Abstract
BRCA1 C-terminal domain (BRCT)-containing proteins are found widely throughout the animal and bacteria kingdoms where they are exclusively involved in cell cycle regulation and DNA metabolism. Whereas most BRCT domains are involved in protein-protein interactions, a small subset has bona fide DNA binding activity. Here, we present the solution structure of the BRCT region of the large subunit of replication factor C bound to DNA and a model of the structure-specific complex with 5′-phosphorylated double-stranded DNA. The replication factor C BRCT domain possesses a large basic patch on one face, which includes residues that are structurally conserved and ligate the phosphate in phosphopeptide binding BRCT domains. An extra α-helix at the N terminus, which is required for DNA binding, inserts into the major groove and makes extensive contacts to the DNA backbone. The model of the protein-DNA complex suggests 5′-phosphate recognition by the BRCT domains of bacterial NAD+-dependent ligases and a nonclamp loading role for the replication factor C complex in DNA transactions.
Keywords: DNA/Protein Interaction, DNA/Repair, DNA/Replication, DNA/Structure, Methods/NMR, Protein/Domains, Protein/Structure
Introduction
Replication factor C (RFC)3 is a five-subunit complex that loads the sliding clamp, PCNA, onto primer-template DNA during synthesis of the daughter strand in DNA replication (1). Human RFC consists of four subunits of 35–40 kDa and a fifth large subunit (p140) of 140 kDa. The C terminus of p140 shares homology with the four small subunits, whereas the unique N-terminal sequence contains a single BRCT domain that is dispensable for its function in PCNA loading (2). The crystal structure of yeast RFC carrying a BRCT-truncated p140 (trRFC) indicated that the five subunits form a spiral complex that precisely matches that of B form DNA (3). Although not required for DNA replication, the BRCT region (residues 375–480) was shown to specifically bind 5′-phosphorylated dsDNA (4, 5). There is currently no structural information available regarding this type of specific DNA recognition.
BRCT domains are small, consisting of roughly 90 amino acids, and are found in more than 900 proteins from all biological kingdoms (6). BRCT domains contain no intrinsic enzymatic activity, rather they appear to play a scaffolding role by mediating primarily protein-protein interactions. Interestingly, all proteins identified so far as containing BRCT domains are strictly involved either directly in DNA transactions or in regulation of the timing of such activities. These proteins, which may contain more than a single copy of the BRCT domain, exhibit functional activities ranging from DNA replication to DNA repair and cell cycle checkpoint regulation (7, 8). The structural information that is available for BRCT domains present in a variety of proteins suggests that the family may be divided into members whose function is contained within a single domain and those that form an obligate tandem repeat. So far, the tandem repeat BRCT domains appear to be specific for binding to phosphopeptide sequences (9, 10) and are exemplified by BRCA1 (11–14) and MDC1 (15). On the other hand, isolated BRCT domains display greater variation in the types of binding in which they participate. For instance, XRCC1 contains two separated copies of the BRCT domain, of which the C-terminal one forms a heterodimer with the BRCT domain of DNA ligase III through residues conserved between the two domains (16). In contrast, the BRCT domain of 53BP1 mediates binding to p53, which does not contain a BRCT domain (17). Finally, there is the distinct class of BRCT domain exemplified by RFC, poly(ADP-ribose) polymerase, and the bacterial NAD+-dependent DNA ligases, some of which mediate DNA binding (18–20). It is clear that despite conservation of the three-dimensional fold of each domain, the mechanism by which BRCT domains execute their function differs significantly within the BRCT superfamily.
Although a limited number of BRCT-DNA interactions are known or have been implied from biochemical data, there is at present no structure of a BRCT-DNA complex. Deletion and mutagenesis data suggest that the region spanning residues 375–480 in RFC p140 (hereafter called p140-(375–480)) is important for DNA binding (5, 21). This portion of RFC p140, which we refer to as the BRCT region, contains the variant BRCT domain and N-terminal sequences, both of which are required for DNA binding. To investigate the molecular basis of this recognition, we employed NMR methods to determine the solution structure of p140-(375–480) bound to dsDNA. Although the data obtained were not sufficient to determine the solution structure of the DNA portion of the complex, the structure of the complexed protein was determined from experimentally derived restraints. The resulting structure of p140-(375–480) consists of a consensus BRCT fold preceded by an α-helix connected to the core domain by a long loop. Here, we present a model of the protein-DNA complex that was generated using HADDOCK (22), an algorithm that docks two molecules using ambiguous interaction restraints based on a variety of experimental data, including mutagenesis, ambiguously assigned intermolecular NOEs, and amino acid conservation. The combination of our p140-(375–480)-dsDNA model and the existing trRFC-PCNA crystal structure reveals a potential function of 5′-phosphate end binding by the p140-(375–480) during Okazaki fragment maturation.
MATERIALS AND METHODS
Sample Preparation
The expression and purification of RFC p140-(375–480) were performed according to published methods (5). The oligonucleotide used in these studies (pCTCGAGGTCGTCATCGACCTCGAGATCA) was produced by standard solid state synthesis and further purified by anion exchange chromatography. For NMR studies, the buffer was exchanged to 25 mm Tris-HCl, pH 7.5, 50 mm NaCl using a PD10 desalting column (Amersham Biosciences). The purity of the DNA was analyzed by mass spectrometry. To form the complex, both the protein and the DNA were diluted to 10 μm in 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 1 mm dithiothreitol to prevent aggregation, mixed in the molar ratio of 1 to 1.2, and concentrated to ∼0.5 mm by vacuum dialysis (Spectrum Labs) using a 10-kDa cutoff membrane. Subsequently, the buffer was exchanged to 25 mm d11-Tris-HCl, pH 7.5, 5 mm NaCl in 95:5 H2O/D2O.
NMR Spectroscopy and Resonance Assignment
The sequential assignment of p140-(375–480) has been described previously (23). An additional three-dimensional [15N,1H] NOESY-HSQC was recorded at 310 K for structure calculation. Spectral data were processed using NMRPipe (24). The assignment and the integration of NOE peaks were performed using the computer program CARA (25). The chemical shift assignments of the protein bound to DNA have been reported (23) and deposited (BMRB accession number 6353). The following half- and double-filtered experiments were acquired: a two-dimensional NOESY (τm = 150 ms) recorded at 900 MHz with heteronuclear multiple quantum correlation purge set to reject 13C- and 15N-coupled protons during t1 and to accept 13C- and 15N-coupled protons during t2, and a two-dimensional NOESY (τm = 150 ms) run at 900 MHz with HMQC purge set to reject 13C- and 15N-coupled protons during both t1 and t2 (26).
