Abstract
Marijuana cannabinoids, the endocannabinoids, and cannabinoid cell receptors have been shown to play important roles in immune regulation particularly as potent modulators of anti-inflammatory cytokines. The predominant cannabinoid receptor involved in this immune regulation is cannabinoid receptor 2 (CB2), which is predominantly expressed in B lymphocytes. However, the promoter region and mechanisms of CB2 gene regulation are unknown in this immune cell type. Utilizing a combination of bioinformatics, 5′ rapid amplification of cDNA ends (5′ RACE), real time RT-PCR, DNA sequencing, and luciferase reporter assays, we show that human B cells express one CB2 transcript while mouse B cells express three CB2 transcripts, with specific transcript selection occurring during B cell activation by LPS. Alignment of our sequenced RACE products to either the mouse or human genome, along with the GenBank submitted mRNA sequences, revealed that the transcripts we isolated contained previously unidentified transcriptional start sites (TSS). In addition, expression construct testing of the genomic region containing the TSSs of the mouse CB2 exon 1 transcripts showed an eight fold increase of promoter activity over baseline. These data show for the first time that human B cells use only one TSS for CB2 while mouse B cells use multiple TSSs and that the mouse TSSs are in a genomic area with promoter activity thus suggesting the location of the gene promoter region. Defining these TSSs also provides clues to the various gene regulatory factors involved in the expression of CB2 during B cell activation.
Keywords: Δ9-tetrahydrocannabinol, cannabinoid, Cannabis sativa, marijuana, endocannabinoid, gene regulation, promoter
INTRODUCTION
The peripheral cannabinoid receptor (CB2) was first identified in 1993 by Munro et al.(Munro et al., 1993) via cloning of a novel G-protein coupled receptor expressed in the human cell line, HL60, having relatively high affinity for cannabinoids. Since its discovery, the expression of CB2 has shown to be almost exclusively on cells of the immune system, with the ranking order of abundance being B cells > NK cells > macrophages > T cells (Galieque et al., 1995; Carayon et al., 1998; Lee et al., 2001a). Though B cells appear to express more CB2 receptor than other immune cell types, little is known about the role this receptor plays in B cell activation and overall biology. There is some evidence that CB2 signaling may be involved in B cell differentiation (Carayon et al., 1998), migration (Jorda et al., 2002; Rayman et al., 2004), proliferation (Massi et al., 1998; Marchand et al., 1999; Jorda et al., 2003., Tanikawa et al., 2007) and antibody class switching (Agudelo et al., 2008), suggesting the receptor is part of the B cell immune activation program. However, the mechanisms involved in the receptor gene (CNR2) regulation and at what stage in B cell activation this process occurs are still unclear. In this regard, several reports suggest that CD40 on B cells as well as stimulation by IL-4, and activation of STAT6, increases CB2 expression (Carayon et al., 1998; Lee et al., 2001a; Schroder et al., 2002; Agudelo et al., 2008); whereas, LPS stimulation reportedly suppresses CB2 mRNA expression in B cells (Lee et al., 2001b). Though these studies provide some clues to regulation of CB2 expression in activated B cells, clearly more studies are needed to define the factors that regulate CNR2 expression and CB2 protein in normal B cells. Identifying the structure of the core promoter and the spectrum of transcription factors involved in CNR2 regulation will help elucidate the role of CB2 in B cell biology.
The core promoter is the minimal region of DNA required for RNA polymerase II (Pol II) to assemble with the general transcription factors and form the pre-initiation complex for initiation of activator-independent (basal) transcription (Gross and Oelgeschlager, 2006). At the center of the core promoter is the initiator (INR) sequence that contains the transcription start site (TSS), which is defined as the most 5′ nucleotide of mRNA transcribed by Pol II (Gross and Oelgeschlager, 2006; Sandelin et al., 2007). Correct identification of the TSS in primary resting B cells will lead to the location of the CNR2 core promoter, including core and cis-acting elements, and provide insights into the molecular mechanisms involved in CB2 expression. In this report, we defined the various CB2 TSSs associated with the core promoter in resting mouse primary splenic and human peripheral B cells, as well as investigated CB2 mRNA transcript production in mouse resting and LPS activated primary B cells.
MATERIALS AND METHODS
Bioinformatics analysis
The bioinformatics programs used for this study include the Genetics Computer Group (GCG) SeqWeb v3.1 software package, Primer3 (Rozen and Skaletsky, 2000), the Database of Transcriptional Start Sites (DBTSS), Ensembl and NCBI databases.
Mice
C57BL/6 mice, 8 to 10 wks old, and of mixed gender where obtained from NCI (Fredericksburg, MD) and housed and cared for in the University of South Florida Health Science Center animal facility, which is fully accredited by the American Association for Accreditation of Laboratory Animal Care.
Isolation of mouse splenocytes, T and B lymphocytes
Mice were euthanized by CO2 asphyxiation, followed by removal of the spleens, which were placed in 12 ml of Hanks balanced buffer saline (HBBS) then pulverized with a Seward Stomacher® 80 (Lab System, England) to release the splenocytes. The splenocytes where collected by centrifugation at 1100 rpm for 10 minutes at 10°C, and washed once with PBS. The T and B cells were then isolated by magnetic negative selection using the EasySep® mouse T or B cell enrichment Kits (StemCell Technologies, Canada) following the manufacturer’s protocol. Total RNA was extracted from the lymphocytes immediately following isolation, except for B cells activated by LPS (5 μg/ml) for up to 8 hrs.
Human subjects, isolation of PBMCs and B lymphocytes
Human subjects recruited for this study were male and female laboratory workers at the University of South Florida, who gave informed consent. Venous blood (25 ml) was drawn into 4 K3 EDTA vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ), then diluted 1:1 or 1:2 with RPMI 1640 medium (Sigma, St Louis, MO). The PBMCs were isolated from blood using Hisopaque®-1077 (Sigma Diagnostics, Inc.) following the manufacturer’s protocol. B cell isolation was performed by magnetic negative selection using the EasySep® human B cell enrichment kit (StemCell Technologies, Canada).
Flow-cytometry analysis of cell populations
Mouse T and B cell subpopulations were analyzed for enrichment by labeling 106 cells with fluorochrome-conjugated anti-mouse mAbs; CD19-PE, CD3-PerCP, NK-pan-FITC and F/480-APC (BD Pharmingen, San Jose, CA). The human B cell populations were analyzed for enrichment by labeling 105 cells with fluorochrome-conjugated anti-human mAbs; CD19-PE, mdCD3-FITC and CD14-APC (BD Pharmingen, San Jose, CA). All flow cytometric analysis was conducted using a FACS Caliber flow cytometer and Cell Quest software (Becton Dickinson, San Diego, CA, USA).
