Abstract
Type II plasmid partition systems utilize ParM NTPases in coordination with a centromere-binding protein called ParR to mediate accurate DNA segregation, a process critical for plasmid retention. The Staphylococcus aureus pSK41 plasmid is a medically important plasmid that confers resistance to multiple antibiotics, disinfectants, and antiseptics. In the first step of partition, the pSK41 ParR binds its DNA centromere to form a superhelical partition complex that recruits ParM, which then mediates plasmid separation. pSK41 ParM is homologous to R1 ParM, a known actin homologue, suggesting that it may also form filaments to drive partition. To gain insight into the partition function of ParM, we examined its ability to form filaments and determined the crystal structure of apoParM to 1.95 Å. The structure shows that pSK41 ParM belongs to the actin/Hsp70 superfamily. Unexpectedly, however, pSK41 ParM shows the strongest structural homology to the archaeal actin-like protein Thermoplasma acidophilum Ta0583, rather than its functional homologue, R1 ParM. Consistent with this divergence, we find that regions shown to be involved in R1 ParM filament formation are not important in formation of pSK41 ParM polymers. These data are also consonant with our finding that pSK41 ParM forms 1-start 10/4 helices very different from the 37/17 symmetry of R1 ParM. The polymerization kinetics of pSK41 ParM also differed from that of R1 ParM. These results indicate that type II NTPases utilize different polymeric structures to drive plasmid segregation.
Keywords: Chromosomes, Chromosomes/Centromeres, Chromosomes/Non-histone Proteins, Methods/Computer Modeling, Methods/X-ray Crystallography, Protein/Molecular Dynamics, Protein/Protein-Protein Interactions, Protein/Protein-Protein Interactions
Introduction
Partition or segregation is the essential process whereby the genetic material is actively distributed into daughter cells (1–3). Prokaryotic plasmid partition systems represent excellent model systems to study DNA segregation at a structural level because they require only three elements: a cis-acting centromere DNA site, an NTPase, and a centromere-binding protein (1–3). In the first step of segregation, multiple centromere-binding proteins bind cooperatively to the centromere to form a partition complex. The partition complex then recruits the NTPase protein, which ultimately drives the physical movement of plasmids to opposite cell poles (1–4). Plasmid-encoded partition (par) systems are typically divided into two main families based on the kind of NTPase present (3). Type I par systems contain NTPases with deviant Walker-A type folds, whereas the type II systems employ NTPases termed ParM (5–8). More recently, type III and IV partition systems have been identified. The type III systems use NTPases with putative tubulin-like folds, whereas the type IV systems carry out partition using a single, non-NTPase protein with a helix-turn-helix motif and coiled-coil domain (9–12).
How the partition proteins mediate the actual physical separation of plasmids has been a central question in partition. Studies on the type II R1 par system have provided key insights into this question. Specifically, experiments showing that the R1 ParM NTPase forms filamentous structures, led to the insertional polymerization model for partition (13–17). According to this model ParM filaments are first captured between paired partition complexes and continued filament growth, made possible by polymer stabilization, leads to plasmid movement to opposite cell poles (13). Remarkably, the R1 partition process can be reconstituted in vitro using only ParM, ParR, and the centromere, parC, which indicates that cellular factors are not required for the general separation mechanism (16). These studies also showed that the filaments grow by insertion of ParM-ATP molecules at the ParR-parC interface (16). Binding to ParR-parC stabilizes the filaments against catastrophic collapse as the filaments show dynamic instability (17). Structural studies showed that R1 ParM belongs to the actin/Hsp70 superfamily of ATPases, which not only includes actin and molecular chaperones but also FtsA, various sugar kinases, and the prokaryotic cell shape-determining protein, MreB (7, 18–20). All members of this superfamily share the same basic architecture (19), but the details vary and additional domains are often present. At the amino acid sequence level the degree of similarity within the family is low. Several actin homologues have also recently been identified in Archaea. The structure of one such archaeal actin-like protein, Ta0583 from Thermoplasma acidophilum, has been solved and it has been suggested that Ta0583 may be an ancestral actin (21, 22).
Recent work has started to shed light on the filamentous structures of actin family proteins (23–25). These studies have revealed the remarkable finding that although these actin-like proteins share similar overall folds, they often form very different filament structures that may be optimized for their specific cellular functions (23, 24, 26). For example, electron microscopy analyses on the R1 ParM filament ultimately showed that it forms a left-handed two-start helix, which is very different from the right-handed F-actin filament (24). More recent work on the DNA segregation protein AlfA has shown that although it forms two-start, left-handed filaments, the filaments adopt structures quite different from those of R1 ParM (25). In these highly twisted filaments, the protomer nucleotide binding pockets are rotated away from the filament axis and the ribbon-like helix is much more open than those of R1 ParM and F-actin. Thus, more work is needed to understand the types of filament structures adopted by actin-like proteins and how these filaments are optimized to carry out their specific cellular functions. In the case of type II partition, only the R1 ParM protein has been studied in any detail in terms of its structure and polymer dynamics and thus it is not clear if all such type II partition NTPases form similar filaments. Here we report studies on the type II partition ParM from the Staphylococcus aureus pSK41 plasmid partition system. pSK41 is representative of a clinically important family of self-transmissible S. aureus multiple resistance plasmids, which confers resistance to several antibiotics, disinfectants, and antiseptics (27).
The pSK41 par operon is similar to the well characterized R1 par system in that it encodes ParR and ParM proteins (28, 29). Our previous studies revealed how multiple pSK41 ParR proteins bind cooperatively to the parC centromere to form a partition complex and also showed that this complex recruits ParM via interactions with the ParR C-terminal domain (29). However, the role of the pSK41 ParM protein in mediating partition once it is recruited to the partition complex has not been characterized. To address these questions we determined the 1.95-Å resolution crystal structure of pSK41 ParM and compared it to other filament forming actin-like proteins. The structure reveals that pSK41 ParM belongs to the actin/Hsp70 superfamily, which includes its functional homologue, R1 ParM. However, the pSK41 ParM structure shows the strongest similarity to the archaeal T. acidophilum Ta0583 protein, which is thought to be involved in cell shape determination (21, 22). Furthermore, the regions shown to be involved in R1 ParM filament formation are not structurally conserved in pSK41 ParM. Mutagenesis experiments revealed that indeed, ParM does not use the same residues in filament formation as R1 ParM, consistent with our electron microscopy (EM)3 and total internal reflection fluorescence microscopy (TIRF) studies, which show that the filament structure and dynamics of the pSK41 ParM protein are very different from that of R1 ParM. These results point to unexpected divergence in the cytoskeletal structures employed by type II plasmid partitioning systems from evolutionarily and morphologically distinct bacterial hosts.
