Abstract
DNA methylation is an epigenetic mark affecting genes and transposons. We screened for mutations that fail to establish DNA methylation, yielding two mutants termed involved in de novo (idn). IDN1 encodes DMS3, an SMC related protein, IDN2 encodes a novel double stranded RNA binding protein with homology to SGS3. IDN1 and IDN2 control de novo methylation and siRNA-mediated maintenance methylation and are components of the RNA-directed DNA methylation pathway.
All known de novo DNA methylation in Arabidopsis is carried out by DOMAINS REARRANGED METHYLASE2 (DRM2)1,2, a homolog of DNMT3 methyltransferases. DRM2 is guided by small interfering RNAs (siRNAs) in a pathway termed RNA-directed DNA methylation (RdDM)3. Current models suggest that the RNA polymerase IV (Pol IV) complex is recruited to a target sequences by an unknown mechanism resulting in the production of single-stranded RNA, which is converted into double-stranded RNA by RNA-DEPENDENT RNA POLYMERASE2 (RDR2) and subsequently processed by DICER-LIKE3 (DCL3) into 24 nucleotide siRNAs. siRNAs are then loaded into ARGONAUTE4 (AGO4) which interacts with the Pol V machinery which transcribes RNA at silent loci 4,5. AGO4-siRNA complexes are then thought to pair with nascent RNA transcripts to guide DRM2 to initial target loci6. In order to better understand RdDM, we performed a screen to identify mutants affecting de novo methylation using FWA silencing as a model.
The endogenous FWA gene is heritably silenced by methylation at two tandem repeats in its upstream region. Unmethylated fwa epialleles cause FWA ectopic expression resulting in a dominant late flowering phenotype that is easily scored7. In wild type plants, methylation of FWA transgenes is efficiently established, while RdDM mutants fail to methylate and silence FWA and therefore flower late. By screening for mutants affecting the establishment of silencing at FWA transgene, but that do not affect pre-existing silencing at the FWA endogene, one can screen for de novo methylation mutants1,2.
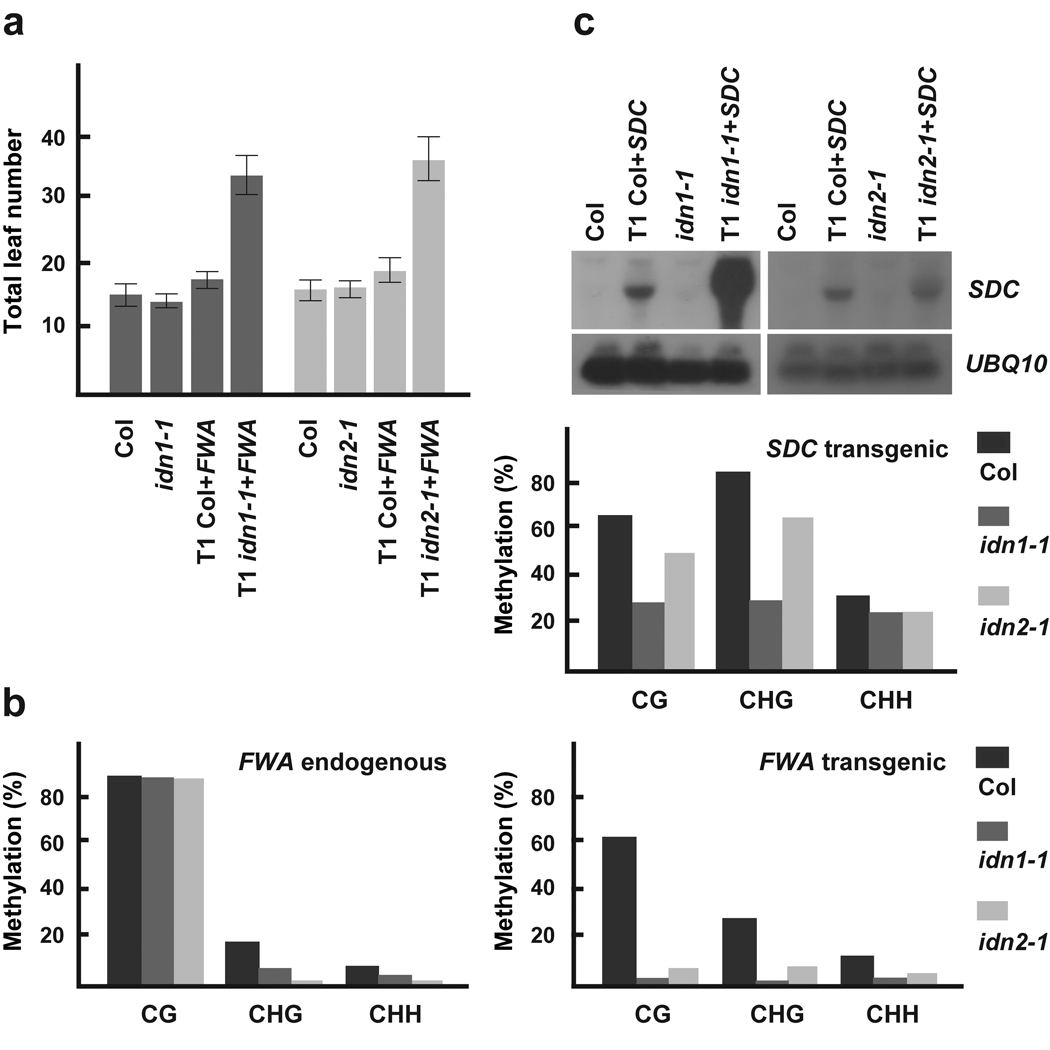
We screened a collection of 429 T-DNA insertion mutants using FWA Agrobacterium-mediated transformation. This collection includes RNA binding proteins, Agenet domain containing proteins, Jumonji domain containing proteins and other chromatin related proteins (Supplementary Table 1). This screen identified two mutants that were late flowering after transformation, but showed normal flowering prior to transformation (Figure 1a). We termed these mutants involved in de novo1 and 2 (idn1-1 and idn2-1). The late-flowering phenotype of these mutants was correlated with reduced methylation of the FWA transgenes as revealed by genomic bisulfite sequencing. Methylation was severely reduced in all DNA sequence contexts (CG, CHG, and CHH, where H = A, T, or C); however, pre-established CG methylation remained unaffected at the FWA endogenous gene (Figure 1b).
Figure 1. De novo methylation phenotype of idn mutants.
