Abstract
Biological macromolecules including DNA, RNA, and proteins, have intrinsic features that make them potential building blocks for the bottom-up fabrication of nanodevices. RNA is unique in nanoscale fabrication due to its amazing diversity of function and structure. RNA molecules can be designed and manipulated with a level of simplicity characteristic of DNA while possessing versatility in structure and function similar to that of proteins. RNA molecules typically contain a large variety of single stranded loops suitable for inter- and intra-molecular interaction. These loops can serve as mounting dovetails obviating the need for external linking dowels in fabrication and assembly.
The self-assembly of nanoparticles from RNA involves cooperative interaction of individual RNA molecules that spontaneously assemble in a predefined manner to form a larger two- or three-dimensional structure. Within the realm of self-assembly there are two main categories, namely template and non-template. Template assembly involves interaction of RNA molecules under the influence of specific external sequence, forces, or spatial constraints such as RNA transcription, hybridization, replication, annealing, molding, or replicas. In contrast, non-template assembly involves formation of a larger structure by individual components without the influence of external forces. Examples of non-template assembly are ligation, chemical conjugation, covalent linkage, and loop/loop interaction of RNA, especially the formation of RNA multimeric complexes. The best characterized RNA multiplier and the first to be described in RNA nanotechnological application is the motor pRNA of bacteriophage phi29 which form dimers, trimers, and hexamers, via hand-in-hand interaction. phi29 pRNA can be redesigned to form a variety of structures and shapes including twins, tetramers, rods, triangles, and 3D arrays several microns in size via interaction of programmed helical regions and loops. 3D RNA array formation requires a defined nucleotide number for twisting and a palindromic sequence. Such arrays are unusually stable and resistant to a wide range of temperatures, salt concentrations, and pH. Both the therapeutic siRNA or ribozyme and a receptor-binding RNA aptamer or other ligands have been engineered into individual pRNAs. Individual chimeric RNA building blocks harboring siRNA or other therapeutic molecules have been fabricated subsequently into a trimer through hand-in-hand interaction of the engineered right and left interlocking RNA loops. The incubation of these particles containing the receptor-binding aptamer or other ligands results in the binding and co-entry of trivalent therapeutic particles into cells. Such particles were subsequently shown to modulate the apoptosis of cancer cells in both cell cultures and animal trials. The use of such antigen-free 20–40 nm particles holds promise for the repeated long-term treatment of chronic diseases. Other potentially useful RNA molecules that form multimers include HIV RNA that contain kissing loop to form dimers, tecto-RNA that forms a “jigsaw puzzle,” and the Drosophila bicoid mRNA that forms multimers via “hand-by-arm” interactions.
Applications of RNA molecules involving replication, molding, embossing, and other related techniques, have recently been described that allow the utilization of a variety of materials to enhance diversity and resolution of nanomaterials. It should eventually be possible to adapt RNA to facilitate construction of ordered, patterned, or pre-programmed arrays or superstructures. Given the potential for 3D fabrication, the chance to produce reversible self-assembly, and the ability of self-repair, editing and replication, RNA self-assembly will play an increasingly significant role in integrated biological nanofabrication. A random 100-nucleotide RNA library may exist in 1.6 × 1060 varieties with multifarious structure to serve as a vital system for efficient fabrication, with a complexity and diversity far exceeding that of any current nanoscale system.
This review covers the basic concepts of RNA structure and function, certain methods for the study of RNA structure, the approaches for engineering or fabricating RNA into nanoparticles or arrays, and special features of RNA molecules that form multimers. The most recent development in exploration of RNA nanoparticles for pathogen detection, drug/gene delivery, and therapeutic application is also introduced in this review.
Keywords: RNA, Nanotechnology, Self-Assembly, RNA Application, phi29 pRNA
1. THE SUITABILITY OF RNA FOR NANOTECHNOLOGY
The emergent field of nanotechnology generally involves the characterization, manipulation, modification, and/or assembly of organized materials on the nanoscale level,1, 2 thereby helping to form supramolecular structures.3–5 These materials can then be used as building blocks for the construction of larger devices and systems.
Living systems contain a wide variety of nanomachines and other such ordered structures,6 including motors,7–14 arrays,15–17 pumps, membrane cores, and valves. The novelty and ingenious design of such machines have helped inspire the development of biomimetics for nanodevices.1, 4 Much current research is being devoted to make these machines as viable and effective as possible outside of their native environment.18 Once this is achieved, these nanodevices could be used in such applications as the delivery of drugs19 and therapeutic macromolecules,20 the gearing of other nanodevices for purposes such as nano-electromechanical systems (NEMS),21 the driving of molecular sorters, the building of intricate arrays and chips for diagnostics, molecular sensors, and novel actuators22 in new electronic and optical devices.
Since biological macromolecules, including DNA, RNA, and proteins, intrinsically have defined features at the nanometer scale, they could serve as unique and powerful building blocks for the bottom-up fabrication of nanostructures and nanodevices. Both DNA and proteins have been much more extensively studied with regard to their potential for nanotechnological applications than has RNA.23–34 What makes RNA particularly useful is the amazing diversity evident in its functionality, which can be directly attributed to the versatility present in its structure.35–39 Most available models that delineate various RNA functions such as transcription, translation, degradation, maturation, and the catalytic activity of ribozymes are based on a specific RNA structure.40 Therefore RNA is a particularly interesting candidate for nanotechnological applications, since RNA molecules can be designed and manipulated with both a level of simplicity characteristic of DNA41, 42 and a versatile flexibility in structure and function similar to that of proteins. Typically, RNA molecule contains large variety of single stranded loops for inter- and/or inter-molecular interaction. These loops can serve as mounting dovetails, thus external linking dowels might not be needed in fabrication and assembly.
RNA molecules are polymers made up of hundreds, thousands, or millions of nucleotides from four groups: adenosine (A), cystidine (C), guanosine (G), and uridine (U) (see Section 3). As to a 100-nucleotide RNA polymer, as many as 4100 (=1.6×1060) different RNA molecules can be designed. Since the size of RNA ranges from the angstrom to the nanometer scale, bottom-up approaches would be reasonable for use with RNA in nanotechnological applications. The first application in RNA nanotechnology is the construction of micrometer-scale RNA arrays derived from the nanometer-scale building block of bacteriophage phi29 motor pRNA.43
2. GENERAL APPROACHES IN BOTTOM UP ASSEMBLY USING RNA AS BUILDING BLOCKS
The self-assembly of nanoparticles from RNA is a prominent bottom-up approach to obtain nanoscale, nanostructured materials, and thus represents an important approach by which biological techniques and biomacromolecules can be successfully integrated into nanotechnology.43–47 Such approaches rely upon the cooperative interaction of individual RNA molecules that spontaneously assemble in a predefined manner to form a larger two- or three-dimensional structure. Within the realm of self-assembly here are two main subcategories: templated assembly and nontemplated assembly. Templated assembly involves the interaction of RNA with one another under the influence of a specific external force, structure, or spatial constraint. RNA transcription, hybridization, replication, annealing of DNA and RNA, molding, or replica, are part of this category. Nontemplated assembly involves the formation of a larger structure by individual components without any influence from external forces. Included in the nontemplated category are ligation, chemical conjugation, covalent linkages, loop/loop interaction of RNA, and the formation of macromolecules by structural interaction in the formation of multimers.41, 43, 46–49
One of the main areas of interest in current nanotechnology is the synthesis of patterned arrays50–53 for technological applications. Arrays can be engineered to serve as chips in the diagnosis of diseases or to function as ultrahigh density data storage systems.21 Ordered nanocrystals have already been assembled into superlattices by techniques such as colloidal crystallization,54–56 macromolecule self-assembly,11, 57–59 complementary interactions,48, 60, 61 and patterned etch pits.62 Ordered biological structural arrays can serve as templates for the further construction of superlattices.11, 57, 58, 63 Bacteriophage phi29 pRNA has been used as building blocks for the construction of RNA arrays with the size in micron.43
Various applications involving replication, molding, embossing, and other related techniques have recently been emerging,45 allowing for the utilization of a variety of materials to improve the diversity and to enhance the resolution of nanomaterials. It may eventually be possible to specifically adapt RNA for use in many of these techniques, which would facilitate the construction of ordered, patterned or preprogrammed arrays or superstructures. It is evident that RNA self-assembly will play an increasingly significant role in integrated biological nanofabrication in the future, given its potential for 3D fabrication, the chance to produce reversible self-assembly, and the ability of self-repairing and replicating. RNA serves as the ultimate model for effective, efficient nanoscale self-assembly, with a complexity far exceeding that of any current nanoscale systems.
3. BASIC CONCEPTS OF RNA STRUCTURE AND FUNCTION
Prior to a detailed description of the wide range of applications of RNA nanotechnology, it is important to provide some basic background information regarding RNA and its specific attributes. Following is a brief discussion of its process of formation and structural characteristics.
Nucleotides form the basic building blocks of ribonucleic acid (RNA). These nucleic acids are composed of three main components: (1) a base, (2) a phosphate group, and (3) a pentose (a five-carbon sugar ribose). In chemical terms, the two most important differences between RNA and DNA are: (1) the presence of an −OH group at the 2′ position of the ribose in RNA and the absence of the oxygen of this group in DNA (hence the deoxy—prefix) and (2) the replacement of the uracil base (U) in RNA by the thymine base (T) in DNA. When there is only a base/pentose composite, this structure is called “nucleoside;” when phosphate is included, a nucleotide is formed. The four nucleosides of RNA are adenosine (A), cystidine (C), guanosine (G), and uridine (U).
RNA can be synthesized chemically or enzymatically by RNA polymerases. In the latter case, nucleoside monophosphates link the hydroxyl group of one nucleotide to the phosphate of another during transcription. A template strand directs insertion of the nucleoside monophosphate. In this manner, a long chain of nucleotides can be formed, with one end of the chain (the 5′ end) having one or up to three phosphates and the other (the 3′ end) a free hydroxyl group. Along the length of the chain, there are two sides. One side is known as the “backbone” and consists of sugar and phosphate groups. The other side is left open for the possibility of the base-pairing of chains. This flexibility allows for the construction of a wide variety of complex structures and provides for a considerable amount of functional diversity.
RNA will self-fold into 2D and 3D structure. A chain’s relative level of stringency determines whether it exists in a double-stranded form under conditions of low stringency or a single-stranded form in a high stringency environment. Salt concentration is generally inversely proportional to stringency, since the presence of only a small amount of salt tends to cause the natural electrostatic repulsion of the two strands to overcome any ionic attraction and thus to manifest a single-stranded, high stringency condition. In contrast, temperature is directly proportional to the level of stringency; for example, a high temperature environment is likely to cause dissociation of dimer, trimer, hexamer into monomer and any double-stranded RNA structures into a single strand. Other conditions affect stringency as well.
One of the RNA functions involves protein synthesis within cells. There are a wide variety of subtypes of RNA with different functions that have been identified. Among the most common types are: (1) ribosomal RNA (rRNA), which serves as the “machinery” in the process of synthesis, (2) messenger RNA (mRNA), which is responsible for the transmission of DNA information, and (3) transfer RNA (tRNA), which performs a variety of specific roles in protein synthesis. These and many other small RNA subtypes, including small interfering RNA (siRNA),64–67 antisense RNA,68, 69 ribozymes,70–73 aptamer RNA,74, 75 pRNA (a viral packaging RNA discussed below),76 and microRNA,77 have potential application in nanotechnology, gene therapy, and other areas. Many of the small RNAs from cells and other microorganisms exhibit novel, emerging and/or unknown functions, and there is still much to be learned in order to fully utilize their differing mechanisms, function, and structure.
4. METHODS FOR THE STUDY OF RNA STRUCTURE
Though nucleotide derivatives have been found in RNA, the primary sequence of the RNA molecule is as simple as DNA, since both are composed of four nucleotides. Both DNA and RNA can form double-helical structure, but RNA has a more diverse structure. Even the smallest RNA molecules, though they contain only the four nucleotides A, G, C, and U, exhibit significant versatility in biological function. Such versatility is ascribed to the flexibility and complexity in RNA structural folding. NMR78–84 and X-ray crystallography85–89 have been used to obtain a physical tertiary structure of RNAs. Yet, the difficulty, uncertainty, and time involved in obtaining a diffractable RNA crystallographic structure, together with the difficulty of using NMR for large RNAs, necessitate the use of alternative approaches to obtain information on RNA structures.
4.1. Genetic Analysis by Truncation, Insertion, Deletion, and Mutation
Mutant RNAs can be easily constructed through the truncation, deletion, insertion, and mutation targeting of any desired position. For example, using an in vitro assembly assay system, dozens of mutant phi29 pRNAs can be produced and tested in one to two weeks.59, 90–93
4.2. Phylogenetic Analysis
Phylogenetic analysis of RNA can be used to compare the sequences of RNA molecules with similar or identical functions from different species.94–97 The supposition that RNA molecules with a similar function possess a common secondary structure has been deduced from such analyses. It has been found that RNA structure plays a critical role in RNA function and that nature most often tends to select the most stable molecule with the best-fit structure or with acceptable base co-variations. Such phylogenetic analysis of species from nature can potentially be expanded into molecules made artificially, such as those made by the use of complementary modification or SELEX (described below).