EXPERIMENTAL PROCEDURES
pSK41 ParM Purification and Crystallization
The pSK41 ParM overexpression plasmid, pSK9019, was constructed by cloning a 1-kb ParM PCR product into the pTTQ18-RGS6H vector, which places a C-terminal RGS6H tag on the protein. To obtain soluble ParM protein, the cells containing the overexpression plasmid were grown at 37 °C and induced for 6 h at 22 °C with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The protein was purified using nickel-nitrilotriacetic acid chromatography and buffer exchanged into 25 mm Tris, pH 7.5, 300 mm NaCl, 5% glycerol, and 1 mm β-mercaptoethanol. For crystallization, the protein was concentrated to 5 mg/ml. Crystals were obtained by hanging drop vapor diffusion at 298 K by mixing equal volumes of ParM and 1.5 m citrate, pH 5.6, which acts as a crystallization reagent. The crystals were monoclinic, P21, a = 53.4 Å, b = 79.2 Å, c = 54.6 Å, and β = 110.2° and contain one ParM monomer in the crystallographic asymmetric unit.
pSK41 ParM Structure Determination
Despite numerous attempts, the pSK41 ParM structure could not be solved by molecular replacement using R1 ParM coordinates. Hence, the structure was solved by the production and crystallization of selenomethionine-substituted pSK41 ParM protein. A multiple wavelength anomalous diffraction (MAD) three-wavelength data set was collected to 2.90-Å resolution and used to solve the structure (30). The selenomethionine sites were located using SOLVE and phases were calculated and density modification carried out using CNS (31, 32). The model was fit using O and refined in CNS (32, 33). The selenomethionine pSK41 ParM structure was refined using the 2.90-Å resolution selenomethionine data until convergence and the model used in molecular replacement with the high resolution 1.95-Å resolution native pSK41 ParM data (34). The 1.95-Å data were used for final refinement of the model, which resulted in an Rwork/Rfree of 19.5/24.6%. Refinement statistics are shown in Table 1. The pSK41 ParM structural topology is: β1, residues 4–12; β2, residues 15–20; β3, residues 23–28; β4, residues 52–55; β5, residues 63–66; α1, residues 70–75; α2, residues 89–106; β6, residues 111–119; α3, residues 126–138; β7, residues 140–145; β8, residues 148–154; β9, residues 157–161; α4, residues 162–171; α5, residues 179–183; β10, residues 185–192; β11, residues 195–205; β12, residues 211–213; α6, residues 218–229; α7, residues 240–246; α8, residues 264–284; β13, residues 282–298; α9, residues 301–311; α10, residues 324–342. SSM (SSM is a Protein structure comparison service SSM at the European Bioinformatics Institute (authors E. Krissinel and K. Hendrick)) was used to identify structural homologues of pSK41 ParM. In addition to root mean square deviation (r.m.s. deviation) similarities, the Z-score was used to judge similarity. The Z-score measures the statistical significance of a match in terms of Gaussian statistics. If the quality of a match may be considered a random value at random picking and if the structures are from a sufficiently large data base, it should obey the Gaussian distribution, with probability that the same or better quality match may be found, expressed as p = erfc(Z/2). The higher the Z-score, the higher the statistical significance of the match (35).
TABLE 1.
Crystallographic data for pSK41 apo-ParM
| Selenomethionine MAD data | |||
| Wavelength (λ) | 0.9797 | 1.0200 | 0.9796 |
| Resolution (Å) | 79.31-2.90 | 68.78-2.90 | 68.84-2.90 |
| Overall Rsym(%)a | 7.6 (14.6)b | 6.2 (14.5) | 7.6 (14.5) |
| Overall I/σ(I) | 10.9 (2.6) | 10.4 (2.3) | 10.4 (2.5) |
| Unique reflections | 10,875 | 10,879 | 10,890 |
| Multiplicity | 3.5 | 3.5 | 3.5 |
| Overall figure of meritc | 0.620 | ||
| Refinement statistics | |||
| Resolution (Å) | 74.53-1.95 | ||
| Overall Rsym (%)a | 6.2 (35.4) | ||
| Overall I/σ(I) | 20.4 (2.1) | ||
| Total reflections | 82,229 | ||
| Unique reflections | 24,845 | ||
| % Complete | 99.6 (99.6) | ||
| Rwork/Rfree (%)d | 19.5/24.6 | ||
| R.m.s. deviations | |||
| Bond angles (°) | 1.37 | ||
| Bond lengths (Å) | 0.009 | ||
| Ramachandran analysis | |||
| Most favored (%/#) | 88.9/240 | ||
| Additional allowed (%/No.) | 10.4/28 | ||
| Generously allowed (%/No.) | 0.7/2 | ||
| Disallowed (%/No.) | 0.0/0 | ||
a Rsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
b Values in parentheses are for the highest resolution shell.
c Figure of merit = 〈|ΣP(α)eiα/ΣP(α)|〉, where α is the phase and P(α) is the phase probability distribution.
d Rwork = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| and Rfree = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|; where all reflections belong to a test set of 5% randomly selected data.
EM Studies
ParM Mutant Studies
ParM filament formation was assayed by using EM. Specifically, negative stain EM studies were carried out using stock solutions of 5–10 μm ParM protein (either wild type or mutant), 1 mm AMP-PCP, and 5 mm MgCl2 in polymerization buffer (25 mm Tris, pH 7.5, 150 mm NaCl, and 1 mm dithiothreitol). Samples at various dilutions were applied to glow discharged, nickel-coated, copper grids, blotted, negatively stained with 1% uranyl acetate, air dried, and examined at the M.D. Anderson High Resolution Electron Microscopy Facility on a Jeol 1010 microscope operating at 80 kV.
Structural Analysis of pSK41 ParM Filaments
Wild type samples in Par buffer (80 mm KCl, 2 mm MgCl2, 25 mm Hepes, pH 7.4, 1 mm dithiothreitol) were applied to carbon-coated copper grids, blot stained with 1% uranyl acetate, and visualized under a Jeol JEM-2010 HC microscope operated at 100 keV at a nominal magnification of 10,000 to 40,000. Films were digitized with Photo Scan 2000 (Z/I Imaging) at 7-μm steps. Optical transforms and filtered images were calculated using the EM software package EOS (36). Otherwise, electron micrographs were scanned at 1200 disintegrations/min on an Epson scanner and displayed by Photoshop.
Total Internal Reflection Fluorescence Microscopy
TIRF experiments were conducted in a similar fashion as previously described (3, 24). pSK41 ParM was diluted to the desired concentration, using Par buffer in the presence of 10 mm dithiothreitol and crowding agents (1% methylcellulose or 8% polyvinyl alcohol). ATP or GTP (2 mm final concentration) were added, directly mixed on the glass slide, covered with a glass slip, and sealed with nail polish. For labeling, ParM was dialyzed against appropriate buffers. Proteins were briefly centrifuged at high speed before use. Lysines were labeled with Alexa 488 carboxylic acid succinimidyl ester 6-isomer or 5-(and-6) carboxyl tetramethylrhodamine succinimidyl ester at a molar ratio of about 1:4 (dye molecule:pSK41 ParM molecule) overnight at 4 °C as previously described (3, 24). Labeled pSK41 ParM filaments were observed with an inverted Nikon microscope as previously described (3, 24). Filament length was analyzed and error estimation determined using the programs Aquacosmos, ImageJ, and KaleidaGraph 3.6, as previously described (37).
