Abstract
RUNX3 is a transcription factor that functions as a tumor suppressor. In some cancers, RUNX3 expression is down-regulated, usually due to promoter hypermethylation. Recently, it was found that RUNX3 can also be inactivated by the mislocalization of the protein in the cytoplasm. The molecular mechanisms controlling this mislocalization are poorly understood. In this study, we found that the overexpression of Src results in the tyrosine phosphorylation and cytoplasmic localization of RUNX3. We also found that the tyrosine residues of endogenous RUNX3 are phosphorylated and that the protein is localized in the cytoplasm in Src-activated cancer cell lines. We further showed that the knockdown of Src by small interfering RNA, or the inhibition of Src kinase activity by a chemical inhibitor, causes the re-localization of RUNX3 to the nucleus. Collectively, our results demonstrate that the tyrosine phosphorylation of RUNX3 by activated Src is associated with the cytoplasmic localization of RUNX3 in gastric and breast cancers.
Keywords: Diseases/Cancer, Diseases/Cancer/Carcinogenesis, Diseases/Cancer/Oncogene, Oncogene, Protein/Post-translational Modification, Protein/Protein-Protein Interactions, Protein/Translocation, Tumor/Suppressor
Introduction
RUNX transcription factors are essential for various cellular developmental and differentiation processes (1). In mammals, there are three family members as follows: RUNX1, RUNX2, and RUNX3. RUNX1 is essential for hematopoiesis (2), and the RUNX1 gene is a frequent target of chromosomal translocation in distinct subtypes of human leukemia (3). RUNX2 is essential for osteogenesis (4, 5) and is involved in the human disease cleidocranial dysplasia, an autosomal dominant bone disorder (6, 7).
Previously, we reported that RUNX3 is closely associated with transforming growth factor-β signaling and functions as a tumor suppressor in gastric carcinogenesis (8). RUNX3 also suppresses the development of colon cancer by forming a ternary complex with β-catenin-TCF4 and attenuating Wnt-mediated signaling activity (9). RUNX3 is frequently inactivated in gastric and colon cancer and in cancers of the lung, bladder, pancreas, liver, prostate, bile duct, breast, larynx, esophagus, and the testicular yolk sac, primarily through promoter hypermethylation (10–26). These studies suggest that RUNX3 functions as a suppressor of various tumors in some cancer contexts (1, 27).
Cytoplasmic mislocalization of RUNX3 and down-regulation of the RUNX3 gene have been observed in human breast tumors and gastric cancers (28, 29) as well as in colon cancers, based on our studies (9). These results suggested that cytoplasmic mislocalization of RUNX3 is another mechanism for inactivating the tumor suppressor activity of RUNX3. However, the molecular mechanisms causing RUNX3 mislocalization in human cancers have not been comprehensively studied.
The Src family of kinases (SFKs)3 includes the largest family of nonreceptor protein kinases. The SFK family is composed of nine members as follows: Blk, Fgr, Fyn, Hck, Lck, Lyn, Src, Yes, and Yrk. Src, Fyn, and Yes are ubiquitously expressed in most tissues, whereas the others are selectively expressed in particular cell lineages (30–33). SFKs are critical components of the signaling cascades initiated by various membrane receptors, including growth factor receptors, integrins, other adhesion receptors, G protein-coupled receptors, cytokine receptors, immunoglobulin-like domain receptors, and ion channels. SFKs are essential for many cellular activities, including proliferation, differentiation, motility, and adhesion. Src is the most extensively studied of the SFKs, and it has been closely associated with tumor development, tumor progression, and distant metastasis by promoting cell proliferation, invasion, and motility (31). It has been found that Src is overexpressed or highly activated in a large number of human cancers (34). It phosphorylates p27Kip1 on tyrosine residues and accelerates p27Kip1 proteolysis (35).
Under normal conditions, the export of signal transducers from the nucleus is important for their recycling in successive rounds of inactivation and reactivation. This signaling process depends on a nuclear export receptor known as chromosome region maintenance 1 (CRM1), which bridges nuclear export signal-containing proteins to the nuclear pore complex (36, 37). In this regard, recent studies have demonstrated that the functions of various regulatory proteins, including APC, p53, p27Kip1, Smad4, and RUNX2, can be modulated via the nuclear export mechanism (38–45).
In this study, we provide evidence that Src phosphorylates RUNX3 at multiple tyrosine residues. This Src-mediated tyrosine phosphorylation results in the cytoplasmic mislocalization of RUNX3. Notably, RUNX3 is localized in the cytoplasm in various cancer cell lines where the Src kinase is highly activated. The knockdown of Src by siRNA facilitates the nuclear localization of RUNX3. Our results suggest that the tyrosine phosphorylation and mislocalization of RUNX3 by the activation of Src could be one of the mechanisms for RUNX3 inactivation in cancer cells.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Reagents were purchased from the following vendors: restriction enzymes and protein phosphatase were from New England Biolabs (Beverly, MA); transfection reagents Lipofectamine Plus reagent, siRNA-Src (12938-109), fetal bovine serum, and medium were from Invitrogen; DNA preparation kit, QIAquick gel extraction/PCR purification kit, and Ni-NTA-agarose were from Qiagen (Hilden Germany); PCR reagents and Pfu DNA polymerase were from Solgent (Seoul, Korea); EDTA-free protease inhibitor was from Roche Applied Science; protein G-Sepharose beads, fast protein liquid chromatography system, and HiLoad Superdex 200 column were from GE Healthcare; ECL solution was from PerkinElmer Life Sciences; polyvinylidene difluoride membranes and Amicon Ultra-15 were from Millipore (Billerica, MA); Abl-Src inhibitor (thiadiazole) was from Calbiochem; and 4,6-diamidino-2-phenylindole and nuclear export inhibitor (leptomycin B) were from Sigma. Mouse monoclonal antibodies against RUNX3 (R3-5G4 and 6E9; Abcam (Cambridge, UK)), phospho-Src and Src (Cell Signaling, Beverly, MA), and phosphotyrosine (4G10; Upstate Biotechnology, Inc.) were used for immunoprecipitation, immunoblotting, or immunocytochemical analysis.