4.3. SELEX
The SELEX (Systematic Evolution of Ligands by Exponential Enrichment) method has permitted the identification of unique RNA/DNA molecules from very large populations of random sequences DNA or RNA libraries. Usually, selections are frequently carried out with RNA pools due to the folding ability of RNAs into complex structures which can be a source of diversity of RNA function.75, 98–101 SELEX allows screening for co-variation of several nucleotides and can be used to reveal non-canonical interaction which is difficult to prove by classic genetic and biochemical approaches.75, 98–101 For example, SELEX has been used to select pRNA sequences that bind procapsids102 and proved that pRNA can bind ATP.103
4.4. Complementary Modification
One approach often utilized to confirm base-pairing in predicted RNA structure is complementary modification. Before the conclusion that “G pairs to C” in an RNA structure can be definitively drawn, at least three mutants should be constructed and analyzed. These mutants, having either one G changed to A (or U) or one C to U (or A) should render the mutant inactive. When both Gs are changed to As (or Us) and both Cs are changed to Us (or As), the mutant should experience restored activity. For example, the complementary modification helps the study of loop/loop interaction in pRNA dimer formation.40, 104, 105
4.5. Chemical Modification and Chemical Modification Interference
Chemical modification is often employed to probe RNA structure.106, 107 The modifying agents utilized include dimethyl sulfate (DMS), which methylates A at N1, G at N7, and C at N3;108, 109 kethoxal, which modifies G at N1 and N2;110, 111 and 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulfonate (CMCT), which attacks U at N3 and G at N1. The locations of modified bases can be identified by primer extension with reverse transcriptase.108, 112 The successful chemical modification of a base is a good indication that the base is unpaired and that the specific functional group is solvent-exposed. Thus this base is a possible candidate for intermolecular interactions. A lack of modification will most likely be due to base pairing, especially in helical regions, but may also be the result of tertiary interactions or non-canonical base–base, base–sugar, or base–phosphate interactions in loop or bulge regions.110 Such chemical modification data can provide information regarding base accessibility, which is helpful in assessing and predicting secondary structures, evaluating 3-D molecular models, and analyzing RNA/protein interactions.
Chemical modification interference113–115 involves the same procedure as chemical modification; the general principle behind the interference method is that an RNA containing an interfering modified base would appear in the fast migrating monomer band, while RNA containing a non-interfering modified base would appear in the slower migrating dimer band.
4.6. Photoaffinity Crosslinking by Psoralen, GMPS/Aryl Azide, Phenphi, or Ultraviolet-Light
The chemical psoralen can intercalate into RNA or DNA helices and after irradiation with 320–400 nm light, freeze (in a helical or pseudoknot form) the uridines of RNA or the thymidines of DNA by covalent attachment116 if they are in close proximity.117–120 The sites of crosslinks can be determined by primer extension121 and/or Mung bean nuclease treatment.122 The psoralen derivative, AMT (4′-aminomethyl-4,5′,8-trimethyl psoralen), is a good choice to crosslink RNA due to its solubility.121 Psoralen crosslinks only RNA or DNA but not protein, which is different from the azido group (see below) which crosslinks non-specifically to both protein and nucleic acids. Psoralen can induce intra-molecular crosslinks within the RNA even in the presence of other proteins and RNA conformations in different environments can therefore be detected.
Aryl azides contain functional groups that are chemically inert in the absence of light, but can be converted to a reactive nitrene after long wavelength UV irradiation which then inserts into nearby bonds resulting in covalent crosslinks.123, 124 Thus, aryl azides can be incorporated into RNA to obtain structural data.125, 126 Specific internal bases of RNA can be uniquely labeled with photoaffinity crosslinking agents to analyze inter-and intra-molecular interactions. Crosslinked RNAs are separated from uncrosslinked RNAs by denaturing gel electrophoresis. Crosslink sites are determined by primer extension.113, 127 Bases identified as crosslink sites by primer extension indicate that these bases are in close proximity to the photoagent-labeled base. Crosslinking is used to determine the proximity in 3D structure determination.
Unlike psoralen, phenphi [(cis-Rh(phen)(phi) (phen = 1,10-phenanthroline; and phi = 9,10-phenanthren-equinone diimine)] induces covalent bonds between guanosine bases upon UV activation. Phenphi has also been shown to crosslink RNA.128
Also, ultraviolet-induced cross-linking of RNA to protein or RNA to RNA has been proved to be an effective methods for the study of RNA protein interaction and RNA structure–function relationships.129
4.7. Cryo-Atomic Force Microscopy (Cryo-AFM)
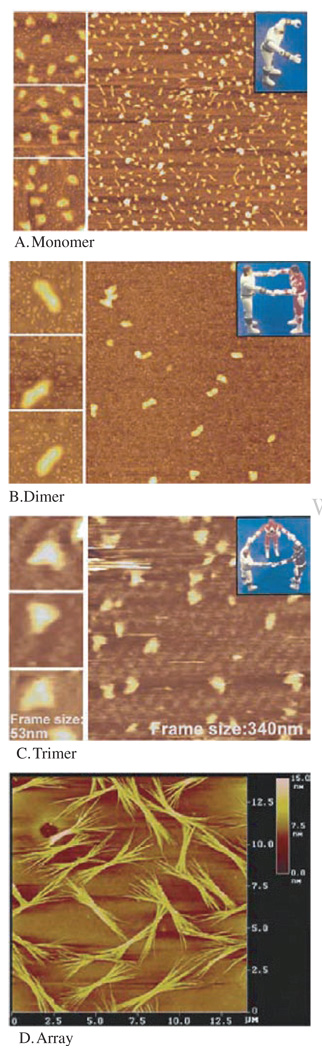
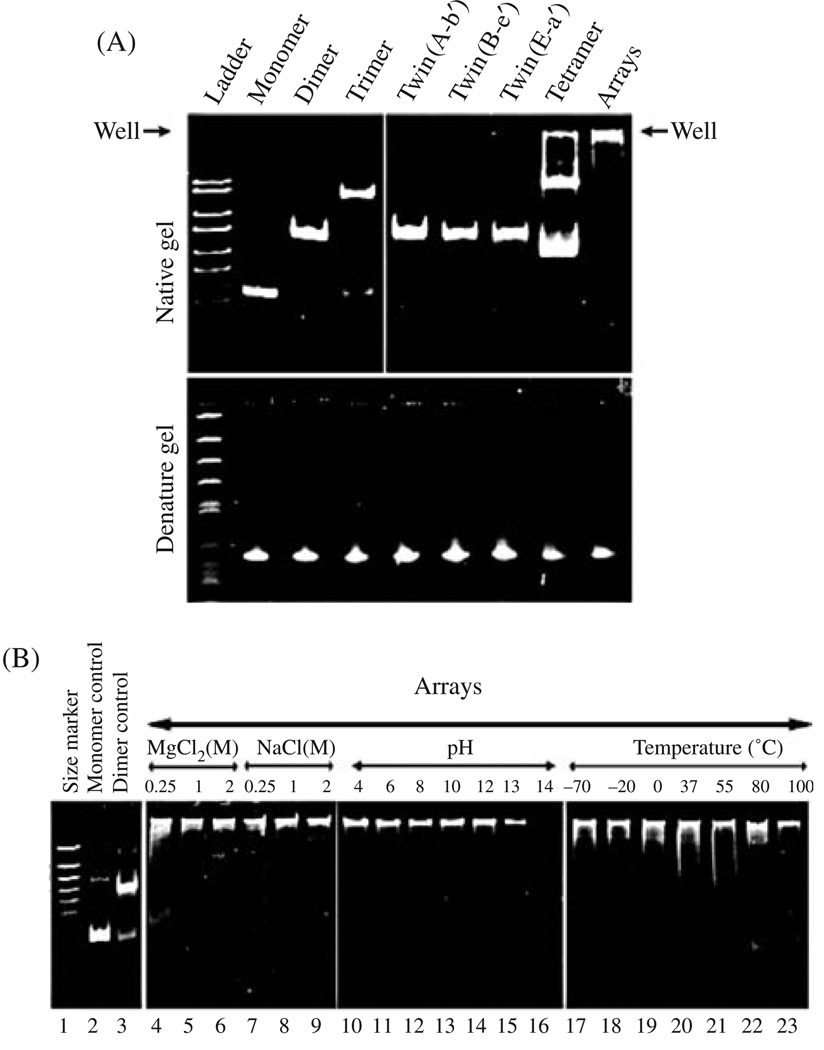
Cryo-AFM130 can be performed to better elucidate the 3-D structure of RNA in native conformation.113, 131, 132 The color indicates the thickness or height of the sample but does not reflect the atomic density observed. The brighter the color indicates the thicker of the sample—the darker the color, the thinner the sample (Fig. 3).
Fig. 3.
Atomic force microscopy (AFM) showing pRNA monomers (A), dimers (B), trimers (C), and arrays (D) of pRNA. The three insets at the left of each panel contain images with higher magnification, as indicated by the size of the frame. The pRNA monomers folded into a checkmark shape, dimers displayed a rod shape, trimer exhibited triangle shape, and arrays displayed as bundles. Formation of dimers requires Mg2+, while the sample on mica was briefly rinsed with water before freezing for cryo-AFM, which resulted in some dissociation of dimers or trimers even when the pRNA was already adsorbed to the activated mica surface. The color within each image reflects the thickness and height of the molecule. The brighter or whiter, the color, the thicker or taller the molecule; the darker the image, the thinner the molecule. Reprinted with permission from [43], D. Shu et al., Nano Lett. 4, 1717 (2004). © 2004, American Chemical Society.
4.8. Computer Modeling of RNA’s Three-Dimensional Structure
Once data is collected from such methods as crosslinking, chemical probing, chemical modification, chemical modification interference, and cryo-AFM, in addition to other methods, computer programs can be used to effectively translate the data into three-dimensional constructions.133–135 Programs such as NAHELIX, MANIP, PRENUC, NUCLIN, and NUCMULT136, 137 can be used to model structures based on such data and constraints. Modified distance geometry and molecular mechanics algorithms using simplified “pseudo atom” representations can be used to generate structures consistent with data from various techniques. A constraint satisfaction algorithm is combined with discrete representations of nucleotide conformations to refine the disturbed area in order to ensure the normal representation of all atoms.138
4.9. Ribonuclease Probing
Some ribonucleases are specific to RNA secondary structure.110, 122, 139 For example, RNases T1 (specific for GpN linkages), U2 (specific for ApN linkages), and S1 prefer to cleave single-stranded RNA. Nuclease V1 is specific for double stranded RNA. End-labeled RNA in various solutions with or without containing Mg++ can be probed by T1, U2, or V1 nucleases.94, 121 T1 and V1 can be used to distinguish the loops and helices. In addition, T1 nuclease can be used to study changes in RNA conformation. Since the activity of RNase T1 is Mg++ independent, this enzyme can also be used to investigate the conformational change of RNA in the presence or absence of Mg++ [Ref. 121], Besides end labeling, the cleavage site can also be identified by primer extension or 2D gel analysis.
4.10. Footprinting
Foot printing is a technique derived from nuclease probing or chemical modification and is particularly useful in probing the interaction of RNA with proteins.140 The procapsid/pRNA complex has been probed with nucleases A, T1, and V1.141 The RNA will be end-labeled, for example, with 32P and the optimal concentration of enzymes is determined empirically to ensure, on the average, one cleavage site per RNA molecule. Alternatively, the results will be analyzed by primer extension or 2D gel analysis.
4.11. Other Methods
Besides, other methods for RNA structure study are available. For example, lead (II) cleavage has previously been used to probe tRNA structure142 and now developed to investigate RNA structure in vivo.143 Hydroxyl radical footprinting can provide information of the solvent-accessible area of a folded RNA molecule.144 Nucleotide analog interference mapping make it possible to determine the chemical basis of RNA function and structure.145 Fluorescence techniques, in particular, fluorescence resonance energy transfer (FRET), have been employed to monitor RNA conformational changes.146, 147 Also, enzymatic footprinting, electrophoresis mobility shift assay (EMSA)37 have been used : for the studies on thermodynamics, RNA– protein interaction or conformational change of RNA.
5. RNA MOLECULES THAT FORM MULTIMERS
The diverse structure and processes of the many types of RNA play a crucial role in determining the functionality of a variety of organisms. Understanding the specific attributes and conformations of these types of RNA will lead to advances in medicine and nanotechnology. Following are some prominent examples of different types of RNA that form multimers by RNA–RNA interaction and their potential applications. Such interaction can be utilized for bottom-up assembly.
5.1. Motor pRNA of Bacteriophages phi29
A striking common feature in the maturation of all linear ds-DNA viruses is that their genome is translocated into a preformed protein shell.148–155 A DNA-translocating motor, powered by ATP hydrolysis, accomplishes this energetically unfavorable DNA transportation process. A RNA molecule called pRNA has been shown to have a novel and indispensable role in phi29 DNA packaging motor.76, 156
pRNA contains two functional domains (Fig. 1).154, 157 The procapsid binding domain is located at the central region91, 132, 141, 158 bases 23–97, while the DNA translocation domain is located at the 5′/3′ paired ends.104 This conclusion comes from different studies including (a) base deletion and mutation;93, 104, 105, 159, 160 (b) ribonuclease probing;94, 121, 141, 159 (c) oligo targeting;161, 162 (d) competition assays to inhibit phage assembly;162–164(e) UV-crosslinking;158 (f) psoralen crosslinking;121 and (g) primer extension.131
Fig. 1.
Sequence and structural elucidation of phi29 motor pRNA and related assemblages. (A) Primary and secondary structure of wild-type pRNA I-i′. The binding domain (shaded area) and the DNA translocation domain (the helical region) are marked with bold lines. The four bases in the right and left loops, which are responsible for inter-RNA interactions, are boxed. (B) Three-dimensional structure of wild-type pRNA I-i′ displayed as ribbon. (C) Diagrams depicting the pRNA monomer A-b′ with unpaired right/left loops. (D) pRNA dimers (A-b′)(B-a′). (E) pRNA trimers (A-b′)(B-e′)(E-a′). (F) pRNA monomer with unpaired right/left loops A-b′ and a 6-nucleotide palindromic sequence. (G) pRNA twin A-b′. (H) Size and three-dimensional computer model of phi29 pRNA trimer. Reprinted with permission from [43], D. Shu et al., Nano Lett. 4, 1717 (2004). © 2004, American Chemical Society.
