Abstract
Dandy-Walker malformation (DWM), the most common human cerebellar malformation, has only one characterized associated locus1,2. Here we characterize a second DWM-linked locus on 6p25.3, showing that deletions or duplications encompassing FOXC1 are associated with cerebellar and posterior fossa malformations including cerebellar vermis hypoplasia (CVH), mega-cisterna magna (MCM) and DWM. Foxc1-null mice have embryonic abnormalities of the rhombic lip due to loss of mesenchyme-secreted signaling molecules with subsequent loss of Atoh1 expression in vermis. Foxc1 homozygous hypomorphs have CVH with medial fusion and foliation defects. Human FOXC1 heterozygous mutations are known to affect eye development, causing a spectrum of glaucoma-associated anomalies (Axenfeld-Rieger syndrome, ARS; MIM no. 601631). We report the first brain imaging data from humans with FOXC1 mutations and show that these individuals also have CVH. We conclude that alteration of FOXC1 function alone causes CVH and contributes to MCM and DWM. Our results highlight a previously unrecognized role for mesenchyme-neuroepithelium interactions in the mid-hindbrain during early embryogenesis.
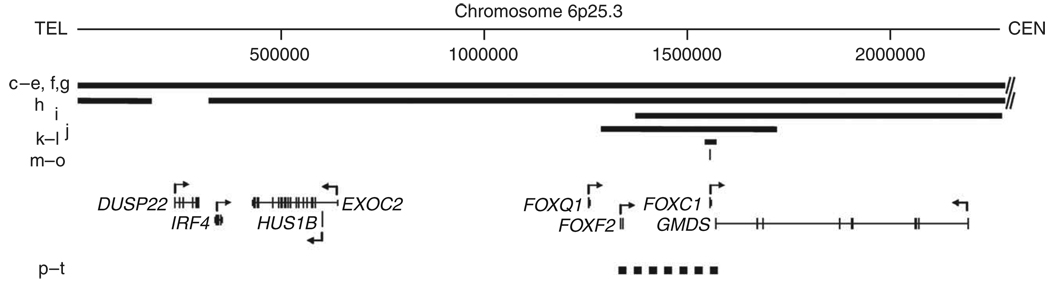
Malformations of the cerebellum are common, heterogeneous human birth defects with chromosomal, single-gene and complex inheritance1–3. CVH, MCM and DWM are generally classified as separate disorders, but they overlap in appearance, and it remains unclear whether they represent distinct entities or share a common pathogenesis1,3,4. We found reports supporting four classic DWM-associated loci, including chromosome 3q24 and 6p25.3, with brain imaging documentation (Supplementary Table 1). We previously identified heterozygous deletion of ZIC1 and ZIC4 on 3q24 as the first molecularly defined cause of classic DWM (MIM no. 220200)2 and mapped a second locus to a 1.8-Mb critical region on 6p25.3 (refs. 5,6).
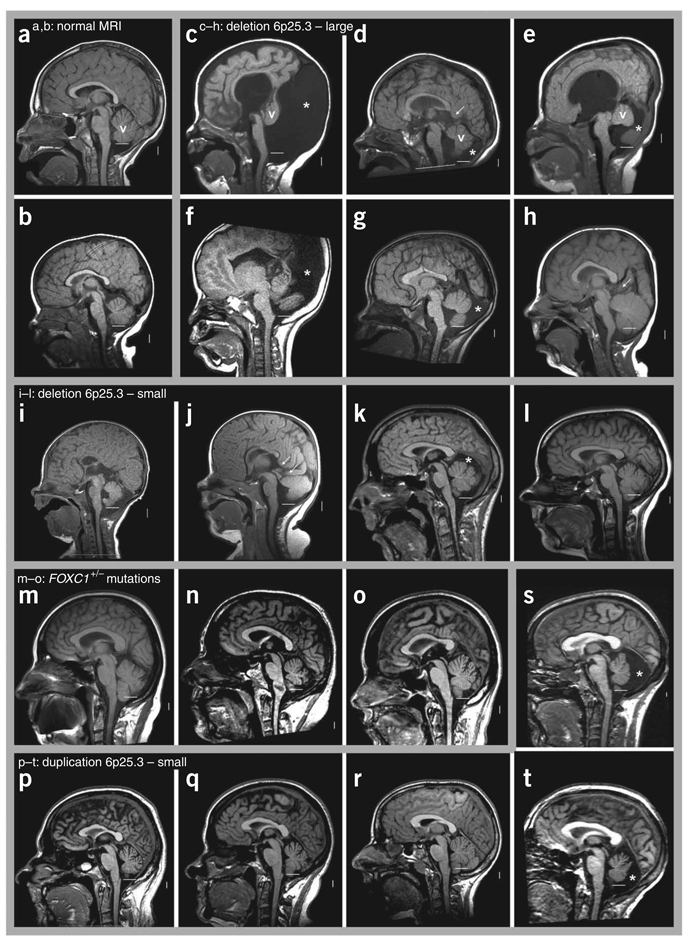
To define this second locus, we obtained brain imaging studies and mapped the breakpoint in 12 individuals with deletion and 6 with duplication of 6p25.3, all ascertained because of multiple congenital anomalies, especially eye malformations consistent with ARS. Anterior chamber dysgenesis was observed in 17/18 affected individuals5,7–9, but too few details were available to compare severity (data not shown). We have even fewer data regarding the retina, although two of the affected individuals had microphthalmia with posterior colobomas. Further, most were examined at a single time point, whereas the ocular abnormalities associated with ARS are known to vary with time (for example, progressive iris alterations or development of glaucoma between 0 and 40 years). Both of these factors limit genotype-phenotype correlation. Brain imaging studies were previously reported for only 5/18 of these individuals5,8. Data illustrated in the first two figures include copy number variants (CNVs), known genes in the region (Fig. 1), brain phenotype details (Table 1) and imaging studies (Fig. 2). One affected individual reported to harbor a telomeric deletion8 proved upon after oligonucleotide array analysis to have a 450-kb segmental deletion (Supplementary Fig. 1). Among the 12 individuals with deletions, 4 were de novo, 3 derived from parental translocations, 2 familial and 3 of unknown origin (Table 1). The duplications were inherited from a common founder9.
