Abstract
Ullrich congenital muscular dystrophy (UCMD) is a disabling and life-threatening disorder resulting from either recessive or dominant mutations in genes encoding collagen VI. Although the majority of the recessive UCMD cases have frameshift or nonsense mutations in COL6A1, COL6A2, or COL6A3, recessive structural mutations in the COL6A2 C-globular region are emerging also. However, the underlying molecular mechanisms have remained elusive. Here we identified a homozygous COL6A2 E624K mutation (C1 subdomain) and a homozygous COL6A2 R876S mutation (C2 subdomain) in two UCMD patients. The consequences of the mutations were investigated using fibroblasts from patients and cells stably transfected with the mutant constructs. In contrast to expectations based on the clinical severity of these two patients, secretion and assembly of collagen VI were moderately affected by the E624K mutation but severely impaired by the R876S substitution. The E624K substitution altered the electrostatic potential of the region surrounding the metal ion-dependent adhesion site, resulting in a collagen VI network containing thick fibrils and spots with densely packed microfibrils. The R876S mutation prevented the chain from assembling into triple-helical collagen VI molecules. The minute amount of collagen VI secreted by the R876S fibroblasts was solely composed of a faster migrating chain corresponding to the C2a splice variant with an alternative C2 subdomain. In transfected cells, the C2a splice variant was able to assemble into short microfibrils. Together, the results suggest that the C2a splice variant may functionally compensate for the loss of the normal COL6A2 chain when mutations occur in the C2 subdomain.
Keywords: Collagen, Electron Microscopy, Extracellular Matrix, Fibroblast, Genetic Diseases, Muscular Dystrophy, Biosynthesis, Mutation
Introduction
Ullrich congenital muscular dystrophy (UCMD,2 OMIM 254090) has recently emerged as one of the most prevalent forms of congenital muscular dystrophy (1, 2). The disorder is characterized by severe muscle weakness in conjunction with proximal joint contractures and co-existing distal joint hypermobility (3). Children afflicted with UCMD may never achieve independent ambulation or may be able to walk for only a limited period of time. They may suffer from respiratory insufficiency, leading to early demise if not treated. UCMD was initially thought to be a recessive condition caused by mutations in the COL6A1, COL6A2, or COL6A3 genes encoding three collagen VI chains (4–6). We have demonstrated that the disease can also result from a dominant mutation in COL6A1 (7). It is now apparent that dominant mutations in the three collagen VI genes are common in UCMD patients (8, 9).
Collagen VI is a ubiquitously expressed extracellular matrix protein composed of α1(VI), α2(VI), and α3(VI) chains encoded by the COL6A1, COL6A2, and COL6A3 genes, respectively (10). Each chain is made up of a triple-helical domain flanked by N- and C-globular regions that consist mainly of von Willebrand factor, type A (vWF-A) modules (11, 12) (Fig. 1A). There are two vWF-A modules in the C-globular region of each chain, named C1 and C2 subdomains, where C2 is more C-terminal. The COL6A2 gene undergoes alternative splicing, resulting in a minor splice variant, in which the C2 subdomain is replaced by a smaller C2a subdomain (13) (see Fig. 1A). After synthesis, the three collagen VI chains associate via the C-globular region and wind together into a triple-helical monomer. The monomers are then assembled in a staggered, anti-parallel manner into dimers, which subsequently associate laterally into tetramers (10) (see Fig. 1B). The tetramers are secreted outside of the cells, where they assemble end-to-end into collagen VI microfibrils. The microfibrils can further undergo lateral association into microfibril bundles. Additional α3(VI)-like chains, two in humans and three in other animal species, have been identified recently (14, 15). Current evidence suggests that these α3(VI)-like chains, named the α4(VI), α5(VI), and α6(VI) chains, most likely can replace the α3(VI) chain in the assembly of collagen VI microfibrils (14, 15). However, the three α3(VI)-like chains show highly restricted tissue distribution patterns compared with the α1(VI), α2(VI), and α3(VI) chains, and therefore their roles in collagen VI assembly in various tissues await further studies.
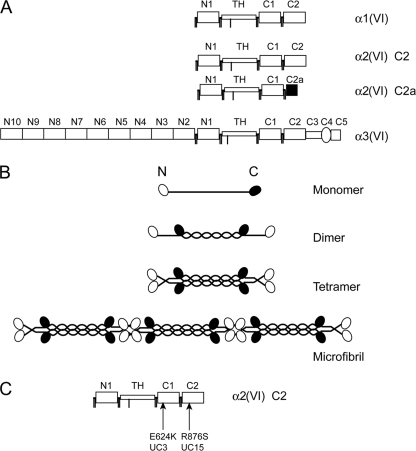
FIGURE 1.
Schematic diagram of collagen VI chains, microfibril assembly, and missense mutations. A, modular structures of the three collagen VI chains. Each chain contains a triple helical domain (TH) flanked by N- and C-globular domains consisting of vWF-A modules, designated N1–N10 and C1–C2. Note that a minor α2(VI) chain variant, named α2(VI) C2a, has a shorter C2a domain at the C-terminal end compared with the major α2(VI) C2 chain, as a result of alternative splicing of the 3′-end of the gene. B, assembly of collagen VI. The three collagen chains form a monomer with a central TH domain flanked by N- and C-globular domains. Monomers assemble into dimers in an anti-parallel manner. Dimers associate laterally into tetramers, which linked end-to-end into a microfibril. C, location of the COL6A2 missense mutations. UC3 has homozygous E624K mutation in the C1 subdomain and UC15 carries a homozygous R876S mutation in the C2 subdomain.
Studies to date indicate that the majority of the recessive cases in UCMD have frameshift or nonsense mutations in either the triple-helical or globular domains of the collagen VI subunits (5, 8, 16). These mutations lead to premature stop codons, which can trigger nonsense-mediated decay of the mutated collagen VI mRNA and thereby result in a complete absence or drastic reduction of the collagen VI protein (17). Recessive amino acid alterations in collagen VI have been reported in two UCMD patients thus far (8, 18). The recessive mutations in these two patients are both located in the C2 subdomain of the COL6A2 gene. One of the patients has a homozygous missense mutation (18), whereas the other patient is compound heterozygous for a missense mutation and a single amino acid deletion (8). The molecular mechanisms underlying these recessive structural mutations are still poorly understood.
Here we report the identification of recessive missense mutations in two additional UCMD patients and characterized the consequences of the mutations using fibroblasts from the patients and cultured cells transfected with the mutant collagen VI chains. Both patients carried homozygous mutations that resided in the C-globular domain of the COL6A2 gene, altering the coding sequences of either the C1 or C2 subdomain. We show that these two mutations have distinct effects on collagen VI biosynthesis and that there is no direct correlation between the collagen VI biosynthetic abnormality and clinical severity of the patients. Significantly, we find that the splice variant COL6A2 C2a with an alternative C2 subdomain is utilized to assemble triple-helical collagen VI molecules in fibroblasts from the patient with the C2 subdomain mutation, which may explain the moderately severe clinical presentation of this patient.