The six pRNAs form a hexagon via intermolecular base-pairing between the right loop (bases 42–45) and the left loop (bases 82–85).163–168 pRNA dimers are the building blocks in hexamer assembly. pRNA has many attributes that make it a novel choice for nano-technological applications (Fig. 1–Fig. 4). Among these are its ATP-binding activity,103 its function in driving the DNA-packaging motor,154 its ability to form dimers, trimers, and hexamers,48, 132, 165–167, 169 its tight and stable folding,91, 93, 105, 135, 160 its two independent domains for structure and function,104, 105, 141, 158 and its functional similarity as compared to protein enzymes.76, 154 Since the size of RNA ranges from the angstrom to the nanometer scale, the bottom-up approach could be reasonably applied to RNA in nanotechnological applications. Larger RNA complexes can be constructed from the following three building blocks:
Fig. 4.
A mixture of two complementary twins, A-b′ and B-a′, assembled into two distinct supramolecular structures. (A) Two complementary twins were able to form a stable tetramer (double-twins) by assembling into a circular structure. (B) Concatemers of alternating twins formed when a twin interacted with two rather than one complementary twin. Reprinted with permission from [43], D. Shu et al., Nano Lett. 4, 1717 (2004). © 2004, American Chemical Society.
Monomer with intramolecularly self-complementary left and right loops.
Monomer with non-complementary left and right loops for intermolecular interaction.
Monomer with intermolecularly self-complementary left and right loops and palindromic 3′ ends.
5.1.1. Nomenclature of RNA Subunits
To simplify the description in the construction of RNA complexes, uppercase and lower case letters are used to represent the right and left hand loops of the pRNA respectively, (Fig. 1A). The matched letters indicate complementarity, whereas different letters indicate non-complementary loops. For example, pRNA(A-b′) contains right hand loop A(5′G45G46A47C48) and left hand loop b′ (3′U85G84C83G82), which can pair with the left hand loop a′(3′C85C84U83G82) and right hand loop B(5′A45C46G47C48) respectively, of pRNA(B-a′).
5.1.2. Construction of Dimers
Dimers are formed by the intermolecular interaction of interlocking right and left loops. Alone, the pRNAs A/b′ and B/a′ were inactive in DNA packaging, but when mixed together, DNA-packaging activity was restored.135, 166 This result can be explained by the trans-complementarity of the interlocking pRNA loops, i.e., the right-hand loop A of pRNA A/b′ can pair with the left-hand loop a′ of pRNA B/a′. Mixing two inactive pRNAs with interlocking loops, such as when pRNA A-b′ and B-a′ are mixed in a 1:1 molar ratio, results in the production of pRNA dimers with up to100% efficiency. The fact that the three inactive pRNAs are fully active when mixed together is not surprising, since the number of pRNAs in the DNA-packaging complex is a multiple of 2 (six copies), in addition to being a multiple of three. The mechanism of dimer formation by interlocking loop/loop interaction will be utilized to construct chimeric pRNA dimers carrying receptor-binding RNA aptamer and/or therapeutic RNA molecules.
5.1.3. Construction of Trimers
Several sets of trimers were constructed, e.g., A-b′, B-c′, and C-a′. Indeed, stable pRNA trimers are formed with a very high efficiency, up to 100%, by using sets of three interlocking pRNA.43, 48, 132, 166, 167 A mechanism similar to that of dimer formation by interlocking loop/loop interaction is utilized to construct chimeric pRNA trimers43 carrying receptor-binding RNA aptamer and/or therapeutic RNA molecules (see below). Nucleotides 23–97 are the central components in the formation of both dimers and trimers. The ability to form dimers or trimers is not affected by 5′ or 3′ end truncation before residue #23 and after residue #97, since one or two truncated or extended pRNAs can be incorporated into dimers while one, two, or three truncated or extended pRNAs can be incorporated into trimers. Specific nucleotide assignment before #29 and after #92 is not required for trimer formation, as long as the complementary helix at the 3′/5′ region is generated.
Trimers are verified by native gel or analyzed by 10–30% glycerol or 5–20% sucrose gradient sedimentation in the presence of 10 mM magnesium. It has been shown that monomers, dimers, and trimers were separated by sedimentation. Plotting the hypothetical molecular weight vs the log of migration distance (the fractional number) in gradient revealed a linear relationship.170 Trimers can be further confirmed by AFM imaging if necessary.
5.1.4. Construction of Hexamers
Recently, an “annealing approach” has been employed to assemble hexamer (unpublished data).
5.2. HIV RNA Dimer
HIV dimerization occurs through loop–loop interactions between the stem loops of two identical RNA molecules (Fig. 5). The resultant “kissing complex” refolds into a more stable complex known as an extended dimer. Still, little is known about the transitional pathway due to difficulties involved in directly observing the intermediate processes.171–173 Understanding this pathway could lead to significant improvements in the treatment of the disease caused by the virus and the application of such RNA–RNA interaction in nanotechnology.
Fig. 5.
Model of HIV-1 RNA and TectoRNA structure. (A) HIV-1 RNA secondary structure models. Model of the structural rearrangements in the TAR hairpin. TAR dimerization induced by the nucleocapsid protein gives rise to a kissing-loop complex and an extended duplex. The following sequences are marked: the central palindrome (red), extended base-pairing (blue), and secondary parallel helix (green; gray dotted lines in extended duplex). Adapted with permission from [216], E. S. Andersen et al., J. Biol. Chem. 279, 22243 (2004). © 2004, American Society for Biochemistry and Molecular Biology. (B) TectoRNAs employing two loop-receptor motifs and their modes of assembly. (a) Two modes of assembly used in this study. (b) Schematic of RNA assembly unit showing elements varied for this study. The tetraloop (L) is shown in red, the tetraloop receptor (R) in green, the linker (or hinge) in blue and the insert [comprising a helix and a second linker (or hinge)] in magenta (refer also to the 2D and 3D models in Fig. 2 and Fig. 3). Adapted with permission from [175], L. Jaeger et al., Nucleic Acids Res. 29, 455 (2001). © 2001, Oxford University Press.
5.3. TectoRNA
RNA has been formed into “jigsaw puzzle” pieces known as tectosquares.174, 175 Their programming allows for the production of nanoscale fabrics with controllable directionality and geometry41 (Fig. 5). In contrast to traditional materials, where materials are selected, rather than designed, for a given application, the next generation of materials will allow for the a priori design of novel building blocks, programmed for assembly and synthesized with particular needs in mind.
5.4. mRNA Dimerization in Drosophila Embryos
The multilization of mRNA is of considerable importance for the localization of ribonucleoprotein particles in Drosophila embryos.176, 177 Dimerization proceeds through a two-step mechanism via a hand-in-arm interaction, which includes a transition from the reversible initiation complex into a very stable one.178, 179 A series of different RNA fragments with the capacity to form a variety of structures, including open dimers and closed dimers and other multimers, were constructed.178–180 The initial open dimer is converted into a stable closed dimer, perhaps due to a kinetically-controlled mechanism.
6. APPLICATIONS OF RNA NANOTECHNOLOGY IN MEDICINE
Nanotechnology has brought about an unprecedented variety of revolutionary approaches for the detection and therapy of diseases. Due to their small size, nanoparticles can readily interact with biomolecules either on the surface or within cells. To take advantage of this, it is desirable to develop multifunctional engineered, targeted complexes capable of bypassing biological barriers to deliver multiple therapeutic agents directly to specific cells or tissues. Due to their easy access to many areas of the body, multivalent nanoparticles could offer a wealth of innovative tools with the potential to combine detection and therapy in ways previously unimaginable.
RNA molecular therapy is widely understood to be one of the most promising applications of modern biological science.181–185 There are many molecules that could potentially be utilized for nanotechnology-based RNA therapy such as small interfering RNA,65–67, 186 ribozymes71, 73, 184 and anti-sense RNA68, 69 which all show significant potentials to down-regulate specific gene expression in cancerous or viral-infected cells.187 The successful application of siRNAs and ribozymes for the treatment of cancer and infectious diseases requires overcoming the following obstacles: (1) the difficulty of entering the cell due to the size limit for membrane penetration; (2) degradation by exonucleases within the cell; (3) trafficking into the appropriate cell compartment; and (4) correct folding of the ribozymes or siRNA in the cell if fused to a carrier. At this time, the development of a safe, efficient, specific, and nonpathogenic nanodevice for the delivery of multiple therapeutic RNAs is highly desirable.
6.1. Small RNA Molecule with Therapeutic Potentials
6.1.1. siRNA188, 189
siRNA (short or small interfering RNA) are 20–25 nucleotide molecules of RNA that are produced when larger pieces of RNA are cleaved. Their function is to interfere with gene expression through the cleavage of mRNA with homologous sequence by a protein/RNA complex named RISC (RNA-induced silencing complex). An enzyme known as Dicer facilitates this process, which, as specified later in the gene therapy section of the manuscript, has already exhibited significant nanotechnological applicability.46, 47
6.1.2. Ribozyme70–73
Ribozymes are RNA molecules that have the capability to catalyze chemical reactions. They have highly significant potential for therapeutic purposes190 since they are capable of regulating gene function by intercepting and cleaving messenger RNA or viral genomic RNA. Even within the RNA subtype of ribozymes, there are significant structural and functional differences between different molecules; hammerhead ribozymes191 and hairpin ribozymes192 are so named because of their appearance when visualized by microscope. These are both categorized as small ribozymes; larger ribozymes include introns and RNase P.193, 194
6.1.3. Antisense RNA68, 69
Antisense RNA molecules function through connecting with particular target RNAs in order to regulate their functionality after transcription. They are called “antisense” because most often they are transcribed in a direction opposite that of the target RNA from the same DNA transcript. This allows for the production of a pair of transcripts that are complementary or nearly complementary. The combination of these two types of RNA produces a stable and easily detectable complex, and thus antisense RNA provides a promising route by which RNA/RNA interactions can be reliably probed. A variety of types of artificially produced antisense RNAs have been developed, and their applications for therapeutic purposes are just beginning to be fully realized.
6.1.4. Aptamer75, 98, 99, 195
Aptamers are a family of RNA- or DNA-based oligonucleotides obtained by in vitro screening that bind to selected targets, including proteins, organic compounds, and nucleic acids.75, 99 Their usefulness is found in their ability to recognize the ligands through the formation of binding pockets analogous to antigen–antibody interaction. SELEX is the method which screens for the aptamers from these randomized RNA pools. Starting with a library containing random RNA sequences with about 1014–17 varieties of RNA molecules, in vitro binding, elution, and reverse PCR amplification techniques allow for the selection of RNA molecules that efficiently bind to a specific receptor or ligand with high affinity.196–199 Using this technique, a number of aptamers that specifically recognize many kinds of targets, such as organic compounds, nucleotides, peptides, proteins, and receptors, have been obtained.103, 199–203
6.2. The Uniqueness of Therapeutic Nanoparticles Derived from phi29 pRNA
Until recently, the use of small RNA in gene therapy was significantly hampered by the difficulties involved in producing a safe and efficient system by which specific cells could be targeted. Recently, phi29 pRNA has been used to harbor small therapeutic RNA molecules46, 47 for the delivery to specific cancer cells (Fig. 6).
Fig. 6.
