Abstract
Justification for article appearing in Angewandte Chemie:
Foldamers are currently being explored by a number of groups as antagonists of protein-protein interactions. Here we report the first high-resolution structure of a foldamer in complex with its target protein, the anti-apoptotic protein Bcl-xL. The structure demonstrates that α/β-peptide foldamers can accurately mimic natural peptide ligands and that the β-amino acid residues can make unique contacts at the binding interface. This structural information provides a basis for the design of foldamers with enhanced Bcl-xL-binding properties and, more generally, encouragement for continued efforts to develop foldameric inhibitors of other protein-protein interactions.
Keywords: foldamer, peptides, peptide mimics, protein-protein interaction, X-ray diffraction
Development of potent small molecule (MW < 500) antagonists of protein-protein interactions is challenging because of the large surface areas (typically > 500 Å2) buried at such interfaces.[1] Unnatural oligomers with strong and predictable conformational propensities, referred to as "foldamers," have shown potential as inhibitors of such interactions.[2] As with α-peptides and proteins, foldamers constitute scaffolds that can display diverse side chains in particular 3D arrangements, enabling mimicry of large protein-recognition surfaces. However, foldameric backbones are typically resistant to enzymatic degradation, an advantage relative to peptide- and protein-based inhibitors. Folding rules for oligomers containing β-amino acid residues exclusively ("β-peptides"[3]) or in combination with α-amino acid residues (e.g., "α/β-peptides" [4]), have been elucidated, and both have been used to block specific protein-protein interactions.[5–9]
Here, we report the first high-resolution structure of a complex between an α/β-peptide foldamer and a protein partner, Bcl-xL. Bcl-xL is a pro-survival member of the Bcl-2 family that inhibits apoptosis signaling.[10] As over-expression of pro-survival Bcl-2 proteins can lead to cancer, strategies that inhibit their action could lead to development of anti-cancer agents.
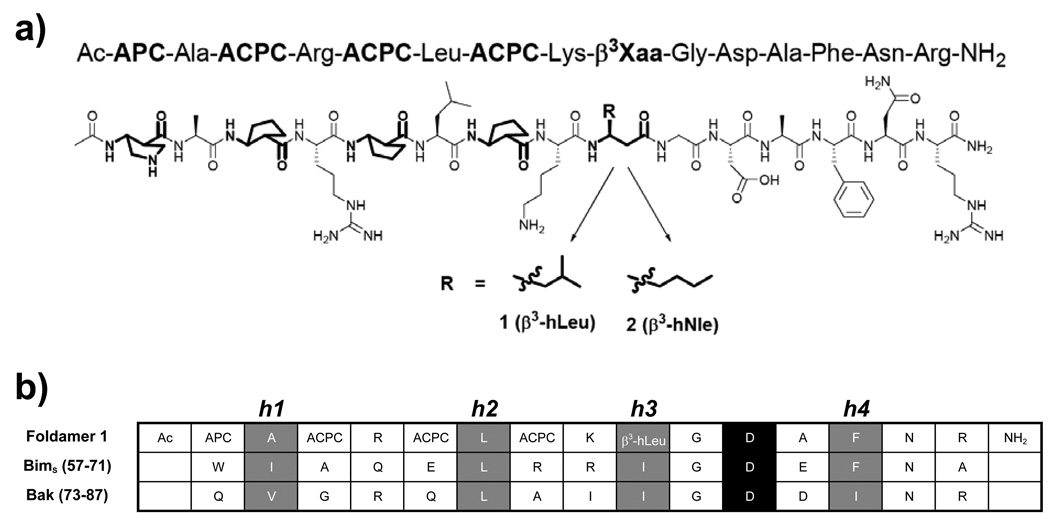
Anti-apoptotic Bcl-2 family members display a hydrophobic cleft that accommodates an α-helical segment from the Bcl-2 homology 3 (BH3) domain of a pro-apoptotic binding partner.[11] Peptide- and non-peptide-based scaffolds have been explored as BH3 domain mimics to antagonize Bcl-2 family interactions, including our efforts to identify helical β- and α/β-peptide foldamer ligands for Bcl-xL.[5, 7, 12] Initial inhibitor designs focused on backbones composed entirely of β-amino acid residues or a 1:1 alternating α:β pattern, but we concluded that more complex patterns of α- and β-residues are necessary for effective BH3 domain mimicry. For example, the isomeric 15-residue α/β-peptides 1 and 2 (Figure 1A), which contain an N-terminal segment with alternating α:β residues and a C-terminal α segment, bind tightly to Bcl-xL (Ki = 2 nM for 1 and 5 nM for 2).[7a] The first four β-residues in 1 and 2 have a five-membered ring constraint that promotes helical folding.[13] The fifth β-residue is the only difference between 1 and 2 (β3-hLeu in place of β3-hNle, respectively). Several side-chains in 1 and 2 (e.g., those of α-Leu6 and α-Asp11) were intended to mimic side chains known to be important for binding of BH3 domains to Bcl-xL (e.g., Leu62 and Asp67 in Bim; Fig. 1b).[14]
Figure 1.