Structure Calculations
Distance restraints were derived from the automated NOE cross-peak assignment of the three-dimensional 15N,1H NOESY-HSQC (recorded at 310 K) and the 13C,1H NOESY-HSQC (recorded at 298 K) using the protocol CANDID implemented in the computer program CYANA 2.0 (27). The chemical shift tolerances used in the automated assignment were 0.02 ppm for protons and 0.1 ppm for heavy atoms. The structures were calculated using the NOE-derived distance restraints and the dihedral angle restraints calculated from the chemical shift values of Cα and Cβ by TALOS (28). One hundred structures were calculated starting from conformers with random dihedral angles using simulated annealing and torsion angle dynamics as implemented in CYANA 2.0. The 24 lowest energy structures with no distance violations greater than 0.3 Å and no angle violations greater than 5° were subjected to water refinement following a previously described scheme (29). The 24 structures with the lowest backbone conformation Z scores (WHATCHECK) (30) were accepted as the final structures representing the solution conformation and deposited in the Protein Data Bank (PDB code 2k6g).
Docking Protocol
The program HADDOCK (31) was used to dock p140-(375–480) to the dsDNA using solvent-accessible residue defined by NACCESS (32) and ambiguous intermolecular restraints defined according to established criteria. Eight amino acids of p140-(375–480) (Tyr-382, Tyr-385, Arg-388, Thr-415, Gly-415, Arg-423, Lys-458, and Lys-461) with >50% solvent accessibility were defined as “active” residues. Ambiguous interaction restraints were then generated between the active residues of p140-(375–480) and the passive residues of dsDNA (Table 3). A more specific ambiguous intermolecular restraint was introduced between Hγ of THR-415 and the 5′-phosphate group of dsDNA (CYT-19). Side chain and backbone flexibility that allow local rearrangement during the semi-flexible simulated annealing step of HADDOCK is confined to the segments around the active and passive residues (377–392 and 414–462 in p140-(375–480) and the entire DNA molecule) (Table 3).
TABLE 3.
Active and passive residues used in the definition of the ambiguous interaction restraints for HADDOCK
| Active residues of p140-(375–480) | Passive residues of dsDNA | Method determined |
|---|---|---|
| Tyr-382, Arg-388, Lys-458, and Lys-461a | Any DNA nucleotide (5′pCTCGAGGTCG3′/ 5′CGACCTCGAGATCA3′) | Mutagenesis data (5) |
| Thr-415 Hγa | O1P, O2P, and O3P of 5′pC19 | Conservation/mutagenesis |
| Tyr-385 Hδ | Any H2, H6, H8 | Intermolecular NOEs (see Table 2) |
| Gly-416 HN | Any H2, H6, H8 | |
| Arg-423 Hϵ | Any H4′, H5′, H5″ | |
| Gly-455 or Gln-456 HN | Any H4′, H5′, H5″ |
a A specific restraint is shown (see text for details).
The starting structures for docking were the 24 NMR structures of p140-(375–480) and 3 models of dsDNA. Because no structure of the DNA portion of the complex was available, a model structure of 5′-phosphorylated dsDNA with a 3′ single-stranded overhang in the standard B-form DNA with three conformations was generated, using the oligonucleotide sequence identical to that used for the NMR studies except that the hairpin was removed (5′pCTCGAGGTCG3′/5′CGACCTCGAGATCA3′). Docking of the p140-(375–480)-dsDNA complex was performed following the protocol of HADDOCK2.0 (22). Inter- and intramolecular energies were evaluated using full electrostatic and van der Waal's energy terms with a distance cutoff using optimized potentials for liquid simulations nonbonded parameters as defined in the default protocol. During rigid body energy minimization, 2400 docking structures were generated (four cycles of orientational optimization for each combination of starting structures were repeated 10 times). The best 200 structures in terms of intermolecular energies were then used for the semi-flexible simulated annealing, followed by explicit water refinement. Finally, the structures were clustered using a 5-Å r.m.s.d. as a cutoff based on the pairwise backbone r.m.s.d. The lowest energy cluster of four structures was chosen as the model of the protein-DNA complex (PDB code 2k7f).
Preparation of the Protein-DNA Complex
The 5′-phosphorylated hairpin oligonucleotide used to form the protein-DNA complex was described previously and shown to bind RFC p140-(375–480) with KD ∼10 nm (5). The protein-DNA complex was formed as described with a starting protein/DNA ratio of 1:1.2 to ensure the formation of a full complex with a 1:1 stoichiometry. Excess DNA eluted through the dialysis membrane. No signals from either unbound protein or DNA could be detected in the NMR spectra. The approach to and extent of the sequential assignment has been described previously (23).
RESULTS AND DISCUSSION
NMR Data Support Binding of p140-(375–480) to 5′-Phosphorylated dsDNA
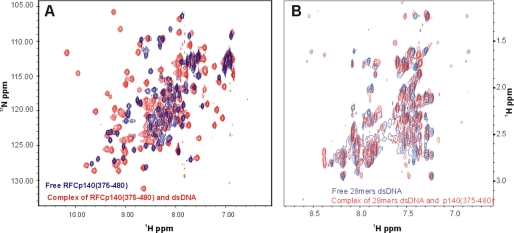
The 15N,1H HSQC spectrum of the free p140-(403–480) was poorly dispersed and exhibited heterogeneous line width and intensity, whereas the spectrum of the DNA-bound protein was clearly better, containing 105 of the 106 expected amide correlations and exhibiting good dispersion with more homogeneous line widths (Fig. 1A) (5). The extensive differences between the spectra of p140-(403–480) in the presence and absence of DNA are strong evidence for tight and intimate binding. Only 60% of backbone resonances of the free protein could be sequentially assigned,4 whereas 99% of the backbone 1H and 15N and 95% of the side chain 1H chemical shifts of the DNA bound p140-(375–480) were assigned (23). The observation of two distinct sets of resonances for the bound and the free protein is characteristic of slow dissociation of the protein-DNA complex on the NMR time scale and is consistent with the previously determined KD of ∼10 nm (5). As a consequence, it was not possible to deduce the DNA-binding site on the protein by chemical shift perturbation analysis.
FIGURE 1.