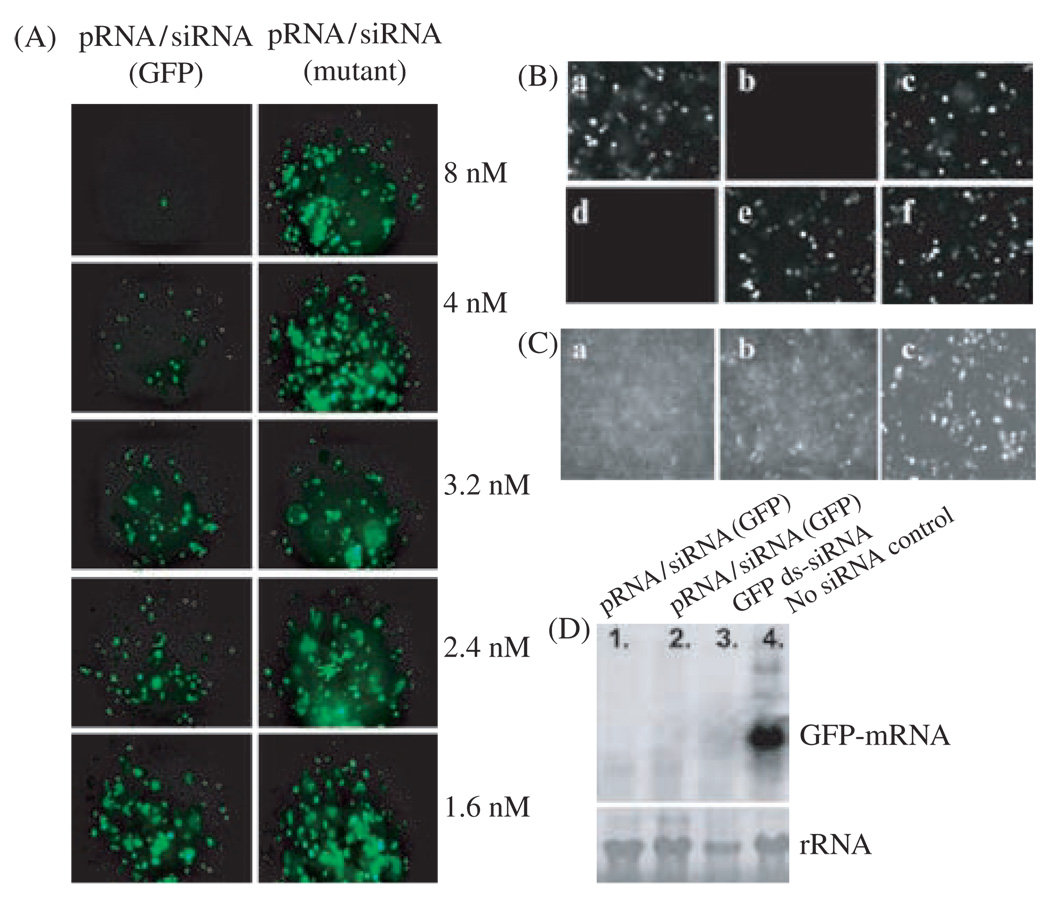
Functional assay of chimeric pRNA/siRNA(GFP) by transfection. (A), (B), and (C) are fluorescence microscopy images to show the silencing of GFP gene by transfection. (A) Dose-dependent silencing of GFP gene by chimeric pRNA/siRNA(GFP) (the left column). A mutant pRNA/siRNA on the right column, served as negative control. (A) GFP expression of cells transfected with various RNA. (a) No RNA; (b) Synthesized double-stranded siRNA(GFP); (c) Double-stranded siRNA(LacZ) control; (d) pRNA/siRNA(GFP); (e) pRNA/siRNA(mutant); (f) pRNA vector alone (C) Comparison of the performance of chimeric pRNA/siRNA(GFP) (a) and the conventional double-stranded -siRNA(GFP) (b) at the same molar concentration. Panel (c) is the control with no siRNA treatment. (D) Northern blot to examine the effect of chimeric pRNA/siRNA(GFP) on GFP mRNA level after transfection. Lanes 1 and 2 show the effects of two different constructs of pRNA/siRNA(GFP). Lane 3 is the double-stranded siRNA and lane 4 is cells without RNA treatment. rRNA was used as loading control. Reprinted with permission from [46], S. Guo et al., Human Gene Therapy 16, 1097 (2005). © 2005, Mary Ann Liebert, Inc.
By utilizing RNA nanotechnology, both therapeutic siRNA and a receptor-binding RNA aptamer was engineered into individual pRNAs of phi29’s motor.46, 47 The RNA building block harboring siRNA or other therapeutic molecules was fabricated subsequently into a trimer through the interaction of engineered right and left interlocking RNA loops.43, 48, 132, 166, 167 The incubation of the protein-free nanoscale particles containing the receptor-binding aptamer or other ligands resulted in the binding and co-entry of the trivalent therapeutic particles into cells, subsequently modulating the apoptosis of cancer cells and leukemia model lymphocytes in cell culture and animal trials. The use of such antigenicity-free 20–40 nm particles holds promise for the repeated long-term treatment of chronic diseases. Such protein-free 30 nm nanoparticles will allow for repeated long-term administration and avoid problems of short retention time of small molecules less than 30 nm and the difficulty of delivery of particles larger than 100 nm.
In addition, since there are six chimeric pRNAs in the hexamer, a polyvalent nanoparticle can be constructed by utilizing six available positions to carry molecules for cell recognition, therapy, and detection (Fig. 12). Besides the small RNA and folate reported, other materials such as heavy metal, quantum dots, fluorescent beads or radioisotopes can also be conjugated for the detection of cancer signatures at different stages of development. The conjugation of endosome-disrupting chemicals could be used to promote the release of internalized therapeutic reagent from the endosome to improve therapeutic efficacy. This polyvalent RNA complex can also potentially be used for treating chronic viral infections through targeting at the specific virus-glycoproteins displayed on the infected cell surface.
Fig. 12.
The potential use of pRNA hexamers as polyvalent gene delivery vectors. Six copies of pRNA have been found to form a hexameric ring to drive the DNA-packaging motor of bacterial virus phi29. There would therefore be six positions available to carry foreign moieties for targeting, therapy, and detection, as shown in the figure. Reprinted with permission from [46], S. Guo et al., Human Gene Therapy 16, 1097 (2005). © 2005, Mary Ann Liebert, Inc.
It is well-established in the scientific community that RNAs do not induce a detectable immune response except when complexed with proteins.204, 205 The use of RNA as a delivery vehicle could avoid the problems of immune response and the rejection of protein vectors after repeated long-term drug administration for chronic diseases.
6.3. Construction of Chimeric pRNA Subunit Harboring Therapeutic Moieties
An important task in constructing chimeric monomers is to protect the delivered therapeutic molecule from exonuclease degradation, a problem commonly encountered in gene therapy. Therapeutic RNA molecules are typically vulnerable to RNase digestion. They can be protected by connecting both ends of the foreign RNA with both ends of the pRNA. Circular permutation strategies were also applied for this purpose.92, 93, 125, 206 The feasibility of constructing circularly permuted RNAs lies in the close proximity of the native 5′ and 3′ ends.104 The feasibility of constructing functional RNA that harbors actual ribozymes has been recently demonstrated.20 The monomer pRNA carrying the foreign daughter RNA molecule can also contain the same designable right and left loops for the formation of dimers, trimers, and hexamers. This section describes the construction of one subunit of the building block of the RNA nano-particles.
6.3.1. Harboring siRNA
Recently, post-transcriptional gene-silencing and RNA interference have been investigated extensively in a wide variety of organisms using double-stranded RNAs.207, 208 This RNA is processed into small interfering RNAs (siRNAs) of 19–25 nucleotides,209, 210 which act as guides for the formation of the silencing enzymatic complex required for cleavage of the target mRNAs.77, 211 These siRNAs specifically suppress the expression of a target mRNA with a sequence identical to the siRNA. Although the detailed mechanism of post-transcriptional gene silencing and RNA interference remains to be elucidated, this powerful new technology for selective inhibition of specific gene expression employing siRNAs has shown great promise in the therapy of cancer and viral infections.64, 67
Bacterial phage phi29 pRNA contains a double-stranded helical domain at 5′/3′ end and an intermolecular binding domain, which fold independently of each other. Complementary modification studies have revealed that altering the primary sequences of any nucleotide of the helical region does not affect pRNA structure and folding as long as the two strands are paired (Fig. 1).104 Extensive studies revealed that siRNA is a double-stranded RNA helix.64, 65, 67, 209 To test whether it is possible to replace the helical region in pRNA with double-stranded siRNA, a variety of chimeric pRNAs with different targets were constructed to carry siRNA connected to nucleotides #29 and 91 of the pRNA (Fig. 1), resulting in pRNA/siRNA(GFP), pRNA/siRNA(Luciferase), and pRNA/siRNA(Survivin).
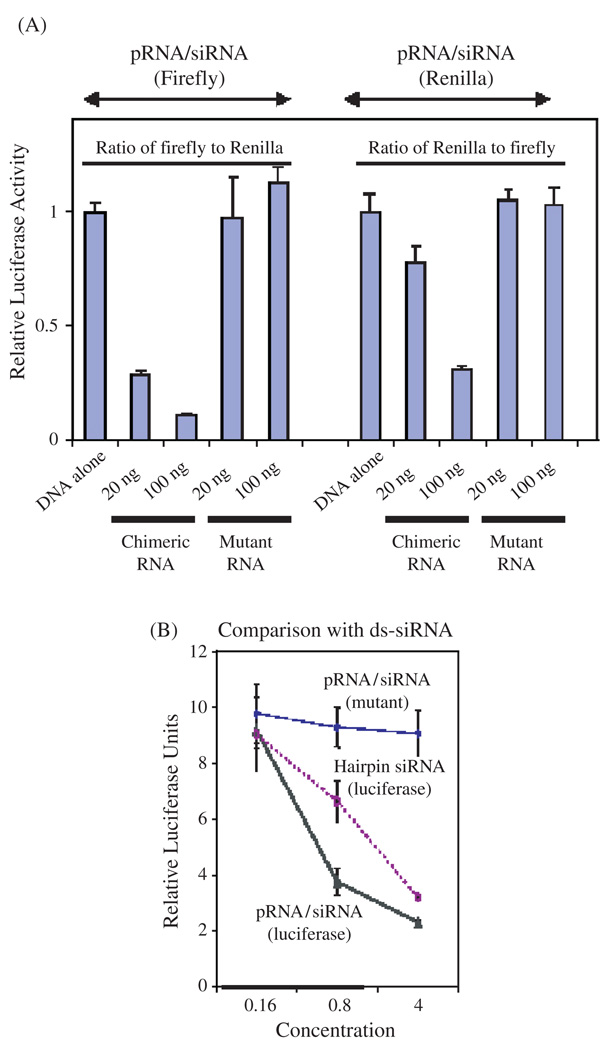
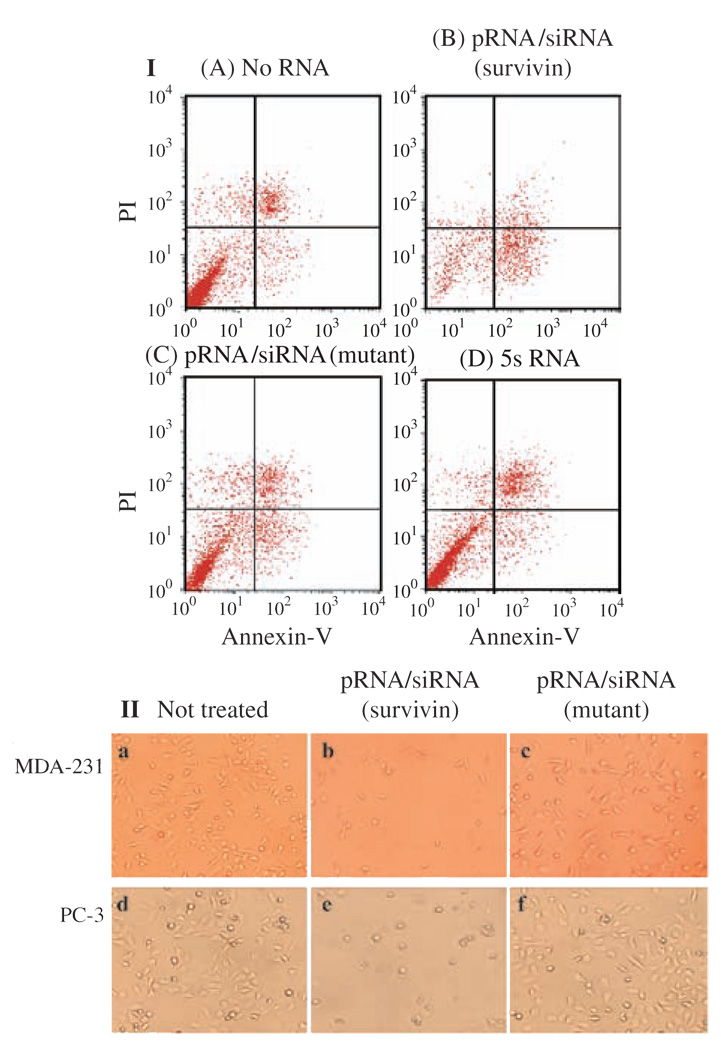
Chimeric pRNA/siRNA(GFP) was constructed and the specific inhibition of GFP expression by this RNA building block was demonstrated in mRNA and protein level (Fig. 6). pRNA/siRNA(Luciferase) was another building block RNA proven to successfully inhibit target gene expression (Fig. 7). Two chimeric pRNA/siRNA constructs targeting either firefly luciferase or Renilla luciferase were found to inhibit target luciferase gene expression without showing non-specific inhibition to internal control luciferase gene expression. In addition, chimeric pRNA/siRNA exhibited equivalent or superior inhibitory effects on target gene expression compared with chemically synthesized double-stranded siRNA or hairpin siRNA (GFP) (Fig. 6). pRNA/siRNA(Survivin) and pRNA/ribozyme(Survivin) targeting an anti-apoptosis factor survivin, which is necessary to down-regulate genes involved in tumor development and progression, was shown to suppress survivin expression, initiate apoptosis and cell death (Fig. 8).
Fig. 7.
Functional assay of chimeric pRNA/siRNA targeting luciferase by transfection. (A) Dual reporter luciferase assay showing the specific knockdown of firefly luciferase or renilla luciferase expression by pRNA/siRNA(Firefly) or pRNA/siRNA(Renilla), respectively in a dose dependent manner. (B) Comparison of the activities of the conventional hairpin siRNA(Luciferase) and pRNA/siRNA(Luciferase). pRNA/ siRNA(mutant) with mutations in siRNA sequences was included as a nonspecific control. Reprinted with permission from [46], S. Guo et al., Human Gene Therapy 16, 1097 (2005). © 2005, Mary Ann Liebert, Inc.
Fig. 8.
Apoptosis and cell death induced by transfection of chimeric pRNA harboring siRNA targeting survivin. (I) Breast cancer MCF-7 cells were transfected with pRNA/siRNA(Survivin) and apoptosis was monitored using PI/annexin V double-labeling followed by flow cytometry. Cells at the lower right quadrant represent apoptotic cells. (II) Breast cancer cells MDA-231 and prostate cancer cells PC-3 were transfected with 20 pmol of pRNA/siRNA(Survivin) in 24-well plates and images were taken in 24 hours after transfection. The mutant pRNA/siRNA was transfected in parallel as a negative control. Reprinted with permission from [46], S. Guo et al., Human Gene Therapy 16, 1097 (2005). © 2005, Mary A nn Liebert, Inc.