RNA extraction and RT-PCR
Total RNA was extracted from the cell populations by standard techniques using Tri-reagent (Sigma; 1 ml per 107 cells) and quantitated using the RiboGreen RNA Quantitation Kit (Molecular Probes, Eugene, OR). Just prior to cDNA synthesis, residual DNA was removed by treatment with Turbo DNA-free™ (Ambion Inc., Austin, TX) following manufacturer’s protocol. To synthesize the cDNA, 1.0μg of the DNAse treated RNA was primed with 1 μl of random primers for 5 minutes at 70°C, then reverse transcribed (RT) at 37°C for 1hr using 15 U avian myeloblastosis virus (AMV), 40 Units RNasin (Promega, Corp., Madison, WI) and 1.25 mM mix of dNTPs (Promega Corp., Madison, WI) in a volume of 20 μl. The PCR reaction was carried out in 25 μl containing 1 μl cDNA, 500 nM of each primer (see Table 2), with 12.5 μl GoTaq Green Master Mix (Promega Corp., Madison, WI) and amplified using the MyCycler™ thermal cycler (Bio-Rad Laboratories, Hercules, CA). The PCR amplification conditions were as follows; for the initial denaturation step, 95°C for 1 min, followed by 32 cycles at 95°C for 20 sec, 55°C for 30 sec, 72°C for 45 sec, with a final elongation at 72°C for 3 min.
Table 2.
Primers and taqman probes used in this study.
| Primer pairsa and probe | Sequence 5′-to-3′b | Assayc | Size of amplicond | |
|---|---|---|---|---|
| mCB2-E3 | F | GCCGTGCTCTATATTATCCTGTCCTC | qRT-PCR | 120 bp |
| R | GACAAAGTTGCAGGCGAAGATCAC | |||
| P | 6FAM-AGAAAGCCCTCGTACCTGTTCATCAGCA-BHQ1 | |||
| mCB2-E1a | F | TCATCTGCGAAAGTGTGA | qRT-PCR | 112 bp |
| R | TTGTCCTGGCTATTCTGTATC | |||
| P | 6FAM-CTGGAGCTGCAGCTCTTGGGAC-BHQ1 | |||
| mCB2-E1b | F | ACACATAGCCTGGCACA | qRT-PCR | 171 bp |
| R | GCGGTTGAATTCTCTCTTC | |||
| P | 6FAM-TCAAGTGAGTTGCAGGACAGCATAC-BHQ1 | |||
| mCB2-E2 | F | TTCTAGAAGGCACCCATGT | qRT-PCR | 189 bp |
| R | CCTCTGCTCATTCAGGTACA | |||
| P | 6FAM-CTTCCTGTTGCTGTGTGCATCCT-BHQ1 | |||
| β-actin | F | GGGAATGGGTCAGAAGGACT | qRT-PCR | 134 bp |
| R | AGGTGTGGTGCCAGATCTTC | |||
| P | ROX-ATGTGGGTGACGAGGCCCAGAGCAA-BHQ2 | |||
| mE1b-DNA | A | ggaggaggcatgaggca | PCR | 189 bp |
| mRNA | B | ACACATAGCCTGGCACA | RT-PCR | 171 bp |
| C | GCGGTTGAATTCTCTCTTC | |||
| G | GACAAAGTTGCAGGCGAAGATCAC | 755 bp | ||
| mE2-DNA | D | atacatcaaacacatccttg | PCR | 224 bp |
| mRNA | E | TTCTAGAAGGCACCCATGT | RT-PCR | 189 bp |
| F | CCTCTGCTCATTCAGGTACA | |||
| G | GACAAAGTTGCAGGCGAAGATCAC | 567 bp | ||
| hE1-DNA | J | gcaagagaaagctggctt | PCR | 99 bp |
| mRNA | I | TCAACAGGTGCTCTGAGTG | RT-PCR | 71 bp |
| H | CTGAGGAGTCCCAGTTGTT | |||
Name of primers; m, mouse; h, human; E3, exon 3; E1a, exon 1a; E1b, exon 1b; E2, exon 2; F, forward primer; R, reverse primer; P, Taqman® probe; A, D, J, DNA forward primers; B, E, I, forward primers for amplification of mRNA derived cDNA; C, F, G, H, reverse primers.
Sequences of the primers and probes. 6FAM, 6-carboxyfluorescein; BHQ1 or 2, Black Hole Quencher®-1 or 2. Primers designed to bind genomic DNA 5′ of the TSSs are in lower case.
The assay the primers were used in. qRT-PCR, quantitative real time, reverse transcription polymerase chain reaction.
Size of the expected amplicons.
SMART-5′-RACE to identify the putative TSS
To identify the TSS we employed the technique; Switching Mechanism At 5′ end of RNA Transcript Rapid Amplification of cDNA Ends (SMART™ RACE cDNA Amplification kit, Clontech Inc., Madison, WI) following manufacturer’s protocol. Two reverse gene specific primers (GSP) were designed, for both mouse and human CB2 using the GCG SeqWeb v3.1 software (see Table 1 for primer sequences). The mouse GSP1 (mCB2-R301) binds within the ORF 301 bp downstream of the ATG, while the human GSP1 (hCB2-R298) binds 298 bp downstream of the ATG. These were used with the universal primer mix (UPM) that anneals to the SMART sequence at the 5′ end of the cDNA and is supplied with the SMART-RACE kit. A second GSP2 (mCB2-R217 and hCB2-R163), located 84 bp upstream of mCB2-R301, and 74 bp upstream of hCB2-R298, was used with UPM in a nested PCR for CB2 amplicon confirmation. RACE products were run on a 2% agarose gel, visualized with ethidium bromide and purified using the Perfectprep® Gel Cleanup kit (Eppendorf, North America) following manufacturer’s protocol, and sent to the Moffitt/USF Molecule Biology Core lab for DNA sequencing. The SeqWeb PileUp program was used to compare the RACE sequences with the GenBank mCnr2 and hCNR2 sequences to confirm CB2 identity, exonal usage, and location of the TSSs.
Table 1.