Kinetics, P1 Release, and Critical Concentration Studies
pSK41 ParM polymerization was initiated by the addition of 2 mm nucleotide in Par buffer. The amount of pSK41 ParM polymer formed was monitored by 90 degree perpendicular light scattering using a Hitachi F-4500 fluorescence spectrophotometer at 300 nm and 24 °C. The data were fitted using the program Dynafit (38). k1 and k−1 describe concentration independent steps of the monomer activation when assembly is initiated by the addition of nucleotide, whereas k2 and k−2 are related to the dimerization step. All subsequent rate constant pairs are equal to ke and k−e and describe the steps of elongation. Without a dimerization step, the fit to the kinetic data were substantially worse.
The release of inorganic phosphate upon nucleotide hydrolysis during pSK41 ParM polymerization was measured at 24 °C using a phosphate assay kit (E-6646) from Molecular Probes (39). To determine the critical concentration of ParM for polymerization, various protein concentrations were tested. Reactions were equilibrated at 24 °C for 1 h. Ninety degree perpendicular light scattering experiments were carried out using a Hitachi F-4500 fluorescence spectrophotometer at 300 nm and 24 °C.
RESULTS AND DISCUSSION
Overall Structure of Apo-pSK41 ParM
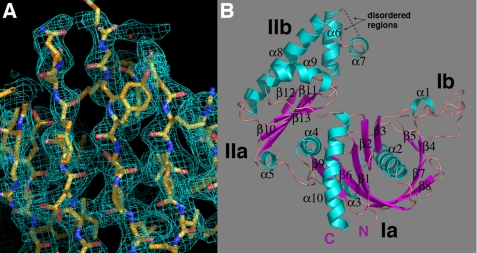
The 346-residue pSK41 ParM structure in its apo form was solved using MAD phasing techniques (Table 1 and Fig. 1A) (30, 31). Specifically, MAD data were collected on a selenomethionine-substituted ParM crystal and used to obtain the phases for structure determination. Once the structure was solved, a high resolution (1.95-Å resolution) native data set was used to carry out the final refinement, which resulted in an Rwork/Rfree of 19.5/24.6%. The final model includes ParM residues 2–230, 239–248, 262–343, and 294 water molecules. The structure shows that pSK41 ParM, which contains an α/β-fold consisting of 13 β-strands and 10 α-helices, is a member of the actin/Hsp70 superfamily of ATP-binding proteins (Fig. 1B).
FIGURE 1.
pSK41 ParM structure. A, representative region of the experimental MAD electron density map (blue mesh), contoured at 1.0 σ, for the selenomethionine apo-pSK41 ParM protein. The protein is shown as sticks with carbon, nitrogen, and oxygen colored yellow, blue, and red, respectively. This figure and B, Figs. 2, A and B, 3, A–C, and 4A were made using PyMOL (55). B, overall structure of the apo-pSK41 ParM protein. β-Strands and α-helices are colored magenta and cyan, respectively, and labeled. Also labeled are the N and C termini and subdomains of the molecule. The disordered regions are shown as dotted lines and their locations labeled.
pSK41 ParM, like other members of actin/Hsp70 superfamily, contains two domains, termed domains I and II, which are further subdivided into subdomains Ia, Ib and IIa, IIb (Fig. 1B). Characteristic of the actin/Hsp70 superfamily is the presence of two similar structural motifs in subdomains Ia and IIa, which are comprised of a 4–5-stranded β-sheet surrounded by three α-helices. Interestingly, construction of this motif in subdomain Ia requires crossover of an α helix, α10 in the pSK41 ParM structure, from domain II into domain I (Fig. 1B). It has been postulated that the actin-like fold evolved through gene duplication of this α/β motif and the acquisition of additional domains modulates the protein function, as members of this superfamily play many diverse roles from filament formation, glycolysis, and chaperone activity to DNA segregation. Movement between the two domains is mediated by a hinge region, which in pSK41 ParM is composed of α4 and α10. The pSK41 ParM nucleotide-binding site is predicted to be located in a cleft between the two domains based on structures of other actin/Hsp70 proteins in complex with nucleotides.
Structural Homology Searches Reveal Strong Homology with Archaeal Actin Ancestor
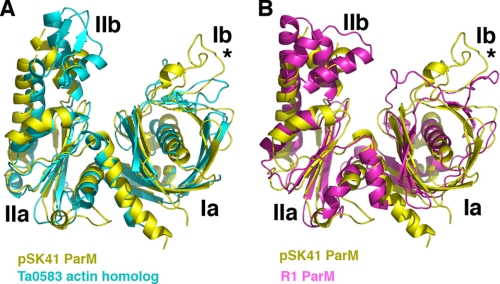
Besides pSK41 ParM, only one other structure of a ParM protein, that of R1 ParM, is available (6). Because both proteins function in plasmid DNA segregation, we anticipated that they would be structurally homologous. Interestingly, however, data base searches reveal that the pSK41 ParM structure shows the strongest structural homology to the archaeal T. acidophilum Ta0583 protein. pSK41 ParM shares 22% sequence identity with Ta0583, which although slightly higher than the sequence identity between the pSK41 and R1 ParM proteins (18%), is still in the “twilight zone” of protein sequence homology. However, the structural correspondence between the pSK41 ParM protein and Ta0583 is notable; the structures can be maximally superimposed (using 76 corresponding Cα atoms) with an r.m.s. deviation of 2.4 Å, resulting in a Z-score of 7.7 (Fig. 2A) (21). The Ta0583-ADP structure shows slightly lower structural homology and superimposes onto pSK41 ParM with an r.m.s. deviation of 2.6 Å for 71 corresponding Cα atoms and a Z-score of 5.1. By contrast, the R1 ParM-GMP-PNP structure (6, 24) can be superimposed onto the pSK41 ParM structure with an r.m.s. deviation of 4.0 Å for 74 corresponding Cα atoms, which provides a Z-score of only 3.1 (“Experimental Procedures”) (Fig. 2B).
FIGURE 2.
Comparison of pSK41 ParM, R1 ParM, and Ta0853 structures. A, superimposition of the pSK41 ParM structure (yellow) onto the T. acidophilum Ta0583 actin-like protein structure (cyan). The structures overlay with a r.m.s. deviation of 2.4 Å for 83 corresponding Cα atoms. B, superimposition of the pSK41 ParM structure (yellow) onto the R1 ParM structure (magenta). The structures overlay with a r.m.s. deviation of 4.0 Å for 74 corresponding Cα atoms. The asterisks in both panels underscore that the loop region in Ib is the most structurally divergent region in all three structures.
Examination of the superimpositions of pSK41 ParM with R1 ParM and Ta0583 reveals that region Ib is the most divergent between all the structures (Fig. 2, A and B). This region contains an extended loop in these proteins and in the pSK41 ParM structure it contains an extra, short helical segment, α1, which is not present in either Ta0583 or R1 ParM. The other major structural difference between pSK41 ParM and Ta0583 is in subdomain IIb. Residues 235 to 248 in this region in the Ta0583 structure forms a short antiparallel β-sheet, whereas in the pSK41 ParM structure the corresponding residues are disordered (Figs. 1B and 2A). Notably, in the R1 ParM structure, the region is helical. However, aside from these two areas in regions Ib and IIb, pSK41 ParM and Ta0583 show strong correspondence in their secondary structural elements. This is not the case in comparing the pSK41 and R1 ParM structures. In addition to structural differences in regions Ib and IIb, the proteins show marked divergence in much of domain I. Both the arrangement and length of the β-strands and loops in subdomains Ia and Ib are different in the two ParM structures. For example, β-strands 2 and 3 are much longer in the R1 ParM structure and as a result, extend from the core of the protein.
