Abstract
The novel ginkgolide analog ginkgolide X was characterized functionally at human glycine and γ-aminobutyric acid type A receptors (GlyRs and GABAARs, respectively) in the fluorescence-based FLIPRTM Membrane Potential assay. The compound inhibited the signaling of all GABAAR subtypes included in the study with high nanomolar/low micromolar IC50 values, except the ρ1 receptor at which it was a significantly weaker antagonist. Ginkgolide X also displayed high nanomolar/low micromolar IC50 values at the homomeric α1 and α2 GlyRs, whereas it was inactive at the heteromeric α1β and α2β subtypes at concentrations up to 300 μm. Thus, the functional properties of the compound were significantly different from those of the naturally occurring ginkgolides A, B, C, J, and M but similar to those of picrotoxin. In a mutagenesis study the 6′ M2 residues in the GlyR ion channel were identified as the primary molecular determinant of the selectivity profile of ginkgolide X, and a 6′ M2 ring consisting of five Thr residues was found to be of key importance for its activity at the GABAAR. Conformational analysis and docking of low-energy conformations of the native ginkgolide A and ginkgolide X into a α1 GlyR homology model revealed two distinct putative binding sites formed by the 6′ M2 residues together with the 2′ residues and the 10′ and 13′ residues, respectively. Thus, we propose that the distinct functionalities of ginkgolide X compared with the other ginkgolides could arise from different flexibility and thus different binding modes to the ion channel of the anionic Cys-loop receptor.
Keywords: Cell Surface Receptor, GABA Receptors, Membrane Proteins, Receptor Structure-Function, Site-directed Mutagenesis, GABA(a) Receptors, Ginkgolide, Glycine Receptors, Ligand-gated Ion Channels, Picrotoxin
Introduction
γ-Aminobutyric acid (GABA)2 and glycine are the predominant inhibitory neurotransmitters in the central nervous system and also maintain important functions in several peripheral tissues (1–5). The ionotropic GABAA and glycine receptors (GABAARs and GlyRs) belong to the Cys-loop receptor superfamily, which also comprises nicotinic acetylcholine receptors (nAChRs) and 5-HT3 receptors (5-HT3Rs) (3–9). The Cys-loop receptors are homomeric or heteromeric assemblies of five subunits, and the pentameric receptor complex consists of three domains: an extracellular domain composed of the N-terminal domains of the five subunits, a transmembrane domain formed by the M1–M4 α-helices of the five subunits (including an ion channel predominantly formed by the five M2 helices), and an intracellular domain composed primarily of the large second intracellular loops of the five subunits (4, 7). Signal transduction through the Cys-loop receptor is initiated by binding of the agonist to orthosteric sites situated at the interfaces between the N-terminal domains of the subunits, and this elicits a conformation change in the pentameric complex leading to flux of ions through the ion channel. Whereas nAChRs and 5-HT3Rs are excitatory Cys-loop receptors mediating the flux of Na+, K+, and Ca2+, GABAARs and GlyRs are inhibitory anionic channels primarily permeable to Cl− ions (4, 6–9).
Native GABAARs are a highly heterogenous population composed of numerous receptor subtypes assembled from a total of 19 subunits (10). The receptors mediate a wide array of important physiological functions and are validated drug targets in the treatment of numerous disorders (10–14). The GlyRs in the spinal cord and brain stem are of key importance for motor functions and sensory signaling in vision and audition (1, 5, 15). The GlyRs are formed from the α1–α4 and β subunits, and both homomeric α and heteromeric αβ subtypes exist (1, 5, 15). To be able to elucidate the composition of the native GABAAR and GlyR populations and delineate the physiological roles governed by specific receptor subtypes, there is a profound need for identification and development of selective ligands targeting one or a subset of subtypes.