SMART 5′ RACE primers used to identify the TSS.
| Primer namea | Sequence 5′-to-3′ | Size (mer) | 5′ end binding siteb |
|---|---|---|---|
| Mouse GSP1 mCB2-R301 | CGACCCCGTGGAAGACGTGGAAGATGACAA | 30 | 301bp downstream ATG |
| Mouse GSP2 mCB2-R217 | TGAACAGGTACGAGGGCTTTCT | 22 | 217bp downstream ATG |
| Human GSP1 hCB2-R298 | GCCAGGAAGTCAGCCCCAGCCAAGCTGCCAA | 31 | 298bp downstream ATG |
| Human GSP2 hCB2-R163 | GCACAGCCACGTTCTCCAGGGCACTTAGCA | 30 | 163bp downstream ATG |
GSP, gene specific primer; 1 designates used for the initial RACE PCR, 2 used for nested PCR.
The number of base pairs from the start of translation in which the 5′ end of the GSP binds for amplification of CB2. The primers were designed to include enough of the coding region for CB2 confirmation of the RACE transcripts.
Primer mapping of the TSS by PCR and RT-PCR
Genomic DNA was extracted from mouse splenic and human peripheral B cells using the Wizard® Genomic DNA Isolation System (Promega Corp., Madison, WI) following manufacturer’s protocol. RNA was extracted, DNAse treated and reverse transcribed from mouse and human B cells as stated in 2.8 above. Using Primer3, forward primers were designed to flank the TSSs identified by 5′RACE (figure 3A) 2 to 10 bp in either direction. Forward primers upstream the TSS should only amplify genomic DNA, whereas forward primers downstream of the TSS should amplify genomic DNA as well as cDNA derived from the CB2 transcripts. The PCR reaction was carried out in 25 μl containing either 1 μl cDNA or 1 μl DNA, 500 nM of each primer (see Table 2), with 12.5 μl GoTaq Green Master Mix (Promega Corp.) and amplified using the MyCycler™ thermal cycler (Bio-Rad Laboratories, Inc). The PCR amplification conditions were as stated in section 2.6 with minor adjustments, the cycle number was increased to 35 and for the mouse samples elongation was increased to 1.5 minutes.
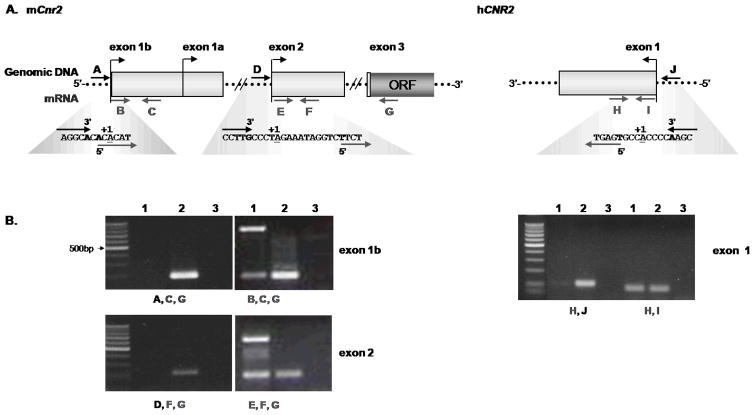
Figure 3. Primer mapping of TSSs.
To confirm the relative position of the TSSs identified by 5′ RACE, primers were designed to map the TSSs of the mouse exon 1b, exon 2 and human exon 1 CB2 transcripts. A. Illustration of the strategy for primer mapping of the mCB2 and hCB2 TSSs. Forward primers were designed to flank the TSSs, in such a way that one forward primer (black arrows A, D, and J) only amplifies genomic DNA and the other forward primer (grey arrows B, E, and I) amplifies both genomic DNA and the CB2 mRNA. The reverse primers (grey arrows C, F, G, and H) are shown. The blown out sequences illustrate where the forward primers bind in relation to the TSSs. The bold letters are the 3′ and 5′ end of the forward primers, and underlined nucleotides are the TSS B. Gel electrophoresis of the mapped CB2 transcripts, where in lane 1 contains cDNA derived from 1 μg of total RNA from mouse splenic and human PBMC B cells, lane 2 is genomic DNA extracted from B cells, and lane 3 contains the no template control. The primers used for each PCR are below the lanes and the panels are labeled with the exon TSSs tested.
Quantitative real time RT-PCR (qRT-PCR) of mouse CB2 transcripts in B cells
mCB2 transcript exonal usage in resting and LPS (5μg/ml) stimulated splenic B cells was measured by qRT-PCR, in which we employed a duplex Taqman PCR strategy; 4 mCB2 exon specific primer sets and probes were designed, one each for the mCB2 exons (1a, 1b, 2, and 3) and 1 primer and probe set for the endogenous β-actin control using Primer3 (see Table 2 for primer/probe sequences). The real-time PCR was carried out in 20 μl containing 1 μl cDNA, 300 nM β-actin and 500 nM CB2 primers, 250 nM fluorescent probe (6-FAM for mCB2 exon, ROX for β-actin), with 10μl IQ™ Multiplex Powermix, and performed in the iCycler IQ™ Real-Time PCR detection system (Bio-Rad Laboratories, Inc). In brief, the reaction was performed in duplicate for each RT cDNA product (see above). Samples were heated for 10 min at 95 °C, followed by 50 cycles of amplification for 15 s at 95 °C and 1 min at 60 °C.
pGL3-clones, transcfection and luciferase reporter assay
Using genomic DNA extracted from mouse B cells (see above) and the pGL3-enhancer vector (Promega), two clones were constructed to test for promoter activity surrounding the TSSs of the exon 1a and 1b variants. The first clone included the region from −359 bp to +205 bp of the TSS (+1) of exon 1a, where as the second clone spanned the region from +68 bp to +205 bp. The DNA regions were PCR amplified and initially cloned into the pBlue TOPO-TA vector (Invitrogen) following manufacturer’s protocol, then sub-cloned by standard methods, into the pGL3-enhancer vector via the Hind III sites. Primary B cells were cultured for 2 days in RPMI medium containing 10% FCS, 10 ng/ml IL-4 and 500 ng/ml anti-CD40, then transfected (107cells/500μl RPMI in 0.4-cm cuvettes) with the pGL3-clones (10μg) by electroporation at 250 V and 800 μFarads using Gene Pulser (BioRad). The transfected B cells were collected 24 hrs after electroporation and lysates analyzed for luciferase activity using Promega’s Luciferase Assay System, following manufacturer’s protocol.