Chemical shift perturbation analysis of RFC p140-(375–80) binding to dsDNA. A, overlay of [15N,1H] HSQC spectra of p140-(375–480) in the absence (blue trace) and in the presence of a 1.2-fold molar excess of a 5′-phosphorylated hairpin dsDNA (red trace). B, overlay of cross-peaks in a region of the two-dimensional [1H,1H] NOESY spectrum of 5′-phosphorylated hairpin dsDNA in the absence (blue trace) and that of two-dimensional F1, F2 [15N/13C] double-filtered NOESY spectrum in the presence of RFC p140-(375–480) (red trace).
The NMR spectra of the 5′-phosphorylated hairpin 28-mer DNA in the presence and absence of p140-(375–480) were also investigated. Two-dimensional NOESY spectra (Fig. 1B) of free DNA were indicative of dsDNA but were not well resolved. Standard isotope-filtered NMR experiments did not yield high quality spectra of the DNA in the presence of p140-(375–480), likely due to dynamic behavior. We therefore tried an alternative approach based on purge pulses (26), which proved to be moderately successful. Two NOESY spectra were obtained by simultaneous suppression of 13C/15N-attached protons in both F1 and F2 or only in F1 (data not shown). The resulting F1, F2 double-filtered spectrum, which contains exclusively resonances from the unlabeled DNA, was substantially different from that of the free oligonucleotide. The differences further support formation of a complex between p140-(375–480) and the dsDNA. Unfortunately, due to poor dispersion of the resonances of the DNA, it was not possible to sequentially assign the majority of resonances. The lack of sequential assignment precludes experimental structure determination of the DNA moiety of the complex. However, comparison of the NOESY spectra listed in Table 2 allowed us to ambiguously assign a few peaks arising from intermolecular magnetization transfer from DNA to protein. Due to the lack of sequence-specific resonance assignments for the DNA, however, the identity of the source proton could not be ascertained.
TABLE 2.
Intermolecular NOEs observed in the p140-(375–480)-DNA complex
| RFCp140-(375–480) | Ambiguously assigned to DNA | Unassigned intermolecular NOE | Experimentsa |
|---|---|---|---|
| ppm | |||
| Tyr-385 QD | CYT H5 or THY H1′ | 5.51 | A and B |
| CYT H6 or THY H6 | 7.72 | ||
| Asn-440 HD21 | CYT H5 or THY H1′ | 5.37 | A, B, and C |
| HB3 | TCH3 | 1.5 | A and B |
| Gly-416 HN | CYT H6, THY H6 | 7.7 | C |
| ADE H2, H8 or | |||
| NH2 of ADE, CYT and GUA | |||
| Arg-423 HE | Ribose H2″ or H2′ | 2.19 | B and C |
| Ribose H4′, H5″ or H5′ | 3.88 | ||
| Gly-455/Gln-456 HNa | Ribose H4′, H5″ or H5′ | 3.94 | A and B |
a A indicates two-dimensional F1-filtered [13C/15N] NOESY. B indicates two-dimensional NOESY. C indicates [15N,1H] NOESY-HSQC.
b Ambiguity in the amide proton resonance was due to overlap in amide proton frequencies between Gly-455 and Gln-456.
Description of p140-(375–480) Bound to dsDNA
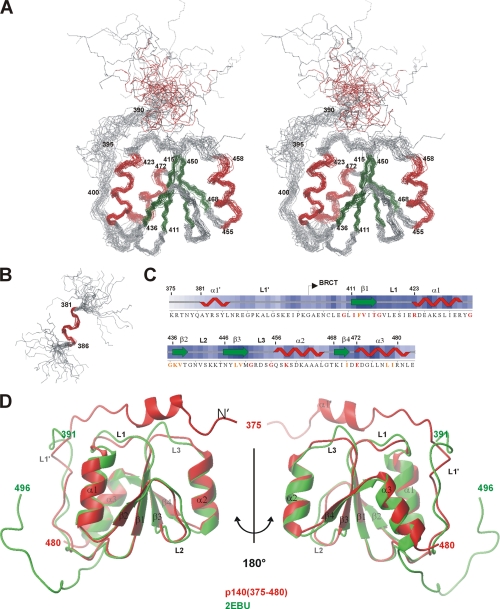
The structure of the protein moiety of the DNA-protein complex was determined primarily from distance restraints derived from NOEs in the three-dimensional [15N,1H] NOESY-HSQC and the three-dimensional [13C,1H] NOESY-HSQC spectra (Table 1). The best fit superposition of the 24 conformers with the lowest backbone Z-scores is depicted in Fig. 2A, left panel, and the quality statistics of the structures are summarized in Table 1. The secondary structure within p140-(403–480) is well defined, with an average r.m.s.d. of 0.98 Å for backbone atoms and 1.66 Å for all heavy atoms (Table 1). The least defined regions are located at the N-terminal helix and the loops that connect the secondary structures and reflect the low number of long range distance restraints. Analysis of the Ramachandran plot for all residues using the program PROCHECK (33) showed that 84% of ϕ and ψ angles lie within the most favored and 12.9% lie in the additionally allowed regions, whereas only 3% are in the generously allowed or disallowed regions (Table 1). The residues that fall into the latter regions are found in the loops.
TABLE 1.
Structural statistics of the best 24 RFCp140-(375–480) conformers
| Restraints used in the calculation | |
| Total no. of NOE upper distance limits | 1812 |
| Intra-residue (|i − j| = 0) | 576 |
| Sequential (|i − j| = 1) | 455 |
| Medium range (1 〈|i − j| < 5) | 295 |
| Long range (|i − j| ≥ 5) | 486 |
| Dihedral angle restraints (TALOS predicted) | 122 |
| CNS 1.0/water refinement | |
| Average no. of distance restraint violations (>0.3 Å) | 0 |
| Average no. of dihedral angle constraint violations (>5°) | 2 |
| WHATCHECK Average backbone conformation Z-score | −3.60 ± 0.4 |
| Average pairwise r.m.s.d. (Å)a | |
| Backbone atoms | 0.98 ± 0.19 |
| Heavy atoms | 1.66 ± 0.24 |
| PROCHECK Ramachandran plot analysis | |
| Most favored regions | 84.1% |
| Additionally allowed regions | 12.9% |
| Generously allowed regions | 1.2% |
| Disallowed regions | 1.8% |
a Residues 403–480 were used for the calculation.
FIGURE 2.