EXPERIMENTAL PROCEDURES
Patient Studies
Muscle and skin biopsies were obtained from patients and controls after informed consent. Dermal fibroblasts were established from skin biopsies and grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (HyClone, Logan, UT) in 5% CO2 at 37 °C. Human subject studies were performed in accordance with protocols approved by the Institutional Review Board of Thomas Jefferson University and the Children's Hospital of Philadelphia. Cryosections of 10-μm thickness were prepared from muscle biopsies and immunostained with a monoclonal antibody against collagen VI (MAB3303, Chemicon) and a polyclonal antibody recognizing the basement membrane protein perlecan as described (7). Mutation analysis of the three collagen VI genes was performed by reverse transcription-PCR amplification of total RNA isolated from the fibroblasts and DNA sequencing as described (7).
HEK293 Cells Expressing α1(VI) and α3(VI) N6-C5 Collagen Chains
Expression constructs for the full-length human α1(VI) collagen chain and the human α3(VI) N6-C5 collagen chain containing a BM40 signal sequence, both in the Rc/CMV vector (Invitrogen, Carlsbad, CA), have been reported previously (19, 20). The constructs were linearized with NruI and co-transfected into human embryonic kidney (HEK) 293 cells (American Type Culture Collection) by electroporation using an ECM 630 electroporator (BTX, Holliston, MA) at 270 V and 800 microfarads. Cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Stably transfected clones were selected by G418 resistance (Invitrogen, 175 μg/ml). Serum-free media were collected from the transfected clones and screened by immunoblotting using antibodies specific for the α1(VI) and α3(VI) collagen chains (19, 21). A cell clone that expressed high levels of both constructs, designated HEK293-α1α3, was used in this study.
HEK293-α1α3 Cells Expressing Normal and Mutant α2(VI) Collagen Chains
A full-length human α2(VI) collagen cDNA, F225 (11), was cloned into the EcoRI site of the pIRESpuro2 expression vector harboring a puromycin resistance gene (Clontech, Mountain View, CA). The F225 cDNA encodes the major α2(VI) collagen chain containing the C2 subdomain, designated α2-C2 (11). An expression vector for the alternatively spliced α2-C2a variant was generated by replacing the C2 subdomain coding region in the α2-C2 expression construct with a fragment from cDNA clone P1 (11), which encodes the α2-C2a form. Expression constructs for the COL6A2 E624K and R876S mutants were generated by replacement of the C1 or C2 coding regions of the α2-C2 cDNA expression constructs with the corresponding cDNA fragments from fibroblasts of the patients, prepared by reverse transcription-PCR. The α2-C2, α2-C2a, α2-E624K, and α2-R876S expression constructs and the empty pIRESpuro2 vector were linearized with NotI and transfected into the HEK293-α1α3 cells using Lipofectamine 2000 (Invitrogen). Stably transfected cells were selected with 175 μg/ml G418 and 0.5 μg/ml puromycin (Sigma).
Northern Blot Analysis, Immunoprecipitation, and Immunoblotting
Northern blotting was performed with total RNA extracted from cells using the RNeasy Mini Kit (Qiagen, Valencia, CA) and hybridized with [32P]dCTP-labeled human COL6A1, COL6A2, and COL6A3 cDNA probes by the procedure described previously (7). For immunoprecipitation, cells were metabolically labeled overnight with [35S]cysteine (MP Biomedicals, Irvine, CA) in the presence of 50 mg/ml sodium ascorbate in serum-free medium as described (5). For the pulse-chase experiments, cells were labeled with [35S]cysteine for 30 min and then chased for 0, 30, 60, and 120 min in the same medium containing unlabeled cysteine. Culture media (0.8 ml) and cell layers were subject to immunoprecipitation with an antibody specific for the α3(VI) collagen chain (21) by the methods described previously (5). The immunoprecipitated material was analyzed after reduction with 25 mm dithiothreitol on 3–8% polyacrylamide gels (Invitrogen) or without reduction on composite 0.5% agarose and 2.4% polyacrylamide gels. Radioactive bands on gels were detected using a Typhoon PhosphorImager (Amersham Biosciences). For immunoblotting, serum-free culture media and cell layers were electrophoresed on 3–8% polyacrylamide gels as described (7). The blots were probed with an antibody specific for the α2(VI) collagen chain, and the blot was developed with the ECL plus reagents (Amersham Biosciences).
Collagen VI Chain-specific Antibodies
The α3(VI) collagen VI-specific polyclonal antibody was generated previously using a recombinant N-globular domain of the human α3(VI) chain produced in HEK293 cells as the antigen (21). A mouse monoclonal antibody against the human α3(VI) collagen was purchased from Chemicon (#3303). The α2(VI) collagen-specific antibody was prepared by the custom antibody service of Proteintech Group Inc. (Chicago, IL), using a glutathione S-transferase fusion protein containing the C-globular domain of the α2(VI) chain produced in Escherichia coli as the antigen. Briefly, a cDNA fragment encoding the C-globular domain coding region of COL6A2 was generated by PCR using a full-length COL6A2 cDNA as the template (11). The PCR primers were 5′-TAT AGG ATC CTT CGT CAT CAA CGT GGT CAA C and 5′-TAT AGA ATT CCT AGC AGA TCC AGC GGA TGA A. The resulting cDNA fragment was digested with BamHI and EcoRI and cloned into the polylinker region of the pGEX-4T-1 vector (Amersham Biosciences). The glutathione S-transferase fusion protein produced in E. coli was purified by affinity chromatography and used to immunize rabbits. The resulting α2(VI)-specific antisera demonstrated minimal cross-reactivity with other proteins in immunoblot analysis of culture medium and cell lysate from human fibroblasts (supplemental Fig. 1S).
Immunostaining and Electron Microscopic Analysis
Fibroblasts and HEK293 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum in the presence of 50 mg/ml sodium ascorbate until 4 days (fibroblasts) and 10 days (HEK293) after confluency, fixed with 3.7% formaldehyde and incubated with a monoclonal antibody against collagen VI (Chemicon 3303, 1:2,500) for 1 h. Immunostaining was detected with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, OR, 1:500 dilution) and observed using a Zeiss Axioskop epifluorescence microscope with a Toshiba 3CCD camera. Rotary shadowing electron microscopy of fibroblasts was performed as described previously (17).
Computer Analysis
Multiple sequence alignment was performed with the ClustalW program. Computer models of the regions spanning the COL6A2 E624K and R876S mutation sites were generated by the Sybyl7.3 program (Tripos, Inc., St. Louis, MO). The models of specific regions were generated by homology modeling with the αXβ2 integrin I domain as a modeling template, a domain belonging to the vWF-A-like family of proteins (Protein Data Base accession number 1N3Y) (22). Molecular density surfaces and the electrostatic potential (physical unit, kilocalories/mole) of the surfaces of the analyzed fragments were calculated using the MOLCAD module of the Sybyl program (23).
RESULTS
Two Patients with Homozygous Missense Mutations in COL6A2
A cohort of patients with typical features of UCMD, including congenital muscle weakness, proximal joint contractures, and distal joint laxity, were screened for collagen VI mutations by DNA sequencing of the genomic DNA and cDNAs (7, 24). Two patients were found to carry homozygous missense mutations in the C-globular domain of the COL6A2 gene. Patient UC3 had a homozygous p.E624K (c.1870G>A) change in the C1 subdomain, whereas patient UC15 carried a homozygous p.R876S (c.2626C>A) substitution in the C2 subdomain (Fig. 1C).