(a) Structures of α/β-peptides 1 and 2. β-Amino acid residues highlighted in bold. (b) Sequence alignment of BH3 domains from Bim and Bak with foldamer 1. The four conserved hydrophobic residues of BH3 domains (h1–h4) are highlighted in grey; the invariant Asp is highlighted in black.
We have now co-crystallized α/β-peptide 1 with Bcl-xL, and solved the structure to 2.3 Å resolution (PDB ID: 3FDM; Supporting Information Table S1). We used a “loop-deleted” form of human Bcl-xL (Δ27–82), which forms an α1 domain-swapped dimer, yet retains BH3 domain binding activity [15] (Supporting Information Figure S1). The asymmetric unit contains three protein molecules (RMSD superposition of Cα atoms over residues 83 – 197 of Bcl-xL molecule C vs. A = 0.51 Å; B vs. A = 0.36 Å; of the foldamer alone that is bound to Bcl-xL molecule C vs. A = 0.32 Å; B vs. A = 0.21 Å). The largest differences between A–C are in the α3 helix of Bcl-xL (Supporting Information Figure S2), which is best ordered in molecule A; we use A for the structural description below.
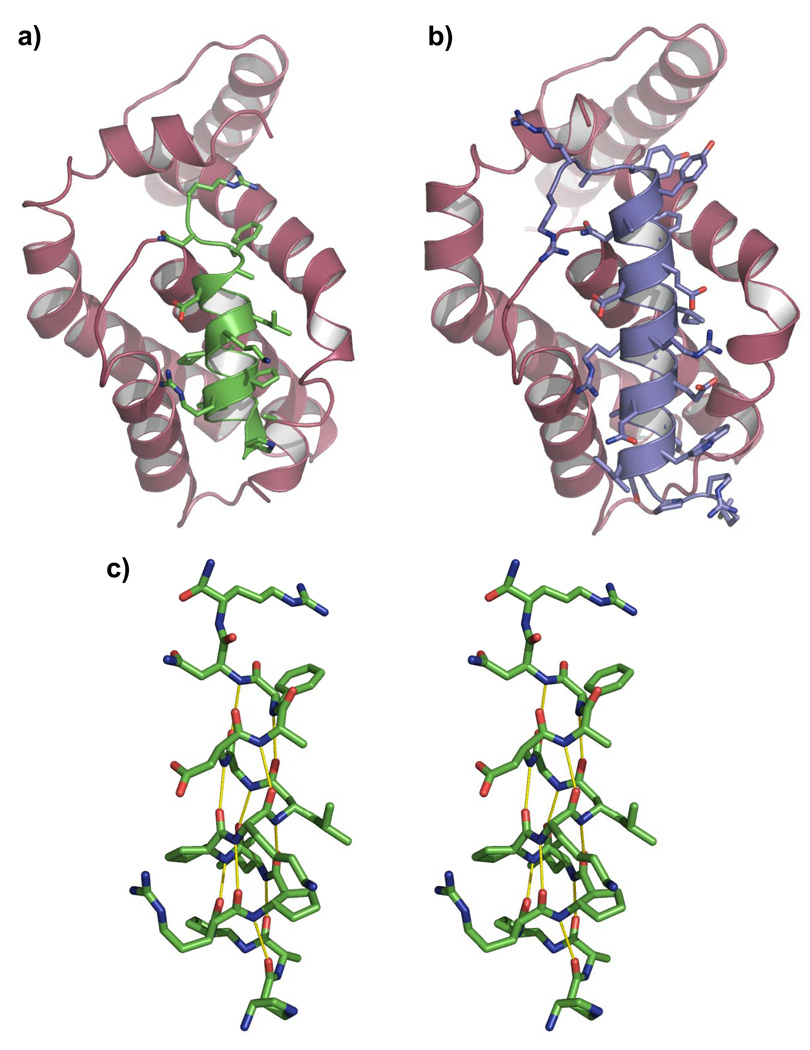
The structure shows that 1 adopts a helical conformation and is oriented within the BH3 recognition groove similarly to natural BH3 domains (Figures 2A,B; the Bim BH3 domain bound to Bcl-xL is shown for comparison). The helix formed by Bcl-xL-bound 1 features a regular pattern of backbone C=O(i)--H-N(i+4) H-bonds over nearly the entire length of the oligomer (the helix is disrupted at the C-terminus; Figure 2C). This H-bond pattern underlies the α-helix formed by pure α-residue peptides, and is characteristic also of the 14/15-helix, a secondary structure favored by 1:1 alternating α:β backbones in which β-residues have a five-membered ring constraint.[16]
Figure 2.