Figure 1.
Physical map of the 6p25.3 DWM locus. Diagram of 6p25.3 drawn to scale shows deletions (solid bars) and duplications (dashed bar) as well as heterozygous mutations of FOXC1 (thin vertical line). Letters to the left of the CNVs refer to the same patients as in Figure 2. RefSeq genes are shown with direction of transcription (arrows). Chromosome position is given in kb.
Table 1.
Genotype-phenotype analysis of affected subjects with CNV of 6p25.3 or mutations of FOXC1
| Subject | Inheritance | MRIg | Sex | CNVh (Mb) | Vermis sizej | Vermis rotk | PFl | DXm | CCn | Men defo | WMp | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Del 6p25.3 | ||||||||||||
| LR04-301 | De novo | 2c | F | >5i | ↓↓ | ↑↑ | ↑↑ | DWM | − | − | − | Subject 17 |
| LR04-302 | De novo | 2d | F | >5i | ↓ | ↑ | ↑ | dwm | − | + | + | Subject 37 |
| LR04-303 | Unknowna | 2e | F | >5i | ↓↓ | ↑↑ | ↑↑ | DWM | − | − | − | Subject 27 |
| LR04-317 | Unknownb | 2f | F | 4.95 | ↓↓ | ↑↑ | ↑↑ | DWM | + | − | − | 001-15 |
| LR04-313a2 | Unknownc | 2g | F | 3.94 | ↓↓ | ↑↑ | ↑↑ | DWM | − | + | + | 002-25,6 |
| LR04-318a1 | Familiald | F | 3.94 | ↓↓ | ↑↑ | ↑↑ | DWM | … | … | … | 003-15 | |
| LR04-318a2 | Familiald | F | 3.94 | ↓↓ | ↑↑ | ↑↑ | DWM | … | … | … | 003-25 | |
| LR07-237 | De novo | 2h | M | 1.9 | − | − | ↑ | Text | − | + | − | Proband 119 |
| LR08-198 | De novo e | 2i | M | 0.45 | ↓↓ | ↑ | − | dwm | + | + | − | Subject8 |
| LR07-072 | De novo | 2j | F | 0.870 | ↓ | ↑ | ↑ | dwm | + | ++ | − | Pedigree 109 |
| LR08-197a1 | Familialf | 2k | M | 0.030 | ↓ | − | ↑ | MCM | − | − | ± | Pedigree 99 |
| LR08-197a2 | Familialf | 2l | F | 0.030 | ↓ | − | CVH | − | − | − | Pedigree 99 | |
| FOXC1 +/− | ||||||||||||
| LR08-027a1 | Familial | 2m | F | p.S82T | ↓ | − | − | CVH | − | + | + | Family 327,28 |
| LR08-027a2 | Familial | 2n | M | p.S82T | ↓ | − | − | CVH | − | − | + | Family 327,28 |
| LR08-075 | Familial | 2o | F | p.S131L | ↓ | − | − | CVH | − | − | + | S131L29 |
| Dup 6p25.3 | ||||||||||||
| LR06-253a1 | Familial | 2p | U | 0.492 | ↓ | − | ↑ | MCM | … | … | … | Pedigree 59 |
| LR06-253a2 | Familial | 2t | U | 0.492 | ↓ | − | ↑ | MCM | … | … | … | Pedigree 59 |
| LR07-230a1 | Familial | M | 0.492 | ↓ | − | CVH | − | − | ++ | Pedigree 69 | ||
| LR07-230a2 | Familial | 2q | M | 0.492 | ↓ | − | − | CVH | − | − | − | Pedigree 69 |
| LR07-230a3 | Familial | 2r | F | 0.492 | ↓ | − | − | CVH | − | − | − | Pedigree 69 |
| LR07-230a4 | Familial | 2s | M | 0.492 | ↓ | − | − | CVH | − | − | − | Pedigree 69 |
Phenotype data are grouped by mutation type.
Mother normal, father not tested.
Parents clinically normal, but chromosomes not tested.
Affected sibling, but parents not tested.
Maternal 6p;16q translocation.
Unbalanced 6p;18p translocation.
Postzygotic mosaicism in one parent.
Indicates the panel in Figure 2 showing the subject’s midline sagittal MRI.