Clinical History
UC3 was the second child of a family without known consanguinity living in Brazil. She was recognized to have hypotonia at 6 months of age, when she started to sit. She was able to ambulate with support at 16 months of age but never achieved the ability to walk independently. She was first evaluated at 3 years and 4 months of age and found to have proximal greater than distal weakness, as well as muscular atrophy that was more prominent proximally than distally. She had contractures at the ankles, knees, and elbows as well as hyperlaxity of the small joints of the fingers and toes. Her contractures were progressive, and her weakness was mostly stable with some mild worsening over the years. At 10 years of age she was able to crawl and pedal a tricycle. She died with an intercurrent pneumonia en route to the hospital at the age of 10 years and 4 months. The child had not shown any signs of overt respiratory insufficiency before that. Serum creatine kinase (CK) levels were normal on multiple occasions. Muscle biopsies at ages of 3.5 years and again at 9 years showed considerable variability in fiber diameter, many rounded and severely atrophic fibers, central nucleation, increased interstitial connective tissue, and some evidence for necrosis of muscle fibers. The variability in fiber diameter, build-up of interstitial connective tissue, and evidence for necrosis were progressive from the first to the second biopsies.
UC15 is of Filipino origin, and the offspring of a consanguineous marriage. Bilateral dislocated hips were noticed at birth. Hyperlaxity of the fingers, hands, and feet was noticed early. He started walking for limited distances without support at 2 years of age. There were increasing contractures predominantly of the shoulders, elbows, hips, and knees, while striking distal hyperlaxity was maintained, in particular in the fingers and toes. Together with slowly increasing weakness, he had increasing difficulties ambulating, and finally was dependent on a wheelchair for ambulation at 11 years of age. CK values were elevated 2–5 times above normal. During the second decade of life, he demonstrated progressive decline in his forced vital capacity with evidence for severe restrictive lung disease. Noninvasive assisted ventilation at night was instituted at 19 years of age. Scoliosis was progressive leading to spinal fusion at the age of 14 years. Now in his third decade of life he continues to be quite stable. He had significant contractures of the neck extensors, elbows, hips, knees, and feet. He had excessive keloid formation as well as follicular hyperkeratoses over the extensor surfaces of the arms and legs. Muscle biopsy revealed a myopathic pattern without specific features. The heterozygous carriers in both families (UC15 and UC3) are clinically unaffected and have no neuromuscular symptoms.
Structural Analysis of the Missense Mutations
The C1 and C2 subdomains in the C-terminal globular region of the α2(VI) collagen chain are vWF-A modules present in a large number of multidomain cell adhesion and extracellular matrix molecules (25). Crystal structures of several vWF-A domains are known, and they adopt the dinucleotide-binding (Rossmann fold) structure consisting of a central β-sheet with mostly parallel β-strands surrounded by α-helices (26–29). Many vWF-A domains contain a metal ion-dependent adhesion site (MIDAS) that elicits divalent metal-dependent ligand binding (27). We performed sequence alignment and molecular modeling of the C1 and C2 subdomains based on the known crystal structures of the vWF-A domains (Fig. 2 and supplemental Fig. 2S). The E624K mutation altered a glutamic acid residue within the consensus MIDAS sequence, DXSXS, in the loop between the β1-strand and α1-helix of the C1 subdomain (Fig. 2A). The R876S mutation affects a different region of the vWF-A module, an arginine residue in the β2 strand of the C2 subdomain (Fig. 2A). Previously reported compound heterozygous COL6A2 mutations, L837P and Asn-897 deletion (8), are located in the β1-strand and α2-helix of the C2 subdomain, respectively (Fig. 2A). Both E624K and R876S mutations result in amino acid charge changes. Computational analysis confirmed that the amino acid substitutions result in significant changes in the electrostatic potentials of the regions surrounding the mutations (supplemental Fig. 2S). The electrostatic potentials (expressed as kilocalories/mole) of the region encompassing the Lys-624 mutation site ranged from (+)11.7 to (+)164.3 compared with the range of (−)90.6 to (−)220.6 for the region spanning the E624 site of the normal C1 subdomain. The electrostatic potential values for the C2 region encompassing the Arg-876 site ranged from (+)92.6 to (+)132.6, whereas the values for the area surrounding the Ser-876 mutation site ranged from (+)17.1 to (+)68.4.
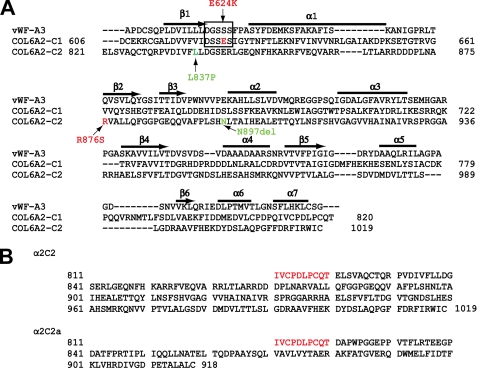
FIGURE 2.
Amino acid sequences of the COL6A2 C-globular domains and the location of missense mutations. A, alignment of the amino acid sequences of the COL6A2 C1 and C2 subdomains with that of the vWF-A3 domain. Locations of α2-E624K and α2-R876S mutations are indicated in red. Previously reported compound heterozygous C2 subdomain mutations, L837P and N897del, are shown in green. Secondary structure assignments of the vWF-A3 domain (seven α-helices and seven β-strands) are shown, and the MIDAS site is boxed. B, amino acid sequences of the COL6A2 splice variants. The COL6A2-C2 and COL6A2-C2a differ in the sequences at the end of C-globular domains due to mutually exclusive use of the most 3′-exons. Sequences shown include 10 amino acids of the constant region (in red) followed by the C2 or C2a subdomains (in black).
Muscle Biopsies and Fibroblasts from the Patients
Muscle biopsies of UC3 and UC15 were co-immunostained with antibodies against collagen VI and the basement membrane marker perlecan (Fig. 3A). There was a substantial increase in endomysial connective tissue in both patients as evidenced by a much wider space between the basement membranes outlining adjacent muscle fibers compared with the control. Collagen VI immunostaining was prominent in the interstitial connective tissue of the UC3 muscle but significantly reduced in the muscle of UC15. This finding was unexpected. Given the recessive nature of the disease in the patients and given that UC3 had a more severe clinical phenotype than UC15, UC3 was predicted to produce less collagen VI than UC15. However, in both patients there was a reduction of collagen VI immunoreactivity in the basement membrane.
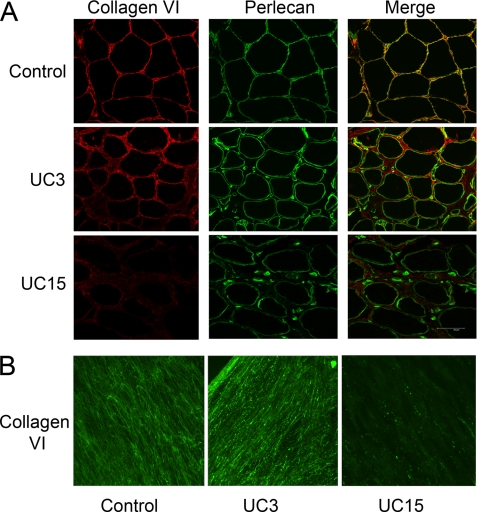
FIGURE 3.