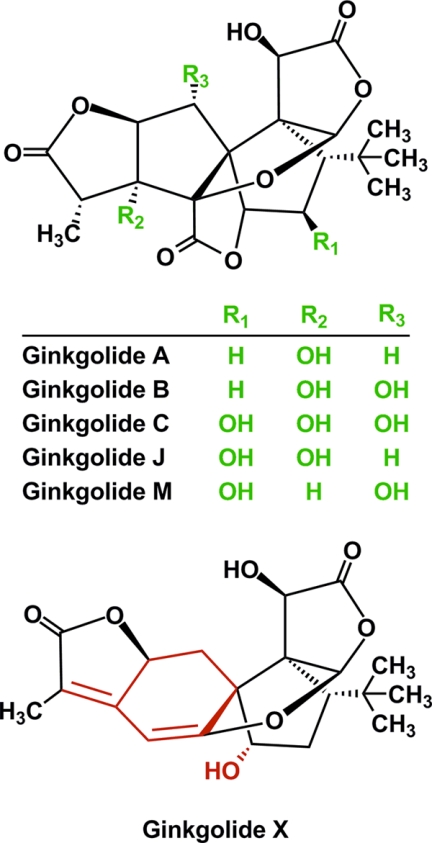
Ginkgolides, natural products from Ginkgo biloba, are diterpenes with a cage structure constituted by 6 five-membered rings and a tert-butyl group (16, 17). The structures of the five naturally occurring ginkgolides A, B, C, J, and M (collectively referred to as “the native ginkgolides”) vary only when it comes to the number and positions of the hydroxyl groups in the molecules (Fig. 1). The ginkgolides are noncompetitive antagonists of GlyRs and GABAARs (18–24). Ginkgolides B, C, and J have exhibited preference for heteromeric over homomeric GlyRs in studies of cloned and native receptors (18, 19, 21). Thus, the compounds display the opposite pharmacological profile compared with that of another natural product, picrotoxin (an equimolar mixture of picrotoxinin and picrotin) (4, 25, 26). In contrast to picrotoxin, however, the native ginkgolides cannot be claimed to be truly selective against some GlyR subtypes over others, as IC50α1/IC50α1β and IC50α2/IC50α2β ratios determined in some studies have been 3–7 and 10–26, respectively, and because other studies have not observed significant differences between their antagonist potencies at different GlyR subtypes (18, 19, 21, 24). The pharmacological properties of the ginkgolides have not been characterized at many recombinant GABAAR subtypes, and thus their selectivity profiles at these receptors are unknown (22, 23).
FIGURE 1.
Chemical structures of native ginkgolides A, B, C, J, and M and ginkgolide X. The structural differences between the native ginkgolides and ginkgolide X are indicated with the color red in the ginkgolide X molecule.
The native ginkgolide has been proposed to bind to the bottom half of the GlyR ion channel, a binding site that overlaps with that of picrotoxin. Both picrotoxin and the native ginkgolides have been shown to form interactions with the 6′ residues in the M2 helices lining the ion channel pore (18–20, 25, 27, 28). Furthermore, the 2′ M2 residue, located one helix turn below, has been proposed to be involved in the binding of picrotoxin (25, 28) and to be involved in the coordination of the ginkgolides to the heteromeric α1β GlyR but not to the homomeric GlyR (19, 20). In structure-activity relationship studies of ginkgolide analogs, the GlyR antagonist activity of the ginkgolide has been demonstrated to be very dependent on its rigid structure, and modifications of the hydroxyl groups in the molecule have been found to have detrimental effects on its activity (24, 29).
Ginkgolide X is a novel ginkgolide analog with a structure distinct from those of the five native ginkgolides (Fig. 1) (30). In a previous study, small amounts of ginkgolide X was isolated from considerable amounts of waste products from the production of the G. biloba extract EGb 761 (30).3 In this study we have characterized the functional properties of ginkgolide X at GlyRs and GABAARs and investigated the molecular basis for its activity at these receptors.