Rigid body rotations of domains I and II have been shown to accompany nucleotide binding in both R1 ParM and Ta0583. However, the magnitude of the structural changes is different. In R1 ParM, the domains rotate ∼25° relative to each other upon nucleotide binding, whereas in Ta0583 the rotation is much smaller, ∼8° (20–21). This structural change facilitates the optimal positioning of residues in the pocket to bind nucleotide. Interestingly, the apo structure of pSK41 ParM shows stronger similarity to the nucleotide bound form of R1 ParM rather than its apo form. This does not seem to be caused by molecules in the crystallization solution mimicking nucleotides, as no such molecules are observed in the binding pocket (supplemental Fig. S1A). As discussed below, we find by EM that pSK41 ParM readily form filaments upon ATP-Mg2+ addition (supplemental Fig. S1B). Thus, to address whether the crystallization solution stabilizes the closed, filamentous state in the absence of nucleotide, we carried out EM on pSK41 ParM in 1.5 m citrate, pH 5.6 (supplemental Fig. S1C). These studies clearly show that the crystallization solution does not force pSK41 ParM into the closed state. Indeed, the EM images are identical to those of the apoprotein.
Examination of the crystal packing shows that there are no crystal contacts to residues in or near the nucleotide binding pocket that might affect the protein conformation. The crystal contacts that are present are fairly equally distributed around the surface of the molecule and are made to both lobes (supplemental Fig. S2). It is possible that these contacts could somehow affect the closure state of pSK41 ParM, but it is not immediately obvious how. Indeed, our EM experiments support the contention that ParM adopts the apo state under these conditions, arguing against this idea. Thus, pSK41 ParM may be similar to Ta0583 in requiring little conformational change to bind nucleotides, but this remains to be determined.
R1 Filament Forming Residues Mapped onto pSK41 ParM: Implications for Polymer Formation
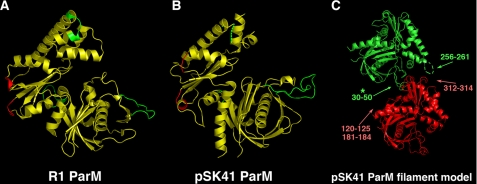
Recently, a three-dimensional model for the R1 ParM filament was deduced using the R1 ParM structure for image reconstruction (24). An important outcome of this work was the determination of the residues involved in intra-strand filament contacts. Three primary regions were found and include residues 33–44, 107–114, and 237–242. In addition, two shorter segments, residues 161–164 and residues 297–299, were also involved in intra-strand interactions. The corresponding regions in the pSK41 ParM protein were 30–50, 120–125, and 256–261 and the shorter segments correspond to residues 181–184 and 312–314.
Analysis of the R1 filament model reveals that the regions containing R1 ParM residues 33–44 and 107–114 provide the most numerous intra-strand contacts and thus, are crucial for filament formation. Interestingly, recent data shows that ParM filaments can contain a small percentage of “open” ParM molecules. Residues 33–44 were shown to be involved in filament formation in the open as well as closed forms of the polymer (40). Consistent with the central role played by these residues in filament formation, it was found that point mutation of residues 33, 34, 36, and 40 completely abrogate R1 ParM filament formation (14). Therefore, if pSK41 ParM forms polymers that are similar to the R1 ParM protein, then residues 30–50 should also be essential for its polymerization. However, when the regions known to be critical for filament formation in R1 are examined in the pSK41 structure, we find that they appear structurally different, in particular pSK41 ParM residues 30–50. As noted, this region forms an extended loop, which adopts very different conformations in the two proteins (Fig. 3, A and B). Indeed, in pSK41 ParM this loop contains an insertion and there are more residues present than in R1 ParM. When a pSK41 ParM filament is modeled based on that of the R1 ParM filament by overlaying the pSK41 structures onto the R1 structures in the polymer, we find that pSK41 ParM residues 30–50 clash with the next subunit within the strand (Fig. 3C). As was mentioned, this region in subdomain Ib also shows the most divergence between the pSK41 ParM and Ta0583 structures. The large differences in the filament forming regions of R1 ParM, pSK41 ParM, and Ta0583 suggest that these proteins may not form the same types of filamentous structures.
FIGURE 3.
Mapping of filament forming regions on ParM. A, regions involved in R1 ParM filament formation. The three key regions involved in intra-filament contacts are colored green and minor regions are colored red. B, mapping of the corresponding filament forming regions from R1 onto the pSK41 ParM structure. Key regions are green and minor regions are red as in A. C, pSK41 ParM filament model derived from the R1 ParM filament structure from Popp et al. (24). To produce the model, pSK41 structures were maximally docked onto the R1 structures. This figure underscores the large structural differences in the regions involved in filament contacts between R1 and pSK41 ParM proteins. Especially note that the large loop in pSK41 ParM subdomain Ib would clash with its interacting neighbor in such a filament, suggesting that pSK41 ParM might not form filaments similar in structure to R1 ParM (indicated by asterisk).
Mutagenesis Studies
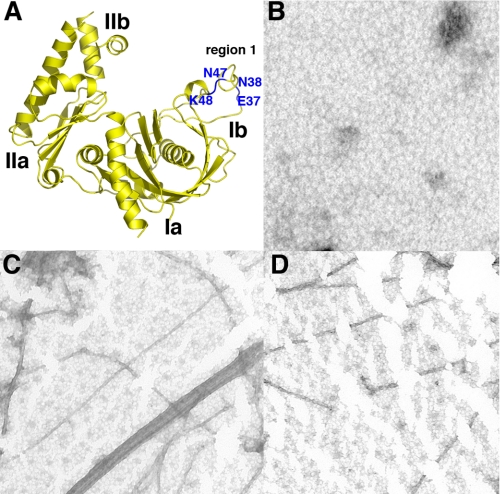
Although the R1 ParM and pSK41 ParM structures show significant differences in the key filament forming region (comprised of residues 33–44 in R1 ParM), it was still possible that this region is important in pSK41 ParM polymerization. For example, structural changes might occur during polymerization and/or nucleotide binding that could influence its structure and hence its ability to engage in polymer forming contacts. Indeed, it has been demonstrated that this region, region Ib, in actin adopts various conformations that are dependent on its nucleotide bound state (41–43). Thus, we next directly tested the importance of this region in pSK41 ParM filamentation. Because it has been shown that point mutations of residues in this region (residues 33, 34, 36, and 40) ablate filamentation by R1 ParM, we created corresponding point mutations in the pSK41 ParM protein and assayed the affect of these substitutions on the ability of ParM to polymerize. Specifically, we made four amino acid substitutions, E37I, N38D, N47T, and K48S (Fig. 4A).
FIGURE 4.