RESULTS
Mouse and human B cells differ in the number of CB2 TSSs
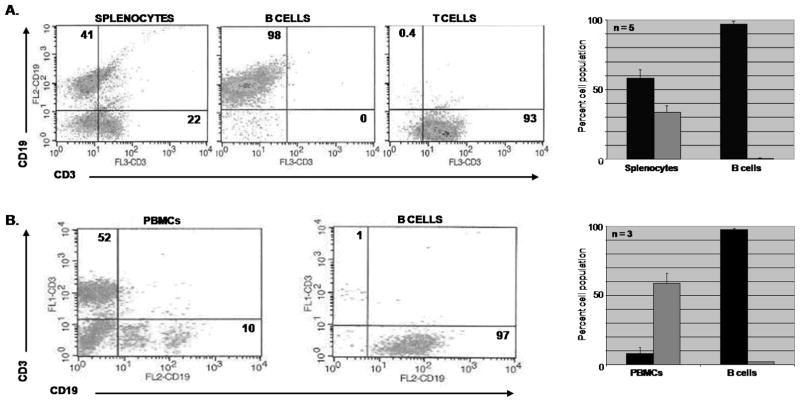
Since the discovery of CB2, several cDNA clones from various mouse and human tissues, as well as the complete gene sequence have been submitted to GenBank and available to researchers. We therefore took advantage of this resource to gain initial insight into CB2 receptor gene expression in B cells. Initially we explored genome databases, such as Ensembl and NCBI, to obtain the location and gene structure of mouse and human CNR2. The mouse Cnr2 (mCnr2) is located on chromosome 4, is 24.7 kb, and produces at least two transcripts containing different 5′ untranslated region (UTR) first exons. On the other hand the human CNR2 (hCNR2) is on the anti-sense strand of chromosome 1, is 39.4 kb, and expresses a single transcript. There is a consensus among the mouse and human clones in which the ORF, encoding CB2 protein, is within a single exon termed exon 3 for mouse and exon 2 for human. Further computational analysis using the GCG SeqWeb package to align the 5′UTR of the GenBank clones, revealed the mouse clones from various immune tissues (GenBank accession nos. NM009924, X86405, and AK037898) share a similar 5′UTR first exon (exon 1) that differ in length at their most 5′ nucleotide. Similarly, the clones reported from bone and liver (GenBank accession nos. BC024052, and AK036658) share a second common 5′UTR first exon (exon 2), yet differ in length at the 5′ nucleotide. Analysis of human data suggested there is only one full length human CB2 clone (GenBank accession no. NM001841) containing a 5′UTR first exon (exon 1). This analysis suggested that mCnr2 utilizes multiple TSSs whereas the human gene utilizes only one. However, none of this existing data provided identification of the TSS in B cells and since studies have shown that CB2 mRNA is most abundant in mouse and human B cells (Galieque et al., 1995; Carayon et al., 1998; Lee et al., 2001a), we began an analysis using purified B cells from mouse spleen and human PBMCs. Purity of the B cell preparations was determined by flow cytometry analysis (FACS) using CD19 and CD3 fluorescent labeled antibodies and demonstrated that the mouse and human B cell populations were enriched to greater than 95% (figure 1).
Figure 1. Phenotypic analysis by flow cytometry of lymphocyte subpopulations isolated from mouse splenocytes and human PBMCs.
A. Mouse B and T lymphocytes isolated from splenocytes by affinity purification (EasySep®) were treated with CD19-PE, CD3-PerCP, NK-pan-FITC and F/480-APC anti-mouse mAbs (FITC and APC data not shown) to confirm lymphocyte enrichment. Graph represents data of 5 independent B cell isolations. B. Human B cells isolated from PBMCs by affinity purification were analyzed with CD19-PE, CD3-FITC and CD14-APC anti-human mAbs (CD14 data not shown) to determine B lymphocyte enrichment. Graph represents data from 3 human donors. Black bars are B cells and grey bars are T cells.
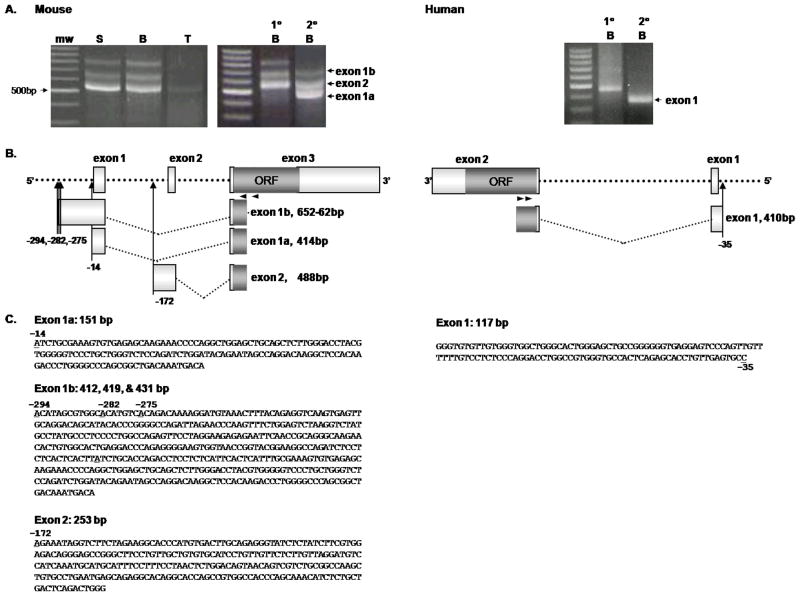
To gain a better understanding of the location of the TSSs, we employed the SMART 5′ RACE technique. Figure 2 shows RACE results of RNA isolated from mouse splenocytes, purified B cells and T cells, as well as from purified human PBMC B cells. For mouse cells we used the GSP1, mCB2-R301, along with the universal primer mix (UPM) to inititate synthesis at the 5′ end and supplied with the kit (see Materials and Methods). RACE PCR yielded three mCB2 transcripts in splenocytes and splenic B cells, as well as one transcript in splenic T cells, respectively (figure 2A, left panel). These three transcripts were confirmed as CB2 RACE products by nested PCR (Figure 2A). RACE was also performed on RNA from human B cells isolated from three different individuals using the hCB2-R298 GSP1 with the UPM, followed by nested PCR using hCB2-R163 GSP2. The results showed only one transcript with the same TSS was obtained from all three donors (Figure 2A, right panel). In order to identify the gene location of the TSSs, the RACE CB2 transcripts were isolated, sequenced and the nucleotides aligned for analysis. Alignment of the sequenced RACE products to either the mouse or human genome, along with the GenBank submitted mRNA sequences revealed several new aspects of CB2 transcript expression in B cells (figure 2B). First the mouse transcripts were homologous to the CNR2 as well as the existing CB2 mRNA data, with the exception that exons 1 and 2 in the transcripts we isolated were longer by 14 to 294 nucleotides, respectively, indicating they contained previously unidentified TSSs. Mouse B cells also expressed and additional transcript, exon 1b, with multiple TSSs (Figure 2B and C). Regarding transcript usage in human B cells, data obtained from three human subjects showed expression of only one transcript containing a single 5′ UTR first exon utilizing the same TSS (figure 2). To our knowledge, this is the first report identifying TSSs in B cells from mouse and human.