EXPERIMENTAL PROCEDURES
Materials
Culture medium, serum, antibiotics, and buffers for cell culture were obtained from Invitrogen. Glycine, GABA, Ach, and serotonin were purchased from Sigma, and picrotoxin, genistein, and epibatidine were obtained from Tocris Cookson (Bristol, UK). The ginkgolide X sample was a generous gift from Dr. Willmar Schwabe Arzneimittel GmbH & Co. KG (Karlsruhe, Germany). The cDNAs encoding for the human α, β, and γ2s GABAAR subunits were kind gifts from Dr. P. J. Whiting and Merck, Sharp and Dohme (Harlow, Essex, UK) and the human ρ1 cDNA was obtained from Dr. D. S. Weiss. The cDNAs for the human GlyR subunits were obtained from Drs. P. R. Schofield (α1 and β) and H. Betz (α2), and the cDNAs for the human α7 nAChR and human Ric-3 were kind gifts from Drs. J. Lindstrom and N. S. Millar, respectively. Finally, the stable cell lines expressing rat α3β4 nAChR, mouse α4β2 nAChR, and human 5-HT3AR were kind gifts from Drs. Y. Xiao and K. J. Kellar, J. A. Stitzel, and J. Egebjerg, respectively (31, 32).
Molecular Biology
The subcloning of α1, α2, and β GlyR cDNAs into pcDNA3.1 and α7 nAChR cDNA into pCI-neo has been described previously (26, 33, 34). The cDNAs for the GABAAR subunits were subcloned into the pcDNA3.1 vector from their original vectors using PCR and subsequent digestion using the unique restriction enzymes NotI and XbaI for β2 and β3 and XbaI and XhoI for α1–α5 and γ2s. The mutations introduced in various plasmids were made using the QuikChange mutagenesis kit according to the manufacturer's instructions (Stratagene, La Jolla, CA). The absence of unwanted mutations in all cDNAs created by PCR was verified by DNA sequencing (Eurofins MWG Operon, Martinsried, Germany).
Cell Culture and Transfections
The tsA-201 cells used for the transient transfections were grown in cell culture medium (Dulbecco's modified Eagle's medium supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% fetal bovine serum). The stable HEK293 cell lines expressing human 5-HT3AR, rat α3β4 nAChR, and mouse α4β2 nAChR were grown in cell culture medium supplemented with 1 mg/ml of G418 (5-HT3AR and α3β4 nAChR) or with 0.5 mg/ml of hygromycin B and 0.1 mg/ml of zeocin (α4β2 nAChR).
For the transient transfections, 8 × 105 tsA-201 cells were split into a 6-cm tissue culture plate and transfected the following day with a total of 4 or 5 μg of cDNA using Polyfect (Qiagen, Hilden, Germany). In the GlyR experiments, cells were transfected with 2 μg of α-subunit cDNA and 2 μg of pCDNA3.1 vector or with 1 μg of α-subunit cDNA and 3 μg of β-pCDNA3.1. In the GABAAR experiments, cells were transfected with 1 μg of α-subunit cDNA and 1 μg of β-subunit cDNA together with 3 μg of pCDNA3.1 vector or 3 μg of γ2s-pCDNA3.1. In the α7 nAChR experiments, cells were transfected with 2 μg of hα7-pCI-neo and 2 μg of hRic3-pRK5. 16–24 h after the transfection, the cells were split into 96-well plates (see below), they were assayed 36–48 h after the transfection.
FMP Blue Assay
Cells were split into poly-d-lysine-coated black 96-well plates with clear bottoms (BD Biosciences). 16–24 h later the medium was aspirated and the cells washed with 100 μl of Krebs buffer (140 mm NaCl, 4.7 mm KCl, 2.5 mm CaCl2, 1.2 mm MgCl2, 11 mm HEPES, 10 mm d-glucose, pH 7.4). 50 μl of Krebs buffer was added to the wells (in the antagonist experiments, various concentrations of the antagonist were dissolved in the buffer) and then an additional 50 μl of Krebs buffer supplemented with the FMP Blue assay dye (1 mg/ml) was added to each well. Then the plate was incubated at 37 °C in a humidified 5% CO2 incubator for 30 min and assayed in a NOVOstarTM plate reader (BMG Labtechnologies, Offenburg, Germany) measuring emission (in fluorescence units) at 560 nm caused by excitation at 530 nm before and up to 1 min after addition of 33 μl of agonist solution. The experiments were performed in duplicate at least three times for each compound at each WT and mutant receptor. Glycine, GABA, and serotonin were used as agonists for GlyRs, GABAARs, and 5-HT3AR, respectively. ACh was used as agonist for the α7 nAChR, whereas epibatidine was used as agonist for the α4β2 and α3β4 nAChRs. EC70–EC95 concentrations of the respective agonists were used at the various WT and mutant receptors. The experiments with α7/Ric3-transfected cells were performed in the presence of 100 μm genistein in the assay buffer.