Plasmids
The Src gene (NM_005417) and a gene encoding a kinase-dead mutant of Src (K298A) and deletion mutants of HA-Src (SH2, SH3, or TyrK) were amplified by PCR and cloned into the pCS4-HA vector to create HA-tagged Src (HA-Src) and HA-Src-KD. The full-length RUNX3 gene (NM_004350) and its deletion mutants were amplified by PCR and subcloned into the pCS4-Myc vector to create FLAG- or Myc-tagged RUNX3 (FLAG-RUNX3 or Myc-RUNX3) and the Myc-tagged deletion constructs. Other SFK family kinases, Myc-tagged Fyn (NM_002037) and Myc-tagged Lck (NM_002037), were used for transfection and immunoprecipitation experiments. For protein purification, the cDNA encoding RUNX3 was subcloned into pCS4–10×His to generate proteins with N-terminal His tags. All the constructs were verified by sequencing.
Cell Culture and Transfection
The HEK293, BOSC23, HeLa, and SYF cells were maintained in Dulbecco's modified Eagle's medium. We used BOSC23 cells and SYF cells because Src is not activated in BOSC23, and c-Src/Yes/Fyn are disrupted in SYF cells. Human gastric cancer cell lines MKN45, SNU16, and human breast cancer cell lines BT549, BT20, and MDA-MB-468 were maintained in RPMI 1640 medium, minimum essential media, or Leibovitz's L-15 medium, respectively, with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin in a humidified atmosphere with 5% CO2 at 37 °C. Transient transfections were carried out using Lipofectamine Plus reagent, and cells were harvested 24–48 h after transfection. Immunoprecipitations, immunoblotting, and immunostaining were performed as reported previously (46).
Immunocytochemistry
Cells grown on microscope coverslips were transfected with siRNA-Src or treated with the Abl-Src inhibitor (thiadiazole). Cells were fixed with 4% paraformaldehyde for 15 min, treated with a serum-free blocking solution (DAKO, Denmark), and incubated with a primary antibody at 4 °C overnight and a secondary antibody at room temperature for 1 h in a diluent solution (DAKO) (28). To visualize the nuclei, cells were counterstained with 4,6-diamidino-2-phenylindole. After mounting, cells were observed with a laser-scanning confocal microscope (Leica, Wetzlar, Germany).
In Vitro Kinase Assay
For the purification of recombinant His10-tagged RUNX3 or His10-tagged Src, HEK293 cells were transfected with 20 μg of the appropriate plasmid and harvested after 36 h. The cell lysates were homogenized in buffer containing 50 mm Tris-HCl (pH 7.8), 150 mm NaCl, and EDTA-free protease inhibitor. The RUNX3 and Src proteins were purified using Ni-NTA-agarose and gel filtration chromatography (Superdex 200). In vitro kinase assays using a fixed concentration of RUNX3 protein and various concentrations of Src kinase were conducted for 30 min at 37 °C in a buffer containing 50 mm Tris-HCl (pH 7.8), 5 mm MgCl2, 1.0 mm dithiothreitol, 5 mm ATP, and 150 mm NaCl. Proteins were separated by SDS-PAGE and analyzed by immunoblotting with antibodies against phosphotyrosine (4G10), RUNX3 (5G4), and Src. In addition, purified His-tagged RUNX3 proteins from Escherichia coli BL21(DE3) and GST-Src protein (Cell Signaling) from E. coli were used in in vitro kinase reactions and pulldown experiments.
Phosphatase Treatment of Cell Extracts
Dephosphorylation of RUNX3 was performed by incubating whole cell extracts with 400 units of λ-phosphatase at 30 °C for 1 h. The reactions were stopped by boiling in SDS sample buffer, followed by SDS-PAGE and immunoblotting with the 4G10 anti-phosphotyrosine antibody.
RESULTS
RUNX3 Protein Is Mislocalized from the Nucleus to the Cytoplasm by Overexpression of Src Kinase
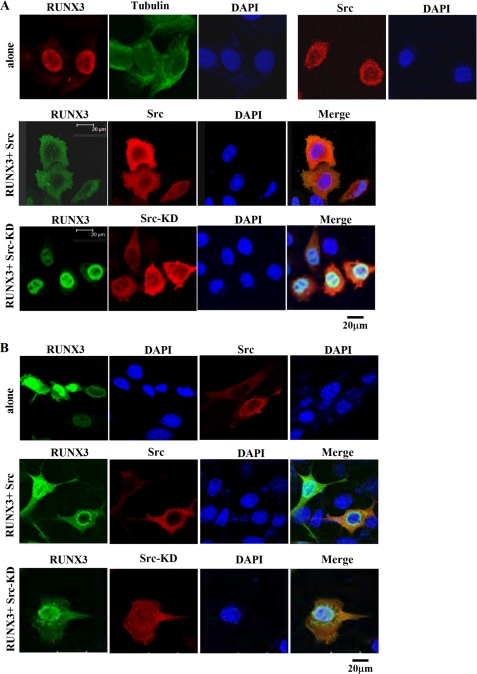
Because Src is frequently activated in various cancers where RUNX3 is mislocalized in the cytoplasm, we hypothesized that the cytoplasmic localization of RUNX3 might be associated with the activation of Src kinase. To investigate this possibility, we examined whether the subcellular localization of RUNX3 is affected by the overexpression of Src. HeLa cells were transiently transfected with an expression plasmid encoding Myc-tagged RUNX3 (Myc-RUNX3) with or without a plasmid encoding HA-tagged wild-type Src (HA-Src). The subcellular localizations of RUNX3 and Src were analyzed by immunocytochemical analysis using corresponding antibodies. In the absence of Src, RUNX3 protein was localized in the nucleus of transfected cells (Fig. 1A, top panel). In contrast, when RUNX3 was coexpressed with wild-type Src, RUNX3 was localized in the cytoplasm (Fig. 1A, 2nd panel). These results suggested that the overexpression of Src is associated with the cytoplasmic localization of RUNX3. We also tried coexpression with a plasmid encoding an HA-tagged kinase-dead mutant of Src (HA-Src-KD). This mutant protein has a K298A substitution in its ATP-binding site that was previously shown to inactivate the tyrosine kinase activity of Src (47). Coexpression with HA-Src-KD failed to induce the cytoplasmic localization of RUNX3 (Fig. 1A, 3rd panel), suggesting that the Src-mediated cytoplasmic localization of RUNX3 is dependent on the tyrosine kinase activity of Src. Essentially the same results were obtained when HeLa cells were replaced by SYF cells (Fig. 1B). Although biochemical interaction experiments were done with HEK293 cells, we have not used HEK293 cells in immunocytochemical analysis, because these cells are easily detached from the plate when staining with antibodies.