Collagen VI immunostaining of muscle biopsies and dermal fibroblasts. A, cryosections of muscle biopsies from the control, UC3, and UC15 were immunostained with anti-collagen VI monoclonal antibody (red) and anti-perlecan polyclonal antibody (green). A merge of green and red fluorescence is shown. Collagen VI immunostaining was readily seen throughout the interstitial connective tissue of UC3 but significantly reduced in the UC15 muscle biopsies. Unlike the control muscle, collagen VI and perlecan did not co-localize in the basement membrane of the muscles from the patients. B, immunostaining of collagen VI deposited by fibroblasts from the control, UC3, and UC15. Cells were grown in the presence of 50 μg/ml ascorbate for 4 days post-confluency and then stained with the anti-collagen VI monoclonal antibody. Abundant but knotty collagen VI microfibrils were deposited by UC3 fibroblasts, whereas UC15 fibroblasts assembled few collagen VI microfibrils in the matrix.
Collagen VI microfibrils deposited by the fibroblasts from the patients were analyzed by immunostaining of post-confluent cultures (Fig. 3B). The UC3 fibroblasts deposited abundant collagen VI microfibrils, but the microfibrils appeared thick and abnormally knotty compared with the control. By contrast, the UC15 fibroblasts produced almost no collagen VI microfibrils in the extracellular matrix. Thus, there was a good correlation between the collagen VI protein expression levels in fibroblast cultures and those in the muscle biopsies.
The collagen VI microfibrils assembled by the UC3 fibroblasts were further analyzed by electron microscopy after rotary shadowing (Fig. 4). In contrast to the evenly dispersed collagen VI network seen in the control fibroblasts (Fig. 4, A–C), the microfibrillar network deposited by the UC3 fibroblasts was scarcely spread out and composed mainly of thick fibrils and spots made of densely packed microfibrils (Fig. 4, D–G). The microfibrils from UC3 preserved the parallel association ability in that thick fibrils were formed and globular domains were in register (Fig. 4G). However, there was disorganization and reduced expansion of the two-dimensional networks with loss or fusion of globular domains.
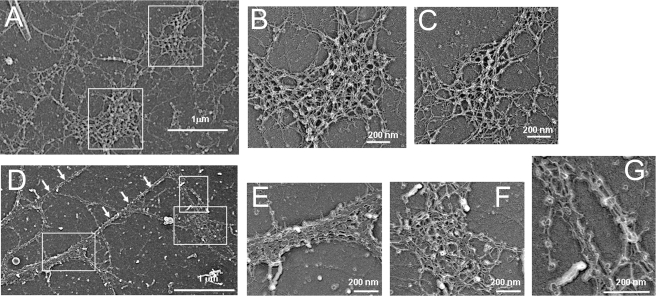
FIGURE 4.
Rotary shadowing electron microscopy of collagen VI microfibrils deposited by the dermal fibroblasts from control (A–C) and UC3 (D–G). A, the collagen VI network in the healthy donor is well spread out. Scale bar = 1 μm. B and C, high magnification images of boxed areas in A, showing a web-like collagen VI network. Scale bars = 200 nm. D, UC3 microfibrillar network is unevenly dispersed and composed mainly of thick fibrils (white arrows) and spots made of densely packed microfibrils (boxed). E, F, and G, high magnification images of boxed areas in D, showing tight lateral association of the UC3 microfibrils with globular domains in register but reduced expansion in two dimensions.
Collagen VI Biosynthesis in Fibroblasts from Patients
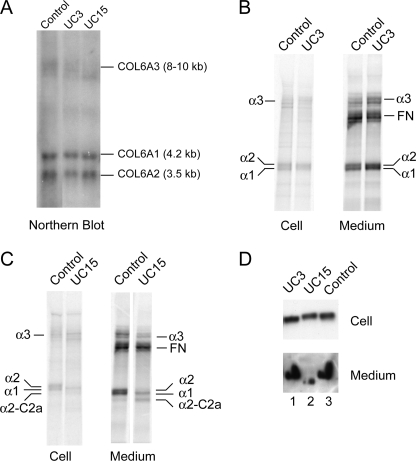
Northern blot analysis showed that the levels and sizes of the COL6A2 mRNAs in fibroblasts from UC3 and UC15 were comparable to the control (Fig. 5A), indicating that the single nucleotide changes did not alter the processing and stability of the COL6A2 mRNA. The biosynthesis of collagen VI was studied by immunoprecipitation of cell layers and culture media from fibroblasts labeled with [35S]cysteine overnight. The antibody used was specific for the α3(VI) collagen chain, which precipitated triple-helical collagen VI consisting of all three chains as well as the free α3(VI) chain. Collagen VI synthesis and secretion in UC3 fibroblasts were comparable to the control (Fig. 5B). In contrast, a marked reduction in collagen VI was observed in the medium of UC15 fibroblasts (Fig. 5C), even though the amount of fibronectin that bound to the Sepharose A beads during immunoprecipitation was similar to that in the control fibroblasts. Strikingly, the secreted collagen VI in UC15 was not composed of normal-sized α2(VI) chains, but was instead made up of a faster migrating band with a size corresponding to that of the C2a splice variant (Fig. 5C). Immunoblotting of culture media and cell lysates (Fig. 5D) confirmed that UC15 fibroblasts secreted only a low level of the faster migrating α2(VI) chain into the medium and that the α2(VI) chain present in the cell lysate was of the normal size. Collectively, the results suggested that the R876S mutant chain in UC15 was synthesized but not assembled into a triple-helical collagen VI molecule and that the secreted collagen VI in UC15 was made up of the C2a splice variant, which contains a shorter alternative C2a subdomain not affected by the missense mutation (Fig. 2B).
FIGURE 5.
Analysis of collagen VI mRNA and protein in dermal fibroblasts of patients. A, Northern blot analysis of total RNAs (10 μg/lane) isolated from dermal fibroblasts hybridized with [32P]dCTP-labeled cDNA probes for COL6A1, COL6A2, and COL6A3. The levels of the three collagen VI mRNAs in UC3 and UC15 were similar to those in the control fibroblasts. B and C, biosynthesis of collagen VI in dermal fibroblasts from patients. Fibroblasts from UC3 (B) and UC15 (C) were labeled with [35S]cysteine overnight, and then serum-free culture media and cell layers were immunoprecipitated with a COL6A3-specific antibody. The amounts and sizes of the three collagen VI chains (α1, α2, and α3) in UC3 were similar to the control fibroblasts. Note that fibronectin (FN), which binds to protein A-Sepharose during immunoprecipitation, was detected in the culture medium. UC15 fibroblasts secreted a polypeptide migrating at the size of C2a splice variant, but there was no normal α2 chain. They also secreted substantially reduced amounts of α3 and α1 chains compared with the control, whereas the levels of FN are similar in UC15 and the control. D, immunoblot analysis of fibroblasts from patients with a COL6A2-specific antibody. Cells were seeded at the same density, grown until 1 day after confluency, and then switched to serum-free medium for 24 h. Cell lysates (30 μg of total protein/lane) and 100-μl aliquots of the serum-free culture media precipitated with 0.9 ml of 100% EtOH were separated on 3–8% polyacrylamide gels. UC15 secreted a faster migrating α2 chain at a markedly reduced level, whereas in the cells the α2 chain produced by UC15 was similar in size and level as the control and UC3.