Mutagenesis of R1 filament forming regions in pSK41 ParM: pSK41 ParM forms filaments different from those of R1 ParM. A, location of the four residues (in blue and labeled) in region Ib of pSK41 ParM that were mutated to test their importance on filament formation. The corresponding residues (location wise) were shown to be essential for filament formation by R1 ParM (14). B, negative stain EM image of wild type pSK41 ParM in the absence of nucleotides. C, negative stain EM image of wild type pSK41 ParM in the presence of 1 mm AMP-PCP, 5 mm MgCl2. D, negative stain EM image of the quadruple mutant (E37I,N38D,N47T,K48S) pSK41 ParM in the presence of 1 mm AMP-PCP, 5 mm MgCl2.
pSK41 single mutant proteins, pSK41 ParM(E37I), ParM(N38D), ParM(N47T), and ParM(K48S), were made first. Subsequently, a protein containing all four substitutions, ParM(E37I,N38D,N47T,K48S) was constructed. All the mutant ParM proteins were expressed and purified similarly to the wild type protein. The proteins were then tested for their ability to form filaments using negative staining, electron microscopy techniques. In the absence of ATP or GTP the wild type protein did not form polymers (Fig. 4B). However, addition of ATP, GTP, or ATP analogs to the wild type protein resulted in the clear formation of filaments as visualized by negative stain EM (Fig. 4C). The proteins with single mutations, E37I, N38D, N47T, and K48S all formed filaments indistinguishable from those formed by the wild type protein. Notably, the protein containing all four mutations also formed filaments nearly identical to those produced by the wild type protein (Fig. 4D).
Filament Structures
Our structural and biochemical data indicate that pSK41 ParM filaments are different from those formed by R1 ParM. To examine the pSK41 ParM filament structure in detail we carried out EM studies. These experiments showed that the pSK41 ParM protein assembled preferentially into well ordered bundles, which were typically several micrometers long, in the presence of ATP, GTP, or the non-hydrolysable nucleotide, AMP-PNP (Fig. 5, A–C). Such filaments were not observed upon addition of ADP and GDP (Fig. 5D). Like R1 ParM, pSK41 ParM does not form filaments in the absence of divalent cations (supplemental Fig. S3, A and B). In addition to bundles, individual filaments (Fig. 4C) and paracrystalline nets were also observed (Fig. 6, A–E). The filaments were found to assemble under a wide range of ionic strengths (supplemental Fig. S3A). Optical diffraction patterns from bundles, in which individual filaments were arranged in parallel, showed a set of layer lines extending to 25 Å, which are typical for a helical structure. For pSK41 ParM-ATP the observed layer lines were spaced at 270, 130, 75, 58, 47, and 40 Å and a meridional reflection arising from the subunit spacing at 26 Å was observed (Fig. 7A). For pSK41 ParM-GTP and pSK41 ParM-AMP-PNP the set of reflections observed was similar: 240, 120, 73, 56, 39, and 25 Å for pSK41 ParM-GTP and 250, 120, 74, 57, and 39 Å for pSK41 ParM-AMP-PNP (Fig. 7, B and C). The slight differences in the observed layer line spacings may indicate a nucleotide-dependent variability in helical twist as observed for F-actin (44).
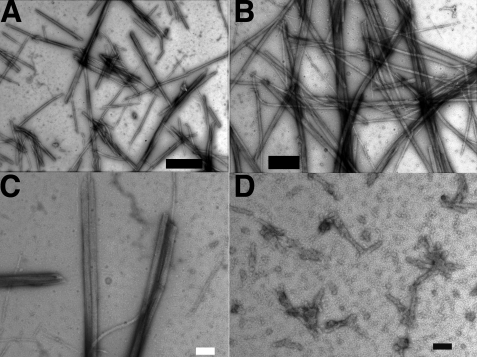
FIGURE 5.
pSK41 ParM filament structure. A, EM image of pSK41 ParM in the presence of 2 mm ATP showing the formation of filaments. B, EM image of the filaments formed by pSK41 ParM in the presence of 2 mm GTP. C, EM image of the filaments formed by pSK41 ParM in the presence of 2 mm AMP-PNP. D, EM image of pSK41 ParM in the presence of 2 mm ADP, scale bars, 200 nm. No filaments are observed with ADP.
FIGURE 6.
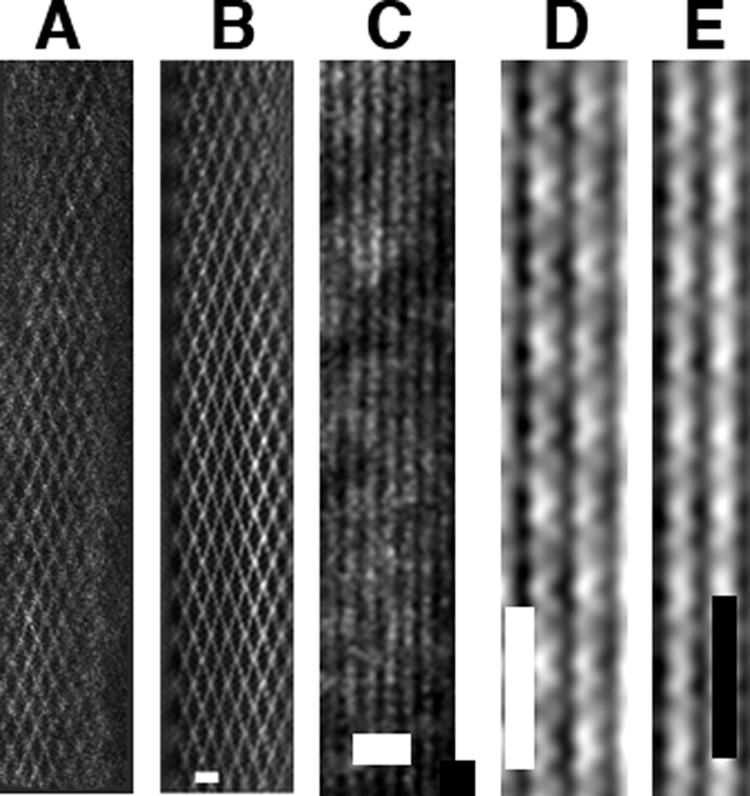
pSK41 ParM filamentous structures. pSK41 ParM forms single-stranded filaments and these filaments further associate into bundles and nets. A, a typical electron micrograph of a pSK41 ParM filament “net.” B, filtered image of the pSK41 ParM net, scale bar, 20 nm. C, typical electron micrograph image of bundles of pSK41 ParM filaments, scale bar, 20 nm. D, filtered image of a thin region of pSK41ParM-AMP-PNP bundles showing the presence of single helical filaments. E, filtered image of a thin region of pSK41ParM-GTP bundles showing single helical filaments, scale bar, 25 nm.
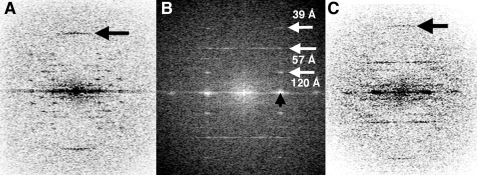
FIGURE 7.
