN IV lesions per genotype. Data presented are mean ± SD. *p < 0.01 and **p < 0.05 (unpaired t test). Scale bar represents 10 µm.
(E) Analysis of cell proliferation in various PIN lesions of 6-month-old Pten+/− and Bub1−/H/Pten+/− males by immunohistochemistry for PCNA (n = 3 mice per genotype). Scale bar is 100 µm.
To explore the possible basis for the tumor suppressive effect of Bub1 insufficiency during the early stages of prostate tumorigenesis, we prepared metaphase spreads of splenocytes of Pten+/−, Bub1−/H and Bub1−/H/Pten+/− mice and performed chromosome counts. We noticed that loss of a single Pten allele was associated with high rates of aneuploidy, with 19% of Pten+/− splenocytes exhibiting aneuploidy compared to 0% of splenocytes of control mice (Figure 7C), indicating that a full complement of Pten is required for accurate chromosome segregation. Strikingly, 80% of aneuploid Pten+/− splenocytes had less than 40 chromosomes, indicating a strong bias toward loss of chromosomes. Importantly, Bub1−/H/Pten+/− splenocytes showed remarkably high rates of chromosome missegregation, with 75–84% of splenocytes being aneuploid, much higher than the percentages of Bub1−/H and Pten+/− single mutants combined (Figure 7C). Notably, 83% of aneuploid splenocytes of double mutant mice contained more than 40 chromosomes, which contrasts the preferential chromosome losses observed in Pten+/− animals. Furthermore, spreads from Bub1−/H/Pten+/− double mutants had a much broader spectrum of abnormal chromosome numbers than spreads from Bub1−/H and Pten+/− single mutants (Figure 7C). These data imply that Bub1 and Pten might act synergistically in preventing chromosome missegregation. Due to the limited sensitivity of interphase FISH on paraffin sections, we were unable to determine the precise ploidy status of neoplastic prostate epithelial cells of Bub1−/H/Pten+/− mice. However, interphase FISH for chromosomes 4 and 7 did indicate that cells of PIN II-IV lesions had no near tetraploid aneuploidies (Figure S5A).
Cell death has previously been linked to chromosomal instability (Dobles et al., 2000; Kops et al., 2004) and might underlie, at least in part, the decrease in prostate tumorigenesis in double mutant males. To test for this possibility, we screened the various PIN lesions of Pten+/−/Bub1−/H and Pten+/− mice for the presence of pyknotic nuclei. These nuclei are indicative of apoptosis and can be easily identified in hematoxylin and eosin-stained paraffin sections. Strikingly, significant increases in pyknotic nuclei were observed in PIN II and III lesions of Pten+/−/Bub1−/H mice, but not in PIN IV lesions (Figure 7D). It has further been suggested that aneuploidy can impair cell proliferation and, as such, inhibit tumorigenesis (Torres et al., 2007; Torres et al., 2008; Williams et al., 2008). To screen for reduced cell proliferation, we stained paraffin sections of Bub1−/H/Pten+/− and Pten+/− prostates for the proliferation marker PCNA. As illustrated in Figure 7E, Bub1−/H/Pten+/− PIN II and PIN III lesions, but not PIN IV lesions, always had fewer PCNA-positive cells than corresponding Pten+/− lesions. To confirm this growth inhibitory effect of Bub1 insufficiency, we performed an in vivo BrdU labeling experiment. We injected 6-month-old Bub1−/H/Pten+/− and Pten+/− males with BrdU, collected their prostates 24 h later and prepared paraffin sections, which we stained for BrdU. As expected, Bub1−/H/Pten+/− PIN II and PIN III lesions, but not PIN IV lesions, contained much less BrdU-positive cells than equivalent lesions of Pten+/− mice (Figures S5B and S5C). Collectively, these data suggest that decreased proliferation and increased cell death may, at least in part, underlie the tumor suppressive effect of Bub1 insufficiency in prostate of Pten+/− mice. Furthermore, they suggest that, during the early stages of tumor evolution, there is a selective pressure to counteract the adverse effect of Bub1 insufficiency on cell growth and survival.