X-Ray structures of human Bcl-xL (magenta) in complex with (a) foldamer 1 (green) or (b) a 26-residue α-peptide from human Bim (DMRPEIWIAQELRRIGDEFNAYYARR) that we recently solved (blue). This structure (PDB ID: 3FDL) is essentially identical to 1PQ1 where a murine Bim peptide was crystallized with murine Bcl-xL (manuscript in preparation).[19] c) Stereo image of foldamer 1 showing that the helix formed by this ligand bound to Bcl-xL adopts a regular pattern of backbone C=O(i)---H-N(i+4) hydrogen bonds, characteristic also of α-helices formed by pure α-residue peptides.
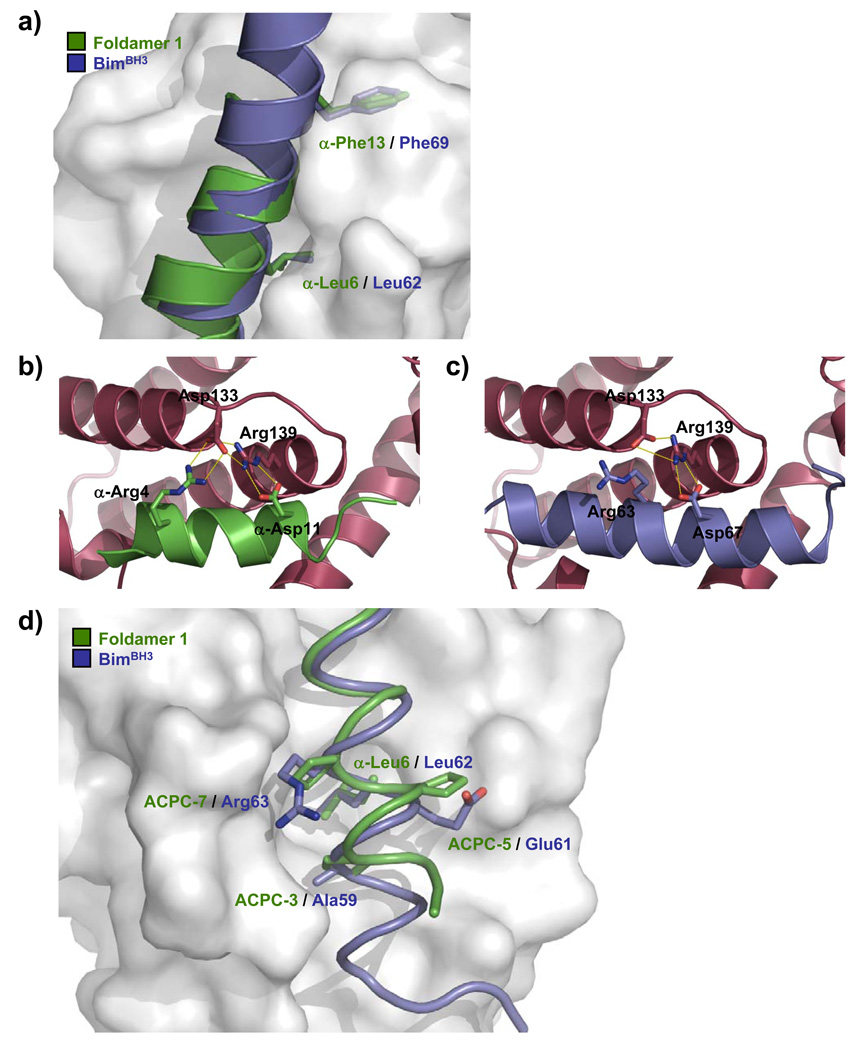
The canonical BH3 domain sequence includes four hydrophobic residues in an i, i+4, i+7, i+11 pattern, designated h1–h4 (Figure 1B), which project from one face of the amphipathic α-helix into the hydrophobic groove of the pro-survival protein. One of the most important of these residues for binding to Bcl-2 family proteins is the h2 residue, conserved as leucine in all known BH3 domains from pro-apoptotic proteins.[14, 17] In the complex with Bcl-xL, the side chain of α-Leu6 in 1 occupies the h2 binding pocket, mimicking the side chain of the conserved BH3 domain leucine (Figure 3A). The h4 binding pocket on Bcl-xL is filled by α-Phe13 in 1 (Figure 3A); some natural BH3 domains, such as BimBH3, have Phe at the analogous position. A critical salt-bridge formed between the aspartate residue strictly conserved in all BH3 domains and the side chain of Arg139 of Bcl-xL is observed also in the foldamer 1/Bcl-xL complex (Figures 3B,C). Interactions involving β-residues include those of the trans-2-aminocyclopentanecarboxylic acid (ACPC) residues at positions 3 and 7 of 1, which make contacts similar to those made by other BH3 domains (e.g., the side chain of Ala59 and the propylene moiety of Arg63, respectively, in BimBH3) (Figure 3D).
Figure 3.
Detailed comparison of the foldamer/Bcl-xL and BimBH3/Bcl-xL interfaces. (a) Overlay of key hydrophobic side chains on foldamer 1 (green) with analogous side chains on BimBH3 (blue). Key electrostatic interactions in the foldamer/Bcl-xL and BimBH3/Bcl-xL interfaces are shown in (b) and (c), respectively (Bcl-xL in magenta ribbon). In (d), ACPC residues of foldamer 1 are overlaid with analogously-positioned side chains on BimBH3.