Structure of RFC p140-(375–480) when bound to dsDNA. A, stereoview of a superposition of the backbone (N, Cα, and C′) atoms of residues 403–480 for the 24 lowest energy structures of RFC p140-(375–480). The α-helices and β-strands are colored red and green, respectively. B, superposition of the backbone of residues 381–386 demonstrating the locally well defined helix α1′ near the N terminus. C, consensus secondary structure found in the ensemble of 24 structures is shown above the amino acid sequence of RFC p140-(375–480). Sequence alignment of RFC p140 from 27 eukaryotic species identified a number of residues with identity or similarity above 90%, which are colored in red and orange, respectively. D, structural superposition of the lowest energy structure of RFC p140-(375–480) and PDB code 2EBU. The average r.m.s.d. of the two structures was 1.3 Å based on superposition of the Cα atoms of residues 400–480. This and other structure figures were generated using MOLMOL (44).
Residues 403–480 of RFC p140-(375–480), which contain weak sequence homology to the BRCT domain family (6), fold into a compact unit consisting of four parallel β-strands surrounded by helices α1 and α3 on one side and by helix α2 on the other (Fig. 2A), thereby forming a canonical BRCT domain. Residues 375–390 form an α-helix (α1′) and a loop (L1′), which separate the helix from the core of the protein. Helix α1′ (residues 381–386) appears consistently in all 24 structures (Fig. 2B, the consensus secondary structure is shown in Fig. 2C); however, it is poorly defined with respect to the rest of the protein. This lack of definition certainly reflects the absence of observable long range NOEs between the helix α1′ and the BRCT domain. Loop L1′ is anchored to helices α1 and α2 through burial of the side chains of residues Leu-399, Pro-400, and Leu-407 between the two helices and through potential salt bridging between the side chains of Lys-397 (L1′) and Glu-472 or Asp-473 (α3) and of Lys-392 (L1′) and Glu-419 (L1).
The BRCT domain of p140-(375–480) belongs to a distinct subclass of the BRCT superfamily (6). One unusual difference from the rest of the superfamily is the presence of a Gly in position 474 in helix α3, where the consensus for the BRCT superfamily is a Trp. The substitution of this Trp by a Leu causes destabilization of the structure of the XRCC1 BRCT domain and may be a possible explanation for the apparent “floppiness” of the present unliganded protein (16). Gly-434 and Gly-435, two of the most conserved residues in the BRCT superfamily (6), form a tight turn between α1 and β2. Substitution of either of these glycines by a larger residue could potentially destabilize the three-dimensional structure. In our own experience, the G435R mutation resulted in a protein prone to precipitation and with reduced DNA binding activity (data not shown), both characteristics suggestive of a decrease in ΔGfold. In the case of BRCA1, the analogous G1788V mutation renders the tandem BRCT repeat more sensitive to proteolytic digestion (34), whereas the G617I mutation in the BRCT domain of the bacterial NAD+-dependent DNA ligase reduces both DNA binding and nick-adenylation activity (20). Interestingly, the G193R mutation in the BRCT domain of Rev1 has been shown to interfere with the in vivo trans-lesion synthesis activity of Rev1 in Saccharomyces cerevisiae (35), but one should perhaps be careful about interpreting such a mutation that leads to a general destabilization of the BRCT domain.
In this structure, the L3 loop displays a high degree of disorder (Fig. 2) because of the limited number of distance restraints found within this region. It is not yet clear whether the disorder reflects actual dynamic motions within the L3 loop or simply a paucity of structural restraints. It is interesting to note that the preceding helix, α2, and loop L3 are the most variable in size and sequence in the BRCT family. Loops L1 and L2 also display some conformational variation in the ensemble, although to a lesser extent than L3 (Fig. 2). In most BRCT domains, loop L1 is more or less flexible as reflected by the high B-factors in x-ray crystal structures and poor definition in NMR structures (12, 36–38). In relation to these other structures, the L1 loop of p140-(375–480) is better defined and buried under loop L1′.
Recently, the NMR structure of RFC p140-(392–496), which lacks the N-terminal amino acids essential for DNA binding, has been reported (PDB code 2EBU). The backbone r.m.s.d. of the conserved BRCT domain in the free and DNA bound state is 1.3 Å, indicating that the core BRCT domain does not undergo major structural changes upon DNA binding (Fig. 2D). The largest deviations in the two structures are seen in the loops.
Comparison with the Phosphopeptide Binding BRCT Domains Reveals Potential 5′-Phosphate DNA Interaction Site on p140-(375–480)
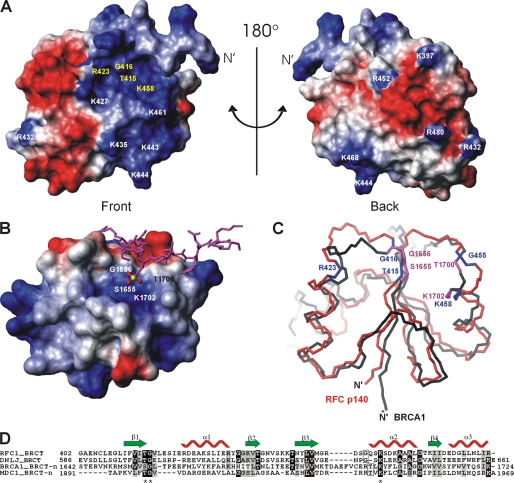
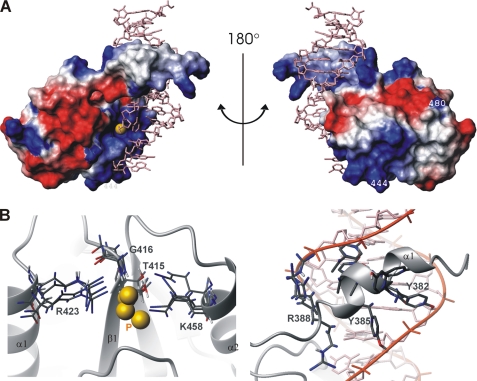
A surface representation of p140-(375–480), colored according to electrostatic potential, is presented in Fig. 3A. Note that the location of helix α1′ relative to the core of the protein in Fig. 3A is arbitrary. The conserved residues (Fig. 3A, yellow) that were identified by sequence alignment of the RFC BRCT domains (Fig. 2C) are distributed mainly within the basic patch of p140-(375–480) (Fig. 3A) and are strictly found within the BRCT domain rather than within the loop L1′ or helix α1′. Because mutation of these conserved residues reduced or abrogated DNA binding activity (5), this basic patch is a likely binding site of either the negatively charged phosphate backbone or the 5′-phosphate of DNA. Negatively charged surfaces, on the other hand, extend from the front to the back of the protein (Fig. 3A).
FIGURE 3.