Figure 2. Transcriptional start sites determined by SMART 5′ RACE.
A. Gel electrophoresis of the mouse (left 2 photographs) 5′ RACE PCR using, mCB2-R301 GSP1 (1°), yielded 3 CB2 transcripts containing either exon 1a, exon 1b or exon 2 in the splenocyte (S) and B lymphocyte (B) subpopulations, whereas the T lymphocyte (T) subpopulation has 1 CB2 transcript containing exon 1a. Nested PCR of the B lymphocyte 5′ RACE PCR product using, mCB2-R217 GSP2 (2°), was done to confirm that the primary PCR products were CB2 transcripts. Human (right photograph) 5′ RACE PCR and nested PCR using, hCB2-R298 (1°) and hCB2-R163 (2°) GSPs, yielded 1 CB2 transcript containing exon 1 in B lymphocytes isolated from PBMCs of the three different donors. All RACE products were visualized on a 2% agarose gel stained with ethidium bromide. mw, 100 bp DNA ladder molecular weight standard, the 500bp band is labeled. B. Diagrams of the mCnr2 (sense strand) and hCNR2 (anti-sense strand) gene illustrating the location of the TSSs (upward arrows) and transcript exonal usage. Numbers below the arrows represent the TSS’s location relative to position 1 of the Genbank™ CB2 first exons; mouse exons 1a & 1b to NM009924 and exon 2 to BC024052; human exon 1 to NM001841. The transcripts are labeled with their corresponding exon along with the number of nucleotides sequenced for each 5′ RACE transcript. Small black arrows mark the location of the GSPs used for the primary (1°) and nested (2°) PCR reactions. C. The 5′UTR exon sequences of the CB2 transcripts identified by 5′ RACE (GenBank accession nos. FJ357033-6). The TSS nucleotides are underlined. Each sequence line has 60bp. Mouse primary RACE fragment sizes; exon 1b 778–788 bp, exon 2 614 bp, exon 1a 543 bp. Nested sizes; exon 1b 697–707 bp, exon 2 533 bp, exon 1a 459 bp. Human RACE fragment sizes; primary PCR 455 bp, nested PCR 381 bp.
To verify the relative location of the TSSs, we designed specific forward primers for PCR of either genomic DNA or cDNA reverse transcribed from 1 μg of total RNA. The strategy for these experiments is illustrated in figure 3A. In brief, the forward primers were designed so that either their 3′ or 5′ ends border the TSSs. Therefore, the forward primer in which the 3′ end borders the TSS will only amplify genomic DNA and not the mRNA transcripts, whereas the forward primer that borders the TSS at the 5′ end will amplify both. There is some limitation with this approach in that it is not as sensitive as 5′ RACE in determining the TSS, but it does confirm the relative location of the TSS and approximate 5′ end of the transcripts. Therefore, using this approach we were able to confirm the TSS location of mCB2 exons 1b and 2, as well as the hCB2 exon 1 transcripts (figure 3B). Another limitation with the assay is mouse exons 1a and 1b share identical sequences with the exception that exon 1b is 280 nucleotides longer at the 5′ end. Consequently, primers designed to border the TSS of exon 1a would not be able to distinguish genomic DNA from cDNA derived from exon 1b and would amplify both. Therefore, we could not use this approach to verify the location of the TSS for transcripts containing exon 1a.
Preferential usage of the CB2 transcript variants in mouse resting and LPS stimulated B Lymphocytes
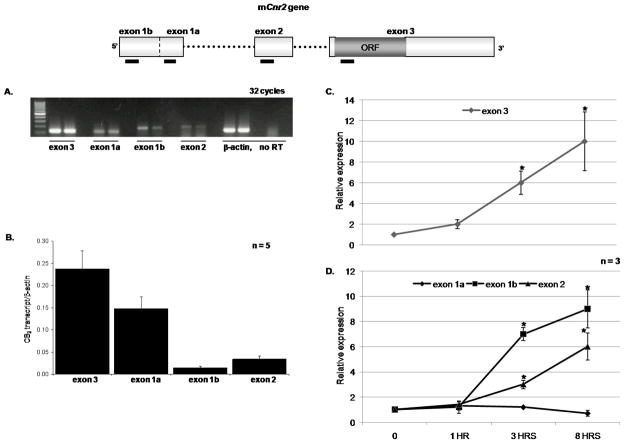
The 5′ RACE data revealed that resting splenic B cells expressed several CB2 transcripts. Therefore, in order to determine which transcript was most abundant we used RT-PCR to quantify the transcripts. Using exon specific primers, the initial results suggested that transcripts containing variants of exon 1 predominated in resting B cells (figure 4A) and to better define this we used qRT-PCR. The primers and CB2 exon specific taqman probes, listed in table 2, were used in conjunction with β-actin primers and taqman probe in a duplex qRT-PCR. Our results showed that the exon 1a transcript variant was the most abundantly expressed transcript in un-stimulated resting splenic B cells (figure 4B). To gain a better understanding as to the role these transcripts may play in B cell biology, we stimulated the primary B cells with LPS (5 μg/ml) for 1, 3 and 8 hrs. We looked at overall CB2 expression (exon 3, figure 4C) and observed a steady increase over time following LPS stimulation. Furthermore, using exon-specific taqman probes revealed that the exon 1b and 2 transcripts were more abundantly expressed than exon 1a with LPS stimulation (figure 4D). These results demonstrate that transcript usage differs in mouse B cells depending upon the state of activation of the cell with exon 1a predominating under resting conditions and exons 1b and 2 under conditions of activation.
Figure 4. Quantitative real time RT-PCR (qRT-PCR) to determine mCB2 mRNA expression in resting and LPS stimulated splenic B cells.
In the diagram of the Cnr2 gene, black boxes under the exons mark the region where the PCR primers and taqman probes bind for cDNA amplification. Since all three transcripts include exon 3, primers designed for this exon will amplify all the transcripts regardless of the first exon. In addition primers designed for exon 1a should amplify all transcripts containing exon 1, whereas primers for exon 1b and 2 were designed to specifically amplify transcripts containing only these exons. A. RT-PCR of the mCB2 transcripts using exon specific primers and 2 separate mouse B cell samples. The samples were collected after 32 cycles of amplification and run on a 2% agarose gel visualized with ethidium bromide. B. Using taqman probes; qRT-PCR was performed to determine the major CB2 transcript utilized in mouse B cells at basal transcription. Results were normalized with βactin and expressed as a ratio of CB2 transcript/β-actin. Data are means ± S.E.M. of five independent experiments. Primary B cells were stimulated with LPS (5μg/ml) for the indicated time points followed by qRT-PCR to establish (C.) total CB2 and (D.) the transcript variants relative expression in activated B cells. Results were obtained by the 2−ΔΔCT method in which βactin is the endogenous control and un-stimulated B cells (time 0) as the calibrator. Data are means ± S.E.M. of three independent experiments. * Significance from time zero at P = 0.05.