Discussion
Whole chromosomal instability, tumor suppressor gene LOH and tumorigenesis
Five observations in p53+/− mice with reduced Bub1 levels provide conclusive experimental evidence for the longstanding hypothesis that whole chromosome instability can drive tumorigenesis by promoting tumor suppressor gene LOH. First, p53+/− mice become highly prone to thymic lymphomas when the rate of chromosome missegregation is accelerated by Bub1 hypomorphism. Second, these tumors consistently show loss of the chromosome 11 copy containing the wild-type p53 allele. Unexpectedly, the loss of this chromosome was always accompanied by a gain of an extra copy of the chromosome harboring the inactivated p53 allele. It has been proposed that loss of an entire chromosome will lead to underexpression of various genes located on that chromosome, which has been linked to cellular stress and reduced proliferation (Torres et al., 2007; Torres et al., 2008; Williams et al., 2008). Thus, the consistent presence of two chromosome copies containing the targeted p53 allele might reflect an early selective pressure to counteract growth inhibitory stress associated with the loss of one copy of chromosome 11. Although the mechanism by which the lymphoma cells obtain two copies of the chromosome containing the targeted p53 allele most likely involves multiple steps, occurring in consecutive cell divisions, it theoretically could also occur in a single division. For instance, due to syntelic microtubule-kinetochore attachment, both chromosome 11 copies containing knockout p53 may be pulled toward one pole and those harboring wild-type p53 toward the other. This scenario cannot be excluded, given that Bub1 functions in microtubule-chromosome attachment and that Bub1 insufficiency increases the rate of congression failure (Jeganathan et al., 2007; Meraldi and Sorger, 2005; Perera et al., 2007). Third, besides two copies of chromosome 11 containing the p53− allele, lymphoma cells from Bub1−/H/p53+/− mice lack recurrent numerical changes of particular chromosomes, supporting the idea that the main tumorigenic effect of chromosomal instability resulting from Bub1 loss is to drive p53 LOH. Fourth, lymphomagenesis in p53+/− mice inversely correlates with Bub1 level of expression. This, combined with the observation that aneuploidy also has an inverse relationship with Bub1 amount (Jeganathan et al., 2007), suggests that the rate with which chromosomes are reshuffled during cell division determines the rate with which cells undergo LOH and become tumor prone. Fifth, sarcomagenesis is the primary neoplastic process observed in p53+/− mice, but when Bub1 levels of these mice drop below a threshold level, tumorigenesis shifts toward development of thymic lymphomas, the most prevalent tumor type seen in p53−/− mice (Jacks et al., 1994). This shift suggests that thymuses of p53+/− mice with low amounts of Bub1 accrue p53−/− thymocytes at high rates as a result of chromosome missegregation, thereby increasing the risk of neoplastic transformation in this organ.
Furthermore, our analysis of colon tumors from Bub1−/H/ApcMin/+ mice provides suggestive evidence that Bub1 drives tumorigenesis through a similar chromosome-reshuffling blueprint in colon epithelial cells, but now involving chromosome 18 containing the Apc gene locus. Due to the pure C57BL/6 genetic background of the Bub1−/H/ApcMin/+ mice, SSLP analysis could not be employed to prove that colon cancer cells had obtained two copies of chromosome 18 containing ApcMin allele through whole chromosome missegregation. However, mitotic recombination, the most plausible alternative mechanism by which these chromosomes could have been obtained, is unlikely because SCE analysis on wildtype and Bub1−/H cells revealed that Bub1 insufficiency does not alter the frequency with which this type of recombination occurs.
Although there are no known functions for Bub1 other than protecting cells against aneuploidy, we cannot formally exclude that insufficiency of this checkpoint protein has other cellular consequences that may in fact be tumor promoting. Bub1 is not only important for faithful chromosome segregation, but also implicated in triggering apoptosis after abnormal mitoses (Jeganathan et al., 2007). Bub1’s dual function in mitosis might diversify the spectrum of chromosome losses and gains tolerated by primary cells, thereby facilitating tumor suppressor gene LOH at the initial stages of cell transformation. MEFs with low amounts of Bub1 respond normally to a broad spectrum of DNA damaging agents (Jeganathan et al., 2007), indicating that pathways that mediate DNA repair or apoptosis following DNA damage are not dependent on Bub1. In addition, these MEFs showed no evidence of increased structural chromosome damage, including chromosome translocations and fusions, large insertions and deletions, and single and double strand breaks (Jeganathan et al., 2007), further arguing against a role for Bub1 outside of mitosis.