COL6A2 Missense Mutants and Splice Variant in Transfected Cells
The biosynthetic studies suggested that the COL6A2 E624K and R876S mutations had distinct effects on collagen VI biosynthesis and assembly. However, it was unclear whether the observed difference resulted from the COL6A2 homozygous missense mutations alone or could also be attributed to other non-synonymous changes of collagen VI genes and/or differences in modifying genes in these two patients. Additionally, it was important to demonstrate that the C2a variant chain indeed could support collagen VI triple-helix formation and microfibrillar assembly. Hence, we established a well defined cell culture system to examine the effects of the COL6A2 missense mutations and C2a splice variant under identical genetic and cellular backgrounds. HEK293 cells were chosen as they do not express endogenous collagen VI chains.
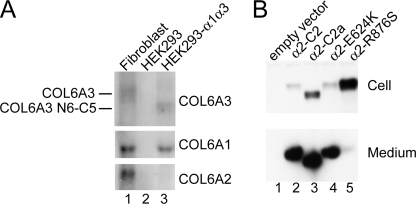
We first generated HEK293 cell clones stably expressing the full-length human α1(VI) and α3(VI) N6-C5 collagen chains. The α3(VI) N6-C5 chain, which lacks the N-terminal N10–N7 domains, was previously shown to be capable of associating with the α1(VI) and α2(VI) chains into a triple-helical collagen VI molecule and then assembling into dimers, tetramers, and microfibrils (20). Northern blot analysis demonstrated that a cell clone, designated HEK293-α1α3, expressed the COL6A1 mRNA and COL6A3 N6-C5 mRNA at similar levels as the corresponding collagen VI mRNAs in normal fibroblasts, but this clone did not express the COL6A2 mRNA (Fig. 6A).
FIGURE 6.
Characterization of the transfected HEK293 cells. A, Northern blot analysis of the HEK293-α1α3 cell clone. Total RNAs (10 μg/lane) were probed with 32P-labeled COL6A1, COL6A2, and COL6A3 cDNAs. The HEK293-α1α3 cell clone (lane 3) expressed the COL6A1 and the COL6A3 N6-C5 mRNAs at comparable levels as the COL6A1 and COL6A3 mRNAs in normal dermal fibroblasts (lane 1). Note that the COL6A3 N6-C5 mRNA is smaller than the COL6A3 mRNA, because it lacks the N10–N7 domains. Parental HEK293 cells (lane 2) did not express the three collagen VI mRNAs. B, immunoblot analysis of the HEK293-α1α3 cells transfected with normal and mutant α2(VI) expression constructs. 30 μg of total protein from cell layers (top panel) and 100-μl aliquots of serum-free culture media (bottom panel) were used for immunoblot analysis with the COL6A2-specific antibody. The normal α2-C2 chain, α2-C2a splice variant, and α2-E624K mutant were secreted into the medium, but the α2-R876S chain was mostly retained inside cells.
The HEK293-α1α3 cells were next transfected with expression constructs of the full-length α2(VI) collagen chain (α2-C2), the mutant α2(VI) chains (α2-E624K and α2-R876S), and the splice variant (α2-C2a). Stably transfected cell clones expressing comparable levels of the normal or mutant COL6A2 mRNAs were selected by Northern analysis and used for the following studies. Immunoblot analysis of the selected cell clones showed that the normal α2-C2 chain, α2-C2a variant, and α2-E624K mutant chain were present at high levels in the culture media and low amounts were detected in the cell lysates, indicating that these chains were efficiently secreted (Fig. 6B). However, only a minute amount of the α2-R876S chain was detected in the medium, and the majority was retained intracellularly.
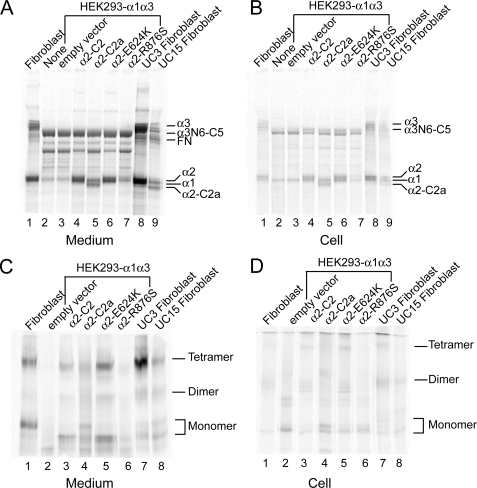
Biosynthesis of collagen VI in the transfected cells was analyzed by immunoprecipitation of cells labeled overnight with the α3(VI)-specific antibody. In the media and cell layers from untransfected and empty vector-transfected HEK293-α1α3 cells (Fig. 7, A and B, lanes 2 and 3), the antibody precipitated the α3(VI) N6-C5 chains and a small amount of the α1(VI) chains that associated with the α3(VI) N6-C5 chains. There were also several bands migrated below the α3(VI)-N6-C5 chain, which were truncated α3(VI)-N6-C5 chains generated by proteolytic processing. In both the medium and cell layers of the transfected cells, the normal α2-C2 chain, α2-C2a variant, and α2-E624K mutant co-immunoprecipitated with the α1(VI) and α3(VI) chains (Fig. 7, A and B, lanes 4–6), indicating that the α2-C2a variant and α2-E624K mutant were able to assemble into triple-helical collagen VI molecules in a manner similar to the α2-C2 chain. On the other hand, only very low amounts of the α2-R876S and α1 chains were detected in the cell layer and medium (Fig. 7, A and B, lanes 7), demonstrating that the mutation impaired the production and/or assembly of collagen VI. The collagen VI biosynthetic patterns in the transfected HEK293 cell clones resemble those observed in the dermal fibroblasts (Fig. 7, A and B, lanes 1, 8, and 9).
FIGURE 7.
Biosynthesis of collagen VI in transfected cells. Transfected HEK293 cells and dermal fibroblasts from patients were labeled overnight with [35S]cysteine. Culture media (A and C) and cell layers (B and D) were immunoprecipitated with a COL6A3-specific antibody. Immunoprecipitated material was analyzed under reduced conditions on 3–8% polyacrylamide gels (A and B) or non-reduced conditions on composite agarose and polyacrylamide gels (C and D). In transfected cells, the α2-R876S chain could not assemble into a triple-helical collagen VI molecule intracellularly and therefore triple-helical collagen VI monomer, dimer, and tetramers were barely seen in the medium. In contrast, the α2-E624K chain and α2-C2a splice variant could be assembled into triple-helical collagen VI monomers, dimers, and tetamers as the normal α2-C2 chain. In UC15 fibroblasts, the α2-C2a variant but not the α2-R876S chain was assembled into a triple-helical collagen VI molecule.
The immunoprecipitated materials were also examined under non-reduced conditions on composite agarose-polyacrylamide gels to assess the assembly of collagen VI dimers and tetramers. In transfected HEK293 cells (Fig. 7, C and D, lanes 2–6), the α2-E624K chain supported tetramer formation as efficiently as the normal α2-C2 chain. The assembled tetramers were barely detected in the cell layers, indicating that most of the tetramers were secreted. The α2-C2a variant also could assemble into tetramers, although less efficiently. By contrast, collagen VI dimers and tetramers were not seen in samples from the α2-R876S-expressing cells. Similarly, in fibroblasts from the patients, collagen VI tetramer assembly was not affected by the E624K mutation in UC3, in contrast to the R876S mutation in UC15 (Fig. 7, C and D, lanes 1, 7, and 8).