6.3.2. Harboring Hammerhead Ribozyme20
Connecting the pRNA 5′/3′ ends with variable sequences did not disturb its folding and function. These unique features, which help prevent two common problems—exonuclease degradation and misfolding in the cell, make pRNA an ideal vector to carry therapeutic RNAs. A pRNA-based vector was designed to carry hammerhead ribozymes that cleave the hepatitis B virus (HBV) polyA signal. The chimeric HBV-targeting ribozyme was connected to the pRNA 5′/3′ ends as circularly permuted pRNA. Two cis-cleaving ribozymes were used to flank and process the chimeric ribozyme. The hammerhead ribozyme including its two arms for HBV targeting was able to fold correctly while escorted by the pRNA. The chimeric ribozyme cleaved the polyA signal of HBV mRNA in vitro almost completely. Cell culture studies showed that the chimeric ribozyme was able to enhance the inhibition of HBV replication when compared with the ribozyme not escorted by pRNA, as demonstrated by Northern blot and e-antigen assays. pRNA could also carry another hammerhead ribozyme to cleave other RNA substrate.
6.3.3. Harboring Antisense RNA68, 69
Antisense RNAs are single-stranded RNA molecules complementary to mRNA. Antisense RNA can inhibit gene expression in the cell.68, 69 This applies to the construction of chimeric RNA monomers carrying single-stranded antisense RNA. All antisense RNA used to block gene function are placed at the 3′-end of the pRNA.
6.3.4. Harboring Receptor-Binding Aptamer
To achieve specific delivery of therapeutic complexes, it is often necessary to incorporate a moiety that recognizes signature molecules on cell surfaces. In comparison to antibodies and phage-displayed peptides, RNA aptamer is an attractive alternative since it avoids the induction of immune responses.204 Using the SELEX approach, a number of RNA aptamers were obtained that specifically recognize a particular cell surface receptor such as CD4.203 One CD4-binding RNA aptamer was chosen to construct chimeric pRNA/aptamer(CD4) via a mutual 5′/3′ end connection (Fig. 1D). The pRNA vector was reorganized into a circularly permuted form, with the nascent 5′ and 3′ ends relocated to residues #71 and 75, respectively, of the original pRNA sequence. The 71/75 end is located in a tightly-folded area135 to bury and protect the ends from exonuclease degradation in vivo.20
6.3.5. Adding Drugs, Folate, or Other Chemical Moieties or Groups to RNA
Some cancer cells overexpress the receptor for folate, which has been shown to be a useful target for the specific delivery of drugs in cancer therapy.212, 213 Other drugs might be incorporated into the therapeutic complex to enhance the therapeutic effect. Chemicals for image detection or endosomal disruption can also be connected to the RNA complex. Therefore, it is desirable to have simple and efficient methods for the conjugation of such groups to RNA. Such additions include labeling with pCp, DIG, phosphate, as well as biotin, amine, sulfhydryl, and carboxyl groups, which are simple and efficient groups for conjugation. NHS can be used to covalently link particles with a primary amine group. NH2-group can be used to link any particles with a COOH group with the help of EDC, or with Cysteine or SH-group using a variety of amine and sulfhydryl reactive heterobifunctional cross-linkers such as BMPS. Sulfhydryl-groups can be used to link NH2-groups via BMPS or with any common chemicals that contain maleimide. Maleimide can be used to link covalently with any particles that contain SH-group. In addition, NH2/NH2 interaction can be achieved via commercially available heterobifunctional crosslinkers as well.
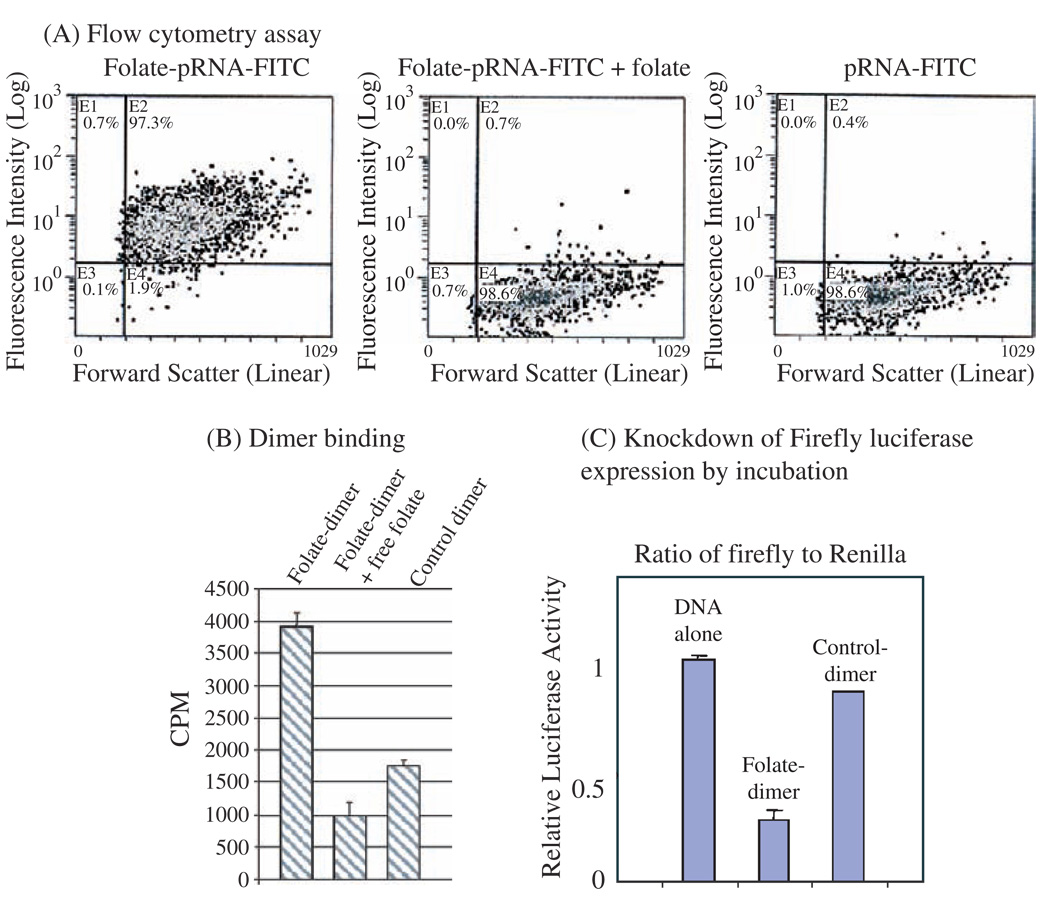
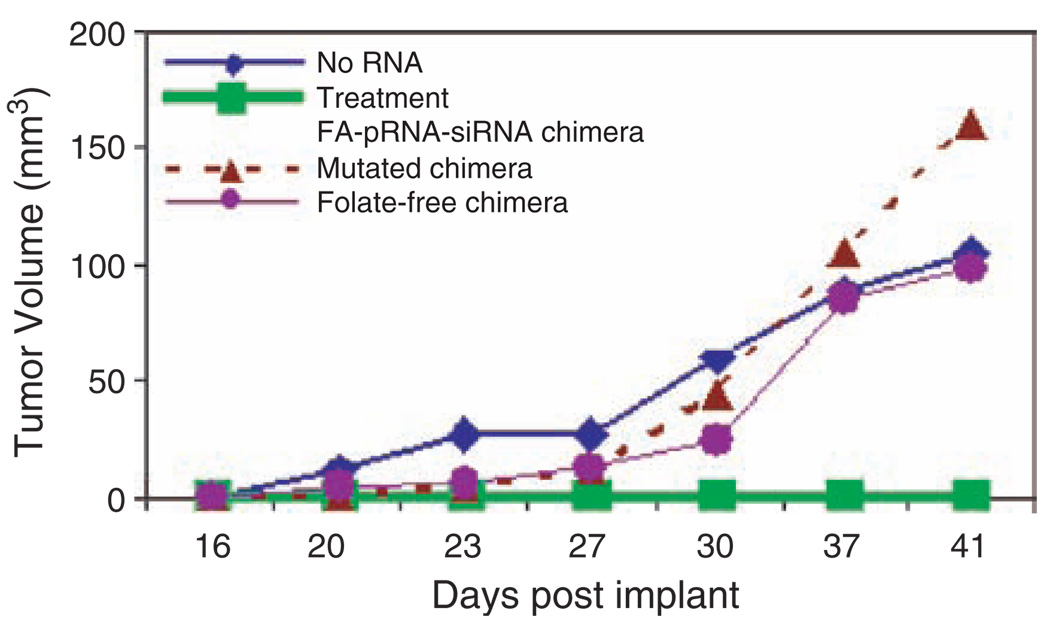
Folate receptors are over-expressed in various types of tumors such as human nasopharyngeal epidermal carcinoma but are generally absent in normal adult tissues. The folate molecule, which binds to folate receptor with high affinity, was incorporated into the 5′ end of RNA by in vitro transcription using folate-AMP. Specific cell binding of pRNA/folate dimer was demonstrated by incubating folate receptor positive KB cells with dimeric pRNA which contains a folate labeled pRNA(A-b′) and a [H3]-pRNA(B-a′). Then the nanoparticles containing both folate-labeling and chimeric pRNA/siRNA were shown to knockdown the firefly luciferase gene expression by a simple incubation process (Fig. 10). Furthermore, animal trials were performed to demonstrate specific suppression of tumorigenicity of cancer cells by ex vivo delivery of the nano-particle containing both folate labeling and chimeric pRNA/siRNA against survivin. The potential of this RNA complex to suppress tumor formation was tested in athymic nude mice. Human nasopharyngeal epidermal carcinoma KB cells were incubated with a chimeric RNA complex with or without folate before being introduced into the nude mice by axilla injection. The mice receiving only cancer cells developed tumors within 3 weeks, while the group of mice that received cancer cells pre-treated with the pRNA complex containing both folate-pRNA and pRNA/siRNA(Survivin) did not develop tumors (Fig. 11). The inhibition of tumor formation is specific since the control RNA complex without folate or the RNA complex containing mutations in survivin siRNA did not affect tumor development in other mice groups.
Fig. 10.
Specific delivery of chimeric pRNA/siRNA by folate-pRNA. (A) Flow cytometry analyses of the binding of folate-pRNA-FITC to KB cells. Cells were incubated with folate-pRNA labeled with FITC (left panel). Cells in the middle panel were pre-incubated with free folate, which served as a blocking agent to compete with folate-pRNA for binding to the receptor. Binding was also tested using folate-free pRNA labeled with FITC (right panel) as a negative control. The percentages of FITC-positive cells are shown in the upper right quadrants. (B) Specific binding of folate-pRNA dimer to KB cells. After incubation of cells with the [3H]-folate-pRNA dimer in the presence (center column) or absence (left column) of free folate, cells were isolated and subjected to scintillation counting. The right column is the [3H]-dimer without folate labeling as a negative control. (C) In the knockdown assay by incubation, folate-chimeric dimer complex containing pRNA(B-a′)/folate and pRNA(A-b′)/siRNA(firefly) was incubated with KB cells for 3 hours to allow the binding and entry of RNA. The luciferase level was measured in the next day by the dual reporter system. The control dimer was identical to the folate dimer except for its lack of folate labeling. Reprinted with permission from [46], S. Guo et al., Human Gene Therapy 16, 1097 (2005). © 2005, Mary Ann Liebert, Inc.
Fig. 11.
Animal trials for cancer therapy using the fabricated RNA nanoparticles. (A) Injection without the pRNA/siRNA chimera (No RNA); (B) Treatment with RNA chimera containing folate-pRNA and siRNA(Survivin); (C) Treatment with RNA chimera containing folate-pRNA and siRNA(Survivin) with mutations in the siRNA sequence; (D) Treatment with pRNA-siRNA chimera that does not contain a folate at its 5′ end. Reprinted with permission from [47], A. Khaled et al., Nano Lett. 5, 1797 (2005). © 2005, American Chemical Society.
6.4. Assembly of Deliverable Dimeric, Trimeric Therapeutic RNA Nano-Particles
RNA nano-particles containing two or three chimeric RNA building blocks were constructed. Nano-particles carry multiple components, including molecules for specific cell recognition, image detection, and therapeutic treatment. One subunit of the deliverable RNA complex (dimer, trimer) was modified to carry an RNA aptamer or other receptor binding ligands that bind a specific cell-surface receptor, thereby inducing receptor-mediated endocytosis. The second subunit carried reporting molecules such as fluorescent beads. The third subunit was altered to carry therapeutic siRNA, ribozyme RNAs, antisense RNA, or other drugs to be delivered.47
6.5. Sequence Requirement of the Hand-in-Hand Loops in Chimeric Trimer Formation
Bases 45–48 and 82–85 of wild type pRNA in the left-and right-hand loops, respectively, were found to engage in pRNA/pRNA interactions.131, 166 Without considering tertiary interaction, in some cases only two G/C pairs between the interacting loops could allow the formation of pRNA multimers. When all four nucleotides were paired, at least one G/C pair was required. The maximum number of base pairings between the two loops to allow optimal polymer formation was five. The minimum number of nucleotides needed for pRNA/pRNA interaction in the right and left loop was five and three, respectively. Our results suggest that a 75-nucleotide RNA segment, nucleotide 23–97, is a self-folded independent domain involved in RNA/RNA interaction in pRNA trimer formation, while nucleotide 1–22 and 98–120 were dispensable for RNA/RNA interaction.