Bub1 loss and tumorigenesis: impact of cellular and genetic context
Although our work reported here demonstrates that Bub1 insufficiency can drive tumorigenesis by eliminating tumor suppressor genes through loss of whole chromosomes, it also reveals that it only does so in the context of select tumor suppressor genes and specific tissues. For instance, in the context of Pten haploinsufficiency, Bub1 loss acts to inhibit the initiation of neoplastic growth in prostate epithelial cells, presumably by increasing apoptosis and reducing cell proliferation in early-stage neoplastic cells. It is tempting to speculate that high aneuploidy rates in prostate epithelial cells underlie both these cellular effects. We find that Pten is different from p53, Apc and Rb in that loss of a single Pten allele causes relatively high percentages of aneuploidy in splenocytes. Pten is further unique in that loss of one Pten allele in combination with Bub1 insufficiency leads to unusually high rates of chromosome missegregation in splenocytes. It has been proposed that extensive gene copy number variation associated with aneuploidy leads to major imbalances in the stoichiometry of protein complex subunits, thereby increasing proteotoxic stress and limiting replicative potential (Luo et al., 2009; Papp et al., 2003; Torres et al., 2007; Torres et al., 2008). However, it should be stressed that due to the experimental restrictions, it was not possible to assess whether the aneuploidy rates observed in splenocytes also apply to prostate epithelial cells. Furthermore, it cannot be excluded that Bub1 insufficiency inhibits formation of PIN lesions in a cell non-autonomous fashion, for instance by altering the tumor microenvironment. Finally, it is important to note that the inhibitory effects of Bub1 insufficiency are restricted to the early stages of prostate tumorigenesis. This is consistent with the idea that aneuploidy creates a selective pressure for mutations that allow cells to neutralize the adverse effects of chromosome imbalances on cell growth and survival (Holland and Cleveland, 2009; Torres et al., 2008). Experimental limitations for detecting near diploid aneuploidies in prostate lesions precluded us from determining whether such adaptation leads to tumors with increased chromosomal instability. Whether Bub1 insufficiency promotes advanced stage prostate tumorigenesis remains unknown, as the number of mice in our study was too small to detect potential differences in the number of PIN IV lesions between Pten+/− and Bub1−/H/Pten+/− males.
Even though Bub1 insufficiency significantly increased the rate of chromosome missegregation in Rb heterozygous mice, it had no effect on pituitary tumorigenesis. There are several possible explanations for this result. First, Bub1 hypomorphism may have little negative impact on the accuracy of chromosome missegregation in the pituitary. Again, due to technical limitations, we were unable to exclude this possibility. However, chromosome counts on various kinds of cultured primary cells from wild-type and Bub1−/H mice indicated that increased aneuploidy is a global rather than a cell-type specific consequence of Bub1 insufficiency in somatic cells (Figure S1). Importantly, of all Bub1−/H cell types tested, only male germ cells showed no increase in aneuploidy (Table 1). Second, Bub1 insufficiency may accelerate Rb LOH through loss of chromosome 14 carrying the residual wild-type Rb allele, but this chromosome might harbor one or more genes whose dosage is critical for cell viability. Third, Rb heterozygous mice may undergo efficient LOH through a mechanism other than whole chromosome instability, such as mitotic recombination. However, involvement of chromosomal instability cannot be fully excluded. For instance, we find that Rb insufficiency induces low, yet significant aneuploidy, at least in splenocytes (Figure 6E), perhaps through aberrant expression E2F target genes with mitotic functions (Bracken et al., 2004). Furthermore, Rb has also been shown to prevent chromosome segregation through its role in regulating pericentric heterochromatin structure and function (Isaac et al., 2006).
A striking finding was that colonic tumor formation in ApcMin/+ mice was dramatically increased when Bub1 was insufficient, whereas tumorigenesis in the adjacent small intestine was unaffected. This might reflect potential differences in chromosomal instability between the two tissues. However, it is also possible that aneuploidy rates are the same but that the resulting mitotic and proteotoxic stress phenotypes are different in epithelial cells of small and large intestine tissues. For instance, chromosomal instability in colon may have no impact on cell proliferation, whereas in the small intestine it may result in delayed cell-cycle progression, senescence and/or apoptosis. The earlier finding that haploinsufficiency of the Bub1-related gene BubR1 in ApcMin/+ mice suppresses polyp formation in the small intestine while promoting tumorigenesis in the large intestine (Rao et al., 2005) supports the idea that the cellular response to aneuploidy resulting from mitotic checkpoint failure is distinct in both tissues.