The DNA region containing the exon 1a and 1b TSSs exhibits promoter activity
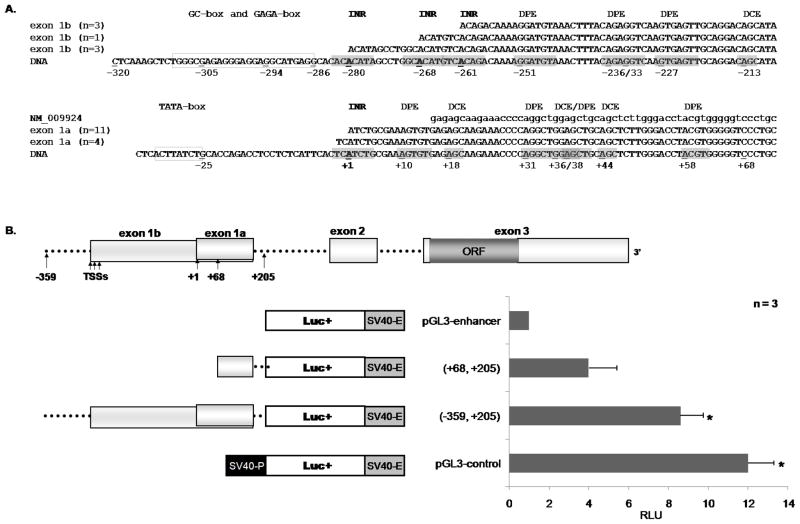
It has been well established that accurate identification of the TSS will lead to the location of the core promoter, which is usually −40 bp upstream to +40 bp downstream of the TSS. The basic elements that comprise the core promoter are the TATA-box, INR (Initiator), DPE (downstream promoter element), and BRE (TFIIB recognition elements)(Sandelin et al., 2007). Therefore, we performed a bioinformatics analysis for the consensus sequences of the core promoter elements in the vicinity of our RACE TSSs to tentatively identify the structure and location of the Cnr2 core promoter. Using GCG SeqWeb, we aligned the 5′ ends of the RACE sequences with that of previously described CB2 mRNA sequences (GenBank accession nos. NM009924 for mouse, and NM001841 for human) as well as the mCnr2 genomic region spanning −320 bp to −208 bp and −36 bp to +73 bp of the TSS (position +1, figure 5A), followed by in silico analysis for core element consensus sequences. For this analysis we used the TSSs of mCB2 exon 1a, the major TSS in resting splenic B cells, and mCB2 exon 1b, the major TSS is LPS stimulated B cells. For exon 1a, we observed an INR-like consensus sequence spanning the area −2bp to +5bp surrounding the TSS (+1), a TATA-like sequence at position −25 bp as well as multiple DPEs and DCEs consensus sequences at positions +10, +18, +31, +36, +38, +44 and +58 bp. The RACE results for the exon 1b transcript identified 3 TSSs, which from this analysis appear to have INR-like sequences. In addition, a GC/GAGA-box spanning from −309 bp to −286 bp is present as well as multiple DPE cis-sites at −251, −236, −233, and −227 bp with two DCE cis-sites at −275 and −213 bp (figure 5A). From this analysis, the mCnr2 region containing exon 1a and 1b has consensus sequences for multiple core promoter elements and therefore we wanted to evaluate this region for promoter activity.
Figure 5. Promoter activity in the region containing the exon 1a and 1b TSSs.
A PileUp analysis was performed using the GCG SeqWeb software of the sequenced CB2 transcripts with that of the Cnr2 gene and GenBank CB2 mRNA clones to determine the location of the 5′ RACE TSS (+1). Once located, the Cnr2 gene region spanning approximately −359 bp to +205 bp of the exon 1a TSS (+1) was analyzed for promoter activity. A. The putative core promoters of the mCB2 exon 1a and 1b transcripts. B. The luciferase reporter assay was used to analyze the relative promoter activity of each construct transfected into IL-4 (10ng/ml) and anti-CD40 (500ng/ml) activated primary B cells. The data are means ± S.E.M expressed as a fold change to that of the pGL3-enhancer vector. RLU, relative light units INR, Initiator (consensus sequence YYANWYY). DPE, Downstream promoter element (consensus sequence RGWCGTG) BREd, TFIIB recognition element downstream the TATA-box (consensus sequence RTDKKKK).
In addition to multiple putative core promoter elements upstream and downstream in the mCnr2 exon 1a and 1b regions, the qRT-PCR results demonstrated that both sets of TSSs are selected either in resting cells (exon 1a) or in LPS-stimulated cells (exon 1b), therefore, we decided to evaluate the region spanning these sites (−359 bp to +205 bp; exon 1a TSS +1) for promoter activity. To do this, we used genomic DNA from mouse B cells to PCR amplify the −359 bp to +205 bp region as well as the region from +68 bp to +205 bp, which excludes the putative core promoters of exon 1a and 1b. The purified DNA fragments were cloned into the pGL3-enhancer vector and evaluated using the luciferase assay. The pGL3-constructs were transfected by electroporation into IL4 (10ng/ml) and anti-CD40 (500ng/ml) activated primary B cells and 24 hrs later luciferase activity was determined for each construct. In total, four constructs were analyzed, two control and two experimental vectors. The pGL3-enhancer vector does not have a promoter and contains only the SV40 enhancer downstream the luciferase gene and therefore served as baseline. The pGL3-control vector contains both the SV40 promoter and enhancer and therefore exhibits full promoter activity. The results demonstrated significant promoter activity when compared to baseline for the pGL3(−359, +205) and pGL3-control vectors (figure 5B), but not for the pGL3(+68, +205) construct. These results demonstrated that the mCnr2 genomic region containing the TSSs for exons 1a and 1b contains strong promoter activity as judged by the luciferase expression studies.
DISCUSSION
The relative robust expression of CB2 in human and mouse B cells suggests that this receptor may have an important role in B cell biology. In spite of this, only a few reports have investigated the function of CB2 in B cells. Furthermore, until this study, examination of the transcript expression and TSSs in B cells had not been done. Therefore, we investigated the genetic sequences involved in transcription of CB2 by identifying the mRNA transcripts and TSSs in the putative core promoter regions in purified resting and activated B cells.