CNV, copy number variants (size listed in Mb) or FOXC1 mutation, listed as the protein (amino acid) change.
Large visible deletions of chromosome 6q25 by clinical karyotype analysis indicated as >5 Mb, although no molecular studies were performed.
Cerebellar vermis size is graded as mild (↓) or moderate to severe (↓↓ ) hypoplasia.
Upward rotation (rot) of the vermis is graded as mild (↑) or moderate to severe (↑↑).
Posterior fossa (PF) enlargement is graded as mild (↑) or moderate to severe (↑↑).
Brain imaging diagnosis (DX) is designated as typical (CVH) or mild (cvh) cerebellar vermis hypoplasia, classic (DWM) or mild (dwm) Dandy-Walker malformation, or mega-cisterna magna (MCM), which also includes mild hypoplasia of the posterior vermis. ‘Text’, see Supplementary Note for diagnosis.
Corpus callosum (CC) size is graded as normal (−) or as short (+) consistent with mild partial agenesis.
Meningeal defects (Men def) present as inappropriate interdigitation of right- and left-sided cerebral gyri secondary to deficiency of the falx cerebri, as medial and anterior displacement of medial parietal gyri into the potential space located between the splenium of the CC and cerebellar vermis secondary to deficiency of the tentorium cerebelli, or both.
Abnormal white matter (WM) signal on T2 or Flair sequences typically correspond to prominent perivascular spaces (+), although one aged subject had more severe changes (++).
Figure 2.
Brain images of affected individuals within the 6p25.3 cohort. (a–t) T1-weighted midsagittal magnetic resonance images in two control subjects (a,b) and 18 subjects with 6p25.3 CNVs that include FOXC1 or intragenic mutations of FOXC1 (c–t). The midline cerebellar vermis (v) is marked only in the top row of images. The lower limit of the vermis typically extends down to the obex, marked by a horizontal white line in each image. In all but one individual (h), the lower border of the vermis does not reach the white line, indicative of cerebellar vermis hypoplasia (CVH). Asterisks (*) indicate an enlarged posterior fossa. In 5/6 individuals with large deletions of 6p25.3 (c–g), brain images show a small and upwardly rotated vermis, cystic dilatation of the fourth ventricle and an enlarged posterior fossa, meeting criteria for classic Dandy-Walker malformation (DWM). The remaining individual (h) unexpectedly has a normal or mildly enlarged vermis. The vermis appears abnormal in all four individuals with small deletions of 6p25.3 (i–l). The first two (i,j) meet criteria for DWM, although the abnormalities are less severe than in the subject with larger deletions. The next (k) has mild CVH with normal vermis position (not rotated upward), a normal fourth ventricle and mildly enlarged posterior fossa, consistent with mega-cisterna magna (MCM). The last individual in this group (l) has mild CVH only. The three individuals with heterozygous intragenic FOXC1 mutations (m–o) all have mild CVH only. Among the five individuals with 6p25.3 duplication, two sibs (p,t) have obvious MCM, whereas three other sibs—probably distantly related to the first two—have mild CVH only.
In general, the brain imaging studies show four overlapping malformation groups: in order of severity, classic DWM, mild DWM, MCM and isolated CVH. The six affected individuals with large deletions involving at least seven genes (IRF4 to GMDS) have the most severe brain phenotypes, with classic DWM in four and mild DWM in another. The sixth has normal vermis size but other subtle defects (Supplementary Note). The four individuals with smaller deletions involving the forkhead transcription factor FOXC1 and all or part of GMDS (GDP-mannose-4,6-dehydratase) have less severe phenotypes, with mild DWM in the two individuals with 450- and 870-kb deletions and even less severe MCM or CVH in the sibs with the 30-kb deletion.
Our combined genotype and phenotype data show consistently more severe phenotypes among individuals with large compared to small deletions, which implicates contributions from more than one causative gene in the region. Further, all 12 affected individuals had deletions of FOXC1 plus at least two exons of GMDS, implicating one or both of these genes as having a previously unrecognized role in cerebellar development. This conclusion is further supported by the results of brain imaging in six affected individuals with small (~492-kb) duplications (FOXF2-FOXC1-GMDS), who also showed MCM or CVH, much like the individuals with the smallest 6p25.3 deletions.
We concurrently evaluated the developmental expression profiles of the eight known genes in the 1.8-Mb initial DWM critical region in mice and examined cerebellar morphology in available mutants (Supplementary Table 2). Neither Gmds nor Foxc1 are expressed in the developing brain, although Foxc1 is expressed in the mesenchyme covering the developing cerebellum (Fig. 3a,b and data not shown). We found that Foxc1-mutant mice have a marked cerebellar malformation (Fig. 3–Fig. 5), supporting our hypothesis that FOXC1 is involved in human 6p25.3 associated DWM.