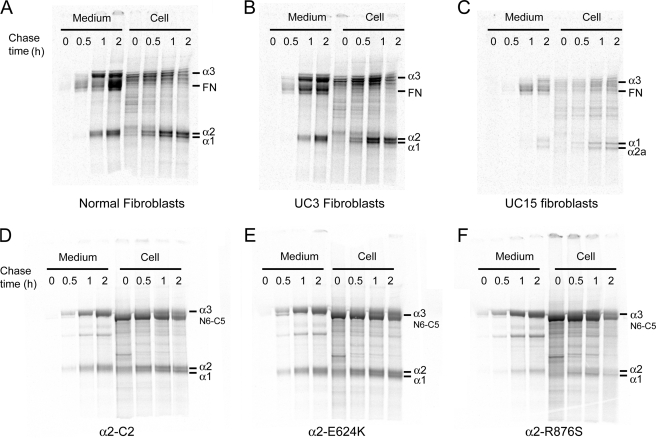
To determine whether the mutations affect the rate of synthesis and secretion of collagen VI, we pulse-labeled both fibroblasts and transfected cells and then immunoprecipitated samples collected at different time points after chase (Fig. 8). In both fibroblasts and transfected HEK293 cells, the rates of collagen VI synthesis and secretion were similar in cells expressing the α2-E624K chain and the normal α2-C2 chain (Fig. 8, A, B, D, and E). In contrast, in UC15 fibroblasts the α2-R876S chain was not seen at all time points and only the α2-C2a chain was produced and secreted (Fig. 8C). The amounts of α2-R876S chain produced in transfected cells were also very low at all time points (Fig. 8F).
FIGURE 8.
Pulse-chase labeling analysis of normal and mutant collagen VI production. Dermal fibroblasts from patients (A–C) and transfected HEK293 cells (D–F) were pulse-labeled with [35S]cysteine for 30 min and then switched to unlabeled medium for 0, 0.5, 1, and 2 h before immunoprecipitation of culture medium and cell layers with the COL6A3-specific antibody. The α2-E624K mutant chains formed triple-helical collagen VI molecules at the same rate as the normal α2-C2 chains in both fibroblasts and HEK293 cells. On the other hand, only a small amount of the α2-R876S mutant chain was co-immunoprecipitated in HEK293 cells at all time points, suggesting that the mutant chain was not efficiently assembled into triple-helical collagen VI molecules. In UC15 fibroblasts, the α2-C2a instead of the α2-R876S chain was incorporated into triple-helical collagen VI molecules and secreted.
Collagen VI Microfibrillar Assembly in Transfected Cells
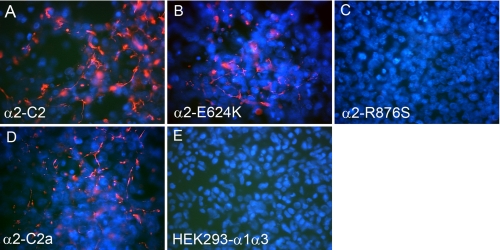
Transfected cells were grown for 10 days after confluency and immunostained with a monoclonal antibody against human collagen VI. Cells expressing the normal α2-C2 chain and the α2-C2a splice variant both assembled short collagen VI microfibrils in the extracellular matrix (Fig. 9, A and D). The α2-E624K-expressing cells could also assemble some short collagen VI microfibrils (Fig. 9B), although the amounts were substantially reduced compared with the normal α2-C2 chain. No collagen VI microfibrils were detected in cells expressing the α2-R876S chains (Fig. 9C).
FIGURE 9.
Immunostaining of collagen VI deposited by the transfected HEK293 cells. HEK293 cell clones expressing the α2-C2 (A), α2-E624K (B), α2-R876S (C), and α2-C2a (D) chains and the parental HEK293-α1α3 cells (E) were grown in the presence of 50 μg/ml ascorbate for 10 days post-confluency and stained with a monoclonal antibody specific for human collagen VI and 4′,6-diamidino-2-phenylindole for nuclei. Short collagen VI microfibrils (red) were deposited by cells expressing the normal α2-C2 chain and the α2-C2a variant. Cells expressing the α2-E624K mutant show reduced amounts of microfibrils compared with those expressing the α2-C2 chain. No microfibrils were deposited by cells expressing the α2-R876S chain.
DISCUSSION
Mutations in the three collagen VI genes are now known to cause overlapping musculoskeletal disease entities with a range of clinical severity. Apart from the severe UCMD, collagen VI defects are found in disorders with intermediate as well as milder clinical manifestations, including Bethlem myopathy, limb-girdle muscular dystrophy, and myosclerosis myopathy (3, 30, 31). Different types of mutations in the three genes have been reported, but the genotype-phenotype relationships remain incompletely understood. In addition to frameshift and nonsense mutations that lead to a decrease or an absence of normal collagen VI chains, structural mutations in collagen VI due to in-frame deletions and missense mutations are frequent. The majority of the structural mutations are dominant acting and located in the triple-helical domains, resulting in a range of manifestations from severe to mild (8, 9, 16, 32, 33). Numerous single amino acid changes in the wWF-A modules of the three collagen VI chain globular domains have also been identified (24). Although most of those are non-pathogenic sequence variations, several heterozygous single amino acid substitutions have been shown to underlie Bethlem myopathy (33, 34). Three of the Bethlem heterozygous amino acid changes in the vWF-A domains have been found to interfere with the folding of the chain, the assembly of the collagen VI microfibrils, or the protein interaction in the extracellular matrix (33, 35). Recessive structural mutations in collagen VI have been reported in two UCMD patients previously (8, 18). One of the patients carries a homozygous COL6A2 R876S mutation in the C2 subdomain like UC15 and is also from a consanguineous family of the Philippines, suggesting a founder effect for the mutation in this ethnic group (18). The other patient has compound heterozygous mutations, also in the C2 subdomain of COL6A2, L837P and delN897, which have been shown to prevent collagen VI assembly (8). Our studies of UC3 and UC15 thus expand the group of UCMD patients with recessive structural mutations in collagen VI and bring novel mechanistic insights into recessive structural mutations.
Of particular interest is that the recessive structural mutations identified in these four UCMD patients to date are all present in the C-globular domain of COL6A2, implying an important role of this region in collagen VI function. The identification of mutations in the C1 and C2 subdomains of COL6A2 in a homozygous state in UC3 and UC15 has provided ideal biological systems to study the role of COL6A2 C1 and C2 subdomains in collagen VI biosynthesis and assembly. Using dermal fibroblasts from the patients, we demonstrate that the COL6A2-E624K mutant mRNA and protein produced by UC3 are stable. Through biosynthetic studies, we show that the UC3 mutation within the MIDAS motif in the C1 subdomain does not affect collagen VI triple-helix formation and dimer/tetramer assembly. The mutant chain does not exert a dominant negative effect as heterozygous carriers in the family are healthy, and ample collagen VI microfibrils are assembled in the extracellular matrix by the patient's fibroblasts. By contrast, both biosynthetic and immunohistochemical studies demonstrate that the homozygous COL6A2-R876S mutation in the C2 subdomain leads to a marked reduction in collagen VI triple-helical monomers, dimers, tetramers, and microfibrils. The distinct effects of these two mutations on collagen VI biosynthesis and assembly are confirmed independently in cells transfected with the mutant COL6A2 constructs. Our studies thus demonstrate that these two missense mutations in the C1 and C2 subdomains of COL6A2 disrupt the proper function of collagen VI through different mechanisms.