a) Flowering time measured as total number of leaves produced by wild type, idn mutants and FWA transformed T1 plants under long day conditions. Error bars depict standard deviation. b) Methylation status of wild type and idn mutants at endogenous and transgenic FWA. c) SDC expression levels in idn mutants before and after SDC transformation. Hybridization with a UBQ10 probe is shown as loading control. Panel below shows methylation status of wild type and idn mutants at transgenic SDC.
Another gene that can be used to test for de novo methylation is SUPPRESSOR OF drm1 drm2 cmt3 (SDC). SDC possesses seven tandem repeats in its promoter region. Hypomethylation at these repeats leads to overexpression of SDC and a characteristic morphological phenotype including curled leaves and short stature. All RdDM mutants tested so far fail to efficiently establish methylation at either SDC or FWA transgenes8. We found that the idn mutants also showed a reduced ability to establish silencing of incoming SDC transgenes. This was especially evident in the case of idn1-1. T1 idn1-1 plants transformed with SDC showed a dramatic increase in SDC expression; on the other hand, idn2-1 T1 transformants displayed nearly wild type SDC expression levels (Figure 1c). SDC overexpression is correlated with moderate hypomethylation at SDC tandem repeats as assayed by bisulfite sequencing (Figure 1c). These data reinforce that IDN1 and IDN2 control de novo methylation.
Thus far all mutations known to block de novo methylation also affect maintenance of methylation at several loci1,2,9. We analyzed the methylation state of representative endogenous targets including the MEA-ISR, 5S, SDC and FWA repeats using methylation sensitive enzymes coupled with Southern blots and/or bisulfite sequencing techniques. The idn1-1 and idn2-1 mutations both caused a reduction of non-CG methylation at MEA-ISR and FWA repeats (Figure 1c, Figure 2a and 2b). Furthermore, they showed reduced CG methylation at the 5S loci and a minor reduction in CHH methylation at the SDC repeats (Figure 2b and 2c). These methylation phenotypes are consistent with those seen in drm2 and other RdDM mutants3 suggesting that IDN1 and IDN2 play an important role in the RdDM pathway.
Figure 2. Maintenance methylation phenotype of idn mutants.
a) Methylation sensitive enzyme Southern hybridization assay at 5S and MEA-ISR loci, and HaeIII cutting assay at AtSN1. HaeIII, is blocked by C methylation in GGCC context, HpaII is blocked by C methylation in CCGG context and MspI is blocked by methylation of the external C in CCGG context. b) Methylation status of wild type, idn and idn cmt3–11 double mutants at endogenous SDC and methylation status of wild type and idn mutants at MEA-ISR. c) Morphological and molecular phenotype of idn cmt3–11 double mutants.
Because SDC silencing is redundantly controlled by both the RdDM and CMT3 pathways8, we crossed the idn mutants to the null CMT3 allele, cmt3–11. As previously reported for drm1 drm2 cmt3 triple mutants8, we observed strong SDC overexpression and the accompanying SDC morphological phenotype in the idn1-1 cmt3–11 double mutant. However, the idn2-1 cmt3–11 double mutant did not overexpress SDC. Consistently idn1-1 cmt3–11 exhibited reduced CG and CHG methylation at SDC, whereas idn2-1 cmt3–11 showed only a partial decrease in CHH methylation (Figure 2b and 2c). These results suggest that IDN1 plays a stronger or more general role in RdDM than IDN2.