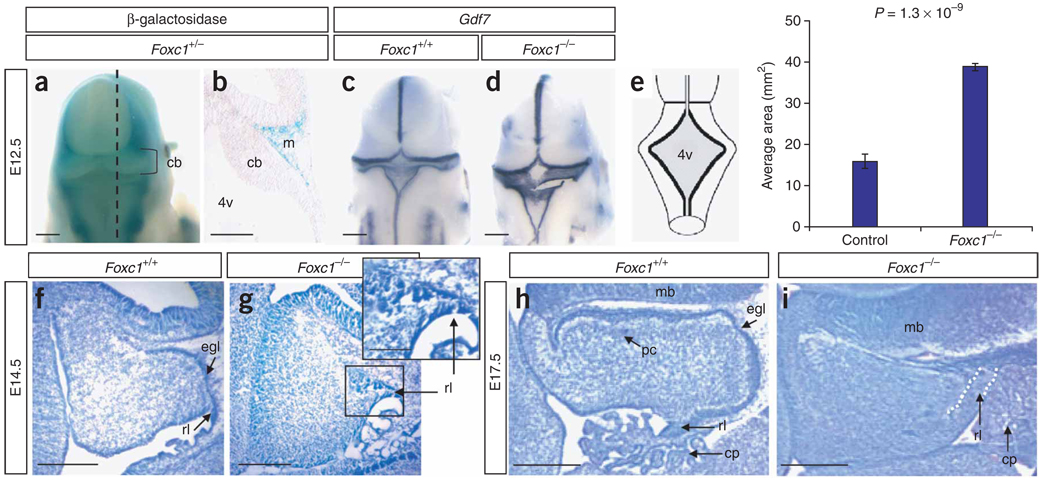
Figure 3.
Morphological cerebellar phenotype of Foxc1 −/− embryos. (a) Dorsal view of the head from an X-gal–stained Foxc1 +/− (Mf1 +/LacZ) embryo at E12.5 showing that Foxc1 expression excludes the developing brain. (b) Midsagittal section through embryo in a (dashed line) showing expression in the mesenchyme (m) adjacent to the cerebellar anlage (cb). (c) Dorsal view of Gdf7 expression in a wild-type embryo. (d) Dorsal view of Gdf7 expression in a Foxc1 −/− embryo revealing an enlarged fourth ventricle roof plate (4v). (e) Schematic of the dorsal view of an E12.5 embryo depicting the 4v area (shaded gray) measured in Foxc1 −/− and littermates. The 4v was 2.5-fold larger in Foxc1 −/− embryos. (f) Cresyl violet–stained midsagittal section of the developing cerebellar anlage in a wild-type E14.5 embryo. (g) Midsagittal section of the developing cerebellar anlage in a Foxc1 −/− E14.5 embryo showing that the rhombic lip (rl) and external granule cell layer (egl) are abnormal and highly disorganized. (h) Midsagittal section through the cerebellar vermis of a wild-type E17.5 embryo showing distinct egl and Purkinje cell (pc) layers. (i) Midsagittal section through the cerebellar vermis of a Foxc1 −/− E17.5 embryo showing a lack of distinct egl and pc layers, an abnormal rl and an expansion of the choroid plexus (cp). Scale bars, 100 (a,c,d), 50 (b,f–i) or 20 µm (g inset).
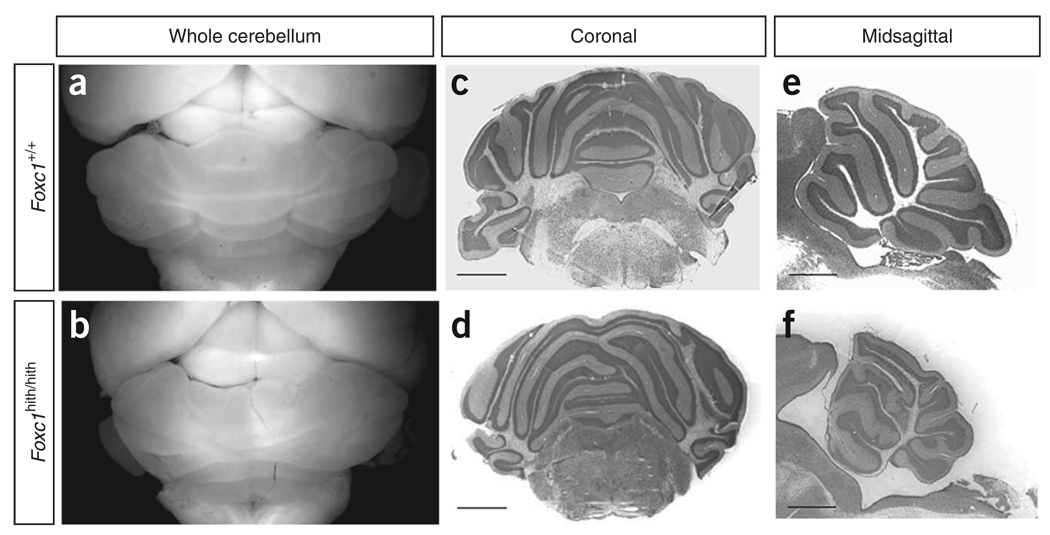
Figure 5.
Cerebellar morphology of Foxc1 hith/hith mice. (a,b) Dorsal views of mouse brains at postnatal day (P) 30. Mean cerebellar area between Foxc1 hith/hith (5.63 arbitrary units (a.u.) ± 0.31 s.e.m.) and littermates (8.60 a.u. ± 0.03 s.e.m.) reveals significant hypoplasia in these adult mutants (P< 0.01). (c,d) Cresyl violet–stained coronal sections through the cerebellum. (e,f) Cresyl violet–stained midsagittal sections through the vermis of brains of mice at P30. The wild-type cerebellum (a) has a stereotypic foliation pattern, which is disrupted in the homozygous mutants. Scale bars, 200 µm.