FIGURE 1.
RUNX3 is mislocalized from the nucleus to the cytoplasm by the overexpression of Src. HeLa cells (A) and SFY cells (B) were transfected with Myc-RUNX3 in the presence or absence of HA-Src or HA-Src-KD. After 48 h, the subcellular localizations of RUNX3 and Src were analyzed by immunofluorescence using a monoclonal anti-Myc antibody and a polyclonal anti-HA antibody. A mouse IgG coupled to green fluorescence and a rabbit IgG coupled to red fluorescence were used to detect the Myc-RUNX3 and HA-Src immune complexes, respectively. Nuclei are visualized by 4,6-diamidino-2-phenylindole staining. In the cells expressing Myc-RUNX3 alone, RUNX3 was almost exclusively localized to the nucleus (top panel). In the cells cotransfected with Myc-RUNX3 and HA-Src, RUNX3 displayed cytoplasmic mislocalization in virtually all of HA-Src coexpressing cells (middle panel). In the cells coexpressing Myc-RUNX3 and HA-Src-KD, RUNX3 was mainly localized in the nucleus (bottom panel), indicating that the cytoplasmic mislocalization of RUNX3 is dependent on the tyrosine kinase activity of Src.
Src Phosphorylates Multiple Tyrosine Residues on RUNX3
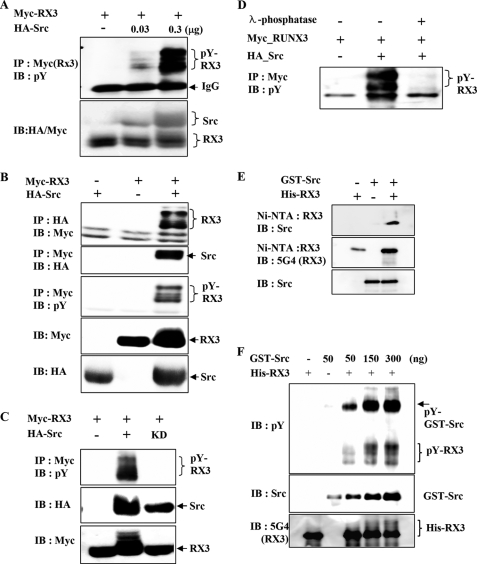
Because the tyrosine kinase activity of Src was required for the cytoplasmic localization of RUNX3, we examined whether RUNX3 is phosphorylated by Src. The coexpression of a fixed amount of Myc-RUNX3 and increasing amounts of HA-Src in HEK293 cells, followed by immunoprecipitation (IP) with anti-Myc (RUNX3) antibody and immunoblotting (IB) with anti-phosphotyrosine antibody (4G10; Tyr(P)), revealed that RUNX3 is phosphorylated at multiple tyrosine residues in an Src concentration-dependent manner (Fig. 2A).
FIGURE 2.
Src kinase phosphorylates RUNX3 on tyrosine residues in vitro and in vivo. A, HEK293 cells were transiently transfected with a fixed amount of Myc-RUNX3 (0.5 μg) and increasing amounts of HA-Src (0, 0.03, and 0.3 μg), and tyrosine phosphorylation of RUNX3 was examined by IP with anti-Myc (RUNX3) antibody and IB with anti-phosphotyrosine antibody (pY; 4G10). B, HEK293 cells were transiently transfected with Myc-RUNX3 and/or HA-Src. Physical interactions between RUNX3 (RX3) and Src were examined by IP and IB using the indicated antibodies (top and 2nd panels). Tyrosine phosphorylation of RUNX3 was analyzed by IP with the anti-Myc antibody and IB with the antibody against phosphotyrosine (4G10; pY) (3rd panel). The levels of Myc-RUNX3 and HA-Src in the transfected cells were measured by IB with the anti-Myc and anti-HA antibodies. C, cells were transfected with Myc-RUNX3 in the presence or absence of HA-Src (wild type) or HA-Src-KD (kinase-dead mutant). Cell lysates were analyzed by IP with the anti-Myc antibody and IB with anti-phosphotyrosine antibody (pY). D, cells were transfected with Myc-RUNX3 in the presence or absence of HA-Src. Cell lysates were treated with λ-phosphatase for 1 h at 30 °C and analyzed by IP and IB with the indicated antibodies. E, GST-Src and His-RUNX3 obtained from E. coli were mixed as indicated, and Ni-NTA pulldown experiments were performed to determine the physical interaction between these two proteins. The interaction was analyzed by IB with the indicated antibodies. F, same proteins were subjected to an in vitro phosphorylation analysis. A fixed amount of His-RUNX3 (500 ng) and increasing amounts of GST-Src (50, 150, and 300 ng) were incubated and analyzed by IB with the anti-RUNX3 (5G4), anti-Src, and anti-phosphotyrosine antibodies. The amount of tyrosine-phosphorylated RUNX3 increased with increasing amounts of Src.
We then examined whether RUNX3 physically interacts with Src. The coexpression of Myc-RUNX3 and HA-Src is followed by IP and IB and revealed that RUNX3 interacts with Src (Fig. 2B, top and 2nd panels). Interestingly, the Src-interacting RUNX3 appeared as multiple bands (Fig. 2A, top panel). A subsequent analysis using IP with the anti-Myc antibody and IB with the anti-phosphotyrosine antibody revealed that the multiple bands represent RUNX3 with various levels of tyrosine phosphorylation (Fig. 2B, 3rd panel). These results demonstrate that Src not only phosphorylates multiple tyrosine residues on RUNX3 but also interacts with the tyrosine-phosphorylated RUNX3. The tyrosine phosphorylation of RUNX3 requires the catalytic activity of the Src kinase, because the kinase-dead Src mutant failed to phosphorylate RUNX3 (Fig. 2C). Src-mediated tyrosine phosphorylation of RUNX3 was dramatically decreased by treatment with a protein phosphatase (λ-phosphatase) in the cell lysates (Fig. 2D).