To determine where the idn mutants act within the RdDM pathway, we examined siRNAs abundance at several loci. Comparison between the idn mutants and other RdDM pathway components showed that both idn mutants display normal (SDC and siR02) or slightly reduced (FWA, AtSN1, MEA-ISR, 5S and siR1003) siRNAs levels, but they did not completely lack siRNAs as observed in rdr2-1 or nrpd1a mutants (Supplementary Fig. 1). This suggests that IDN1 and IDN2 likely act downstream of initial siRNA biogenesis.
To gain further insight into the molecular mechanism of de novo methylation, we cloned both IDN1 and IDN2. Since neither of the T-DNAs insertion co-segregated with the respective mutant phenotypes, we isolated both IDN1 and IDN2 genes using a map-based approach. The idn phenotype at MEA-ISR segregated in a Mendelian fashion in F2 Ler×idn populations allowing us to follow the mutant phenotypes. Fine mapping strategies localized each mutation to a small genomic interval on Chromosome III (Supplementary Fig. 2a). We identified IDN1 and IDN2 initially by DNA sequence mutations in these regions. The idn1-1 allele carries a point mutation, whereas idn2-1 allele carries a point mutation and a 24 base pair deletion. To confirm gene identification, we complemented idn1-1 and idn2-1 with genomic fragments containing wild type IDN1 or IDN2 respectively (Supplementary Fig. 2b and 2c). In addition, we characterized additional alleles of IDN1 and IDN2 obtained from available T-DNA collections (Supplementary Fig. 2b) and found similar methylation defects at the 5S and MEA-ISR loci (Figure 2a).
IDN1 encodes DEFECTIVE IN MERISTEM SILENCING3 (DMS3=At3g49250), a previously described protein that shares homology with structural maintenance of chromosome (SMC) proteins10. The dms3 alleles were show to be required for RdDM, and for the production of secondary siRNAs10 as well as Pol V non-coding transcripts6, consistent with a role of IDN1/DMS3 in a downstream part of the RNA directed de novo methylation pathway.
IDN2 (At3g48670) encodes a predicted protein of 648 amino acids containing a zinc finger domain, and XS and XH domains separated by a coiled-coil region. An XS/XH protein family has been recently defined based on homology with the rice gene X and the protein SGS3 involved in posttranscriptional gene silencing11,12. In addition to IDN2, there are related homologous genes in the Arabidopsis genome containing XS/XH domains including At1g15910, At4g00380, At4g01780, At1g13790, At3g12550, At1g80790, At5g59390, and At4g01180, and one or more of these might act redundantly with IDN2 and explain why the idn2 loss-of-function phenotype is not as strong as that of idn1.
The XS domain has been predicted to be an RNA binding domain and regions around residues N16 and F64 are conserved between XS and RNP1/RNP2 functional motifs in RBPs13. Recently, it has been shown that SGS3 XS domain can bind 5’ overhanging double stranded (dsRNA) species14. To test IDN2 RNA binding abilities we performed gel mobility shift assays using the XS and coiled-coiled domains, (IDN2ΔZnFΔXH), and several RNA species previously described14. Interestingly, IDN2ΔZnFΔXH showed specificity for dsRNAs possessing 5’ overhangs (Supplementary Figure 3). Mutations at N16 and F64 abolished this binding, indicating that XS is the domain responsible for the RNA binding. While it is unclear what double stranded 5’ overhang substrate might be involved in RdDM it is tempting to speculate that IDN2 could recognize an siRNA which is bound to its target non-coding RNA, facilitating the downstream targeting of chromatin factors such as DRM2 to methylation targets. While future studies will be required to determine the precise function of both IDN1 (DMS3) and IDN2, it is clear that they are critical factors in the RdDM pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jacobsen lab members for supportive discussions. Jacobsen lab research was supported by NIH grant GM60398. I.A. was supported by a postdoctoral fellowship from the Ministerio de Educacion y Ciencia. S.E.J. and J. C. are investigators of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
T.C.M. and J. C. provided the majority of the insertion mutagenized population. I.A. and S.E.J. designed the experiments. I.A. performed the experiments and wrote the manuscript.
REFERENCES
- 1.Chan SW, et al. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 2.Cao X, Jacobsen SE. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 3.Henderson IR, Jacobsen SE. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 4.Li CF, et al. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Pontes O, et al. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 6.Wierzbicki AT, Haag JR, Pikaard CS. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soppe WJ, et al. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 8.Henderson IR, Jacobsen SE. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao X, Jacobsen SE. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanno T, et al. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 11.Bateman A. BMC Bioinformatics. 2002;3:21. doi: 10.1186/1471-2105-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mourrain P, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D, Trudeau VL. Cell Cycle. 2008;7:2268–2270. doi: 10.4161/cc.7.14.6306. [DOI] [PubMed] [Google Scholar]
- 14.Fukunaga R, Doudna JA. Embo J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.