The Foxc1 gene is widely expressed in mesenchyme and its protein product participates in a broad range of developmental processes including somatic, cardiovascular, kidney, eye, skull and cortical development, but no cerebellar phenotypes have been associated with mutations in this gene10–15. It is never expressed in the cerebellum itself, but it is expressed in the adjacent posterior fossa mesenchyme starting from embryonic day (E) 11.5 (data not shown). The earliest defect we observed in Foxc1-null (Foxc1 −/−) mouse embryos was an enlarged fourth ventricle roof plate at E12.5 (Fig. 3c – e). By E14.5, the cerebellar rhombic lip induced by the rhombomere 1 roof plate was disorganized (Fig. 3g), with the presumptive external granule layer (EGL) forming a clump rather than a thin layer covering the nascent cerebellum. By E17.5, the Foxc1 −/− cerebellum was small, lacked posterior foliation and showed deficient midline fusion (Fig. 3i). Later stages could not be examined due to perinatal lethality.
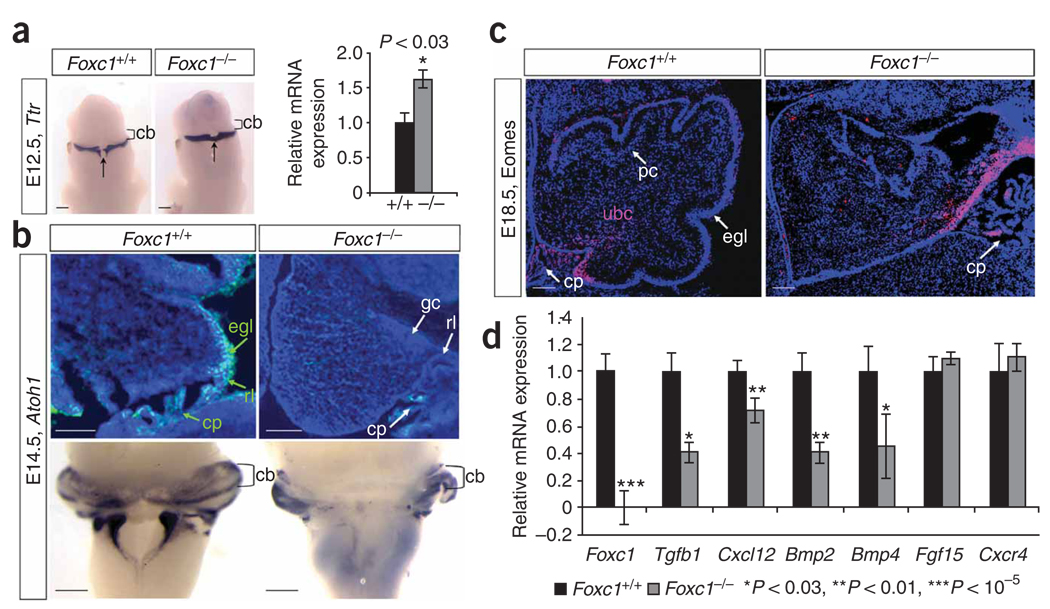
The roof plate and rhombic lip are transient embryonic structures required for proper cerebellar development16–18. Results from molecular analyses of early roof plate and rhombic lip specification, induction (Supplementary Fig. 2 and 3) and anlage proliferation (data not shown) in Foxc1 −/− embryos were normal. We next examined markers of differentiating cell types that arise in successive waves from the rhombic lip1,19. At E12.5, ectopic Ttr+ choroid plexus epithelium spanned the midline roof plate in Foxc1 −/− embryos (Fig. 4a), correlating with the roof plate morphologic change. At E14.5, Tbr1+ deep cerebellar nuclei cells were normal (Supplementary Fig. 4), an expected finding as these cells are generated and migrate from the rhombic lip before the E11.5 onset of posterior fossa Foxc1 expression. Expression of Atoh1 (previously Math1), a marker of rhombic lip and EGL cells, was normal at E12.5, but medial expression was lost by E14.5 (Fig. 4b). The Atoh1− cells in the medial EGL domain were Zic1+ and Pax6+, and they continued to proliferate, confirming their EGL identity (Supplementary Fig. 4 and data not shown). The failure of EGL migration was not accounted for by substrate loss, as the basement membrane at the pial surface of the cerebellum was intact at E14.5. Eomes+ (previously Tbr2+) unipolar brush cells, the last derivative to leave the rhombic lip, were present in Foxc1 −/− embryos at E18.5 but did not migrate away from the rhombic lip (Fig. 4c). Many of these phenotypes can be attributed to loss of medial Atoh1 expression20. Together, our data demonstrate that Foxc1 is required to direct the normal differentiation and migration of roof plate and rhombic lip derivatives but is not required for the progression of cell fate specification in the rhombic lip.
Figure 4.