To investigate whether Src directly interacts with RUNX3, bacterially expressed His-RUNX3 and GST-Src were analyzed by Ni-NTA pulldown assays followed by IB. The results revealed that His-RUNX3 interacts with GST-Src (Fig. 2E). In vitro kinase assays performed with GST-Src and His-RUNX3 proteins showed tyrosine phosphorylation of RUNX3 by Src in an Src concentration-dependent manner (Fig. 2F). Together, these results demonstrate that RUNX3 is a direct target of Src.
Src Kinase Interacts with the Runt Domain of RUNX3
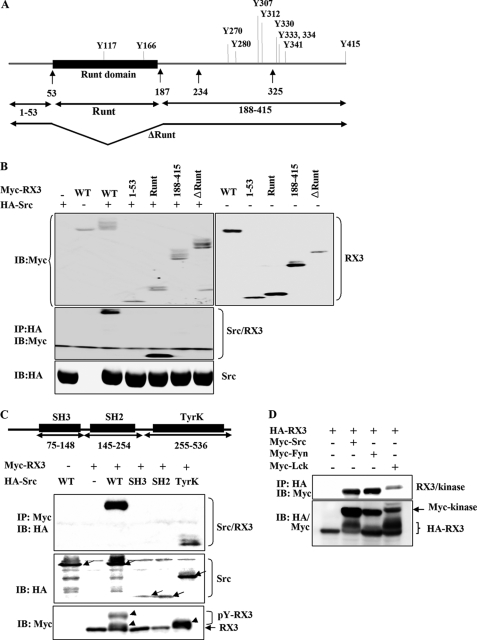
To identify the Src-interacting region within RUNX3, the full-length Myc-RUNX3 construct (which encodes a protein containing 11 tyrosine residues) or truncated versions of Myc-RUNX3 were transiently coexpressed with HA-Src in HEK293 cells (Fig. 3, A and B). Both the full-length RUNX3 protein and the Runt domain alone physically interacted with Src (Fig. 3B). However, the Runt domain deletion mutant (ΔRunt; deleted between amino acids 54 and 187), the N-terminal region of the protein (amino acids 1–53), and the C-terminal region (amino acids 188–415) failed to interact with Src (Fig. 3B). These results suggest that Src interacts with RUNX3 at the Runt domain. Interestingly, however, multiple shifted RUNX3 bands were observed in cells transfected not only with full-length RUNX3 but also with the mutant encoding the RUNX3 C-terminal region (188–415) and the ΔRunt mutant (ΔRunt). This result suggests that the amino acids 188–415 region of RUNX3 may also interact with Src weakly. Although we have evidence that RUNX3 is a target of Src (Fig. 2), our results do not rule out the possibility that another tyrosine kinase, activated by Src, could also phosphorylate RUNX3.
FIGURE 3.
Mapping of the interacting regions within RUNX3 and Src. A, schematic representations of wild-type RUNX3 and deletion constructs (1–53, Runt, 188–415, and ΔRunt). Positions of 11 tyrosine residues are shown on the representation of the wild-type gene (top). B, Myc-RUNX3 (WT) or various truncated constructs of RUNX3 were transiently coexpressed with or without full-length HA-Src in HEK293 cells. Expression of each construct was examined by IB with the anti-Myc (top panels) or anti-HA (bottom panel) antibody. Physical interactions between Src and the WT RUNX3 or the deletion mutants (collectively indicated as RX3) were analyzed by IP with the anti-HA antibody and IB with the anti-Myc antibody (middle panel). C, full-length HA-Src (WT) or deletion mutants of HA-Src (SH2, SH3, or TyrK) were transiently coexpressed with Myc-RUNX3 in HEK293 cells. Physical interactions between RUNX3 and Src (or one of its deletion mutants) were analyzed by IP with the anti-Myc antibody and IB with the anti-HA antibody (top panel). Expression of each construct was examined by IB with the anti-HA or anti-Myc antibody. Full-length Src and its fragments are indicated by arrows (middle panel). Full-length and fragments of Src and tyrosine-phosphorylated RUNX3 are indicated by arrows and arrowheads, respectively. D, cells were transiently transfected with HA-RUNX3 and/or SYF kinases (Myc-Src, Myc-Fyn, and Myc-Lck). Physical interactions between RUNX3 and SYF kinases were examined by IP and IB using the indicated antibodies. The levels of HA-RUNX3 and Myc-SYF kinases in the transfected cells were measured by IB with the anti-Myc and anti-HA antibodies.
The physical interaction between RUNX3 and Src was further examined by IP and IB experiments following cotransfections with full-length RUNX3 and full-length (WT) or truncated versions of Src (SH2, SH3, and TyrK). WT Src and the kinase domain of Src (TyrK) were capable of binding RUNX3. In contrast, the SH2 and SH3 domains of Src did not interact with RUNX3 (Fig. 3C). These results demonstrate that the kinase domain of Src is essential for its physical binding to RUNX3. Notably, tyrosine phosphorylation of RUNX3 was detected in cells that were cotransfected with either the full-length Src or the kinase domain construct (TyrK). However, complete phosphorylation of RUNX3 only occurred in cells transfected with full-length Src, suggesting that the SH2 or SH3 domain may also play a role in fully phosphorylating RUNX3 (Fig. 3C, bottom panel).
Because RUNX3 interacts with the kinase domain of Src, we investigated whether other tyrosine kinases also interact with RUNX3. The results revealed that Fyn and Lck also interact with and phosphorylate RUNX3 as shown by the coimmunoprecipitation and the induced band shift of RUNX3 (Fig. 3D), suggesting that RUNX3 could be a target of other tyrosine kinases.