Experimental Procedures
Mouse strains
Bub1−/H mice (Jeganathan et al., 2007) were bred to p53−/− (Jacks et al., 1994) to generate Bub1+/H/p53+/− and Bub1+/−/p53+/− mice. These mice were then intercrossed to generate p53+/−, Bub1+/H/p53+/−, Bub1+/−/p53+/−, Bub1H/H/p53+/− and Bub1−/H/p53+/− mice. Bub1+/H, Bub1+/−, Bub1H/H and Bub1−/H cohorts were as described (Jeganathan et al., 2007). Bub1−/H mice were crossed to Rb+/− mice (Jacks et al., 1992) and the resulting Bub1+/H/Rb+/− and Bub1+/−/Rb+/− mice were interbred to establish Bub1−/H/Rb+/−, Bub1−/H and Rb+/− experimental cohorts. A similar breeding strategy was used to generate Bub1−/H/Pten+/−, Bub1−/H, and Pten+/− mice. Both Rb+/− and Pten+/− (Podsypanina et al., 1999) mutant mice were acquired from the Mouse Models of Human Cancers Consortium, at the National Cancer Institute. All the above mice were on a C57BL/6 × 129/Sv mixed genetic background. Bub1−/H mice on a C57BL/6 × 129/Sv mixed genetic background were first backcrossed to C57BL/6 mice for ten generations and then to ApcMin/+ C57BL/6 inbred mice (Moser et al., 1990; Jackson Laboratory). The resulting Bub1+/H/ApcMin/+ and Bub1+/−/ApcMin/+ mice were interbred to generate Bub1+/H/ApcMin/+, Bub1−/H/ApcMin/+, Bub1+/−/ApcMin/+, Bub1+/−, Bub1−/H and ApcMin/+ experimental cohorts. Animals were housed in a pathogen free environment for the duration of the study and monitored daily for tumor formation or ill health. All mouse experiments were conducted after approval of the Mayo Clinic Committee on Animal Care and Use. Prism software (GraphPad Software) was used for the generation of survival curves and statistical analysis.
Tumor analysis in p53 and Rb mutant mice
Moribund animals were sacrificed and subjected to a complete autopsy. Questionable areas were removed and processed for routine histopathology. Board certified pathologists assisted in the characterization of tumor samples. A Fisher’s exact test was used to compare the incidence of tumors between the various genotypes. For p53 LOH analysis, genomic DNA was extracted from thymic lymphoma and sarcoma samples and analyzed by Southern blotting or PCR as described (Jacks et al., 1994). For Rb LOH analysis, fresh tumor samples were micro-dissected and DNA was extracted. PCR analysis was as previously described (Keramaris et al., 2008). Lysates of pituitary tumors and normal flanking brain tissue were prepared for western blotting as previously described (Jeganathan et al., 2007). Antibodies used were as follows: rabbit anti-human Bub1(25–165) (Jeganathan et al., 2007), rabbit anti-human BubR1, and mouse anti-alpha tubulin (Sigma, T-9026).
Analysis of intestinal tumors
Analysis of intestinal tumors was as described (Rao et al., 2005). For LOH analysis, fresh tumor samples were isolated and subjected to a previously reported PCR-based method for detection of Apc+ and ApcMin (Luongo et al., 1994).
Prostatic intraepithelial neoplasia analysis
At 6 months of age, five Bub1−/H/Pten+/− and five Pten+/− mice were sacrificed by CO2 asphyxiation. Whole prostates were collected, fixed in 10% formalin and processed for histological evaluation according to standard methods. Five-µm paraffin sections were prepared of the entire prostate. One of every 10 sections was stained with hematoxylin and eosin and screened for PIN II, III and IV lesions using light microscopy. Our classification of PIN lesions was based on previously published criteria (Park et al., 2002). Pyknotic nuclei were determined as described (Wang et al., 1998). Immunohistochemistry for PCNA (Zymed, 93–1143) and Pten (Millipore, 04–035) were performed according to instructions provided by the manufacturers. In vivo BrdU labeling experiments were performed as described (Baker et al., 2008).
Karyotype, FISH, SSLP and SCE analyses
Metaphase spreads of mouse splenocytes were prepared and analyzed for numerical and structural chromosome abnormalities as described (Jeganathan et al., 2007). Chromosome counts on cultured primary cells from various non-hematopoietic mouse tissues are described in the Supplemental Data. Spectral karyotypic analysis was carried out using the protocol, reagents, instrumentation and software from Applied Spectral Imaging. SSLP and FISH analysis for chromosomes 4, 7, 9, 11,12 and 18 and SSLP were as described in the Supplemental Data. SCE analysis on MEFs were performed as previously described (German and Alhadeff, 2001).