Loss of Foxc1 from hindbrain mesenchyme disrupts rhombic lip–derived cerebellar cellular populations. (a) Ttr expression in the differentiating choroid plexus epithelium of E12.5 WT and Foxc1 −/− littermate embryos assayed by in situ hybridization (ISH) shows midline expansion of choroid plexus (arrow) adjacent to the cerebellar anlage (cb) in the mutant. Increased Ttr expression confirmed by qRT-PCR (relative to Gapdh expression) is presented as mean ± s.e.m. *P < 0.03 (b) DAPI-counterstained (blue) parasagittal section shows Atoh1 expression (green) in the cerebellar anlage of E14.5 W Tand Foxc1 −/− littermate embryos. No Atoh1+ cells are observed in the mutant egl. Whole-mount ISH of E14.5 brains confirms that loss of Atoh1 in the mutant is restricted to the developing cerebellar vermis. (c) Eomes expression (pink) in the cerebellar anlage of E18.5 WT and Foxc1 −/− littermate embryos. The Eomes+ unipolar brush cells (ubc) remain in the rl of the mutant, rather than migrating through the developing white matter as in WT. DAPI staining (blue) shows egl and pc in WT cerebellum. (d) Expression of six known Foxc1 downstream targets in eye mesenchyme and endothelial development in the mid-hindbrain of E12.5 WT (black) and Foxc1−/− (gray) littermate embryos, assayed by qRT-PCR. Loss of Foxc1 significantly decreases expression of genes secreted from hindbrain mesenchyme (Tgfb1, Cxcl12, Bmp2 and Bmp4) but not genes expressed in the neural tube (Fgf15 and Cxcr4). Data presented as mean ± s.e.m. represent four independent experiments. *P < 0.03, **P < 0.01, ***P< 0.00001. Scale bars, 100 µm.
Foxc1 expression in mesenchyme influences the proliferation and differentiation of eye and endothelial development by regulating secreted signaling molecules that include Bmp2, Bmp4, Cxcl12 and Tgfb1 (refs. 21–23). We examined expression of these signaling molecules in E12.5 Foxc1 −/− hindbrain (posterior fossa) to test whether the same molecules direct cerebellar development. All four were significantly reduced in posterior fossa mesenchyme (Fig. 4d and Supplementary Fig. 5). These data provide a possible mechanism by which nonautonomous loss of Foxc1 leads to a severe cerebellar phenotype, as Bmp2, Bmp4 and Cxcl12 are essential for normal EGL development at later postnatal stages18,24 and Bmp signaling is known to be critical for Atoh1 expression16,18,25,26. It remains possible that additional signaling pathways influence Atoh1 expression. Because our data show that Foxc1-dependent signaling from head mesenchyme also regulates roof plate development, it is possible that some defects in granule cell migration may be mediated by abnormal roof plate signaling.
Although complete loss of Foxc1 in mice causes perinatal lethality and a severe cerebellar phenotype, homozygous Foxc1 hypomorphs (Foxc1 hith/hith), carrying a protein-destabilizing point mutation, survive to adulthood15. We observed that Foxc1 hith/hith mice were ataxic and had CVH combined with defects in midline vermis fusion and folial patterning (Fig. 5). These changes resemble the cerebellar phenotypes seen in individuals with 6p25.3-associated DWM, prompting us to ask whether intragenic mutations of FOXC1 in humans also result in a cerebellar phenotype. We therefore assessed brain imaging studies from three affected individuals, originally ascertained because of anterior eye defects, who harbored heterozygous FOXC1 point mutations in the coding region (resulting in the amino acid substitutions S82T27,28 and S131L29). Both mutations were previously shown to disrupt DNA binding in vitro 30. All three of these individuals have mild CVH with normal vermis position and posterior fossa size (Fig. 1 and Fig. 2m–o), proving that partial loss of FOXC1 alone results in a cerebellar malformation in humans.
Our mouse data provide the first evidence that altered gene expression in mesenchyme can result in cerebellar malformation. Because cranial mesenchyme differentiates into skull and meninges surrounding the brain, we also examined the brain imaging studies from our cohort of subjects with 6p25.3 mutations for developmental defects of these structures. We found meningeal defects leading to abnormal positioning of cerebral gyri in 6/17 (Supplementary Fig. 6 and Supplementary Note) and enlarged posterior fossae (MCM plus DWM) in 12/21 affected individuals. These findings led us to hypothesize that the enlarged posterior fossa characteristic of MCM and DWM, and the meningeal deficiency, result from defective mesenchymal signaling rather than from increased intracranial pressure or other previously proposed mechanisms3,4.
In examining our entire cohort of affected individuals, including those with intragenic FOXC1 mutations, we found complete penetrance and variable expressivity, although with a consistent genotype-phenotype correlation. The cerebellar malformations were most severe with large 6p25.3 deletions, intermediate with small deletions or duplications, and least severe with intragenic FOXC1 point mutations (Fig. 1 and Fig. 2). This suggests that loss (or gain) of FOXC1 alone is sufficient to cause isolated CVH and contribute to the more severe MCM and DWM phenotypes, whereas loss (or gain) of additional genes increases the likelihood of a more severe phenotype. The phenotypes of intermediate severity found with small deletions or duplications suggest that the nearby genes FOXF2 or GMDS contribute to the more severe phenotype, but other genes may also be involved, given the consistently greater severity that accompanies the very large deletions. We examined the expression of all eight genes within the 6p25.3 DWM locus in control human lymphoblastoid cell lines (LBLs) and found that most genes were expressed at very low or undetectable levels (Supplementary Fig. 7). Therefore, evaluation of gene expression in LBLs derived from individuals with 6p25.3 CNVs is unlikely to aid our understanding of which additional 6p25.3 genes contribute to the severity of cerebellar phenotypes.