The mechanism of pRNA trimer formation by interlocking loop/loop interaction was utilized for the fabrication of the trimer of chimeric pRNA harboring receptor-binding RNA aptamer and/or therapeutic siRNA (Fig. 9). Individual chimeric pRNA building block was engineered to carry one daughter RNA molecule such as siRNA or receptor-binding aptamer. Each building block was intentionally designed to have specific right or left loops, such as A-b′ (right–left), to interact with other building block. The appropriate folding of pRNA and their competency in forming trimers were confirmed. Mixing of individual chimeric pRNAs with counterpart partners with appropriate interlocking loops resulted in the efficient formation of the desired trimer, as documented by gel electrophoresis, AFM imaging, and sucrose gradient sedimentation. This suggests that RNA trimers were generated from the monomeric building block despite the replacement of the 5′/3′ helix with ds-siRNA or the connection of the 5′/3′ end to a CD4-binding aptamer.
Fig. 9.
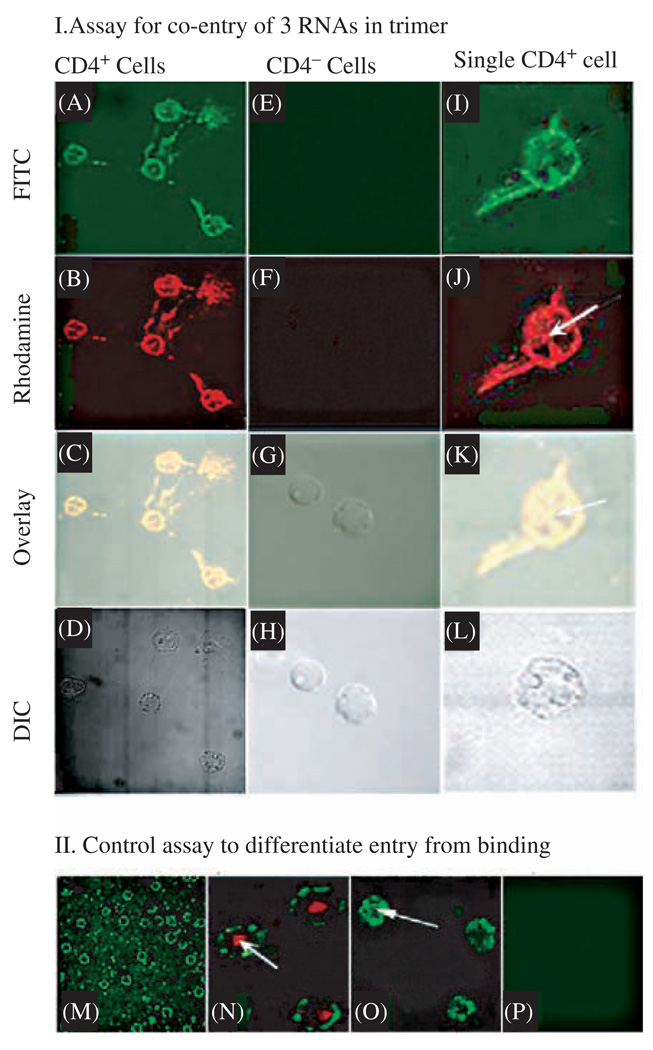
Confocal microscopy showing the specific and simultaneous delivery of three components to CD4 over-expressing cells. (I) Assay for the binding of pRNA trimer containing pRNA(A-b′)/aptamer(CD4), pRNA(B-e′)-FITC, and pRNA(E-a′)-Rhodamine to CD4 over-expressing T cells (A–D of left column, and I–L of right column) and CD4 negative cells T cells (E–H of middle column). A, E, and I were imaged with an FITC filter; whereas B, F, and J were viewed with a Rhodamine filter; C, G, and K are overlays; and D, H, and L are DIC images. The right column represents a close-up view of CD4 over-expressing cells. Arrows point to the complexes that had entered the cell. (II) Section of confocal microscopy images to differentiate between binding (M) and cell entry (arrows in N and O) as well as negative control (P). Binding of FITC-labeled pRNA trimer containing CD4-binding aptamer to lymphocytes was shown as a circle and entry was shown as a green spot inside cell (arrow in O). The red color in N is a positive entry control of transferrin labeled with Texas red. Reprinted with permission from [47], A. Khaled et al., Nano Lett. 5, 1797 (2005). © 2005, American Chemical Society.
6.6. Confirmation of Binding and Entry of the Nano-Particles into Cells
To determine whether RNA can serve as an effective vehicle for the concurrently delivery of multiple therapeutic particles, a trimeric pRNA/aptamer(CD4) complex was constructed and additional components were added to it in order to demonstrate the successful binding and entry of the multivalent complex into cells. In this trimer, one of the subunits was the pRNA(A-b′) chimera harboring the CD4-binding aptamer. The other two subunits carry the FITC (green) molecule and Rhodamine (orange label) molecule, respectively. To test the binding and entry of the trimer, a confocus section approach was attempted with the use of difference filters (Fig. 9). Binding of the trimer to the cells was demonstrated by the finding that fluorescence color appeared on the cell surface as a circle, which is one layer of the cell from the confocal microscope. Entry of the trimer into the cell via CD4-mediated endocytosis was demonstrated by the appearance of a fluorescence spot within the cell (Fig. 9A, B, K, and L). Such binding and entry was specific as long as CD4 was present, since no fluorescence was observed on CD4-negative cells (Fig. 9-I, lower panel). The appearance of both types of fluorescence on the cell surface suggests that pRNA(B-c′) and pRNA(C-a′) had been conjugated onto CD4-binding aptamer A-b′, which had no fluorescence label but had the capacity for CD4 binding. Figure 9K–L, when compared with Texas Red-labeled transferrin as the positive control for entry (Fig. 9J), further illustrates the success in entry. The similarity and overlap of the FITC image with the rhodamine image in Figures 9K – L strongly indicates that the FITC-labeled pRNA(B-c′) had combined with the rhodamine-labeled pRNA(C-a′) and been co-delivered to the cell. All of these results indicate that RNA trimers can serve for the delivery of multiple therapeutic components.
7. PERSPECTIVES
Phi29 pRNA has a strong tendency to form dimers, trimers, and hexamers due to the interaction of the interlocking right- and left-hand loops, which are maneuverable and controllable. It has been shown that insertion of a ribozyme or siRNA at pRNA’s 3′/5′ paired ends does not interfere with the folding of the pRNA itself, the ribozyme or the siRNA. In addition, it also does not affect the formation of dimers, trimers, and hexamers. Instead, the pRNA can escorte the ribozyme or siRNA to destroy Hepatitis B virus or cancer cells. These features enable the development of several new approaches for prostate cancer detection and treatment. The formation of defined pRNA multimers, which are controllable through the design of complementary interlocking loop sequences, will allow for the construction of detection and delivery vehicles. These vehicles will carry multiple components, including molecules for specific cell recognition, image detection, endosome disruption, and therapeutic treatment. For example, in hexameric complex, one subunit of the deliverable RNA complex (dimer, trimer, or hexamer) will be modified to carry an RNA aptamer that binds a specific cell-surface receptor, thereby inducing receptor-mediated endocytosis. The second subunit of the hexamer will carry reporting molecules like heavy metal complexes, quantum dots, fluorescent beads, or radioisotopes for prostate cancer detection. The third subunit of the hexamer will be altered to carry components that will be used to enhance endosome disruption so that the therapeutic molecules are released. The fourth and fifth subunits of the RNA complex will carry therapeutic siRNA, ribozyme, antisense RNA, or other drugs to be delivered. The sixth subunit of the hexamer will be designed to allow for the detection of apoptosis, the planned outcome of this treatment. Although phi29 motor pRNA is unusually stable and the formation of hexamer makes the complex more resistant to RNAase digestion due to the intertwining nature, further procedure to enhance the stability of the RNA complex in blood stream is still needed. Nucleotide derivatives, such as 2′-F-pyrimidine214 or spiegelmer,215 will be incorporated into the RNA to produce stable in vitro RNA transcripts that are resistant to RNase digestion. Other nucleotides with lower cell toxicity will be further developed to enhance safely of this therapeutic reagent produced by RNA nanotechnology.
Fig. 2.
Polyacrylamide gel showing monomers, dimers, trimers, twins, tetramers, and arrays. (A) Native and denatured gel. (B) Test of the stability of pRNA dimers under different conditions. Reprinted with permission from [43], D. Shu et al., Nano Lett. 4, 1717 (2004). © 2004, American Chemical Society.
Acknowledgments
The work in the author’s laboratory was supported by NIH Grants RO1-GM59944 and R01-EB003730 (Nanosciences and Nanotechnology in Biology and Medicine), as well as DOD Breast Cancer Concept award DAMD17-03-1-0589 and DOD Prostate Cancer Grant W81XWH-05-1-0158. I thank Drs. Larry Glickman and Chengde Mao for insightful comments on the manuscript and Jeremy Hall, Jane Kovach, and Songchuan Guo for their assistance in manuscript preparation.
Biography

Dr. Guo is a Professor of Molecular Virology and Biomedical Engineering at Purdue University where he serves as the Director of the Lab of Gene Therapy. His research expertise lies in viral DNA packaging and RNA biochemistry. He has been working on bottom-up assembly of biomacromolecules in nanoscale for more than twenty years. For example, he constructed an in-vitro phi29 DNA packaging motor (the most powerful biomotor studied to date) (PNAS, 1986); discovered the motor pRNA (the viral RNA that binds ATP) (Science, 1987); and served as the guest editor of the Journal Viral Assembly in 1994. However, he has not used the word “nanotechnology” until recently. Other pioneering work includes his demonstration that RNA can serve as building blocks for the construction of devices in nanotechnology. His latest contribution is the application of RNA nanotechnology for gene or drug delivery to treat cancers and viral-related diseases and for pathogen detection. His lab is the first to assemble infectious dsDNA virions in the test tube using synthetic components and purified recombinant nanomotors. His finding that pRNA binds ATP and forms a hexamer to gear the phi29 DNA-packaging motor suggests that RNA might play a role that protein enzymes (such as helicase) play. His research effort has led to an NIH “First Award” in 1992, the “Pfizer Distinguished Faculty Award for Research Excellence” in 1995, the “Purdue Faculty Scholar” in 1998, and the “Seed Award” in 2004. Dr. Guo came to the United States at the end of 1983. He finished his Ph.D. training in Microbiology and Genetics in Dr. Dwight Anderson’s lab at the University of Minnesota/School of Dentistry in May of 1987. He was a postdoctoral fellow with Dr. Enzo Paoletti in Wadsworth Center and later a visiting scientist with Dr. Bernard Moss—a member of National Academy of Sciences at NIH. He joined Purdue in 1990 and became a full Professor in 1997. His lab has been affiliated with a number of interdisciplinary programs including Genetics, Biochemistry and Molecular Biology, Virology, the Cancer Research Center, Veterinary Pathobiology, Nanotechnology, and Biomedical Engineering. He also founded the Purdue Graduate Program in Viral Research. He has also chaired and been the keynote speaker at many international conferences. He is an editor or editorial board member for six journals, including four in nanotechnology and bionanotechnology. He published 70 original papers in high impact refereed journals including Science, PNAS, and Molecular Cell.