Potential DNA-binding site in p140-(375–480) revealed by amino acid conservation and structure comparison. A, electrostatic potential of the accessible surface of 140(375–480) is shown. Negative potential is colored in red and positive in blue. The absolutely conserved amino acids in the eukaryotic RFC family are in yellow. The orientation of the protein in A is equivalent to Fig. 2D. B, electrostatic surface presentation of the N-terminal BRCT domain (BRCT-n) of BRCA1 (PDB code 1T29) in complex with a phosphoserine peptide (in magenta) colored as in A. The C-terminal BRCT domain is not directly involved in phosphate binding and therefore has been omitted from this figure for clarity. The amino acid residues forming the pocket to accommodate the phosphate moiety (P in yellow and O in red) of phosphoserine are indicated on the surface. C, superposition of p140-(375–480) (red) and the BRCT-n from BRCA1 (black). The orientation of the BRCT-n is identical to that of B. The conserved residues of p140-(375–480) are shown in blue and the phosphate-moiety recognition residues of BRCA1 BRCT-n bound to the phosphoserine peptide are shown in magenta. D, sequence alignment of BRCT domains that bind DNA or phosphopeptides as generated by ClustalW. The sequence alignment was adjusted based on three-dimensional structure alignment using DALILITE (45). The secondary structure of the BRCT domain of p140-(375–480) is depicted. Residues that are >70% identical are shaded black, whereas residues that are >50% similar are shaded gray. The asterisks indicate the three amino acids that bind the phosphate moiety of the bound phosphoserine in the BRCA1 BRCT-n and the MDC1 BRCT-n structures (see the text for the references).
The crystal structure of the complex of the BRCA1 tandem BRCT repeat with a phosphoserine peptide shows that the phosphate moiety of the bound peptide is hydrogen-bonded to the three residues of the N-terminal BRCT domain (BRCT-n) (11–13) (residues indicated in white, Fig. 3B). Our structure-based superposition of p140-(375–480) with BRCA1 BRCT-n revealed a striking similarity between the binding site for the phosphate moiety of the phosphoserine on BRCA1 and the conserved basic patch on RFC p140-(375–480) (Fig. 3C), a relationship that had been anticipated (5). This similarity is further underlined by the crystal structure of the analogous complex between the tandem BRCTs of MDC1 and a phosphoserine peptide (39). Whereas in the case of BRCA1, the phosphate moiety of the bound peptide is hydrogen-bonded to the trio of Ser, Gly, Lys (Fig. 3B), the analogous residues in MDC1 are Thr, Gly, and Lys. Despite the overall low level of conservation between the N-terminal BRCTs of BRCA1 and MDC1 on the one hand and p140-(375–480) on the other, both the chemical nature and the three-dimensional structure of the phosphate-binding triad is exactly maintained, corresponding to Thr-415, Gly-416, and Lys 458 in p140 (Fig. 3D). This analysis suggests that the positive patch present on p140 is important for interaction with the 5′-phosphate of dsDNA.
The BRCT domain of RFC p140 belongs to a distinct subgroup of the BRCT superfamily (6). Within the distinct subgroup, there is increasing evidence to suggest that the BRCT domain from the bacterial NAD+-dependent ligase binds to DNA (18–20). The BRCT domain is located at the C terminus of the multidomain enzyme and is responsible for stable association of protein and DNA (18). Amino acid sequence analysis of the distinct subgroup of BRCT domains indicates that the potential DNA-binding residues, including Thr-415, Gly-416, Arg-423, Gly-455, and Lys-458, are absolutely conserved between the NAD+-dependent DNA ligases and RFC p140 (Fig. 3D). As mutations in these residues severely affect the DNA binding as well as the 5′-phosphate adenylate moiety transfer activities of this class of ligases (20), it may be inferred that the 5′-phosphate could also be the specific target for DNA binding by the BRCT domain of the bacterial DNA ligases.
Experimentally Based Protein-DNA Docking by HADDOCK
Because the sequential assignment of the DNA was not available, it was not possible to calculate the structure of the protein-DNA complex based upon the usual restraints such as NOEs. To generate a model of the p140-(375–480)-DNA complex, the data-driven docking program HADDOCK (22) was employed. HADDOCK can make use of a broader array of restraints, including those derived from biochemical and biophysical data. The mutagenesis (5), intermolecular NOEs (Table 2), and structural conservation (Fig. 3) clearly indicate at least some of the residues that interact with the dsDNA. In the docking procedure, ambiguous distance restraints maybe introduced between residues with at least 50% solvent-accessible surface and biochemical or conservation data supporting interaction with the DNA and the 5′-PO4 or any specific nucleotides of the dsDNA (Table 3). In addition to the ambiguous restraints, a specific restraint was generated between the hydroxyl of Thr-415 and the 5′-phosphate of the DNA on the basis of the following three observations. 1) The resonance of the γ-1H of Thr-415 has been tentatively assigned on the basis of NOEs (at 9.22 ppm) indicating that this 1H is in slow exchange with the solvent. Both the reduced exchange rate and the large downfield shift are indications of the involvement of Thr-415 γ-1H in a strong hydrogen bond, whereas inspection of the protein structure indicates that there are no neighboring residues within sufficiently close distance to form such a hydrogen bond. 2) Residue Thr-415 is structurally equivalent to Ser-1655 of BRCA1 and Thr-1898 of MDC1, which form hydrogen bonds to the phosphate moiety of phosphoserine (Fig. 3), and mutation of this residue resulted in reduced DNA binding (5). 3) The specificity of binding to 5′-phosphorylated dsDNA is conserved across the BRCT region of RFC from different species (4), but the absolutely conserved amino acids can only be found in the BRCT domain itself and not in N-terminal α1′-helix or in the L1′ loop.
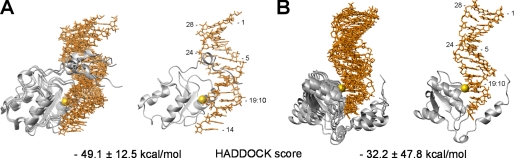
As no structure of the DNA portion of the complex was available, a model structure of 5′-phosphorylated dsDNA with a 3′ single-stranded overhang in the standard B-form conformation was generated using the sequence of the oligonucleotide used in the NMR studies. The model DNA structure, the experimentally determined protein structure, and the intermolecular restraints described in Table 3 were used as input to HADDOCK. To optimize interaction at the protein-DNA interface, the N-terminal residues 377–392 of p140-(375–480) were allowed to move freely during the docking procedure. As a result, the docking calculations generated 200 solutions that were sorted into clusters using a pairwise backbone r.m.s.d. of 5 Å as a cutoff criterion. This procedure resulted in 10 clusters, which were then ranked according to their HADDOCK scores calculated on the basis of the intermolecular energy. The top two clusters, 1 and 4, had HADDOCK scores of −49 ± 12 and −32 ± 48, respectively, whereas the next best cluster scored −13 ± 32. The ensembles of the four best structures of the top two clusters are depicted in Fig. 4. The definition of both clusters is moderate, with a pairwise r.m.s.d. of 2.4 and 2.9 Å over all the backbone atoms of the complex for clusters 1 and 4, respectively (Fig. 4, A and B).