FOX genes exist mostly in clusters throughout the genome and often act synergistically31. Three FOX genes (FOXQ1, FOXF2 and FOXC1) occur in tandem within 6p25.3. In mice, Foxf2 briefly shares the Foxc1 hindbrain mesenchyme expression domain32, and Foxc1 and Foxq1 are each required for proper endothelial development13,23,33,34. No brain phenotype has been reported in Foxq1−/− or Foxf2−/− mice, and our examination did not reveal morphological changes in the cerebellum of either mutant (Supplementary Table 2 and data not shown). However, the regulation of cerebellar development via genetic interaction among Foxq1, Foxf2 and Foxc1 has not been evaluated and remains a plausible cause for the severe cerebellar phenotype observed among humans with large 6p25.3 deletions.
Considered together, our data demonstrate that specific loss of FOXC1 and general defects in mesenchymal signaling may result in cerebellar malformations and that the clinical diagnoses of isolated CVH, MCM and DWM belong to a single spectrum with shared etiology in some affected individuals. Our observations have several important implications beyond the developmental biology of these phenotypes. First, the clinical difficulty in distinguishing between isolated CVH, MCM and DWM becomes understandable, and our results should thus lead to more flexible diagnostic criteria. Second, classic DWM is frequently associated with hydrocephalus caused by an unknown mechanism. Our data demonstrate a consistent and unequivocal expansion of the choroid plexus epithelium in Foxc1−/− mice. We propose that increased cerebrospinal fluid production by an enlarged choroid plexus contributes to hydrocephalus in humans, but this hypothesis requires further analysis in mouse models. Finally, mutations of other genes such as OPHN1 and CASK are associated with CVH or diffuse cerebellar hypoplasia but not with MCM or DWM35,36. Our current working hypothesis is that genes with primary cerebellar expression will be associated with CVH or diffuse cerebellar hypoplasia, whereas genes with posterior fossa mesenchymal expression will be associated with the complete CVH-MCM-DWM spectrum of malformations.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgments
We thank the affected individuals and their families for their participation; C. Beaulieu for assistance with MRI; T. Kume for providing Foxc1 +/− mice; S. Pleasure and A. Peterson for Foxc1 +/hith mice; H. Sing, H. Hang, R. Arkell, N. Miura, J. Belmont, Q. Ma, B. Hogan, W. Duan, J. Johnson, M.E. Hatten, R. Miller, E. Grove, M. German, T. Jessell and R. Hevner for providing mutant brains, in situ probes or antibodies; and M. Walter for helpful discussions. This work was supported by the following awards: an Autism Speaks predoctoral fellowship to K.A.A., Alberta Heritage for Medical Research and Canadian Institute for Health Research grants to O.J.L., US National Institutes of Health grants KO8-NS48174-01A to A.G.B., R01-NS050375 to W.B.D. and R01-NS050386 to K.J.M., and March of Dimes Birth Defects Foundation award 6-FYO7-334 to K.J.M.
Footnotes
AUTHOR CONTRIBUTIONS
K.A.A. contributed to study design and performed most human and mouse studies and manuscript writing. O.J.L. provided brain scans for multiple individuals and contributed to data analysis and manuscript writing. V.V.C. contributed to mouse immunohistochemical studies. L.H., A.G.B. and L.C.A. provided key phenotype data on one or more individuals. I.D.K. provided much of the initial ideas and data used to start this project, as well as new phenotype data on one subject. W.B.D. contributed to the initial design of the study, patient ascertainment, clinical evaluation including interpretation of all brain scans, delineation of the phenotype and manuscript writing. K.J.M. designed the study, supervised the mouse studies and contributed to manuscript writing.