Phosphorylation of RUNX3 Tyrosine Residues Occurs in Src-activated Tumor Cell Lines
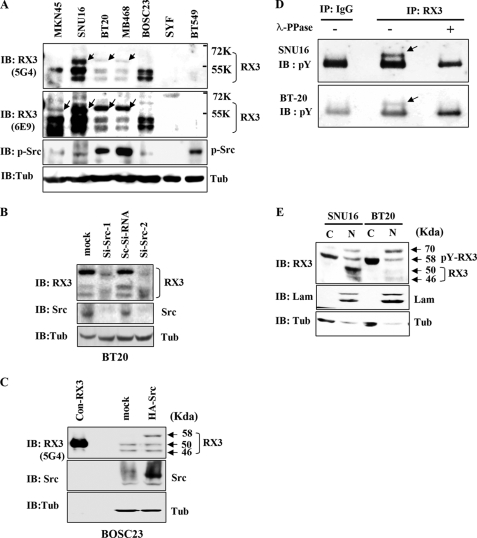
To address the question of whether tyrosine phosphorylation of endogenous RUNX3 occurs in tumor cell lines with cytoplasmic mislocalization of RUNX3 (MKN45 and SNU16 gastric cancer cell lines) or lines with activated Src (BT20 and MB468 breast cancer cell lines (14, 48)), we performed IB experiments with the anti-RUNX3 antibody (5G4). Among the cell lines with cytoplasmic mislocalization of RUNX3, SNU16 also bears activated Src (49). The antibody detected not only two normal RUNX3 bands (46 and 50 kDa) but also a shifted band (58 kDa) in the Src-activated cell lines (Fig. 4A, top panel, shifted bands are indicated by arrows). IB with another anti-RUNX3 antibody (6E9) showed essentially the same result except for an enhanced sensitivity to the 58-kDa protein (Fig. 4A, 2nd panel). In this case, the 58-kDa band was also detected in the MKN45 cell line. These results indicated that the anti-RUNX3 antibodies detect a shifted band in Src-activated cell lines and in lines with cytoplasmic mislocalization of RUNX3. On the other hand, the shifted band was not detected in the BOSC23 cells (Src-nonactivated), SYF cells (c-Src/Yes/Fyn triple knock-out), or BT549 cells (RUNX3 down-regulated) (29) (Fig. 4A).
FIGURE 4.
Tyrosine phosphorylation of endogenous RUNX3 in cancer cell lines. A, immunoblotting analysis of whole cell lysates from the indicated cell lines using two different anti-RUNX3 (RX3) antibodies (5G4 and 6E9) and an antibody against phosphorylated-Src (p-Src). An anti-tubulin (Tub) antibody was used as a loading control. Both the anti-RUNX3 antibodies detected a 58-kDa protein (indicated by arrows) in Src-activated cells and in cells with mislocalized RUNX3 (see text for further details). This phosphorylated RUNX3 is larger than the normally detected bands (46 and 50 kDa). Activated Src was detected by the anti-p-Src antibody. B, BT20 cells were grown for 48 h after treatment with scrambled Si-RNA (Sc-Si-RNA) or Src specific siRNA (Si-Src-1, 5′-GCCUCUCAGUGUCUGACUUCGACAA-3′; Si-Src-2, 5′-GCCUCUCAGUGUCUGACUUCGACAA-3′). Cell lysates were immunoblotted with the indicated antibodies. C, BOSC23 cells were transiently transfected with or without HA-Src, and the cell lysates were immunoblotted with the indicated antibodies. Cell lysates were also obtained from BOSC23 cells transfected with RUNX3 (without tag), and small amounts (1:20 compared with other samples) were loaded as molecular weight markers (con-RX3, 50 kDa). D, SNU16 and BT20 cell lysates were treated with or without λ-phosphatase (λ-PPase) for 1 h at 30 °C, then immunoprecipitated with the anti-RUNX3 antibody (6E9; RX3) or IgG as a control, and immunoblotted with the anti-phosphotyrosine antibody (pY). E, nuclear (N) and cytoplasmic (C) fractions were obtained separately from each of the Src-activated cell lines and analyzed by immunoblotting with the anti-RUNX3 antibody (6E9; RX3). Laminin (Lam) and tubulin (Tub) were used as fractionation controls. The 58-kDa protein (tyrosine-phosphorylated RUNX3) was detected mainly in the cytoplasmic fractions.
To determine whether the shifted RUNX3 band found in lysates from the cancer cell lines might represent tyrosine phosphorylation of RUNX3 by Src kinase, we analyzed cell lysates after the siRNA-mediated knockdown of Src or the overexpression of Src. Interestingly, the knockdown of Src in BT20 cells (Src activated) resulted in a significant decrease in the intensity of the shifted (58 kDa) RUNX3 band (Fig. 4B; Si-Src). Consistent with this, the overexpression of Src in BOSC23 cells (Src-nonactivated) resulted in the appearance of the 58-kDa RUNX3 band (Fig. 4C; HA-Src).
To further confirm that the shifted (58 kDa) RUNX3 band represents the tyrosine-phosphorylated form of RUNX3, SNU16 and BT20 cell lysates were incubated with or without λ-phosphatase and then analyzed by IP with the anti-RUNX3 antibody and IB with the anti-phosphotyrosine antibody. The results revealed that endogenous RUNX3 is phosphorylated on the tyrosine residues in these cancer cell lines (Fig. 4D). Taken together, our results suggest that the shifted (58 kDa) band represents endogenous RUNX3 that has been phosphorylated at the tyrosine residues by Src kinase.