Concentration-response curves for agonists and concentration-inhibition curve antagonists obtained in the FMP Blue assay were constructed based in the difference in the fluorescence units (ΔFU) between the maximal fluorescence recording made before and after addition of agonist obtained for different concentrations of the respective ligands. The curves were generated by nonweighted least-squares fits using the program KaleidaGraph 3.6 (Synergy Software).
Conformational Analysis
The conformational space of ginkgolide X, picrotin, picrotoxinin, and ginkgolide A was sampled using a high temperature quenched molecular dynamics approach with Impact version 50207 (35) as the molecular dynamics engine. Each compound was simulated for 1 ns using the OPLS2005 force field, 0.001-ps time steps, GB/SA implicit solvent model, and a target temperature of 1000 K. Every fifth frame was extracted and subsequently energy minimized using the multiple minimization protocol in MacroModel version 96207 (36) (MMFF94s force field, GB/SA solvent model) with elimination of redundant conformers and keeping only low energy conformations less than 3 kcal/mol above the global energy minimum. The procedure was repeated 5 times using different starting geometries to ensure complete sampling.
Homology Modeling
A homology model of the transmembrane domain (M1–M4) of the α1 GlyR homopentamer was constructed using the 4-Å resolution electron microscopy structure of the nAChR from Torpedo marmorata as template (Protein Data Bank code 2bg9) (37, 38). Initially, a structure-based sequence alignment of the five template subunits (two α1, one β, one γ, and one δ) was established using the structural alignment feature of PyMOL (39). Amino acid sequences for all known human Cys-loop receptor subtypes (GABAAR, GlyR, nAChR, and 5-HT3R) were retrieved from the UniProt data base (62). With ClustalX (40) a “profile alignment” of these sequences to the previously established template alignment was performed. A profile alignment keeps the original (template) alignment fixed while aligning the new sequences to it one by one, thus preserving the structural alignment. The final sequence alignment of M1–M4 of the α1 GlyR subunit to the template is given in supplemental Fig. S1. Note that the long intracellular M3–M4 loop was excluded due to missing structural information of this segment.
Using the program MODELLER (41), 120 models were built and subsequently evaluated using the built-in molpdf scoring functions. The best scoring model was inspected visually to ensure the model was as expected, and a Ramachandran plot confirmed the overall quality (five violations, neither in M2 nor close to the putative ginkgolide binding site). After performing a standard protein preparation (protonation, tautomerization, and geometry optimization) in Maestro version 8.5.207 (42), the resulting α1 GlyR model was used for subsequent docking studies. Additionally, a standard PASS analysis (43) was performed on the model to visualize the size and shape of the putative ginkgolide and picrotoxin binding site.
Induced Fit Docking
All unique ring conformers of ginkgolide X, picrotonin, picrotoxinin, and ginkgolide A were docked using the induced fit docking protocol (44, 61, 63) implemented in Maestro version 85207. Compounds were initially docked to the receptor using scaling factors of 0.70 for both ligand and receptor generating 20 poses for each unique ring conformation of the ligands. Side chains of the M2 helices (−1′ to 14′) were then sampled and minimized and the compounds were finally docked to the optimized receptor using scaling factors of 1.00 and 0.80 for receptor and ligand, respectively. The obtained poses were scored using the Glide-XP scoring function and the IFD-score (Glide G-score + 0.05 × prime energy).