FIGURE 4.
Models of the p140-(375–480)-dsDNA complex generated by HADDOCK. The ensemble of four structures from the two clusters, cluster 1 (A) and cluster 4 (B), which resulted in the lowest HADDOCK scores (indicated at the bottom), are displayed. The structures of p140-(375–480) are in gray, and the DNA is in orange. The 5′-phosphate is indicated by the yellow sphere. The structure with the lowest HADDOCK score in the ensemble is shown to the right of each bundle. The nucleotide positions in the dsDNA are indicated.
The four best structures from cluster 1, which had the lowest HADDOCK score of any cluster, were accepted as the representative model of the complex over the structures from cluster 4 based on a number of observations. The most critical problem with cluster 4 is that helix α1′ binds to the 3′ ssDNA overhang. We previously demonstrated that the 3′ ssDNA is not critical for binding, although helix α1′ is (5). Furthermore, the protein in cluster 4 only interacts with the first 3 bp of DNA, although it was shown that 9 bp are required for high affinity binding. Finally, Lys-444 is close enough to the DNA to interact, although our mutagenesis data suggested it did not. In contrast, the structures of cluster 1 are consistent with these and other observations (see below).
Model of p140-(375–480)-dsDNA Complex
The DNA-binding surface of p140-(375–480) is composed of residues in the α1′-helix and in the BRCT domain; the former is inserted into the major groove making extensive contacts with bases and phosphate backbone of the DNA, whereas the latter accommodates the 5′-phosphate (CYT-19, Fig. 5A) against the positively charged surface. The model of the complex is also consistent with previous mutagenesis data (5) of R480A and K444A, which had suggested that those residues do not participate in DNA contacts (Fig. 5A). A number of interactions with the 5′-phosphate are observed. In addition to Thr-415, the 5′-phosphate is primarily ligated by the conserved residues, Arg-423 and Lys-458, whose side chains, along with the backbone amide of Gly-416, are all within hydrogen bonding or salt-bridging distance to the oxygen atoms of the phosphate (Fig. 5B, left). The constraints introduced for these residues were to the bases of the DNA; thus, the interaction with the 5′-phosphate is not a simple result of the input data.
FIGURE 5.
Model of the RFC p140-(375–480)-dsDNA complex generated by HADDOCK. A, electrostatic surface presentation. The positive and negative charged surfaces are colored in blue and red, respectively. Exposed hydrophobic residues are in white. Residues 444 and 480 had been previously shown not to interact with the dsDNA (see text). B, left, detail of residues involved in 5′-phosphate binding in the four structures of cluster 1. The 5′-phosphate is depicted as a yellow sphere. Right, detail of side chain-DNA interaction in the N-terminal α1′ helix.
A variety of additional interactions with the phosphate backbone of the DNA are also observed in the calculated model structures. For example, hydrogen bonds involving Hϵ of Arg-452 are found in all four model structures even though no constraint was introduced in the calculation. Furthermore, although no intermolecular NOE was observed between Arg-452 and the DNA, the resonance of Hϵ of Arg-452 is clearly visible at 9.3 ppm in the [1H,15N] HSQC spectrum of the p140-(375–480)-dsDNA complex. This large downfield shift (the random coil chemical shift of Hϵ is 7.75 ppm) is suggestive of hydrogen bonding (40). On the other hand, our previous mutagenesis data show a dramatic reduction of DNA binding for the K461E mutant (5), whereas in the present model of the complex, the side chain of Lys-461 is alternatively about 8 Å from the closest phosphate of the DNA backbone or the 5′-phosphate. Although neither of these distances is very close, the introduction of negative charge would still perturb the positively charged patch of Fig. 3A.
The orientation of helix α1′ relative to the BRCT domain is better defined in the complex with DNA than in the free protein and lies in the major groove of the dsDNA. In the model structures helix α1′ is clearly separated from the core of the protein, which explains the lack of long range NOEs between the helix and the core BRCT domain. There are extensive contacts between the side chains of residues in helix α1′ and the backbone of the DNA (Fig. 5B, right). Bearing in mind that p140-(375–480) binds 5′-phosphorylated dsDNA in a nonsequence-specific manner, the model may reflect that the amino acids in α1′ are capable of various interactions. The α-helix is a commonly used structural element for recognition of bases as well as backbone phosphates in sequence-specific and nonsequence-specific DNA binding. In the nonspecific complex of DNA-lac headpiece-62 (41), many of the side chains that confer direct interactions with the base pairs in the major groove of the sequence-specific complex shift and participate in hydrogen bonds and electrostatic interactions with the backbone phosphates that are similar to those observed here. In the nonspecific DNA-lac headpiece-62 complex, residues located at the protein-DNA interface were clearly shown to undergo exchange dynamics on the micro- to millisecond time scale indicating that they sample different base pair environments (41). Such dynamic behavior is also suggested to exist in the p140-(375–480)-dsDNA complex by a variety of NMR data. For example, the transverse relaxation rate of magnetization was abnormally fast for a complex of this size as evidenced by the critical need to reduce the length of the period required for filtering heteronuclear correlated 1H. An experiment based on purge pulses (26), which reduces the amount of time required to perform the magnetization filter, yielded moderate results where more traditional approaches that would normally be effective failed. This observation, in conjunction with the previously reported missing correlations in the three-dimensional [13C,1H] NOESY-HSQC spectrum (23) and the low number of intermolecular NOEs, likely reflects the nature of the complex, in which the residues making contact with DNA undergo intermediate exchange on the NMR time scale between conformations leading to loss of resonance signals due to efficient relaxation of the transverse magnetization. In addition to dynamic behavior, the nature of the nonspecific protein-DNA interactions likely provides a further explanation for the small number of inter-molecular NOEs that were observed. Because these interactions mostly involve the phosphate backbone of the DNA and may well be bridged by water molecules (42), the 1H-1H distances would be beyond the 5-Å limit detectable by NMR.