Tyrosine-phosphorylated RUNX3 Is Localized in the Cytoplasm
The experimental results presented thus far have shown the following: 1) Src overexpression induces alteration of subcellular localization of RUNX3 from the nucleus to the cytoplasm; and 2) Src phosphorylates RUNX3 at the tyrosine residues. To understand the relationship between RUNX3 tyrosine phosphorylation and RUNX3 cytoplasmic localization, we analyzed the subcellular localization of endogenous RUNX3 protein in tumor cell lines where the Src kinase was activated. Nuclear and cytoplasmic protein fractions were separated from SNU16 and BT20 cell lysates, and the levels of RUNX3 were then assessed by IB with the anti-RUNX3 antibody. In each cell line, 46-, 50-, 58-, and 70-kDa proteins were detected by anti-RUNX3 antibody in the nuclear fraction. The majority of the tyrosine-phosphorylated RUNX3 (58 kDa) was detected in the cytoplasmic fraction, whereas the tyrosine phosphorylation-free forms of RUNX3 (46 and 50 kDa) were exclusively detected in the nucleus (Fig. 4E). So far, the identity of the 70-kDa protein remains unknown. This result suggests that the cytoplasmic localization of RUNX3 is associated with its tyrosine phosphorylation.
RUNX3 Is Relocalized to the Nucleus by Treatment with si-Src or an Src Inhibitor
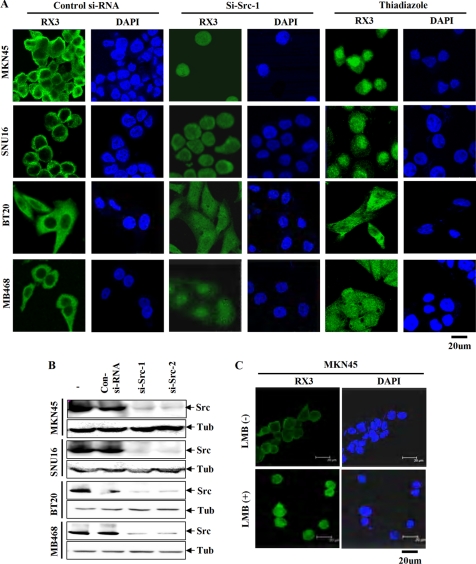
It has been reported previously that RUNX3 is localized in the cytoplasm in the MKN45 and SNU16 gastric cancer cell lines (28), and this was confirmed in our study (Fig. 5A). Notably, BT20 and MB468 breast cancer cell lines, which contain activated Src, also showed cytoplasmic localization of RUNX3 (Fig. 5A).
FIGURE 5.
Relocalization of endogenous RUNX3 by the knockdown of Src or the inhibition of Src activity in Src-activated cancer cell lines. A, two gastric cancer cell lines (MKN45 and SNU16) and two breast cancer cell lines (BT20 and MB468) were grown on coverslips for 24 h and then treated with Src siRNA (si-Src-1) or Src inhibitor (thiadiazole). Forty eight hours after siRNA transfection or 24 h later after Src inhibitor, the cells were fixed, permeabilized, and analyzed by immunofluorescence using the anti-RUNX3 monoclonal antibody (6E9) and a mouse IgG coupled to green fluorescence to detect the subcellular localization of endogenous RUNX3. Nuclei are visualized by 4,6-diamidino-2-phenylindole staining. For each cell line, RUNX3 was detected mainly in the cytoplasm when cells were treated with a control siRNA construct. Treatment with si-Src-1 or the Src inhibitor resulted in an increase in nuclear localization and a decrease in cytoplasmic localization of RUNX3 in all four cell lines. B, cell lines were treated with Src siRNAs (si-Src-1 or si-Src-2) for 48 h, and the expression level of Src was analyzed by immunoblotting with the anti-Src antibody. The results show that both siRNAs effectively knock down Src expression. Tub, tubulin. C, MKN45 cells, which show cytoplasmic localization of RUNX3, were treated with or without leptomycin B (LMB) for 24 h, and then the subcellular localization of RUNX3 was analyzed by immunocytochemical staining. Treatment with leptomycin B resulted in the nuclear localization of RUNX3.
We then asked whether the localization of endogenous RUNX3 in Src-activated cell lines could be counteracted by the siRNA-mediated knockdown of Src. By using two kinds of Src-specific siRNAs (si-Src-1 and si-Src-2), we confirmed that these siRNAs efficiently knocked down Src (Fig. 5B). In all four lines, treatment with si-Src-1 resulted in an increase in the nuclear localization of the RUNX3 protein (Fig. 5A). Similar results were obtained with si-Src-2 (data not shown). Furthermore, treatment with the Src kinase inhibitor thiadiazole also resulted in the relocalization of RUNX3 to the nucleus (Fig. 5A).
The nuclear re-localization of endogenous RUNX3 was also observed after MKN45 cells were treated with 10 ng/ml CRM1 inhibitor leptomycin B for 4 h (Fig. 5C). This suggests that tyrosine-phosphorylated RUNX3 is localized in the cytoplasm through a CRM1-dependent nuclear export mechanism.
DISCUSSION
RUNX3 is a transcription factor that functions as a tumor suppressor regulating cell proliferation, apoptosis, and differentiation. In various cancers, RUNX3 is frequently inactivated by promoter methylation. Recently, it has been shown that RUNX3 is inactivated not only by promoter methylation, but also by mislocalization of the protein to the cytoplasm in gastric cancer and breast cancer cells. However, the molecular mechanisms controlling this mislocalization have been poorly understood. To address this question, we investigated the possibility that the oncogenic protein Src kinase might be responsible for the cytoplasmic localization of RUNX3, because Src is frequently activated in various cancers where RUNX3 is mislocalized in the cytoplasm. In this study, we found the following. 1) Overexpression of Src results in cytoplasmic localization of RUNX3. 2) Src phosphorylates RUNX3 at multiple tyrosine residues, both in vitro and in vivo. 3) Tyrosine residues of endogenous RUNX3 are phosphorylated in Src-activated cell lines. 4) Tyrosine-phosphorylated RUNX3 is localized in the cytoplasm. 5) siRNA-mediated knockdown of Src, or inhibition of Src by an Src inhibitor, results in the relocalization of RUNX3 to the nucleus. Collectively, our results demonstrate that RUNX3 is a target of the Src kinase and that the tyrosine phosphorylation of RUNX3 by Src is associated with the cytoplasmic localization of RUNX3.