Note: Supplementary information is available on the Nature Genetics website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Millen KJ, Gleeson JG. Cerebellar development and disease. Curr. Opin. Neurobiol. 2008;18:12–19. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grinberg I, et al. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat. Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- 3.Parisi MA, Dobyns WB. Human malformations of the midbrain and hindbrain: review and proposed classification scheme. Mol. Genet. Metab. 2003;80:36–53. doi: 10.1016/j.ymgme.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Patel S, Barkovich AJ. Analysis and classification of cerebellar malformations. AJNR Am. J. Neuroradiol. 2002;23:1074–1087. [PMC free article] [PubMed] [Google Scholar]
- 5.DeScipio C, et al. Subtelomeric deletions of chromosome 6p: molecular and cytogenetic characterization of three new cases with phenotypic overlap with RitscherSchinzel (3C) syndrome. Am. J. Med. Genet. A. 2005;134:3–11. doi: 10.1002/ajmg.a.30573. [DOI] [PubMed] [Google Scholar]
- 6.DeScipio C, et al. Fine-mapping subtelomeric deletions and duplications by comparative genomic hybridization in 42 individuals. Am. J. Med. Genet. A. 2008;146A:730–739. doi: 10.1002/ajmg.a.32216. [DOI] [PubMed] [Google Scholar]
- 7.Lin RJ, et al. Terminal deletion of 6p results in a recognizable phenotype. Am. J. Med. Genet. A. 2005;136:162–168. doi: 10.1002/ajmg.a.30784. [DOI] [PubMed] [Google Scholar]
- 8.Maclean K, et al. Axenfeld-Rieger malformation and distinctive facial features: clues to a recognizable 6p25 microdeletion syndrome. Am. J. Med. Genet. A. 2005;132:381–385. doi: 10.1002/ajmg.a.30274. [DOI] [PubMed] [Google Scholar]
- 9.Chanda B, et al. A novel mechanistic spectrum underlies glaucoma-associated chromosome 6p25 copy number variation. Hum. Mol. Genet. 2008;17:3446–3458. doi: 10.1093/hmg/ddn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kume T, et al. The forkhead/winged helix gene Mf1 is disrupted in the pleiotropic mouse mutation congenital hydrocephalus. Cell. 1998;93:985–996. doi: 10.1016/s0092-8674(00)81204-0. [DOI] [PubMed] [Google Scholar]
- 11.Kume T, Deng K, Hogan BL. Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development. 2000;127:1387–1395. doi: 10.1242/dev.127.7.1387. [DOI] [PubMed] [Google Scholar]
- 12.Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo S, et al. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev. Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Wilm B, James RG, Schultheiss TM, Hogan BL. The forkhead genes, Foxc1 and Foxc2, regulate paraxial versus intermediate mesoderm cell fate. Dev. Biol. 2004;271:176–189. doi: 10.1016/j.ydbio.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 15.Zarbalis K, et al. Cortical dysplasia and skull defects in mice with a Foxc1 allele reveal the role of meningeal differentiation in regulating cortical development. Proc. Natl. Acad. Sci. USA. 2007;104:14002–14007. doi: 10.1073/pnas.0702618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chizhikov VV, et al. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- 17.Wingate R. Math-Map(ic)s. Neuron. 2005;48:1–4. doi: 10.1016/j.neuron.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Alder J, Lee KJ, Jessell TM, Hatten ME. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat. Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- 19.Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci. Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Arie N, et al. Atoh1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 21.Rice R, Rice DP, Thesleff I. Foxc1 integrates Fgf and Bmp signalling independently of twist or noggin during calvarial bone development. Dev. Dyn. 2005;233:847–852. doi: 10.1002/dvdy.20430. [DOI] [PubMed] [Google Scholar]
- 22.Sommer P, Napier HR, Hogan BL, Kidson SH. Identification of Tgf beta1i4 as a downstream target of Foxc1. Dev. Growth Differ. 2006;48:297–308. doi: 10.1111/j.1440-169X.2006.00866.x. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi H, Kume T. Forkhead transcription factors regulate expression of the chemokine receptor CXCR4 in endothelial cells and CXCL12-induced cell migration. Biochem. Biophys. Res. Commun. 2008;367:584–589. doi: 10.1016/j.bbrc.2007.12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Q, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc. Natl. Acad. Sci. USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krizhanovsky V, Ben-Arie N. A novel role for the choroid plexus in BMP-mediated inhibition of differentiation of cerebellar neural progenitors. Mech. Dev. 2006;123:67–75. doi: 10.1016/j.mod.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Qin L, Wine-Lee L, Ahn KJ, Crenshaw EB., III Genetic analyses demonstrate that bone morphogenetic protein signaling is required for embryonic cerebellar development. J. Neurosci. 2006;26:1896–1905. doi: 10.1523/JNEUROSCI.3202-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears AJ, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am. J. Hum. Genet. 1998;63:1316–1328. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould DB, Mears AJ, Pearce WG, Walter MA. Autosomal dominant Axenfeld-Rieger anomaly maps to 6p25. Am. J. Hum. Genet. 1997;61:765–768. [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura DY, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat. Genet. 1998;19:140–147. doi: 10.1038/493. [DOI] [PubMed] [Google Scholar]
- 30.Saleem RA, Banerjee-Basu S, Berry FB, Baxevanis AD, Walter MA. Analyses of the effects that disease-causing missense mutations have on the structure and function of the winged-helix protein FOXC1. Am. J. Hum. Genet. 2001;68:627–641. doi: 10.1086/318792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development, disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 32.Ormestad M, Astorga J, Carlsson P. Differences in the embryonic expression patterns of mouse Foxf1 and −2 match their distinct mutant phenotypes. Dev. Dyn. 2004;229:328–333. doi: 10.1002/dvdy.10426. [DOI] [PubMed] [Google Scholar]
- 33.Hong HK, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29:163–171. doi: 10.1002/gene.1020. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi H, Kume T. Foxc transcription factors directly regulate Dll4 and Hey2 expression by interacting with the VEGF-Notch signaling pathways in endothelial cells. PLoS One. 2008;3:e2401. doi: 10.1371/journal.pone.0002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philip N, et al. Mutations in the oligophrenin-1 gene (OPHN1) cause X linked congenital cerebellar hypoplasia. J. Med. Genet. 2003;40:441–446. doi: 10.1136/jmg.40.6.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Najm J, et al. Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat. Genet. 2008;40:1065–1067. doi: 10.1038/ng.194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.