References and Notes
- 1.Niemeyer CM. Trends Biotechnol. 2002;20:395. doi: 10.1016/s0167-7799(02)02022-x. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt OG, Eberl K. Nature. 2001;410:168. doi: 10.1038/35065525. [DOI] [PubMed] [Google Scholar]
- 3.Baneyx G, Baugh L, Vogel V. Proc. Natl. Acad. Sci. USA. 2002;99:5139. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyman P, Valluzzi R, Goldberg E. Proc. Natl. Acad. Sci. USA. 2002;99:8488. doi: 10.1073/pnas.132544299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberger J, He R, Zhang Y, Lee S, Yan H, Choi HJ, Yang P. Nature. 2003;422:599. doi: 10.1038/nature01551. [DOI] [PubMed] [Google Scholar]
- 6.Zandonella C. Nature. 2003;423:10. doi: 10.1038/423010a. [DOI] [PubMed] [Google Scholar]
- 7.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 8.Serwer P. J. Struct. Biol. 2003;141:179. doi: 10.1016/s1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 9.Oster G, Wang H. Nature. 2003;396:279. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 10.Berry RM. Philos Trans. R. Soc. Lond. B Biol. 2003;355:503. doi: 10.1098/rstb.2000.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoriev DN, Moll W, Hall J, Guo P. Encyclopedia of Nanoscience and Nanotechnology. 2003;1:361. [Google Scholar]
- 12.Inoue A, Saito J, Ikebe R, Ikebe M. Nat. Cell Biol. 2002;4:302. doi: 10.1038/ncb774. [DOI] [PubMed] [Google Scholar]
- 13.Moore SD, Prevelige PE., Jr Curr Biol. 2002;12:R96. doi: 10.1016/s0960-9822(02)00670-x. [DOI] [PubMed] [Google Scholar]
- 14.Sablin EP, Fletterick RJ. Curr. Opin. Struct. Biol. 2001;11:716. doi: 10.1016/s0959-440x(01)00265-2. [DOI] [PubMed] [Google Scholar]
- 15.Shenton W, Pum D, Sleytr UB, Mann S. Nature. 1997;389:585. [Google Scholar]
- 16.Carazo J, Santisteban A, Carrascosa J. J. Mol. Biol. 1985;183:79. doi: 10.1016/0022-2836(85)90282-7. [DOI] [PubMed] [Google Scholar]
- 17.Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD. Nat. Biotechnol. 2002;20:353. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- 18.Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. Nano Lett. 2003;3:413. [Google Scholar]
- 19.Soong RK, Bachand GD, Neves HP, Olkhovets AG, Craighead HG, Montemagno CD. Science. 2000;290:1555. doi: 10.1126/science.290.5496.1555. [DOI] [PubMed] [Google Scholar]
- 20.Hoeprich S, ZHou Q, Guo S, Qi G, Wang Y, Guo P. Gene Therapy. 2003;10:1258. doi: 10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- 21.Craighead HG. Science. 2000;290:1532. doi: 10.1126/science.290.5496.1532. [DOI] [PubMed] [Google Scholar]
- 22.Fennimore AM, Yuzvinsky TD, Han WQ, Fuhrer MS, Cumings J, Zettl A. Nature. 2003;424:408. doi: 10.1038/nature01823. [DOI] [PubMed] [Google Scholar]
- 23.Mao C, LaBean TH, Relf JH, Seeman NC. Nature. 2000;407:493. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 24.Moll D, Huber C, Schlegel B, Pum D, Sleytr UB, Sara M. Proc. Natl. Acad. Sci. USA. 2002;99:14646. doi: 10.1073/pnas.232299399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nulf CJ, Corey DR. Nucleic Acids Res. 2002;30:2782. doi: 10.1093/nar/gkf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soukup GA, Breaker RR. Trends Biotechnol. 1999;17:469. doi: 10.1016/s0167-7799(99)01383-9. [DOI] [PubMed] [Google Scholar]
- 27.H g Yan, Zhang X, Shen Z, Seeman NC. Nature. 2002;415:62. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Bergstrom DE. Angewandte Chemie. 1997;36:111. [Google Scholar]
- 29.Seeman NC, Belcher AM. Proc. Natl. Acad. Sci. USA. 2002;99:6451–5. doi: 10.1073/pnas.221458298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess H, Vogel V. Rev. Mol. Biotechnol. 2001;82:67. doi: 10.1016/s1389-0352(01)00029-0. [DOI] [PubMed] [Google Scholar]
- 31.Yan H, LaBean TH, Feng L, Reif JH. Proc. Natl. Acad. Sci. USA. 2003;100:8103. doi: 10.1073/pnas.1032954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner MG, Hutchison JE. Nat. Mater. 2003;2:272. doi: 10.1038/nmat853. [DOI] [PubMed] [Google Scholar]
- 33.Keren K, Krueger M, Gilad R, Ben Yoseph G, Sivan U, Braun E. Science. 2002;297:72. doi: 10.1126/science.1071247. [DOI] [PubMed] [Google Scholar]
- 34.Gerion D, Parak WJ, Williams SC, Zanchet D, Micheel CM, Alivisatos AP. J. Am. Chem. Soc. 2002;124:7070. doi: 10.1021/ja017822w. [DOI] [PubMed] [Google Scholar]
- 35.Studnicka GM, Rahn GM, Cummings IW, Salser WA. Nucleic Acids Res. 1978;5:3365. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. Science. 1989;244:48. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 37.Jaeger JA, SantaLucia JJ, Tinoco IJ. Ann. Rev. Biochem. 1993;62:255. doi: 10.1146/annurev.bi.62.070193.001351. [DOI] [PubMed] [Google Scholar]
- 38.Pleij CWA, Bosch L. Meth. Enzymol. 1989;180:289. doi: 10.1016/0076-6879(89)80107-7. [DOI] [PubMed] [Google Scholar]
- 39.Correll CC, Freeborn B, Moore PB, Steitz TA. Cell. 1997;91:705. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 40.Chen C, Zhang C, Guo P. RNA. 1999;5:805. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaeger L, Leontis NB. Angew. Chem. Int. Ed. Engl. 2000;39:2521. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 42.Hansma HG, Oroudjev E, Baudrey S, Jaeger L. J. Microscopy. 2003;212:273. doi: 10.1111/j.1365-2818.2003.01276.x. [DOI] [PubMed] [Google Scholar]
- 43.Shu D, Moll D, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glotzer SC. Science. 2004;306:419. doi: 10.1126/science.1099988. [DOI] [PubMed] [Google Scholar]
- 45.Gates BD, Xu Q, Stewart M, Ryan D, Willson CG, Whitesides GM. Chem. Rev. 2005;105:1171. doi: 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 46.Guo S, Tschammer N, Mohammed S, Guo P. Human Gene Therapy. 2005;16:1097. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khaled A, Guo S, Li F, Guo P. Nano Lett. 2005;5:1797. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shu D, Huang L, Hoeprich S, Guo P. J. Nanosci. Nanotechnol. (JNN) 2003;3:295. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 49.Prats AC, Roy C, Wang PA, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL. J. Virol. 1990;64:774. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan YNC, Schrock RR, Cohen RE. J. Am. Chem. Soc. 1992;114:7295. [Google Scholar]
- 51.Spatz JP, Roescher A, Möller A. Adv. Mater. 1996;8:337. [Google Scholar]
- 52.Burkett SL, Mann S. Chem. Commun. 1996;3:321. [Google Scholar]
- 53.Braun PV, Osenar P, Stupp S. Nature. 1996;380:325. [Google Scholar]
- 54.Murray CB, Kagan CR, Bawendi MG. Science. 1995;270:1335. [Google Scholar]
- 55.Vossmeyer T, Reck G, Katsikas L, Etk Haupt, Schulz B, Weller H. Science. 1995;267:1476. doi: 10.1126/science.267.5203.1476. [DOI] [PubMed] [Google Scholar]
- 56.Motte L, Billoudet F, Lacaze E, Pileni M. Adv. Mater. 1996;8:1018. [Google Scholar]
- 57.Mao C, Flynn CE, Hayhurst A, Sweeney R, Qi J, Georgiou G, Iverson B, Belcher AM. Proc. Natl. Acad. Sci. USA. 2003;100:6946. doi: 10.1073/pnas.0832310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SW, Mao C, Flynn CE, Belcher AM. Science. 2002;296:892. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 59.Lee CS, Guo P. J. Virol. 1995;69:5018. doi: 10.1128/jvi.69.8.5018-5023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 61.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Nature. 1996;382:609. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 62.Heath JR, Williams RS, Shiang JJ, Wind SJ, Chu J, DEmic C, Chen W, Stanis CL, Bucchignano JJ. J. Phys. Chem. 1996;100:3144. [Google Scholar]
- 63.Mao C, W Sun, Seeman N. J. Am. Chem. Soc. 1999;121:5437. [Google Scholar]
- 64.Li H, Li WX, Ding SW. Science. 2002;296:1319. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 65.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 66.Jacque JM, Triques K, Stevenson M. Nature. 2002;418:435. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carmichael GG. Nature. 2002;418:379. doi: 10.1038/418379a. [DOI] [PubMed] [Google Scholar]
- 68.Coleman J, Hirashima A, Inocuchi Y, Green PJ, Inouye M. Nature. 1985;315:601. doi: 10.1038/315601a0. [DOI] [PubMed] [Google Scholar]
- 69.Knecht DA, Loomis WF. Science. 1987;236:1081. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- 70.Forster AC, Symons RH. Cell. 1987;50:9. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- 71.Sarver NA, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, Rossi JJ. Science. 1990;247:1222. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 72.Chowrira BM, Berzal-Herranz A, Burke JM. Nature. 1991;354:320. doi: 10.1038/354320a0. [DOI] [PubMed] [Google Scholar]
- 73.Ojwang Joshua O, Hampel Arnold, Looney David J, Wong-Staal Flossie. Proc. Natl. Acad. Sci. USA. 1992;89:10802. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shu D, Guo P. Manuscript in preparation. 2004 [Google Scholar]
- 75.Gold L. Harvey Lect. 1995;91:47. [PubMed] [Google Scholar]
- 76.Guo P, Erickson S, Anderson D. Science. 1987;236:690. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 77.Hutvagner G, Zamore PD. Science. 2002;297:2056. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 78.Ayback P, Sandstrom A, Yamakage S, Sund C, Glemarec C, Chattopadthyaya J. J. Biochem. Biophys. Meth. 1993;27:229. doi: 10.1016/0165-022x(93)90006-a. [DOI] [PubMed] [Google Scholar]
- 79.Kearns DR, Wong YP. J. Mol. Biol. 1974;87:755. doi: 10.1016/0022-2836(74)90083-7. [DOI] [PubMed] [Google Scholar]
- 80.Jones CR, Kearns DR. Proc. Natl. Acad. Sci. USA. 1974;71:4237. doi: 10.1073/pnas.71.10.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. J. Mol. Biol. 2005;351:371. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 82.Furtig B, Richter C, Wohnert J, Schwalbe H. Chembiochem. 2003;4:936. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 83.Varani G, Tinoco I., Jr Q. Rev. Biophys. 1991;24:479. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 84.Reid BR. Ann. Rev. Biochem. 1981;50:969. doi: 10.1146/annurev.bi.50.070181.004541. [DOI] [PubMed] [Google Scholar]
- 85.Scott WG. Curr Opin. Struct. Biol. 1998;8:720. doi: 10.1016/s0959-440x(98)80091-2. [DOI] [PubMed] [Google Scholar]
- 86.Cruse WB, Saludjian P, Biala E, Strazewski P, Prange T, Kennard O. Proc. Natl. Acad. Sci. USA. 1994;91:4160. doi: 10.1073/pnas.91.10.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ennifar E, Walter P, Dumas P. Nucleic Acids Res. 2003;31:2671. doi: 10.1093/nar/gkg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ke A, Doudna JA. Methods. 2004;34:408. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 89.Holbrook SR, Kim SH. Biopolymers. 1997;44:3. doi: 10.1002/(SICI)1097-0282(1997)44:1<3::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 90.Lee CS, Guo P. Virology. 1994;202:1039. doi: 10.1006/viro.1994.1434. [DOI] [PubMed] [Google Scholar]
- 91.Reid RJD, Zhang F, Benson S, Anderson D. J. Biol. Chem. 1994;269:18656. [PubMed] [Google Scholar]
- 92.Zhang CL, Trottier M, Guo PX. Virology. 1995;207:442. doi: 10.1006/viro.1995.1103. [DOI] [PubMed] [Google Scholar]
- 93.Zhang CL, Tellinghuisen T, Guo P. RNA. 1997;3:315. [PMC free article] [PubMed] [Google Scholar]
- 94.Bailey S, Wichitwechkarn J, Johnson D, Reilly B, Anderson D, Bodley JW. J. Biol. Chem. 1990;265:22365. [PubMed] [Google Scholar]
- 95.Lane DJ, Pace B, Olsen GL, Stahl DA, Sogin ML, Pace NR. Proc. Natl. Acad. Sci. USA. 1985;82:6955. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma Y, Mathews MB. J. Virol. 1996;70:5083. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pace NR, Smith DK, Olsen GJ, James BD. Gene. 1989;82:65. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- 98.Marshall KA, Ellington AD. Methods Enzymol. 2000;318:193. doi: 10.1016/s0076-6879(00)18053-x. [DOI] [PubMed] [Google Scholar]
- 99.Ellington AD, Szostak JW. Nature. 1992;355:850. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 100.Tuerk G, Gold L. Science. 1990;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 101.Conrad RC, Baskerville S, Ellington AD. Mol Divers. 1995;1:69. doi: 10.1007/BF01715810. [DOI] [PubMed] [Google Scholar]
- 102.Zhang F, Anderson D. J. Biol. Chem. 1998;273:2947. doi: 10.1074/jbc.273.5.2947. [DOI] [PubMed] [Google Scholar]
- 103.Shu D, Guo P. J. Biol. Chem. 2003;278:7119. doi: 10.1074/jbc.M209895200. [DOI] [PubMed] [Google Scholar]
- 104.Zhang CL, Lee C-S, Guo P. Virology. 1994;201:77. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 105.Zhang CL, Tellinghuisen T, Guo P. RNA. 1995;1:1041. [PMC free article] [PubMed] [Google Scholar]
- 106.Cammarano P, Londei P, Biagini R, De RM, Gambacorta A. Eur. J. Biochem. 1982;128:297. doi: 10.1111/j.1432-1033.1982.tb06965.x. [DOI] [PubMed] [Google Scholar]
- 107.Browner MF, Lawrence CB. Anal. Biochem. 1988;168:206. doi: 10.1016/0003-2697(88)90030-9. [DOI] [PubMed] [Google Scholar]
- 108.Moazed U, Stern S, Noller HF. J. Mol. Biol. 1986;187:399. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 109.Yap LP, Musier-Forsyth K. RNA. 1995;1:418. [PMC free article] [PubMed] [Google Scholar]