We previously reported that RUNX3 is stabilized as an inactive form and localized in the cytoplasm after phosphorylation by the PIM1 serine/threonine kinase (46), which is often hyper-activated in hematopoietic malignancies and prostate carcinomas (50). Our finding that Src inactivates RUNX3 by cytoplasmic sequestration in gastric cancer and breast cancer suggests that the inactivation of RUNX3 may be a key function of oncogenic kinases. Further studies are needed to understand the physiological conditions under which this RUNX3 inactivation occurs and the biological meaning of RUNX3 inactivation by the Src and PIM1 oncogenic kinases.
Cell fractionation followed by immunoblotting analyses indicated that most of the tyrosine-phosphorylated RUNX3 was detected in the cytoplasm, although substantial levels of nonphosphorylated RUNX3 were detected in the nuclear fractions of Src-activated cancer cell lines (Fig. 4D). On the other hand, immunocytochemical staining with the same antibody detected RUNX3 mainly in the cytoplasm (Fig. 5A). This discrepancy may be explained by epitope masking following the incorporation of RUNX3 into macromolecular transcriptional complexes that are nuclear matrix-associated (51).
In conclusion, we found that Src phosphorylates RUNX3 and localizes the protein in the cytoplasm of gastric and breast cancer cells. Although it is generally difficult to restore the function of a lost tumor suppressor gene, it might be feasible to pharmacologically restore the function of the mislocalized RUNX3, because RUNX3 can be relocalized in the nucleus by an Src inhibitor. Our results not only demonstrate a new mechanism for RUNX3 inactivation but also provide an additional rationale for developing Src inhibitors as anti-cancer drugs.
This work was supported by Research Grant R16-2003-002-01001-02006 (to S.-C. B.) from the Korean Science and Engineering Foundation.
- SFK
- Src family kinase
- HA
- hemagglutinin
- Ni-NTA
- nickel-nitrilotriacetic acid
- siRNA
- small interfering RNA
- SH
- Src homology
- WT
- wild type
- IP
- immunoprecipitation
- IB
- immunoblot.
REFERENCES
- 1.Blyth K., Cameron E. R., Neil J. C. (2005) Nat. Rev. Cancer 5, 376–387 [DOI] [PubMed] [Google Scholar]
- 2.Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A. H., Speck N. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speck N. A., Gilliland D. G. (2002) Nat. Rev. Cancer 2, 502–513 [DOI] [PubMed] [Google Scholar]
- 4.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 5.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 6.Lee B., Thirunavukkarasu K., Zhou L., Pastore L., Baldini A., Hecht J., Geoffroy V., Ducy P., Karsenty G. (1997) Nat. Genet. 16, 307–310 [DOI] [PubMed] [Google Scholar]
- 7.Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 8.Li Q. L., Ito K., Sakakura C., Fukamachi H., Inoue K., Chi X. Z., Lee K. Y., Nomura S., Lee C. W., Han S. B., Kim H. M., Kim W. J., Yamamoto H., Yamashita N., Yano T., Ikeda T., Itohara S., Inazawa J., Abe T., Hagiwara A., Yamagishi H., Ooe A., Kaneda A., Sugimura T., Ushijima T., Bae S. C., Ito Y. (2002) Cell 109, 113–124 [DOI] [PubMed] [Google Scholar]
- 9.Ito K., Lim A. C., Salto-Tellez M., Motoda L., Osato M., Chuang L. S., Lee C. W., Voon D. C., Koo J. K., Wang H., Fukamachi H., Ito Y. (2008) Cancer Cell 14, 226–237 [DOI] [PubMed] [Google Scholar]
- 10.Goel A., Arnold C. N., Tassone P., Chang D. K., Niedzwiecki D., Dowell J. M., Wasserman L., Compton C., Mayer R. J., Bertagnolli M. M., Boland C. R. (2004) Int. J. Cancer 112, 754–759 [DOI] [PubMed] [Google Scholar]
- 11.Kang G. H., Lee S., Lee H. J., Hwang K. S. (2004) J. Pathol. 202, 233–240 [DOI] [PubMed] [Google Scholar]
- 12.Kang S., Kim J. W., Kang G. H., Park N. H., Song Y. S., Kang S. B., Lee H. P. (2005) Gynecol. Oncol. 96, 173–180 [DOI] [PubMed] [Google Scholar]
- 13.Kato N., Tamura G., Fukase M., Shibuya H., Motoyama T. (2003) Am. J. Pathol. 163, 387–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T. Y., Lee H. J., Hwang K. S., Lee M., Kim J. W., Bang Y. J., Kang G. H. (2004) Lab. Invest. 84, 479–484 [DOI] [PubMed] [Google Scholar]
- 15.Ku J. L., Kang S. B., Shin Y. K., Kang H. C., Hong S. H., Kim I. J., Shin J. H., Han I. O., Park J. G. (2004) Oncogene 23, 6736–6742 [DOI] [PubMed] [Google Scholar]
- 16.Li Q. L., Kim H. R., Kim W. J., Choi J. K., Lee Y. H., Kim H. M., Li L. S., Kim H., Chang J., Ito Y., Youl Lee K., Bae S. C. (2004) Biochem. Biophys. Res. Commun. 314, 223–228 [DOI] [PubMed] [Google Scholar]
- 17.Mori T., Nomoto S., Koshikawa K., Fujii T., Sakai M., Nishikawa Y., Inoue S., Takeda S., Kaneko T., Nakao A. (2005) Liver Int. 25, 380–388 [DOI] [PubMed] [Google Scholar]