Typical Fourier transforms of pSK41 ParM in complex with NTPs. A, Fourier transform of a pSK41 ParM-ATP bundle. The arrow indicates the meridional reflection at 26 Å. B, Fourier transform of the ParM-AMP-PNP bundle. The strongest layer lines in the transform are indicated by white arrows. The black arrow points out the equatorial reflection arising from interparticle spacing of the filament. C, Fourier transform of a pSK41 ParM-GTP bundle. Arrow points to the 25-Å meridional reflection.
The helical parameters of pSK41 ParM were determined from the Fourier transforms. In general, the symmetry of a helical structure can be defined in terms of three parameters, the subunit repeat (h), the repeat of the helix (C), and the radius of the helix (r). If one is dealing with a helix with S subunits in T turns of the helix, S equals C/h and T corresponds to a strong off-meridional reflection observed in the “first helix” cross (44, 45). One also expects to see reflections at T layer lines up and down from the meridional reflection in the “second helix” cross (44, 45). From both pSK41 ParM-ATP and pSK41 ParM-GTP we observed strong meridional reflections at ∼25 Å (Fig. 7, A and C), arising from the subunit repeat, h. The first layer line at about 250 Å arises from helical repeat C (Fig. 7A). pSK41 ParM filaments can therefore be described as 10/4 helices (10 subunits in 4 turns of the helix) with a repeat of about 250 Å. This assignment is consistent with the strong reflections at ∼57 Å (4th layer line), and on the 6th (10-4) layer line at ∼39 Å in the first helix cross (Fig. 7B).
The radius of pSK41 ParM filaments was ∼50 Å as measured from filtered images, where the bundle width was only a single layer thick (Fig. 6, D and E), consistent with the interfilament distance in bundles determined from the equatorial reflections of Fourier transforms (Fig. 7B). This radius appears smaller than the radii observed for F-actin or R1 ParM filaments, indicating that pSK41 ParM filaments may, in fact, be single-stranded (24, 46). Indeed, filtered images of thin bundles clearly showed that pSK41 ParM formed single-stranded helices unlike the two-start double helical polymers formed by R1 ParM (24).
Filament Dynamic Properties
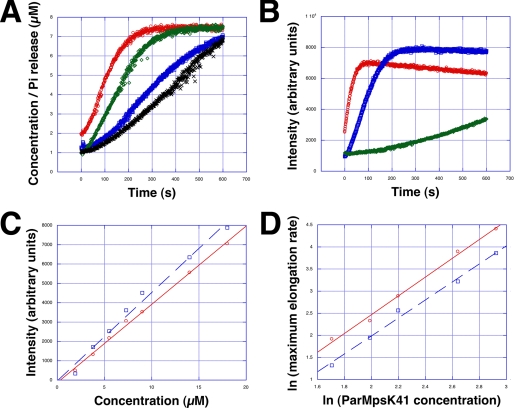
Next we investigated the polymerization kinetics of pSK41 ParM using light scattering. Assembly was substantially slower, ∼10 times, than R1 ParM and ∼3 times faster than F-actin under similar protein concentrations (24, 47). Polymerization kinetics were nucleotide dependent and fastest for ATP, slightly slower for GTP, although AMP-PNP formed polymers substantially slower (Fig. 8, A and B). At physiological protein concentrations of ∼7 μm, steady state was reached about 300 s after adding either ATP or GTP. Phosphate release lagged behind polymerization, as observed for R1 ParM and F-actin (Fig. 8A) (24, 47). The critical concentration of polymerization was about 0.4 μm and similar for both pSK41 ParM-ATP and pSK41 ParM-GTP (Fig. 8C). This value is comparable with the critical concentration of actin but lower than R1 ParM, which was ∼2 μm (16, 47, 48). Polymerization of F-actin required a nucleation step prior to elongation (49). In actin polymerization experiments, a lag phase prior to the intensity increase of light scattering was characteristic for the period of nucleation. In the case of pSK41 ParM no lag phase for pSK41 ParM-ATP or ParM-GTP was detected, whereas for pSK41 ParM-AMP-PNP, due to the slow kinetics, a considerable lag phase was observed (Fig. 8B). We therefore estimated if and how large the initial nucleus size may be from the slope of double logarithmic plots of the maximal rate of apparent elongation versus the protein concentration, as applied before for F-actin (49, 50). The slopes of pSK41 ParM-ATP and pSK41 ParM-GTP were similar and the size of the nucleus was calculated to be 2–2.2 consistent with a dimer nucleus. By contrast, the filament nucleus of actin is a trimer (47, 49, 50). Assembly kinetic data could be fit using a minimal kinetic model similar as described for the linear protofilaments of Escherichia coli FtsZ (51) involving three steps: monomer activation, dimer formation, and elongation (supplemental Fig. S4). The rate constants are summarized in Table 2.
FIGURE 8.
pSK41 ParM polymerization kinetics. A, relationship between the time course of pSK41 ParM polymerization and Pi release. Light scattering units were scaled to the concentration of pSK41 ParM (7.3 μm). Red, pSK41 ParM polymerization in the presence of 2 mm ATP. Green, pSK41 ParM polymerization in the presence of 2 mm GTP. Blue, Pi release from pSK41 ParM-ATP. Black, Pi release from pSK41 ParM-GTP. B, polymerization time courses at 18.6 μm pSK41 ParM. Red, ATP; blue, GTPl; green, AMP-PNP. C, critical concentration of pSK41 ParM. Red, ATP. Blue, GTP. Each data point is the average of three individual experiments. D, the maximal apparent elongation rates. The maximal rates were determined using the light scattering time courses at different pSK41 ParM concentrations as shown for the two examples in A and C. Each data point is the average of three individual experiments.
TABLE 2.
Rate constants for pSK41 ParM filament formation
| ATP | GTP | |
|---|---|---|
| k1 (s−1) | 0.04453 | 0.01973 |
| k−1 (s−1) | 0.04986 | 10−7 |
| k2 (μm−1 s−1) | 0.0621 | 0.0411 |
| k−2 (μm−1 s−1) | 0.0136 | 0.01014 |
| ke (μm−1 s−1) | 1.536 | 0.8597 |
| k−e (μm−1 s−1) | 0.09325 | 0.04852 |
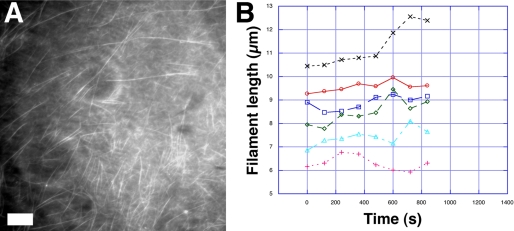
To investigate the dynamics of pSK41 ParM filaments at steady state, we analyzed fluorescently labeled pSK41 ParM filaments using TIRF microscopy (Fig. 9A). Labeled pSK41 ParM filaments were identical to those formed by unlabeled protein as ascertained from EM images and their Fourier transforms (supplemental Fig. S5, A and B). Interestingly, time lapse TIRF images revealed no significant dynamic instability for pSK41 ParM filaments (Fig. 9B). Indeed, pSK41 ParM filaments displayed a form of treadmilling more like F-actin rather than the marked dynamic instability exhibited by R1 ParM filaments.