In complexes of Bcl-xL with BH3 domain peptides, a hydrophobic side chain on the BH3 domain (e.g., Ile58 in BimBH3) invariably occupies the h1 pocket on Bcl-xL. In the complex of Bcl-xL with 1, by contrast, the h1 pocket is not formed since Leu108 of Bcl-xL occludes the site. This is but one of several significant changes in the shape of the BH3-recognition cleft of Bcl-xL in complex with 1 relative to the shape of this cleft in complex with BH3 domain α-peptide ligands (e.g., BimBH3). Interestingly, these changes arise primarily from structural differences in the α3 helix segment of Bcl-xL, which contacts the α/β segment of 1 (Supporting Information Figure S2).
Specific aspects of foldamer-protein recognition observed in the crystal structure are consistent with previously described sequence/affinity relationships established with modified α/β-peptide ligands, suggesting that the crystal structure faithfully reflects foldamer-protein recognition in solution.[7a] For example, Ala-scanning results showed that side chain truncation at α-Leu6, α-Asp11 or α-Phe13 caused substantial decreases in foldamer binding. As discussed above, each of these residues in 1 makes a close contact with Bcl-xL. The side chain of β3-hLeu at position 9 is largely solvent exposed, consistent with the observation that replacing this residue with either β3-hNle or β3-hAla has little effect on affinity. However, moving this side chain one atom toward the C-terminus (β3-hNle → β2-hNle) caused a large decrease in binding, indicating the importance of side chain packing at position 9. Indeed, the structure shows that the backbone methylene of β3-hLeu is very close to Tyr101 of Bcl-xL, which suggests low tolerance for a side chain at this position (i.e., a β2-residue). Replacing a hydrophobic ACPC residue with the hydrophilic analogue APC was deleterious at position 3 or 7, but not position 5. The structure rationalizes these observations because ACPC5 is the most solvent-exposed of these β-residues in the complex.