RESULTS
Functional Characterization of Ginkgolide X at Cys-loop Receptors
The functional properties of ginkgolide X were characterized at a wide range of Cys-loop receptors in the FMP Blue assay. Cys-loop receptor signaling has been investigated in this assay in numerous previous studies, including references (26, 34, 45–47). In supplemental Fig. S2, sample experimental data from the assay are given for anionic and a cationic Cys-loop receptor, the α2 GlyR, and the 5-HT3AR, respectively.
GABAARs
Several human GABAAR subtypes made up from the ρ1 subunit, from α and β subunits or from α, β, and γ2s subunits were expressed in tsA-201 cells by transient transfection of the respective subunit cDNAs, and the functional properties of GABA and ginkgolide X were determined at these receptors in the FMP Blue assay (Table 1 and Fig. 2). Because transfection of cells with α and β subunits gives rise to functional GABAARs, the incorporation of the γ2s subunit in pentamers formed at the cell surface of cells transfected with α, β, and γ2s subunits was verified in two different ways. First, the receptors formed in these cells were shown to be sensitive to allosteric potentiation by diazepam, which is known to bind to the α/γ interface of the αβγ GABAAR complex (data not shown) (3, 6). Second, cells transfected with α5, β2, and γ2sT6′F cDNAs form functional receptors insensitive to picrotoxin and ginkgolide X, a characteristic not shared by WT α5β2 nor WT α5β2γ2s receptors (see below). This further suggests that at least the majority of receptors formed at the surface of cells transfected with α, β, and γ2s cDNAs actually do contain the γ2s subunit.
TABLE 1.
Functional properties of ginkgolide X at various Cys-loop receptors in the FMP Blue assay
Stable cell lines expressing rat α3β4 nAChR, mouse α4β2 nAChR, and human 5-HT3AR, and tsA-201 cells transiently transfected with cDNAs encoding human GlyRs, human GABAARs, and the human α7 nAChR (co-expressed with human Ric-3) were used for the experiments. EC50 values for agonists and IC50 values for ginkgolide X are given in micromolar with pEC50 ± S.E. and pIC50 ± S.E. values in parentheses, respectively. The experiments were performed as described under “Experimental Procedures,” and the data are based on three to five individual experiments.
| Receptor | Transf. ratio | Agonist EC50a | Ginkgolide X IC50b |
|---|---|---|---|
| GlyRs | |||
| α1 | 1α:1 vector | 92 (4.04 ± 0.03) | 0.76 (6.12 ± 0.05) |
| α1β | 1α:3β | 64 (4.19 ± 0.03) | >300 (<3.5) |
| α2 | 1α:1 vector | 83 (4.08 ± 0.03) | 2.8 (5.56 ± 0.05) |
| α2β | 1α:3β | 95 (4.02 ± 0.04) | >300 (<3.5) |
| GABAARs | |||
| α1β2 | 1α:1β:3 vector | 7.4 (5.13 ± 0.05) | 1.1 (5.97 ± 0.04) |
| α1β2γ2s | 1α:1β:3γ | 42 (4.37 ± 0.05) | 2.7 (5.57 ± 0.01) |
| α2β2γ2s | 1α:1β:3γ | 7.2 (5.14 ± 0.06) | 0.71 (6.15 ± 0.04) |
| α2β3γ2s | 1α:1β:3γ | 3.5 (5.46 ± 0.02) | 3.3 (5.48 ± 0.05) |
| α3β2γ2s | 1α:1β:3γ | 9.7 (5.01 ± 0.06) | 3.9 (5.41 ± 0.05) |
| α3β3γ2s | 1α:1β:3γ | 4.6 (5.33 ± 0.05) | 6.1 (5.21 ± 0.04) |
| α4β2 | 1α:1β:3 vector | 1.2 (5.93 ± 0.02) | 0.60 (6.22 ± 0.05) |
| α5β2γ2s | 1α:1β:3γ | 0.72 (6.14 ± 0.06) | 2.2 (5.65 ± 0.02) |
| ρ1 | 0.64 (6.19 ± 0.05) | ∼100 (∼4.0) | |
| nAChRs | |||
| α7/Ric-3 | 1:1 | 9.1 (5.05 ± 0.04) | >300 (<3.5) |
| α4β2 | 0.015 (7.82 ± 0.04) | >300 (<3.5) | |
| α3β4 | 0.031 (7.51 ± 0.05) | >300 (<3.5) | |
| 5-HT3Rs | |||
| 5-HT3A | 0.32 (6.49 ± 0.07) | >300 (<3.5) | |
a Glycine, GABA, and serotonin were used as agonists for GlyRs, GABAARs, and 5-HT3AR, respectively. ACh was used as agonist for the α7 nAChR, whereas epibatidine was used as agonist for the α4β2 and α3β4 nAChRs.