The observation that UC3 can assemble collagen VI microfibrils agrees with previous findings in a cell-free coupled transcription-translation system and further support the notion that alterations in the COL6A2 C1 subdomain do not interfere with collagen VI dimer formation (36). In the transfected cells, the COL6A2-E624K mutant chain was less efficient in supporting microfibrillar assembly than the normal chain (Fig. 9). Although abundant collagen VI microfibrils are deposited by the UC3 fibroblasts, light and electron microscopic analyses indicate that the microfibrils are not able to expand in two dimensions to form the web-like collagen VI network. Instead, they tend to associate laterally into thick fibers and densely packed aggregates. The results thus highlight the importance of the C1 subdomain in the formation of a proper microfibrillar network. The E624K mutation is located in the vWF-A modules, which often mediate protein-protein interactions. For instance, the vWF-A domains in the von Willebrand factor and integrins bind collagens and other ligands (37, 38). Ample studies demonstrate that point mutations in the MIDAS motif abrogate cation or ligand binding (39, 40). Molecular modeling indicates that the change from an acidic to a basic amino acid in the MIDAS site in UC3 profoundly alters the electrostatic environment surrounding the MIDAS region. We speculate that the missense mutation in UC3 may disrupt an important ligand binding interaction that hampers the supramolecular assembly of collagen VI network. Although the effects of the mutation highlight the biologic importance of the motif for collagen VI function, the specific binding interaction of the MIDAS motif in the COL6A2-C1 domain is currently unknown and remains to be investigated. It is noteworthy that large collagen VI microfibrillar aggregates have been reported in another patient with clinical manifestations of UCMD but no mutations in the three collagen VI genes (8). Interestingly, this patient was homozygous for COL6A2-R680H amino acid substitution located in the C1 subdomain. Although the R680H change was thought to be a non-pathogenic polymorphism, our observation in UC3 raises the possibility that in the homozygous state this amino acid substitution may result in dysfunctional collagen VI microfibrils.
The UC15 fibroblasts bearing the homozygous COL6A2 R876S mutation in the C2 subdomain produce normal amounts of COL6A2 mRNA. However, pulse-chase biosynthetic studies indicate that both intracellular and extracellular collagen VI levels are significantly reduced at all time points. Immunoblotting of the transfected cells shows that the R876S mutant chain is mostly retained within the cells (Fig. 6B). Together, the results suggest that the R876S mutant chain may be misfolded due to a significant change in electrostatic interactions and thereby unable to support collagen VI triple-helix formation. This in turn leads to markedly reduced amounts of dimers, tetramers, and microfibrils, as demonstrated by the composite polyacrylamide-agarose gel and immunofluorescence studies. Of note is that the intracellular accumulation of the mutant chain was not apparent in the UC15 fibroblasts. This is probably because the level of collagen VI synthesis is substantially lower in fibroblasts than in transfected cells. Interestingly, fibroblasts from the patient with the compound heterozygous COL6A2 C2 subdomain mutation reported previously exhibit a similar collagen VI biosynthetic defect as the UC15 fibroblasts, in that only a minute amount of collagen VI is secreted (8). The COL6A2 C2 subdomain has previously been shown to be critical for collagen VI dimer formation in the cell-free transcription-translation system (36). The biosynthetic studies of the two patients with the C2 mutations imply that the C2 subdomain may also contain sequences that facilitate the association of the three collagen VI chains into a triple-helical molecule.
An unexpected finding is that, even though UC15 fibroblasts assemble few collagen VI microfibrils, the patient was able to ambulate until 11 years of age and hence had a moderately severe disease manifestation compared with the severe clinical presentation of many UCMD cases with similar absence of collagen VI microfibrils (5, 16). Because the mutation in UC15 resides in the COL6A2 C2 subdomain that is subject to alternative splicing, the C2a splice variant remains wild type. Our studies show that UC15 produces a small amount of collagen VI, which is composed of the C2a splice variant exclusively. The amount of α2-C2a splice variant in UC15 fibroblasts appears to be increased compared with in the control fibroblasts. It is possible that increased utilization of the C2a splice variant could partially compensate for the total loss of the normal COL6A2 chain, thus explaining the less severe clinical manifestation of UC15. We demonstrate that in the transfected cells collagen VI triple-helical molecules and short microfibrils can be assembled with the α2-C2a splice variant. Although few collagen VI microfibrils were detected in UC15 fibroblasts under the experimental conditions used in this study, some collagen VI protein could be assembled in vivo over time as evidenced by the immunostaining of the muscle biopsy. It is noteworthy that the two patients with recessive COL6A2 mutations in the C2 subdomain reported previously, including the Filipino patient who is also homozygous for the identical mutation, also presented with a moderately severe UCMD disease progression (8, 18), which is consistent with our hypothesis of functional compensation by the C2a splice variant. Whether the C2a splice variant is utilized in those two patients reported previously remains to be investigated. On the other hand, the possibility that other genetic factors may modify the clinical phenotypes cannot be ruled out.
In conclusion, our studies demonstrate that the biochemical mechanisms underlying these two homozygous missense mutations in COL6A2 C-globular domains are distinct and that the clinical severity of patients cannot be dictated exclusively by the degree of collagen VI biosynthetic abnormalities. Our observations of the C2a splice variant in UC15 raises an intriguing possibility that up-regulation of the C2a splice variant expression may be a potential treatment strategy for UCMD with mutations in the COL6A2 C2 subdomain.
Supplementary Material
Acknowledgment
We thank Patrizia Sabatelli for helping with the ultrastructural analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants AR053251 (to M. L. C.), AR049537 (to A. F.), and AR051999 (to C. G. B.). This work was also supported by Muscular Dystrophy Association USA (Grant 3896 to C. G. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- UCMD
- Ullrich congenital muscular dystrophy
- vWF-A
- von Willebrand factor, type A
- HEK
- human embryonic kidney cell
- MIDAS
- metal ion-dependent adhesion site
- CK
- creatine kinase.