The hypothesis that foldamer-protein interactions observed in the crystal structure mirror interactions in solution is further supported by the impact of the A142L mutation to Bcl-xL on the binding of foldamer 2. This mutation does not disrupt Bcl-xL folding, but the modified side chain occupies more space within the BH3-recognition cleft, and the mutant protein displays diminished affinity for natural BH3 domains (Supporting Information Figure S3).[18] We used fluorescence polarization to compare the binding affinities of wild-type BclxL and the A142L mutant for a fluorescein-labeled derivative of α/β-peptide 2 (Flu-Ahx-2). The mutant protein showed at least 13-fold weaker binding for Flu-Ahx-2 relative to the wild-type protein, consistent with the crystal structure because α-Leu6 of 1 is in close contact with Ala142 of wild-type Bcl-xL.
These results contribute significantly to our understanding of foldamer-protein recognition. The crystal structure of Bcl-xL bound to 1 shows that this foldamer effectively mimics natural α-helical ligands. This finding validates our "chimeric" design strategy,[7c] since the N-terminal segment of 1 adopts the 14/15-helical conformation documented for simpler α/β-peptides,[16] while the C-terminal segment replicates the analogous segments of BH3 domain α-peptides bound to Bcl-xL (e.g., the Bak BH3 domain).[17] The data suggest that the foldamer achieves high affinity in part by mimicking the three-dimensional display of the canonical side chains projected by natural BH3 domains. However, inspection of the foldamer-protein interface at atomic resolution indicates that β-residue contacts may also contribute significantly to foldamer affinity. These conclusions are supported by effects of mutations to the protein or to the foldamer on binding affinity. Overall, our results demonstrate the value of characterizing protein-foldamer interfaces at high-resolution. Elucidation of additional structures should enhance our ability to design foldamers that target specific protein surfaces.
Methods
Bcl-xL Δ27–82 and A142L (both without the membrane anchor) were produced as described previously.[7a, 15a] The synthesis of 1 has been described.[7a] Crystals of Bcl-xL Δ27–82 and 1 were obtained by mixing at a molar ratio of 1:1.3 then concentrating the sample to 10 mg/ml. Crystals were grown by the sitting drop method at room temperature in 0.2 M lithium nitrate, 20% (w/v) polyethylene glycol 3350. Prior to flash freezing in liquid N2, crystals were equilibrated into cryoprotectant consisting of reservoir solution plus increasing concentrations of ethylene glycol to a final concentration of 20%. Data collection and refinement methods are detailed in Table S1.
Supplementary Material
Footnotes
This work was supported by NIGMS grant GM-56414 (SHG), JDS was supported in part by an NSF Graduate Fellowship, KJP-K was supported in part by a Chemistry-Biology Interface Training Grant from NIGMS (T32 GM008505), EFL, WDF, PMC were supported by grants and fellowships from the Cancer Council of Victoria, the NHMRC of Australia (WDF, PMC), Australian Cancer Research Foundation (PMC) and the Leukemia and Lymphoma Society (PMC). Crystallization trials were performed at the Bio21 Collaborative Crystallisation Centre.
Contributor Information
Erinna F. Lee, The Walter and Eliza Hall Institute of Medical Research (Australia)
Jack D. Sadowsky, University of Wisconsin
Brian J. Smith, The Walter and Eliza Hall Institute of Medical Research (Australia)
Peter E. Czabotar, The Walter and Eliza Hall Institute of Medical Research (Australia)
Kimberly J. Peterson-Kaufman, University of Wisconsin
Peter M. Colman, The Walter and Eliza Hall Institute of Medical Research (Australia)
Samuel H. Gellman, Department of Chemistry, University of Wisconsin, 1101 University Avenue, Madison, WI 53706 (USA), Fax: (+1) 608-265-4534, gellman@chem.wisc.edu, Homepage: http://www.chem.wisc.edu/~gellman/
W. Douglas Fairlie, Structural Biology Division, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3050 (Australia), Fax: (+61) 3-9345-2686, fairlie@wehi.edu.au.