FIGURE 9.
Steady state dynamics of pSK41 ParM filaments. A, typical TIRF microscopy image of labeled ParM filaments, scale bar, 5 μm. B, typical pSK41 ParM filament length fluctuations, as measured by TIRF at steady state.
ParM Filament Formation and Implications for Partition
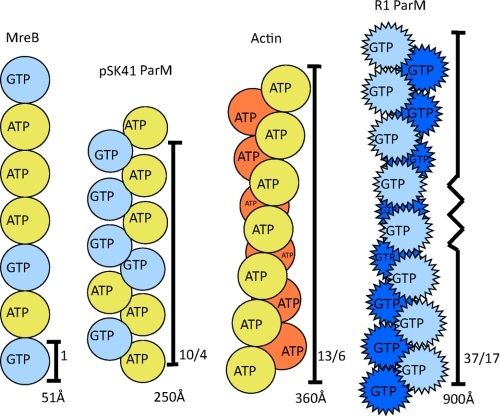
Our structural, mutagenesis, and filament data demonstrate that pSK41 ParM forms filaments that are distinct from those formed by R1 ParM. Specifically, pSK41 ParM forms helical filaments that contain a single strand. This differs significantly from the two-stranded helical filaments of F-actin (13/6) (3), R1 ParM (37/17) (46), and from the linear protofilaments of MreB (20) (Fig. 10). In addition, pSK41 ParM filaments are more stable than R1 ParM filaments, which display markedly dynamic instability (17, 24). These large distinctions between pSK41 and R1 filament structures and dynamics have important ramifications for the multistep partition processes mediated by these proteins.
FIGURE 10.
Actin-like filament systems. Schematic representations of MreB, pSK41 ParM, actin, and R1 ParM filaments. The pointed protomers of R1 ParM refer to the dynamic instability exhibited by its filaments. Notably, pSK41 ParM filaments do not display such dynamic instability.
The first step of partition is mediated by the centromere-binding protein, ParR. This protein binds multiple tandem repeats that comprise the DNA centromere site, giving rise to a superhelical partition complex. Our recent pSK41 ParR-centromere structure showed that the partition complex has a large diameter of ∼180 Å (29). We found that the highly acidic C-terminal domains of ParR, which are flexible, fill the central pore of the superhelix, which ranges from 50 to 90 Å. The structure of pB171 ParR, which also revealed that, at high protein concentrations the ParR protein alone forms a superhelix similar to that of pSK41 ParR-centromere complex (52), suggests that all type II ParR proteins form similar partition complex superstructures, which function to capture the requisite ParM filament. Indeed, the next step in partition is recruitment of the ParM filament to the paired partition complexes of two plasmids. In the case of R1, data indicate that the filament binds the center of the pore of each partition complex (14). ParR binding to ParM activates its NTPase activity. The next step is NTP hydrolysis of bound nucleotide by ParM. Once ParM hydrolyzes the NTP, it no longer binds ParR with high affinity. As a result the filament disengages briefly from the partition complex. The presence of a two-start helix in R1 ParM allows the second strand to remain bound, whereas the first has been disengaged allowing time for insertion of a new ParM-NTP. This continued binding, NTP hydrolysis, release, and ParM-NTP binding at the interface ultimately leads to growth of the filament between the bound partition complexes and hence separation of the plasmids bound at each end.
We have shown that several differences exist between pSK41 and R1 ParM filaments. Instead of a two-stranded ParM helix, the pSK41 ParM helix consists of a single strand, composed of ParM molecules ∼50 Å in diameter. This single-stranded helix must either bind to the partition complex pore as does the R1 filament or at the edges of the filaments that are undergoing treadmilling motion. The latter could ultimately move plasmids to distinct locations in the cell. However, the former model, binding at the center of the pore, is also plausible as, unlike the R1 ParM filaments, the pSK41 filaments appear more stable. Hence, after NTP hydrolysis, even if the pSK41 ParM filament is momentarily released from its contact from the partition complex, the polymer remains stable long enough to permit insertion of a fresh ParM-NTP subunit at the growing interface.
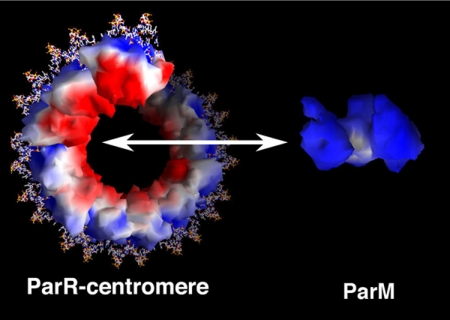
Examination of the electrostatic potential of ParM reveals that its outside surface faces or “sides” are quite positively charged (Fig. 11). This is interesting in light of the data showing that pSK41 ParM forms a single-stranded filament because it suggests that the sides of the filament would contain a positive charge, whereas the region of ParR known to bind and recruit ParM is highly negatively charged (Fig. 11). Hence, charge complementarity between the surfaces of ParR and ParM could play some role in binding between the ParM filament to the center of the ParR-centromere pore. Specifically, each end of a ParM filament could engage or interact with a partition complex. Alternatively, the partition complexes could bind on the outside surface of filament bundles undergoing treadmilling.
FIGURE 11.
Electrostatic surface potential of pSK41 partition proteins. Red and blue surfaces correspond to electronegative and electropositive regions, respectively. The ParR molecules in the partition complex are shown bound to the centromere as surfaces, the DNA as sticks. The electrostatic surface potential of the ParM molecule is also shown. The range for electrostatic potential is ±5 kT/e. This figure was made using GRASP (56).
In conclusion, our combined data show that despite their commonality of ancestry and function, the pSK41 and R1 type II plasmid segregation actin-like proteins have diverged to adopt distinct filament properties in disparate bacterial hosts. Existing models of type II segregation rely on filament assembly along the long axis of a rod-shaped cell as a basis for effective plasmid distribution into future daughter cells (53). However, such a simple explanation seems inadequate to account for efficient plasmid partition in spherical S. aureus cells. The extent to which such a fundamental difference in bacterial host morphology may have contributed to the evolution of the observed filament diversity is an open question at this time. Nonetheless, our study highlights and further strengthens the hypothesis that the only common features between all actin-like polymers are their ability to form lateral protofilament contacts (54).
Supplementary Material
Acknowledgments
We thank the Advanced Light source (A.L.S.) and support staff. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, Material Sciences Division of the United States Department of Energy under contract No. DE-AC03-76SF00098 at the Lawrence Berkeley National Laboratory. R. C. R. thanks the Biomedical Research Council of A*STAR for support.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI080687 (to M. A. S.), Australian Research Council Discovery Grant DP0346013 (to N. F. and R. A. S.), and National Health and Medical Research Council of Australia Project Grant 571028 (to N. F., M. A. S., and R. A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (code 3JS6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- EM
- electron microscopy
- MAD
- multiple wavelength anomalous diffraction
- AMP-PCP
- adenosine 5′-(β,γ-methylenetriphosphate)
- NTPase
- nucleoside 5′-triphosphatase
- Hsp70
- heat shock protein 70
- TIRF microscopy
- total internal reflection fluorescence microscopy
- r.m.s.