b In the ginkgolide X experiments, assay concentrations of 200 μm glycine, EC70–EC95 concentrations of GABA, and 1 μm serotonin were used as agonists for the GlyRs, GABAARs, and 5-HT3AR, respectively. 20 μm ACh and 50 nm epibatidine were used as agonist concentrations for α7 nAChR and α4β2 and α3β4 nAChRs, respectively.
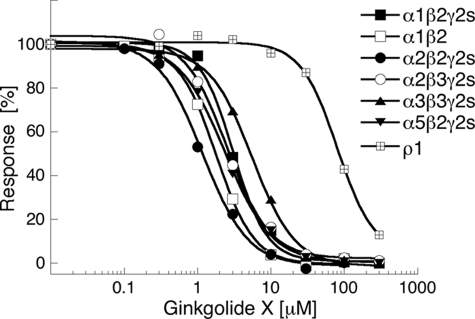
FIGURE 2.
Concentration-inhibition curves of ginkgolide X at tsA-201 cells transiently transfected with selected human GABAARs in the FMP Blue assay. The FMP Blue assay was performed as described under “Experimental Procedures” using EC70–EC95 concentrations of GABA as final agonist concentrations in the assays. The figures depict data from single representative experiments, and error bars are omitted for reasons of clarity.
For the characterization of ginkgolide X at GABAARs in the FMP Blue assay, EC70–EC95 concentrations of GABA were used as the agonist concentration at the respective subtypes. Ginkgolide X displayed IC50 values in the high nanomolar/low micromolar range at all heteromeric GABAARs, whereas its antagonist potency was significantly lower at the homomeric ρ1 receptor (Fig. 2 and Table 1).
GlyRs
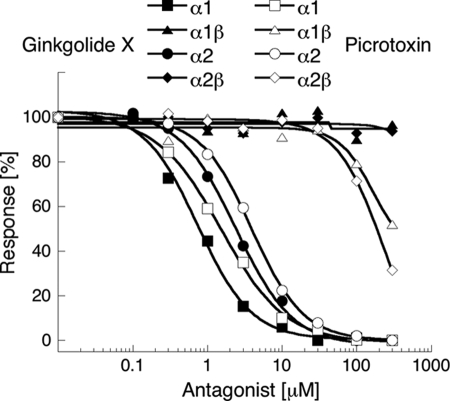
For characterization of ginkgolide X at human GlyRs in the FMP Blue assay, 200 μm glycine was used as agonist, corresponding to EC70–EC95 concentrations at the different receptors (Table 1). Ginkgolide X displayed high nanomolar/low micromolar IC50 values as antagonist of the homomeric α1 and α2 GlyRs (Fig. 3 and Table 1). In contrast, the compound was completely inactive at the heteromeric α1β and α2β subtypes at concentrations up to 300 μm (Fig. 3 and Table 1). Thus, ginkgolide X displayed a selectivity profile at GlyRs similar to that of picrotoxin (4). In the FMP Blue assay, picrotoxin displayed IC50 values of 2.9 μm (pIC50 ± S.E.: 5.54 ± 0.04, n = 3) and 2.4 μm (pIC50 ± S.E.: 5.62 ± 0.05, n = 3) at α1 GlyR and α2 GlyR, respectively, and IC50 values of ∼300 μm at the α1β and α2β subtypes (n = 3 for both). Thus, with IC50α1β/IC50α1 and IC50α2β/IC50α2 ratios of >400 and >100, ginkgolide X exhibited an even higher selectivity for homomeric over heteromeric GlyRs than picrotoxin with its IC50α1β/IC50α1 and IC50α2β/IC50α2 ratios of ∼140 and ∼130 (Table 1).