References
- 1.Arkin MR, Wells JA. Nat. Rev. Drug Discov. 2004;3:301. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.a) Hecht S, Huc I, editors. Foldamers: Structure, Properties, and Applications. Weinheim: Wiley-VCH; 2007. [Google Scholar]; b) Gellman SH. Acc. Chem. Res. 1998;31:173. [Google Scholar]; c) Goodman CM, Choi S, Shandler S, DeGrado WF. Nat. Chem. Biol. 2007;3:252. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 4.Horne WS, Gellman SH. Acc. Chem. Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) English EP, Chumanov RS, Gellman SH, Compton T. J. Biol. Chem. 2006;281:2661. doi: 10.1074/jbc.M508485200. [DOI] [PubMed] [Google Scholar]; b) Horne WS, Boersma MD, Windsor MA, Gellman SH. Angew. Chem. Int. Ed. Engl. 2008;47:2853. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J. Am. Chem. Soc. 2004;126:9468. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 7.a) Sadowsky JD, Fairlie WD, Hadley EB, Lee HS, Umezawa N, Nikolovska-Coleska Z, Wang S, Huang DC, Tomita Y, Gellman SH. J. Am. Chem. Soc. 2007;129:139. doi: 10.1021/ja0662523. [DOI] [PubMed] [Google Scholar]; b) Sadowsky JD, Murray JK, Tomita Y, Gellman SH. Chembiochem. 2007;8:903. doi: 10.1002/cbic.200600546. [DOI] [PubMed] [Google Scholar]; c) Sadowsky JD, Schmitt MA, Lee HS, Umezawa N, Wang S, Tomita Y, Gellman SH. J. Am. Chem. Soc. 2005;127:11966. doi: 10.1021/ja053678t. [DOI] [PubMed] [Google Scholar]
- 8.Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J. Am. Chem. Soc. 2005;127:13126. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werder M, Hauser H, Abele S, Seebach D. Helv. Chim. Acta. 1999;82:1774. [Google Scholar]
- 10.Cory S, Adams JM. Nat. Rev. Cancer. 2002;2:647. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 11.Petros AM, Olejniczak ET, Fesik SW. Biochim. Biophys. Acta. 2004;1644:83. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 12.a) Fu X, Apgar JR, Keating AE. J. Mol. Biol. 2007;371:1099. doi: 10.1016/j.jmb.2007.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee EF, Czabotar PE, Smith BJ, Deshayes K, Zobel K, Colman PM, Fairlie WD. Cell Death Differ. 2007;14:1711. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]; c) Lessene G, Czabotar PE, Colman PM. Nat. Rev. Drug Discov. 2008;7 doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]; d) Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Horne WS, Price JL, Gellman SH. Proc. Natl. Acad. Sci. USA. 2008;105:9151. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schmitt MA, Choi SH, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2005;127:13130. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]
- 14.Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, Willis SN, Puthalakath H, Bouillet P, Colman PM, Huang DC, Fairlie WD. J. Cell Biol. 2008;180:341. doi: 10.1083/jcb.200708096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Kvansakul M, Yang H, Fairlie WD, Czabotar PE, Fischer SF, Perugini MA, Huang DC, Colman PM. Cell Death Differ. 2008;15:1564. doi: 10.1038/cdd.2008.83. [DOI] [PubMed] [Google Scholar]; b) Oberstein A, Jeffrey PD, Shi Y. J. Biol. Chem. 2007;282:13123. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 16.a) Choi SH, Guzei IA, Gellman SH. J. Am. Chem. Soc. 2007;129:13780. doi: 10.1021/ja0753344. [DOI] [PubMed] [Google Scholar]; b) Choi SH, Guzei IA, Spencer LC, Gellman SH. J. Am. Chem. Soc. 2008;130:6544. doi: 10.1021/ja800355p. [DOI] [PubMed] [Google Scholar]
- 17.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Science. 1997;275:983. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 18.Manion MK, O'Neill JW, Giedt CD, Kim KM, Zhang KY, Hockenbery DM. J. Biol. Chem. 2004;279:2159. doi: 10.1074/jbc.M306021200. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. Immunity. 2003;19:341. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.