Our data provide the first evidence that resting splenic B cells in mice use multiple TSSs and express at least three CB2 transcript variants. Based on present models of transcription initiation it is possible that two mechanisms of transcription could be involved in the generation of these variants: 1) alternative splicing of the 5′UTRs in the case of exons 1 or 2, and in fact donor-acceptor sites occur in these regions; and 2) alternative transcription initiation (dispersed initiation, see below) generating exon 1 variants that differ in the length of their 5′ ends. The latter event may have occurred in the case of exon 1 in that we observed different lengths of the 5′ ends ranging over 295 bp and containing a cluster of four TSSs. Interestingly, a cluster of TSSs was predicted by the database, DBTSS, in the 5′ flanking regions of exons 1 and 2; furthermore, multiple TSSs were reported in GenBank CB2 clones from various tissues in these same regions. Our RACE products from B cells identified new TSSs for exons 1 and 2 that were not only different than reported in other tissues but for the most part longer at the 5′ ends. Because of these many TSSs spread over hundreds of bps, we analyzed for core promoter sequences in these areas using an in silico approach. Interestingly, we found consensus core promoter sequences such as INR, DPE, DCE along with either TATA or GC boxes in abundance and in proximity to all of the TSSs expressed in mouse and the one TSS expressed in human B cells (see Figures 5A and 6). However, although present, these sequences were in different numbers and relative distances to the TSS position suggesting a heterogeneity in core promoter activity under resting conditions. Although the functional significance of multiple TSSs and core promoters is unknown, previous studies suggested this heterogeneity relates to cell type and/or cell activation state. This was observed in studies on the control of alternative first exons of the glucocorticoid receptor (GR) which are under the control of specific transcription factors that control both tissue specific and cell activation state specific GR expression (Turner et al., 2006). This was also observed with adenosine A2A receptor (A2AR) 5′UTR splice variants wherein the long 5′UTR A2AR variants were observed in resting PMNs, whereas the short 5′UTRs were expressed to a greater extent in LPS- stimulated cells suggesting short 5′UTR variants were more efficiently translated (Kreth et al., 2008) and suggesting the length of the 5′UTR can be a factor in determining tissue specificity and cell activation state.
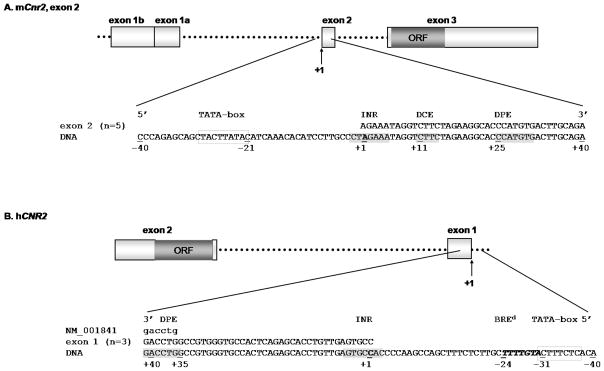
Figure 6. Putative core promoter elements near the TSSs.
A PileUp analysis was performed using the GCG SeqWeb software of the sequenced CB2 transcripts with the GenBank CB2 mRNA clones and gene sequence to determine the location of the 5′ RACE TSS (+1). Once identified, the gene region spanning approximately −40bp to +40bp of the TSSs was analyzed for core promoter elements, which is blown-up here in order to view the sequences. A. The putative core promoter of the mCB2 exon 2 transcript. B. The hCNR2 gene putative core promoter. INR, Initiator (consensus sequence YYANWYY). DPE, Downstream promoter element (consensus sequence RGWCGTG) BREd, TFIIB recognition element downstream the TATA-box (consensus sequence RTDKKKK).
In our mouse CB2 studies, different TSSs and transcript expression were observed in different cell types. For example, resting T cells expressed only the exon 1a variant (figure 2A) and variants of this have been reported in other studies in thymocytes, splenocytes, and the macrophage like cell line, NFS107 (GenBank accession nos. AK037898, X86405, and NM009924). Furthermore, bone and liver tissue expressed exon 2 variants shorter than those we observed in B cells, and only B cells expressed the exon 1b variant (Figure 2A). This variation in transcript expression among the various cell types may be accounted for by variations in core promoter activity surrounding the different TSSs.
In contrast to the multiple TSSs and transcript variants we saw in mouse cells, human peripheral B cells collected from the three different donors expressed a single CB2 transcript and TSS. Interestingly, our observations in mouse and human are in line with those showing that two different strategies are employed by Pol II for transcription initiation. The hCNR2 appears to utilize the more common strategy termed “focused initiation” in which a single TSS and the core promoter contains a TATA-box, BREd, INR, and DPE. On the other hand, the mCnr2 is more like the second strategy that involves multiple weak TSSs dispersed over DNA regions of approximately 50 to 150 bp, thereby dubbed “dispersed initiation” (Juven-Gershon et al., 2006). The mechanisms of dispersed initiation are not clear but probably involve selective usage of multiple upstream and downstream recognition and promoter elements similar to what we observed surrounding the mouse TSSs.
Different mCB2 transcripts are not only associated with different cell types but also with different cell activation states. This is because, in addition to core promoter activity, cell activation can lead to gene transcription through enhancer elements on the DNA either 5′ or 3′ to the core promoter region (Birney et al., 2007). Using qRT-PCR, we showed that the mouse exon 1a transcript was the predominant transcript expressed in resting splenic B cells (Figure 4B) but that exons 1b and 2 were more enhanced in LPS-activated B cells (Figure 4D). In addition, reporter plasmid transfected mouse B cells containing genomic DNA constructs spanning −359 to +205 bp for exon 1 only showed significant promoter activity when stimulated with IL-4 and anti-CD40 antibodies (Figure 5B); non-stimulated cells showed little in the way of gene expression. Enhancer sites proximal to the TSSs for the mCB2 exon 1 and exon 2 contain consensus sequences for binding NF-kB at +52 bp of exon 1a and −82 bp of exon 2. These sites may contribute to increases in mCB2 exon 1 and 2 transcripts in our LPS and/or IL-4/anti-CD40 stimulated cells (Figures 4 and 5) because all of these stimuli activate B cells through an increase in NF-kB (Snapper et al., 1996; Dadgostar et al., 2002; Thieu et al., 2007). In addition to NF-kB, STAT6 binding might also be involved because a putative STAT6 site is located mid-way between the TSSs for mCB2 exon 1a and 1b and also located at the TSS of exon 2; furthermore, increased STAT6 activity has been reported in B cells in response to IL-4 stimulation (Schroder et al., 2002).