Significance
Aneuploidy is a hallmark of human cancers, but the central question as to whether chromosomal instability gene mutations contribute to cancer by eliminating tumor suppressor genes through loss of whole chromosomes remains unresolved. By crossing Bub1 mutant mice with high rates of chromosome missegregation onto heterozygous null backgrounds for various prominent tumor-suppressor genes, we provide evidence that cancer can be caused by a mechanism of tumor suppressor gene inactivation that involves whole chromosome loss. Remarkably, cells that lose the chromosome that contains the wild-type tumor suppressor gene copy, consistently gain an extra copy of the chromosome harboring the inactive allele, implying a strong selective pressure for tumor suppressor gene loss of heterozygosity without losing the other genes on the chromosome.
Supplementary Material
Acknowledgements
We thank Liviu Malureanu, Robin Ricke, Paul Galardy and Rick Bram for helpful discussions and critical evaluation of this manuscript, the Mayo Clinic Cytogenetics shared resource for SKY/FISH analysis and the Genotyping shared resource for SSLP analysis. This work was supported by National Institutes of Health Grant CA126828 and the Mayo Clinic Robert and Arlene Center on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Chi YH, Ward JM, Cheng LI, Yasunaga J, Jeang KT. Spindle assembly checkpoint and p53 deficiencies cooperate for tumorigenesis in mice. Int J Cancer. 2009;124:1483–1489. doi: 10.1002/ijc.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, Clevers H. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- German J, Alhadeff B. Analysis of sister-chromatid exchanges. Curr Protoc Hum Genet Chapter 8. 2001 doi: 10.1002/0471142905.hg0806s02. Unit 8 6. [DOI] [PubMed] [Google Scholar]
- Hawkins CA, Bergstralh EJ, Lieber MM, Zincke H. Influence of DNA ploidy and adjuvant treatment on progression and survival in patients with pathologic stage T3 (PT3) prostate cancer after radical retropubic prostatectomy. Urology. 1995;46:356–364. doi: 10.1016/S0090-4295(99)80220-9. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Gutsmann A, Herbert DC, Bradley A, Lee WH, Lee EY. Heterozygous Rb-1 delta 20/+mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene. 1994;9:1021–1027. [PubMed] [Google Scholar]
- Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, Binne UK, Harrington L, Sicinski P, Berube NG, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Griffiths B, Earle E, Chow CW, Jamsen K, Choo KH. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer. 2005;44:29–36. doi: 10.1002/gcc.20215. [DOI] [PubMed] [Google Scholar]
- Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- Keramaris E, Ruzhynsky VA, Callaghan SM, Wong E, Davis RJ, Flavell R, Slack RS, Park DS. Required roles of Bax and JNKs in central and peripheral nervous system death of retinoblastoma-deficient mice. J Biol Chem. 2008;283:405–415. doi: 10.1074/jbc.M701552200. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo C, Moser AR, Gledhill S, Dove WF. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994;54:5947–5952. [PubMed] [Google Scholar]
- Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. Embo J. 2005;24:1621–1633. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haploinsufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol. 2005;15:43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, Cardiff RD. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161:727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D, Tilston V, Hopwood JA, Barchi M, Boot-Handford RP, Taylor SS. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, Jhanwar-Uniyal M, Dai W. Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci U S A. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliekelman M, Cowley DO, O’Quinn R, Oliver TG, Lu L, Salmon ED, Van Dyke T. Impaired Bub1 Function In vivo Compromises Tension-Dependent Checkpoint Function Leading to Aneuploidy and Tumorigenesis. Cancer Res. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichiri M, Yoshinaga K, Hisatomi H, Sugihara K, Hirata Y. Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival. Cancer Res. 2002;62:13–17. [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LD, Zhou Q, Wei JP, Yang WC, Zhao X, Wang LX, Zou JX, Gao SS, Li YX, Yang C. Apoptosis and its relationship with cell proliferation, p53, Waf1p21, bcl-2 and c-myc in esophageal carcinogenesis studied with a high-risk population in northern China. World J Gastroenterol. 1998;4:287–293. doi: 10.3748/wjg.v4.i4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KW, Montpetit B, Hieter P. The kinetochore and cancer: what’s the connection? Curr Opin Cell Biol. 2005;17:576–582. doi: 10.1016/j.ceb.2005.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.