A dramatic reduction in DNA binding of p140-(375–480) was observed when the size of the DNA duplex becomes less than 7 bp long or when the +6 nucleotide position (G24:C5) from the 5′-phosphate end contains a non-Watson-Crick base pair (T24:C5) (5). The model of the complex nicely explains these observations because there are close contacts with both of the base pairs that were not introduced as constraints. Furthermore, the side chain of Ser-384 in helix α1′ is oriented toward the solvent in the model, which is consistent with the mutagenesis data that clearly showed Ser-384 was not essential for DNA binding. Finally, in the model of the protein-DNA complex, the 3′ single-stranded DNA tail (nucleotides CYT-13 and ADE-14) interacts via the bases with the side chains of Thr-438 and Asn-440 as well as the amide proton of Gly-439, although no explicit constraints were included for any of these residues. This interaction explains the earlier observation that p140-(375–480) binds a 5′-recessed dsDNA with higher affinity than blunt ended DNA (4, 5). In support of this observation, the side chain amide resonance of Asn-440 is shifted away from the random coil value suggesting involvement in some interactions.
Although constraints were used to maintain the overall structure of B-form DNA, the minor groove of the DNA in the best cluster becomes progressively compressed moving in the direction of the 5′-phosphate. At this point, it is not possible to say whether this is an artifact of the calculation or a real result of protein binding.
Potential Role of the BRCT Region of RFC p140 in DNA Replication
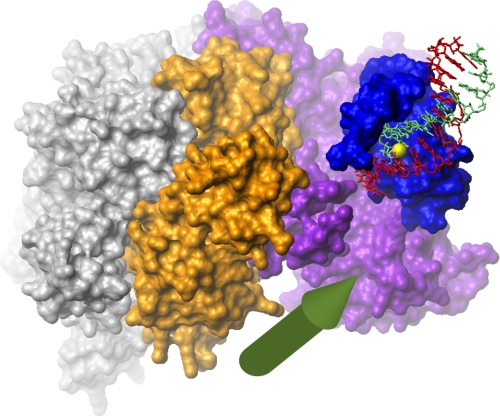
At present, a potential cellular role of 5′-phosphate DNA binding by the BRCT region of RFC remains elusive. In contrast, the cellular role of binding of the pentameric RFC complex at the 3′ end of primer-template DNA, where it directs PCNA loading and subsequent recruitment of PCNA-associated DNA-transacting enzymes, is well documented. The crystal structure of the five-subunit complex of N-terminally truncated RFC1 (p140) with RFC2-5 from yeast and PCNA (3) demonstrated that the five subunits of trRFC form a cap at the primer-template junction that defines the relative orientation of the DNA and trRFC. Our structure orients the C terminus of p140-(375–480) toward the upstream 3′ DNA terminus. By connecting the C terminus of our model to the N terminus of the crystal structure of trRFC, it is possible to ascertain an approximate relative orientation of the BRCT region to the pentameric clamp-loading complex (Fig. 6). In both yeast and humans, the connection between the two structures is about 40 amino acids long and is predicted to be flexible. By placing the C terminus of p140-(375–480) within a reasonable distance of the N terminus of the p140 subunit of trRFC (here 35 Å) and the 3′ end of the template strand as close to the predicted exit of the 5′ end of the template strand from the trRFC-PCNA complex (here 25 Å), it is possible to generate a reasonable model of the relative orientation of the two complexes. This model suggests that binding of the BRCT region to 5′-phosphorylated dsDNA terminus would orient the trRFC complex upstream toward an encroaching 3′ terminus. An important implication of this combined model is that binding by the BRCT region of a 5′ dsDNA terminus of a previously synthesized Okazaki fragment, for instance, would place the clamp loader portion of the complex in the correct position to interact with proteins at the 3′ terminus of an Okazaki fragment that is currently being synthesized or compete with them for PCNA binding. However, the structures in cluster four are inconsistent with this model. Interestingly, Levin et al. (43) demonstrated binding of ligase I to both the N-terminal portion of p140 and p38 and showed that this interaction was inhibitory to ligase I but was abrogated by the presence of PCNA. In the proposed structure of the complex, the BRCT region of p140 and p38 is in close proximity resulting in a potential binding surface for ligase I consistent with the biochemical data. This observation suggests the possibility of a handoff mechanism whereby 5′-phosphate binding by p140-(375–480) serves to localize ligase I whose activity is subsequently enabled when FEN1 is released from PCNA. Efficient completion of the Okazaki fragment maturation requires coordinated activities of DNA polymerase δ, FEN1, DNA ligase I, and PCNA. Our structure suggests that RFC plays an important, yet subtle, role in this process because yeast missing the BRCT region exhibit no obvious phenotype under normal growth conditions (21).
FIGURE 6.
Schematic model of the complex of the RFC BRCT region, the clamp loader, and DNA. The model was created by combining the present NMR-based model of the RFC p140-(375–480) bound to 5′-phosphorylated dsDNA and the trRFC-PCNA complex (PDB code 1SXJ). The five-subunit RFC clamp loader complex is shown as a surface representation in gray (subunits p40, p37, and p36) and purple (p140) and orange (p38). The collar domains are at the top, and the PCNA-binding surface is at the bottom. The RFC BRCT region is shown in blue, and the DNA is in red (template strand) and light green (daughter strand) with the 5′-phosphate in yellow. The predicted exit point of the template strand is indicated by the green arrow (3). The distance between the C terminus of the BRCT region and the N terminus of the p140 clamp loader subunit is 35 Å. The distance between the 3′ end of the template strand of the BRCT region complex and the predicted exit point of the clamp loader complex is 25 Å. Biochemical data (43) suggest that DNA ligase I binds the proximally located BRCT region and p38.
Acknowledgments
We thank Prof. Rolf Boelens for recording the isotope-filtered NOESY spectra and Marc van Dijk for initiating the early part of the HADDOCK calculations.
The atomic coordinates and structure factors (codes 2k6g and 2k7f) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
M. Kobayashi, E. AB, A. M. J. J. Bonvin, and G. Siegal, unpublished data.
- RFC
- replication factor C
- trRFC
- truncated RFC
- BRCT
- BRCA1 C-terminal domain
- dsDNA
- double-stranded DNA
- PCNA
- proliferating cell nuclear antigen
- NOE
- nuclear Overhauser effect
- NOESY
- nuclear Overhauser effect spectroscopy
- HSQC
- heteronuclear single quantum coherence
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank.