REFERENCES
- 1.Muntoni F., Voit T. (2004) Neuromuscul. Disord. 14, 635–649 [DOI] [PubMed] [Google Scholar]
- 2.Peat R. A., Smith J. M., Compton A. G., Baker N. L., Pace R. A., Burkin D. J., Kaufman S. J., Lamandé S. R., North K. N. (2008) Neurology 71, 312–321 [DOI] [PubMed] [Google Scholar]
- 3.Lampe A. K., Bushby K. M. (2005) J. Med. Genet. 42, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higuchi I., Shiraishi T., Hashiguchi T., Suehara M., Niiyama T., Nakagawa M., Arimura K., Maruyama I., Osame M. (2001) Ann. Neurol. 50, 261–265 [DOI] [PubMed] [Google Scholar]
- 5.Camacho Vanegas O., Bertini E., Zhang R. Z., Petrini S., Minosse C., Sabatelli P., Giusti B., Chu M. L., Pepe G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7516–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demir E., Sabatelli P., Allamand V., Ferreiro A., Moghadaszadeh B., Makrelouf M., Topaloglu H., Echenne B., Merlini L., Guicheney P. (2002) Am. J. Hum. Genet. 70, 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan T. C., Zhang R. Z., Sudano D. G., Marie S. K., Bönnemann C. G., Chu M. L. (2003) Am. J. Hum. Genet. 73, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker N. L., Mörgelin M., Peat R., Goemans N., North K. N., Bateman J. F., Lamandé S. R. (2005) Hum. Mol. Genet. 14, 279–293 [DOI] [PubMed] [Google Scholar]
- 9.Lampe A. K., Zou Y., Sudano D., O'Brien K. K., Hicks D., Laval S. H., Charlton R., Jimenez-Mallebrera C., Zhang R. Z., Finkel R. S., Tennekoon G., Schreiber G., van der Knaap M. S., Marks H., Straub V., Flanigan K. M., Chu M. L., Muntoni F., Bushby K. M., Bönnemann C. G. (2008) Hum. Mutat. 29, 809–822 [DOI] [PubMed] [Google Scholar]
- 10.Timpl R., Chu M. L. (1994) in Extracellular Matrix Assembly and Structure (Yurchenoco P. D., Birk D. E., Mecham R. P. eds) pp. 207–242, Academic Press, San Diego, CA [Google Scholar]
- 11.Chu M. L., Pan T. C., Conway D., Kuo H. J., Glanville R. W., Timpl R., Mann K., Deutzmann R. (1989) EMBO J. 8, 1939–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu M. L., Zhang R. Z., Pan T. C., Stokes D., Conway D., Kuo H. J., Glanville R., Mayer U., Mann K., Deutzmann R. (1990) EMBO J. 9, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitta B., Stokes D. G., Vissing H., Timpl R., Chu M. L. (1990) J. Biol. Chem. 265, 6473–6480 [PubMed] [Google Scholar]
- 14.Fitzgerald J., Rich C., Zhou F. H., Hansen U. (2008) J. Biol. Chem. 283, 20170–20180 [DOI] [PubMed] [Google Scholar]
- 15.Gara S. K., Grumati P., Urciuolo A., Bonaldo P., Kobbe B., Koch M., Paulsson M., Wagener R. (2008) J. Biol. Chem. 283, 10658–10670 [DOI] [PubMed] [Google Scholar]
- 16.Giusti B., Lucarini L., Pietroni V., Lucioli S., Bandinelli B., Sabatelli P., Squarzoni S., Petrini S., Gartioux C., Talim B., Roelens F., Merlini L., Topaloglu H., Bertini E., Guicheney P., Pepe G. (2005) Ann. Neurol. 58, 400–410 [DOI] [PubMed] [Google Scholar]
- 17.Zhang R. Z., Sabatelli P., Pan T. C., Squarzoni S., Mattioli E., Bertini E., Pepe G., Chu M. L. (2002) J. Biol. Chem. 277, 43557–43564 [DOI] [PubMed] [Google Scholar]
- 18.Petrini S., Tessa A., Stallcup W. B., Sabatelli P., Pescatori M., Giusti B., Carrozzo R., Verardo M., Bergamin N., Columbaro M., Bernardini C., Merlini L., Pepe G., Bonaldo P., Bertini E. (2005) Mol. Cell Neurosci. 30, 408–417 [DOI] [PubMed] [Google Scholar]
- 19.Tillet E., Wiedemann H., Golbik R., Pan T. C., Zhang R. Z., Mann K., Chu M. L., Timpl R. (1994) Eur. J. Biochem. 221, 177–185 [DOI] [PubMed] [Google Scholar]
- 20.Lamandé S. R., Sigalas E., Pan T. C., Chu M. L., Dziadek M., Timpl R., Bateman J. F. (1998) J. Biol. Chem. 273, 7423–7430 [DOI] [PubMed] [Google Scholar]
- 21.Specks U., Mayer U., Nischt R., Spissinger T., Mann K., Timpl R., Engel J., Chu M. L. (1992) EMBO J. 11, 4281–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vorup-Jensen T., Ostermeier C., Shimaoka M., Hommel U., Springer T. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1873–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warwicker J., Watson H. C. (1982) J. Mol. Biol. 157, 671–679 [DOI] [PubMed] [Google Scholar]
- 24.Lampe A. K., Dunn D. M., von Niederhausern A. C., Hamil C., Aoyagi A., Laval S. H., Marie S. K., Chu M. L., Swoboda K., Muntoni F., Bonnemann C. G., Flanigan K. M., Bushby K. M., Weiss R. B. (2005) J. Med. Genet. 42, 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittaker C. A., Hynes R. O. (2002) Mol. Biol. Cell 13, 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu A., Leahy D. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10277–10281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J. O., Rieu P., Arnaout M. A., Liddington R. (1995) Cell 80, 631–638 [DOI] [PubMed] [Google Scholar]
- 28.Emsley J., Cruz M., Handin R., Liddington R. (1998) J. Biol. Chem. 273, 10396–10401 [DOI] [PubMed] [Google Scholar]
- 29.Huizinga E. G., Martijn van der Plas R., Kroon J., Sixma J. J., Gros P. (1997) Structure 5, 1147–1156 [DOI] [PubMed] [Google Scholar]
- 30.Merlini L., Martoni E., Grumati P., Sabatelli P., Squarzoni S., Urciuolo A., Ferlini A., Gualandi F., Bonaldo P. (2008) Neurology 71, 1245–1253 [DOI] [PubMed] [Google Scholar]
- 31.Scacheri P. C., Gillanders E. M., Subramony S. H., Vedanarayanan V., Crowe C. A., Thakore N., Bingler M., Hoffman E. P. (2002) Neurology 58, 593–602 [DOI] [PubMed] [Google Scholar]
- 32.Pace R. A., Peat R. A., Baker N. L., Zamurs L., Mörgelin M., Irving M., Adams N. E., Bateman J. F., Mowat D., Smith N. J., Lamont P. J., Moore S. A., Mathews K. D., North K. N., Lamandé S. R. (2008) Ann. Neurol. 64, 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker N. L., Mörgelin M., Pace R. A., Peat R. A., Adams N. E., Gardner R. J., Rowland L. P., Miller G., De Jonghe P., Ceulemans B., Hannibal M. C., Edwards M., Thompson E. M., Jacobson R., Quinlivan R. C., Aftimos S., Kornberg A. J., North K. N., Bateman J. F., Lamandé S. R. (2007) Ann. Neurol. 62, 390–405 [DOI] [PubMed] [Google Scholar]
- 34.Pan T. C., Zhang R. Z., Pericak-Vance M. A., Tandan R., Fries T., Stajich J. M., Viles K., Vance J. M., Chu M. L., Speer M. C. (1998) Hum. Mol. Genet. 7, 807–812 [DOI] [PubMed] [Google Scholar]
- 35.Sasaki T., Hohenester E., Zhang R. Z., Gotta S., Speer M. C., Tandan R., Timpl R., Chu M. L. (2000) FASEB J. 14, 761–768 [DOI] [PubMed] [Google Scholar]
- 36.Ball S., Bella J., Kielty C., Shuttleworth A. (2003) J. Biol. Chem. 278, 15326–15332 [DOI] [PubMed] [Google Scholar]
- 37.Jenkins P. V., Pasi K. J., Perkins S. J. (1998) Blood 91, 2032–2044 [PubMed] [Google Scholar]
- 38.Takagi J., Springer T. A. (2002) Immunol. Rev. 186, 141–163 [DOI] [PubMed] [Google Scholar]
- 39.González-Manchón C., Butta N., Larrucea S., Arias-Salgado E. G., Alonso S., López A., Parrilla R. (2004) Thromb. Haemost. 92, 1377–1386 [DOI] [PubMed] [Google Scholar]
- 40.Chen J., Yang W., Kim M., Carman C. V., Springer T. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.