- 18.Nakase Y., Sakakura C., Miyagawa K., Kin S., Fukuda K., Yanagisawa A., Koide K., Morofuji N., Hosokawa Y., Shimomura K., Katsura K., Hagiwara A., Yamagishi H., Ito K., Ito Y. (2005) Br. J. Cancer 92, 562–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osaki M., Moriyama M., Adachi K., Nakada C., Takeda A., Inoue Y., Adachi H., Sato K., Oshimura M., Ito H. (2004) Eur. J. Clin. Invest. 34, 605–612 [DOI] [PubMed] [Google Scholar]
- 20.Oshimo Y., Oue N., Mitani Y., Nakayama H., Kitadai Y., Yoshida K., Ito Y., Chayama K., Yasui W. (2004) Pathobiology 71, 137–143 [DOI] [PubMed] [Google Scholar]
- 21.Sakakura C., Hagiwara A., Miyagawa K., Nakashima S., Yoshikawa T., Kin S., Nakase Y., Ito K., Yamagishi H., Yazumi S., Chiba T., Ito Y. (2005) Int. J. Cancer 113, 221–228 [DOI] [PubMed] [Google Scholar]
- 22.Schulmann K., Sterian A., Berki A., Yin J., Sato F., Xu Y., Olaru A., Wang S., Mori Y., Deacu E., Hamilton J., Kan T., Krasna M. J., Beer D. G., Pepe M. S., Abraham J. M., Feng Z., Schmiegel W., Greenwald B. D., Meltzer S. J. (2005) Oncogene 24, 4138–4148 [DOI] [PubMed] [Google Scholar]
- 23.Tamura G. (2004) Histol. Histopathol. 19, 221–228 [DOI] [PubMed] [Google Scholar]
- 24.Wada M., Yazumi S., Takaishi S., Hasegawa K., Sawada M., Tanaka H., Ida H., Sakakura C., Ito K., Ito Y., Chiba T. (2004) Oncogene 23, 2401–2407 [DOI] [PubMed] [Google Scholar]
- 25.Xiao W. H., Liu W. W. (2004) World J. Gastroenterol. 10, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagawa N., Tamura G., Oizumi H., Takahashi N., Shimazaki Y., Motoyama T. (2003) Cancer Sci. 94, 589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balmain A. (2002) Nature 417, 235–237 [DOI] [PubMed] [Google Scholar]
- 28.Ito K., Liu Q., Salto-Tellez M., Yano T., Tada K., Ida H., Huang C., Shah N., Inoue M., Rajnakova A., Hiong K. C., Peh B. K., Han H. C., Ito T., Teh M., Yeoh K. G., Ito Y. (2005) Cancer Res. 65, 7743–7750 [DOI] [PubMed] [Google Scholar]
- 29.Lau Q. C., Raja E., Salto-Tellez M., Liu Q., Ito K., Inoue M., Putti T. C., Loh M., Ko T. K., Huang C., Bhalla K. N., Zhu T., Ito Y., Sukumar S. (2006) Cancer Res. 66, 6512–6520 [DOI] [PubMed] [Google Scholar]
- 30.Martin G. S. (2001) Nat. Rev. Mol. Cell Biol. 2, 467–475 [DOI] [PubMed] [Google Scholar]
- 31.Summy J. M., Gallick G. E. (2003) Cancer Metastasis Rev. 22, 337–358 [DOI] [PubMed] [Google Scholar]
- 32.Thomas S. M., Brugge J. S. (1997) Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 33.Yeatman T. J. (2004) Nat. Rev. Cancer 4, 470–480 [DOI] [PubMed] [Google Scholar]
- 34.Kopetz S., Shah A. N., Gallick G. E. (2007) Clin. Cancer Res. 13, 7232–7236 [DOI] [PubMed] [Google Scholar]
- 35.Chu I., Sun J., Arnaout A., Kahn H., Hanna W., Narod S., Sun P., Tan C. K., Hengst L., Slingerland J. (2007) Cell 128, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fornerod M., Ohno M., Yoshida M., Mattaj I. W. (1997) Cell 90, 1051–1060 [DOI] [PubMed] [Google Scholar]
- 37.Kudo N., Wolff B., Sekimoto T., Schreiner E. P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. (1998) Exp. Cell Res. 242, 540–547 [DOI] [PubMed] [Google Scholar]
- 38.Blain S. W., Massagué J. (2002) Nat. Med. 8, 1076–1078 [DOI] [PubMed] [Google Scholar]
- 39.Boyd S. D., Tsai K. Y., Jacks T. (2000) Nat. Cell Biol. 2, 563–568 [DOI] [PubMed] [Google Scholar]
- 40.Geyer R. K., Yu Z. K., Maki C. G. (2000) Nat. Cell Biol. 2, 569–573 [DOI] [PubMed] [Google Scholar]
- 41.Inman G. J., Nicolás F. J., Hill C. S. (2002) Mol. Cell 10, 283–294 [DOI] [PubMed] [Google Scholar]
- 42.Kastan M. B., Zambetti G. P. (2003) Cell 112, 1–2 [DOI] [PubMed] [Google Scholar]
- 43.Rosin-Arbesfeld R., Townsley F., Bienz M. (2000) Nature 406, 1009–1012 [DOI] [PubMed] [Google Scholar]
- 44.Xu L., Massagué J. (2004) Nat. Rev. Mol. Cell Biol. 5, 209–219 [DOI] [PubMed] [Google Scholar]
- 45.Pockwinse S. M., Rajgopal A., Young D. W., Mujeeb K. A., Nickerson J., Javed A., Redick S., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S., Doxsey S. J. (2006) J. Cell Physiol. 206, 354–362 [DOI] [PubMed] [Google Scholar]
- 46.Kim H. R., Oh B. C., Choi J. K., Bae S. C. (2008) J. Cell Biochem. 105, 1048–1058 [DOI] [PubMed] [Google Scholar]
- 47.Kaplan K. B., Bibbins K. B., Swedlow J. R., Arnaud M., Morgan D. O., Varmus H. E. (1994) EMBO J. 13, 4745–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biscardi J. S., Belsches A. P., Parsons S. J. (1998) Mol. Carcinog. 21, 261–272 [DOI] [PubMed] [Google Scholar]
- 49.Kim H. P., Lee M. S., Yu J., Park J. A., Jong H. S., Kim T. Y., Lee J. W., Bang Y. J. (2004) Biochem. J. 379, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah N., Pang B., Yeoh K. G., Thorn S., Chen C. S., Lilly M. B., Salto-Tellez M. (2008) Eur. J. Cancer 44, 2144–2151 [DOI] [PubMed] [Google Scholar]
- 51.Pande S., Ali S. A., Dowdy C., Zaidi S. K., Ito K., Ito Y., Montecino M. A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2009) J. Cell Physiol. 218, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]