REFERENCES
- 1.Waga S., Stillman B. (1994) Nature 369, 207–212 [DOI] [PubMed] [Google Scholar]
- 2.Uhlmann F., Cai J., Gibbs E., O'Donnell M., Hurwitz J. (1997) J. Biol. Chem. 272, 10058–10064 [DOI] [PubMed] [Google Scholar]
- 3.Bowman G. D., O'Donnell M., Kuriyan J. (2004) Nature 429, 724–730 [DOI] [PubMed] [Google Scholar]
- 4.Allen B. L., Uhlmann F., Gaur L. K., Mulder B. A., Posey K. L., Jones L. B., Hardin S. H. (1998) Nucleic Acids Res. 26, 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi M., Figaroa F., Meeuwenoord N., Jansen L. E., Siegal G. (2006) J. Biol. Chem. 281, 4308–4317 [DOI] [PubMed] [Google Scholar]
- 6.Bork P., Hofmann K., Bucher P., Neuwald A. F., Altschul S. F., Koonin E. V. (1997) FASEB J. 11, 68–76 [PubMed] [Google Scholar]
- 7.Caldecott K. W. (2003) Science 302, 579–580 [DOI] [PubMed] [Google Scholar]
- 8.Koonin E. V., Altschul S. F., Bork P. (1996) Nat. Genet. 13, 266–268 [DOI] [PubMed] [Google Scholar]
- 9.Yu X., Chini C. C., He M., Mer G., Chen J. J. (2003) Science 302, 639–642 [DOI] [PubMed] [Google Scholar]
- 10.Manke I. A., Lowery D. M., Nguyen A., Yaffe M. B. (2003) Science 302, 636–639 [DOI] [PubMed] [Google Scholar]
- 11.Botuyan M. V., Nominé Y., Yu X., Juranic N., Macura S., Chen J., Mer G. (2004) Structure 12, 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams R. S., Lee M. S., Hau D. D., Glover J. N. (2004) Nat. Struct. Mol. Biol. 11, 519–525 [DOI] [PubMed] [Google Scholar]
- 13.Shiozaki E. N., Gu L., Yan N., Shi Y. (2004) Mol. Cell 14, 405–412 [DOI] [PubMed] [Google Scholar]
- 14.Clapperton J. A., Manke I. A., Lowery D. M., Ho T., Haire L. F., Yaffe M. B., Smerdon S. J. (2004) Nat. Struct. Mol. Biol. 11, 512–518 [DOI] [PubMed] [Google Scholar]
- 15.Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 16.Dulic A., Bates P. A., Zhang X., Martin S. R., Freemont P. S., Lindahl T., Barnes D. E. (2001) Biochemistry 40, 5906–5913 [DOI] [PubMed] [Google Scholar]
- 17.Derbyshire D. J., Basu B. P., Serpell L. C., Joo W. S., Date T., Iwabuchi K., Doherty A. J. (2002) EMBO J. 21, 3863–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson A., Smith A., Bullard D., Lavesa-Curto M., Sayer H., Bonner A., Hemmings A., Bowater R. (2005) Biochim. Biophys. Acta 1749, 113–122 [DOI] [PubMed] [Google Scholar]
- 19.Jeon H. J., Shin H. J., Choi J. J., Hoe H. S., Kim H. K., Suh S. W., Kwon S. T. (2004) FEMS Microbiol. Lett. 237, 111–118 [DOI] [PubMed] [Google Scholar]
- 20.Feng H., Parker J. M., Lu J., Cao W. G. (2004) Biochemistry 43, 12648–12659 [DOI] [PubMed] [Google Scholar]
- 21.Gomes X. V., Gary S. L., Burgers P. M. (2000) J. Biol. Chem. 275, 14541–14549 [DOI] [PubMed] [Google Scholar]
- 22.van Dijk M., van Dijk A. D., Hsu V., Boelens R., Bonvin A. M. (2006) Nucleic Acids Res. 34, 3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi M., Siegal G. (2005) J. Biomol. NMR 31, 183–184 [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 25.Keller R. C. (2004) The Computer Aided Resonance Assignment Tutorial, 1st Ed., pp. 15–54, Cantina Verlag, Goldau, Switzerland [Google Scholar]
- 26.Ikura M., Bax A. (1992) J. Am. Chem. Soc. 114, 2433–2440 [Google Scholar]
- 27.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 28.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 29.Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Guntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. J. J. (2005) Proteins Struct. Funct. Bioinformat. 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 30.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 31.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 32.Hubbard S. J., Thornton J. M. (2008) NACCESS, Department of Biochemistry and Molecular Biology, University College London, London [Google Scholar]
- 33.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 34.Ekblad C. M., Wilkinson H. R., Schymkowitz J. W., Rousseau F., Freund S. M., Itzhaki L. S. (2002) J. Mol. Biol. 320, 431–442 [DOI] [PubMed] [Google Scholar]
- 35.Nelson J. R., Gibbs P. E., Nowicka A. M., Hinkle D. C., Lawrence C. W. (2000) Mol. Microbiol. 37, 549–554 [DOI] [PubMed] [Google Scholar]
- 36.Krishnan V. V., Thornton K. H., Thelen M. P., Cosman M. (2001) Biochemistry 40, 13158–13166 [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., Moréra S., Bates P. A., Whitehead P. C., Coffer A. I., Hainbucher K., Nash R. A., Sternberg M. J., Lindahl T., Freemont P. S. (1998) EMBO J. 17, 6404–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaiser O. J., Ball L. J., Schmieder P., Leitner D., Strauss H., Wahl M., Kühne R., Oschkinat H., Heinemann U. (2004) Biochemistry 43, 15983–15995 [DOI] [PubMed] [Google Scholar]
- 39.Lee M. S., Edwards R. A., Thede G. L., Glover J. N. (2005) J. Biol. Chem. 280, 32053–32056 [DOI] [PubMed] [Google Scholar]
- 40.Pervushin K., Billeter M., Siegal G., Wüthrich K. (1996) J. Mol. Biol. 264, 1002–1012 [DOI] [PubMed] [Google Scholar]
- 41.Kalodimos C. G., Biris N., Bonvin A. M., Levandoski M. M., Guennuegues M., Boelens R., Kaptein R. (2004) Science 305, 386–389 [DOI] [PubMed] [Google Scholar]
- 42.Viadiu H., Aggarwal A. K. (2000) Mol. Cell 5, 889–895 [DOI] [PubMed] [Google Scholar]
- 43.Levin D. S., Vijayakumar S., Liu X., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2004) J. Biol. Chem. 279, 55196–55201 [DOI] [PubMed] [Google Scholar]
- 44.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 45.Holm L., Park J. (2000) Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]