- root mean square.
REFERENCES
- 1.Schumacher M. A. (2008) Biochem. J. 412, 1–18 [DOI] [PubMed] [Google Scholar]
- 2.Hayes F., Barillà D. (2006) Nat. Rev. Microbiol. 4, 133–143 [DOI] [PubMed] [Google Scholar]
- 3.Gerdes K., Møller-Jensen J., Bugge Jensen R. (2000) Mol. Microbiol. 37, 455–466 [DOI] [PubMed] [Google Scholar]
- 4.Funnell B. E. (2005) Plasmid 53, 119–125 [DOI] [PubMed] [Google Scholar]
- 5.Dunham T. D., Xu W., Funnell B. E., Schumacher M. A. (2009) EMBO J. 28, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Ent F., Møller-Jensen J., Amos L. A., Gerdes K., Löwe J. (2002) EMBO J. 21, 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin E. V. (1993) J. Mol. Biol. 229, 1165–1174 [DOI] [PubMed] [Google Scholar]
- 8.Barillà D., Rosenberg M. F., Nobbmann U., Hayes F. (2005) EMBO J. 24, 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen R. A., Cusumano C., Fujioka A., Lim-Fong G., Patterson P., Pogliano J. (2007) Genes Dev. 21, 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M., Bideshi D. K., Park H. W., Federici B. A. (2007) J. Bacteriol. 189, 8053–8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tinsley E., Khan S. A. (2006) J. Bacteriol. 188, 2829–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson A. E., Skurray R. A., Firth N. (2003) J. Bacteriol. 185, 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Møller-Jensen J., Borch J., Dam M., Jensen R. B., Roepstorff P., Gerdes K. (2003) Mol. Cell 12, 1477–1487 [DOI] [PubMed] [Google Scholar]
- 14.Salje J., Löwe J. (2008) EMBO J. 27, 2230–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell C. S., Mullins R. D. (2007) J. Cell Biol. 179, 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. (2007) Science 315, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garner E. C., Campbell C. S., Mullins R. D. (2004) Science 306, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 18.Erickson H. P. (2007) Bioessays 29, 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch W., Holmes K. C. (1995) FASEB J. 9, 167–174 [DOI] [PubMed] [Google Scholar]
- 20.van den Ent F., Amos L. A., Löwe J. (2001) Nature 413, 39–44 [DOI] [PubMed] [Google Scholar]
- 21.Roeben A., Kofler C., Nagy I., Nickell S., Hartl F. U., Bracher A. (2006) J. Mol. Biol. 358, 145–156 [DOI] [PubMed] [Google Scholar]
- 22.Hara F., Yamashiro K., Nemoto N., Ohta Y., Yokobori S., Yasunaga T., Hisanaga S., Yamagishi A. (2007) J. Bacteriol. 189, 2039–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlova A., Garner E. C., Galkin V. E., Heuser J., Mullins R. D., Egelman E. H. (2007) Nat. Struct. Mol. Biol. 14, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popp D., Narita A., Oda T., Fujisawa T., Matsuo H., Nitanai Y., Iwasa M., Maeda K., Onishi H., Maéda Y. (2008) EMBO J. 27, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polka J. K., Kollman J. M., Agard D. A., Mullins R. D. (2009) J. Bacteriol. 191, 6219–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rommelaere H., Waterschoot D., Neirynck K., Vandekerckhove J., Ampe C. (2003) Structure 11, 1279–1289 [DOI] [PubMed] [Google Scholar]
- 27.Firth N., Skurray R. A. (2006) in Gram-Positive Pathogens (Fischetti V. A., Novick R. P., Ferretti J. J., Portnoy D. A., Rood J. I. eds) 2nd Ed., pp. 413–426, American Society for Microbiology, Washington, DC [Google Scholar]
- 28.Dam M., Gerdes K. (1994) J. Mol. Biol. 236, 1289–1298 [DOI] [PubMed] [Google Scholar]
- 29.Schumacher M. A., Glover T. C., Brzoska A. J., Jensen S. O., Dunham T. D., Skurray R. A., Firth N. (2007) Nature 450, 1268–1271 [DOI] [PubMed] [Google Scholar]
- 30.Hendrickson W. A. (1991) Science 254, 51–58 [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 33.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 34.Kissinger C. R., Gehlhaar D. K., Fogel D. B. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 484–491 [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E., Henrick K. (2004) Acta Crystallogr. D 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 36.Yasunaga T., Wakabayashi T. (1996) J. Struct. Biol. 116, 155–160 [DOI] [PubMed] [Google Scholar]
- 37.Popp D., Gov N. S., Iwasa M., Maéda Y. (2008) Biopolymers 89, 711–721 [DOI] [PubMed] [Google Scholar]
- 38.Kuzmic P. (1996) Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 39.Webb M. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galkin V. E., Orlova A., Rivera C., Mullins R. D., Egelman E. H. (2009) Structure 17, 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belmont L. D., Orlova A., Drubin D. G., Egelman E. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otterbein L. R., Graceffa P., Dominguez R. (2001) Science 293, 708–711 [DOI] [PubMed] [Google Scholar]
- 43.Sablin E. P., Dawson J. F., VanLoock M. S., Spudich J. A., Egelman E. H., Fletterick R. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10945–10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egelman E. H., Francis N., DeRosier D. J. (1982) Nature 298, 131–135 [DOI] [PubMed] [Google Scholar]
- 45.Holmes K. C., Blow D. M. (1965) Methods Biochem. Anal. 13, 113–239 [DOI] [PubMed] [Google Scholar]
- 46.Holmes K. C., Popp D., Gebhard W., Kabsch W. (1990) Nature 347, 44–49 [DOI] [PubMed] [Google Scholar]
- 47.Iwasa M., Maeda K., Narita A., Maéda Y., Oda T. (2008) J. Biol. Chem. 283, 21045–21053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popp D., Yamamoto A., Iwasa M., Narita A., Maeda K., Maéda Y. (2007) Biochem. Biophys. Res. Commun. 353, 109–114 [DOI] [PubMed] [Google Scholar]
- 49.Cooper J. A., Buhle E. L., Jr., Walker S. B., Tsong T. Y., Pollard T. D. (1983) Biochemistry 22, 2193–2202 [DOI] [PubMed] [Google Scholar]
- 50.Nishida E., Sakai H. (1983) J. Biochem. 93, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Bjornson K., Redick S. D., Erickson H. P. (2005) Biophys. J. 88, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Møller-Jensen J., Ringgaard S., Mercogliano C. P., Gerdes K., Löwe J. (2007) EMBO J. 26, 4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebersbach G., Gerdes K. (2005) Annu. Rev. Genet. 39, 453–479 [DOI] [PubMed] [Google Scholar]
- 54.Löwe J., Amos L. A. (2009) Int. J. Biochem. Cell Biol. 41, 323–329 [DOI] [PubMed] [Google Scholar]
- 55.Delano W. L. (2002) The PyMOL, Delano Scientific, San Carlos, CA [Google Scholar]
- 56.Nicholls A., Sharp K. A., Honig B. (1991) Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.