- 110.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Nucleic Acids Res. 1987;15:9109. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miura K, Tsuda S, Ueda T, Harada F, Kato N. Biochim. Biophys. Acta. 1983;739:281. doi: 10.1016/0167-4781(83)90102-1. [DOI] [PubMed] [Google Scholar]
- 112.Stern S, Moazed D, Noller HF. Meth. Enzymol. 1988;164:481. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 113.Mat-Arip Y, Garver K, Chen C, Sheng S, Shao Z, Guo P. J. Biol. Chem. 2001;276:32575. doi: 10.1074/jbc.M100045200. [DOI] [PubMed] [Google Scholar]
- 114.Conway L, Wickens M. Meth. Enzymol. 1989;180:369. doi: 10.1016/0076-6879(89)80112-0. [DOI] [PubMed] [Google Scholar]
- 115.Rymond BC, Rosbash M. Genes Dev. 1988;2:428. doi: 10.1101/gad.2.4.428. [DOI] [PubMed] [Google Scholar]
- 116.Wassarman DA. Mol. Biol. Rep. 1993;17:143. doi: 10.1007/BF00996222. [DOI] [PubMed] [Google Scholar]
- 117.Tyc K, Steitz JA. Nucleic Acids Res. 1992;20:5375. doi: 10.1093/nar/20.20.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brereton HM, Chen J, Rychkov G, Harland ML, Barritt GJ. Biochim. Biophys. Acta. 2001;1540:107. doi: 10.1016/s0167-4889(01)00124-0. [DOI] [PubMed] [Google Scholar]
- 119.Wassarman DA, Steitz JA. Science. 1992;257:1918. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 120.Cantor CR, Wollenzien PL, Hearst JE. Nucleic Acids Res. 1980;8:1855. doi: 10.1093/nar/8.8.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen C, Guo P. J. Virol. 1997;71:495. doi: 10.1128/jvi.71.1.495-500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hui CF, Cantor CR. Proc. Natl. Acad. Sci USA. 1985;82:1381. doi: 10.1073/pnas.82.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hixson SH, Hixson SS. Biochemistry. 1975;14:4251. doi: 10.1021/bi00690a016. [DOI] [PubMed] [Google Scholar]
- 124.Burgin AB, Pace NR. EMBO J. 1990;9:4111. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nolan JM, Burke DH, Pace NR. Science. 1993;261:762. doi: 10.1126/science.7688143. [DOI] [PubMed] [Google Scholar]
- 126.Demeshkina N, Repkova M, Ven’yaminova A, Graifer D, Karpova G. RNA. 2000;6:1727. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Garver K, Guo P. J. Biol. Chem. 2000;275:2817. doi: 10.1074/jbc.275.4.2817. [DOI] [PubMed] [Google Scholar]
- 128.Mohammad T, Chen C, Guo P, Morrison H. Bioorg. Med. Chem. Lett. 1999;9:1703. doi: 10.1016/s0960-894x(99)00265-6. [DOI] [PubMed] [Google Scholar]
- 129.Quarless SA, Cantor CR. Anal. Biochem. 1985;147:296. doi: 10.1016/0003-2697(85)90275-1. [DOI] [PubMed] [Google Scholar]
- 130.Han W, Mou J, Sheng J, Yang J, Shao Z. Biochemistry. 1995;34:8215. doi: 10.1021/bi00026a001. [DOI] [PubMed] [Google Scholar]
- 131.Trottier M, Mat-Arip Y, Zhang C, Chen C, Sheng S, Shao Z, Guo P. RNA. 2000;6:1257. doi: 10.1017/s1355838200992501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen C, Sheng S, Shao Z, Guo P. J. Biol. Chem. 2000;275:17510. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 133.Babcock MS, Pednault EPD, Olson WK. J. Mol. Biol. 1994;237:125. doi: 10.1006/jmbi.1994.1213. [DOI] [PubMed] [Google Scholar]
- 134.Gautheret D, Cedergren R. FASEB-J. 1993;7:97. doi: 10.1096/fasebj.7.1.7678567. [DOI] [PubMed] [Google Scholar]
- 135.Hoeprich S, Guo P. J. Biol. Chem. 2002;277:20794. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- 136.Massire C, Westhof E. J. Mol. Graph. Model. 1998;16:197. doi: 10.1016/s1093-3263(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 137.Westhof E, Dumas P, Moras D. J. Mol. Biol. 1985;184:119. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 138.Gautheret D, Major F, Cedergren R. J. Mol. Biol. 1993;20:1049. doi: 10.1006/jmbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- 139.Marciniec T, Ciesiolka J, Krzyzosiak W. Acta Biochim. Pol. 1989;36:123. [PubMed] [Google Scholar]
- 140.Wang XD, Padgett RA. Proc. Natl. Acad. Sci. 1989;86:7795. doi: 10.1073/pnas.86.20.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reid RJD, W Bodley J, Anderson D. J. Biol. Chem. 1994;269:5157. [PubMed] [Google Scholar]
- 142.Behlen LS, Sampson JR, DiRenzo AB, Uhlenbeck OC. Biochemistry. 1990;29:2515. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- 143.Lindell M, Romby P, Wagner EG. RNA. 2002;8:534. doi: 10.1017/s1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tullius TD, Greenbaum JA. Curr Opin. Chem. Biol. 2005;9:127. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 145.Strobel SA. Curr. Opin. Struct. Biol. 1999;9:346. doi: 10.1016/S0959-440X(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 146.Walter NG, Harris DA, Pereira MJ, Rueda D. Biopolymers. 2001;61:224. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]
- 147.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 148.Guo P. Seminars in Virology (Editor’s Introduction) 1994;5:1. [Google Scholar]
- 149.Bazinet C, Benbasat J, King J, Carazo J, Carrascosa J. Biochemistry. 1988;27:1849. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- 150.Black LW. BioEssays. 1995;17:1025. doi: 10.1002/bies.950171206. [DOI] [PubMed] [Google Scholar]
- 151.Catalano CE, Cue D, Feiss M. Mol. Microbiol. 1995;16:1075. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 152.Fujisawa H, Shibata H, Kato H. Virology. 1991;185:788. doi: 10.1016/0042-6822(91)90550-u. [DOI] [PubMed] [Google Scholar]
- 153.Grimes S, Jardine PJ, Anderson D. Adv. Virus Res. 2002;58:255. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- 154.Guo P. Prog. in Nucl. Acid. Res. Mol. Biol. 2002;72:415. doi: 10.1016/s0079-6603(02)72076-x. [DOI] [PubMed] [Google Scholar]
- 155.Feiss M, Fogarty S, Christiansen S. Mol. Gen. Genet. 1988;212:142. doi: 10.1007/BF00322457. [DOI] [PubMed] [Google Scholar]
- 156.Guo P, Bailey S, Bodley JW, Anderson D. Nucleic Acids Res. 1987;15:7081. doi: 10.1093/nar/15.17.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo P. Acta Biochimica et Biophysica Sinca. 2002;34:533. [PubMed] [Google Scholar]
- 158.Garver K, Guo P. RNA. 1997;3:1068. [PMC free article] [PubMed] [Google Scholar]
- 159.Reid RJD, Bodley JW, Anderson D. J. Biol. Chem. 1994;269:9084. [PubMed] [Google Scholar]
- 160.Wichitwechkarn J, Johnson Darrin, Anderson Dwight. Mol. Biol. 1992;223:991. doi: 10.1016/0022-2836(92)90257-k. [DOI] [PubMed] [Google Scholar]
- 161.Zhang CL, Garver K, Guo P. Virology. 1995;211:568. doi: 10.1006/viro.1995.1439. [DOI] [PubMed] [Google Scholar]
- 162.Trottier M, Garver K, Zhang C, Guo P. Nucleic Acids Symposium Series. 1997;36:187. [Google Scholar]
- 163.Trottier M, Guo P. J. Virol. 1997;71:487. doi: 10.1128/jvi.71.1.487-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Trottier M, Zhang CL, Guo P. J. Virol. 1996;70:55. doi: 10.1128/jvi.70.1.55-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen C, Guo P. J. Virol. 1997;71:3864. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guo P, Zhang C, Chen C, Trottier M, Garver K. Mol. Cell. 1998;2:149. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 167.Zhang F, Lemieux S, Wu X, St.-Arnaud S, McMurray CT, Major F, Anderson D. Mol. Cell. 1998;2:141. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 168.Hendrix RW. Cell. 1998;94:147. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 169.Chen C, Trottier M, Guo P. Nucleic Acids Symposium Series. 1997;36:190. [Google Scholar]
- 170.Shu D, Huang L, Guo P. J. Virol Meth. 2003;115:19. doi: 10.1016/j.jviromet.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 171.Clever JL, Wong ML, Parslow TG. J. Virol. 1996;70:5902. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. J. Mol. Biol. 1997;270:36. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 173.Chang KY, Tinoco I., Jr J. Mol. Biol. 1997;269:52. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 174.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 175.Jaeger L, Westhof E, Leontis NB. Nucleic Acids Res. 2001;29:455. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stephenson EC, Pokrywka NJ. Curr. Top. Dev. Biol. 1992;26:23. doi: 10.1016/s0070-2153(08)60438-x. [DOI] [PubMed] [Google Scholar]
- 177.Brunel C, Ehresmann C. Biochimie. 2004;86:91. doi: 10.1016/j.biochi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 178.Wagner C, Palacios I, Jaeger L, St. Johnston D, Ehresmann B, Ehresmann C, Brunel C. J. Mol. Biol. 2001;313:511. doi: 10.1006/jmbi.2001.5057. [DOI] [PubMed] [Google Scholar]
- 179.Wagner C, Ehresmann C, Ehresmann B, Brunel C. J. Biol. Chem. 2004;279:4560. doi: 10.1074/jbc.M306511200. [DOI] [PubMed] [Google Scholar]
- 180.Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. EMBO J. 1997;16:1751. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hao D, Hidalgo M. Clin. Lung Cancer. 2002;4:111. doi: 10.3816/clc.2002.n.021. [DOI] [PubMed] [Google Scholar]
- 182.Gadi VK, Alexander SD, Waud WR, Allan PW, Parker WB, Sorscher EJ. J. Pharmacol. Exp. Ther. 2003;304:1280. doi: 10.1124/jpet.102.044743. [DOI] [PubMed] [Google Scholar]
- 183.Friedrich I, Shir A, Klein S, Levitzki A. Semin. Cancer Biol. 2004;14:223. doi: 10.1016/j.semcancer.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 184.Jung HS, Kwon BS, Lee SW. Biotechnol. Lett. 2005;27:567. doi: 10.1007/s10529-005-2883-6. [DOI] [PubMed] [Google Scholar]
- 185.Torresi J, Locarnini SA. Expert. Opin. Investig. Drugs. 1999;8:289. doi: 10.1517/13543784.8.3.289. [DOI] [PubMed] [Google Scholar]
- 186.Ryther RC, Flynt AS, Phillips JA, III, Patton JG. Gene Ther. 2005;12:5. doi: 10.1038/sj.gt.3302356. [DOI] [PubMed] [Google Scholar]
- 187.Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Cancer Res. 2003;63:8791. [PubMed] [Google Scholar]
- 188.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;19:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 189.Hannon GJ, Rossi JJ. Nature. 2004;431:371. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 190.Kashani-Sabet M. J. Investig. Dermatol. Symp. Proc. 2002;7:76. doi: 10.1046/j.1523-1747.2002.19642.x. [DOI] [PubMed] [Google Scholar]
- 191.Bratty J, Chartrand P, Ferbeyre G, Cedergren R. Biochim. Biophys. Acta. 1993;1216:345. doi: 10.1016/0167-4781(93)90001-t. [DOI] [PubMed] [Google Scholar]
- 192.Hampel A. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:1. doi: 10.1016/s0079-6603(08)60032-x. [DOI] [PubMed] [Google Scholar]
- 193.Cech TR. Ann. Rev. Biochem. 1990;59:543. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 194.Pace NR, Brown JW. J. Bacteriol. 1995;177:1919. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Joshi PJ, Fisher TS, Prasad VR. Curr. Drug Targets. Infect. Disord. 2003;3:383. doi: 10.2174/1568005033481060. [DOI] [PubMed] [Google Scholar]
- 196.Klug SJ, Famulok M. Mol. Biol. Rep. 1994;20:97. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 197.Gold M, Law D, DeFranco A. Nature. 1990;345:810. doi: 10.1038/345810a0. [DOI] [PubMed] [Google Scholar]
- 198.Ellington AD, Szostak JW. Nature. 1990;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 199.Ciesiolka J, Gorskl J, Yarus M. RNA. 1995;1:538. [PMC free article] [PubMed] [Google Scholar]
- 200.Clark S, Remcho V. Electrophoresis. 2002;23:1335. doi: 10.1002/1522-2683(200205)23:9<1335::AID-ELPS1335>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 201.Bouvet P. Methods Mol. Biol. 2001;148:603. doi: 10.1385/1-59259-208-2:603. [DOI] [PubMed] [Google Scholar]
- 202.Wang C, Jin YX, Wang DB, Wu Hua Xue Sheng, Sheng Yu, Wu Wu Li, Xue Bao. (Shanghai) Vol. 30. 1998. p. 402. [PubMed] [Google Scholar]
- 203.Kraus E, James W, Barclay AN. J. Immunol. 1998;160:5209. [PubMed] [Google Scholar]
- 204.Goldsby RA, Kindt TJ, Osborne BA, Kuby J. Immunology. New York: W H. Freeman and Company; 2002. Chap. 3; p. 57. [Google Scholar]
- 205.Madaio MP, Hodder S, Schwartz RS, Stollar BD. J. Immunol. 1984;132:872. [PubMed] [Google Scholar]
- 206.Pan T, Gutell RR, Uhlenbeck OC. Science. 1991;254:1361. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- 207.Zilberman D, Cao X, Jacobsen SE. Science. 2003;299:716. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 208.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. Nature. 2002;418:38. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 209.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 210.Coburn GA, Cullen BR. J. Virol. 2003;76:9225. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Science. 2002;297:1833. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 212.Mathias CJ, Wang S, Lee RJ, Waters DJ, Low PS, Green MA. J. Nucl. Med. 1996;37:1003. [PubMed] [Google Scholar]
- 213.Lee RJ, Low PS. J. Biol. Chem. 1994;269:3198. [PubMed] [Google Scholar]
- 214.Monia BP, Lesnik EA, Gonzalez C. Biol. Chem. 1993;268:14514. [PubMed] [Google Scholar]
- 215.Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Erfurth J, Burgstaller P, Klussmann S. Chem. Biol. 2002;9:351. doi: 10.1016/s1074-5521(02)00111-4. [DOI] [PubMed] [Google Scholar]
- 216.Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. J. Biol. Chem. 2004;279:22243. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]