In conclusion, we have characterized for the first time multiple TSSs that define alternative CB2 transcripts in mouse splenic B cells as well as a single TSS and transcript in human PBMC B cells. We were able to confirm by RT-PCR primer mapping, the relative location of the TSS for mouse exons 1b and 2, as well as the human exon 1. These experimentally defined TSSs directed further in silico analysis and showed that these regions contain consensus sequences for multiple elements such as TATA-box, INR and DPE. These elements were found at the expected distances from the TSSs and by reporter assay experiments these segments contained significant promoter activity inferring that we correctly identified several of the TSSs in mouse B cells. However, further research needs to be done to verify the individual core promoter elements that are important for CB2 transcription in mouse and human B cells and also to determine the extent to which transcript selection changes as the cell is activated. We report here a change in selection between resting and LPS activated B cells; this difference could explain why B cells in mice have the potential to express multiple transcripts and could provide alternate therapeutic target sequences for controlling CB2 expression in B cell diseases. However, more research on transcript selection and expression during B cell activation needs to be done in order to determine the potential therapeutic benefit of targeting specific CB2 transcripts.
Acknowledgments
Financial support: National Institute on Drug Abuse grant DA019824.
List of abbreviations
- aa
amino acid(s)
- BRE
TFIIB recognition element
- CB2
cannabinoid receptor 2
- hCB2
human CB2
- mCB2
mouse CB2
- CNR2
human cannabinoid receptor 2 gene
- Cnr2
mouse CB2 gene
- DCE
downstream core element
- DPE
downstream promoter element
- IL-4
interleukin 4
- INR
initiator
- LPS
lipopolysaccharide
- Pol II
RNA polymerase II
- SMART™ 5′ RACE
switching mechanism at 5′ end of RNA transcript-rapid amplification of cDNA ends
- STAT6
signal transducer and activator of transcription 6
- TSS
transcription start site
- UTR
untranslated region
References
- Agudelo M, Newton C, Widen R, Sherwood T, Nong L, Friedman H, Klein T. Cannabinoid Receptor 2 (CB2) Mediates Immunoglobulin Class Switching from IgM to IgE in Cultures of Murine-Purified B Lymphocytes. Journal of NeuroImmune Pharmacology. 2008;3:35–42. doi: 10.1007/s11481-007-9088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carayon P, Marchand J, Dussossoy D, Derocq JM, Jbilo O, Bord A, Bouaboula M, Galiegue S, Mondiere P, Penarier G, LeFur G, Defrance T, Casellas P. Modulation and functional involvement of CB2 preipheral cannabinoid receptors during B-cell differentiation. Blood. 1998;92:3605–3615. [PubMed] [Google Scholar]
- Dadgostar H, Zarnegar B, Hoffmann A, Qin XF, Truong U, Rao G, Baltimore D, Cheng G. Cooperation of multiple signaling pathways in CD40-regulated gene expression in B lymphocytes. Proc Natl Acad Sci U S A. 2002;99:1497–1502. doi: 10.1073/pnas.032665099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galieque S, Mary S, Marchand J, Dussossoy D, Carriere D, Carayon P, Bouaboula M, Shire D, Le Fur G, Casellas P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- Gross P, Oelgeschlager T. Core promoter-selective RNA polymerase II transcription. Biochem Soc Symp. 2006:225–236. doi: 10.1042/bss0730225. [DOI] [PubMed] [Google Scholar]
- Jorda MA, Verbakel SE, Valk PJ, Vankan-Berkhoudt YV, Maccarrone M, Finazzi-Agro A, Lowenberg B, Delwel R. Hematopoietic cells expressing the peripheral cannabinoid receptor migrate in response to the endocannabinoid 2-arachidonoylglycerol. Blood. 2002;99:2786–2793. doi: 10.1182/blood.v99.8.2786. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Biochem Soc Trans. 2006;34:1047–1050. doi: 10.1042/BST0341047. [DOI] [PubMed] [Google Scholar]
- Kreth S, Ledderose C, Kaufmann I, Groeger G, Thiel M. Differential expression of 5′-UTR splice variants of the adenosine A2A receptor gene in human granulocytes: identification, characterization, and functional impact on activation. Faseb J. 2008;22:3276–3286. doi: 10.1096/fj.07-101097. [DOI] [PubMed] [Google Scholar]
- Lee SF, Newton C, Widen R, Friedman H, Klein TW. Differential expression of cannabinoid CB2 receptor mRNA in mouse immune cell subpopulations and following B cell stimulation. Eur J Pharmacol. 2001a;423:235–241. doi: 10.1016/s0014-2999(01)01122-0. [DOI] [PubMed] [Google Scholar]
- Lee SF, Newton C, Widen R, Friedman H, Klein TW. Downregulation of cannabinoid receptor 2 (CB2) messenger RNA expression during in vitro stimulation of murine splenocytes with lipopolysaccharide. Adv Exp Med Biol. 2001b;493:223–228. doi: 10.1007/0-306-47611-8_26. [DOI] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Rayman N, Lam KH, Laman JD, Simons PJ, Lowenberg B, Sonneveld P, Delwel R. Distinct expression profiles of the peripheral cannabinoid receptor in lymphoid tissues depending on receptor activation status. J Immunol. 2004;172:2111–2117. doi: 10.4049/jimmunol.172.4.2111. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Schroder AJ, Pavlidis P, Arimura A, Capece D, Rothman PB. Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL-4-stimulated B lymphocytes. J Immunol. 2002;168:996–1000. doi: 10.4049/jimmunol.168.3.996. [DOI] [PubMed] [Google Scholar]
- Snapper CM, Rosas FR, Zelazowski P, Moorman MA, Kehry MR, Bravo R, Weih F. B cells lacking RelB are defective in proliferative responses, but undergo normal B cell maturation to Ig secretion and Ig class switching. J Exp Med. 1996;184:1537–1541. doi: 10.1084/jem.184.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieu VT, Nguyen ET, McCarthy BP, Bruns HA, Kapur R, Chang CH, Kaplan MH. IL-4-stimulated NF-kappaB activity is required for Stat6 DNA binding. J Leukoc Biol. 2007;82:370–379. doi: 10.1189/jlb.1106707. [DOI] [PubMed] [Google Scholar]
- Turner JD, Schote AB, Macedo JA, Pelascini LP, Muller CP. Tissue specific glucocorticoid receptor expression, a role for alternative first exon usage? Biochem Pharmacol. 2006;72:1529–37. doi: 10.1016/j.bcp.2006.07.005. [DOI] [PubMed] [Google Scholar]