FIGURE 3.
Concentration-inhibition curves of ginkgolide X and picrotoxin at tsA-201 cells transiently transfected with human GlyRs in the FMP Blue assay. The FMP Blue assay was performed as described under “Experimental Procedures” using 200 μm glycine as the final agonist concentration in the assay. This figures depict data from a single representative experiment, and error bars are omitted for reasons of clarity.
nAChRs and 5-HT3AR
The functional properties of ginkgolide X were also characterized at four cationic Cys-loop receptors: the three major neuronal nAChR subtypes α4β2, α3β4, and α7, and the 5-HT3AR. The compound was inactive at these receptors at concentrations up to 300 μm (Table 1).
The Molecular Basis for Subtype Selectivity of Ginkgolide X at GlyRs
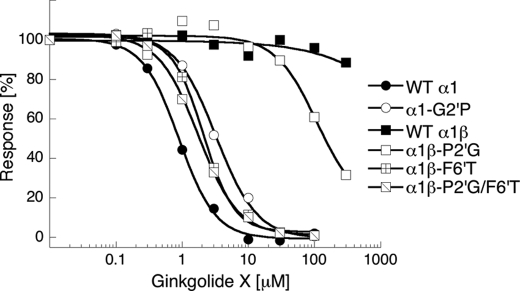
The 6′ residue in the bottom half of the M2 α-helices forming the ion channel of the GlyR have been shown to be involved in binding of native ginkgolides as well as picrotoxin and its two components picrotin and picrotoxinin (19, 20, 25, 27, 28). The 6′ residue is highly conserved through the Cys-loop receptor family, being a Thr or a Ser residue in almost all subunits (Fig. 4). A notable exception is the β GlyR subunit, which contains a Phe residue in this position (Fig. 4). To investigate the role of this residue for ginkgolide X and picrotoxin inhibition of GlyR signaling, the F6′ residue in the β GlyR subunit (Phe282) was mutated to the corresponding Thr residue in α1 and the reverse T6′F mutation (Thr258 → Phe) was introduced in the α1 subunit, and the activities of the two antagonists at GlyRs incorporating these mutant subunits were characterized in the FMP Blue assay.
FIGURE 4.
Alignment of the amino acid sequences of the M2 regions in selected Cys-loop receptor subunits. The positions of the 2′, 6′, 10′, and 13′ residues are indicated, and the residues are highlighted in bold in the individual subunits. Human, mouse, and rat subunits are indicated by the prefixes h, m, and r, respectively.
Mutation of the F6′ residue in the β subunit to Thr converted the inactivity of ginkgolide X at the WT α1β GlyR to an inhibitory activity very similar to that displayed by the compound at the homomeric WT α1 GlyR (Table 2 and Fig. 5). In agreement with a previous study (25), introduction of the F6′T mutation into the β subunit also converted the weak antagonist activity of picrotoxin at the WT α1β to almost the same antagonist activity as it displayed at the WT α1 GlyR (Table 2).
TABLE 2.
Functional properties of ginkgolide X and picrotoxin at WT and mutant GlyRs transiently expressed in tsA-201 cells in the FMP Blue assay
For the antagonist experiments, EC70–EC95 concentrations of glycine at different WT and mutant receptors were used as agonist. The experiments were performed as described under “Experimental Procedures,” and the data are based on three to nine individual experiments.
| Receptor | Transfection ratio | Glycine | Ginkgolide X | Picrotoxin |
|---|---|---|---|---|
| EC50 (pEC50 ± S.E.) | IC50 (pIC50 ± S.E.) | IC50 (pIC50 ± S.E.) | ||
| WT α1 | 1α:1 vector | 99 (4.00 ± 0.03) | 1.4 (5.84 ± 0.02) | 3.4 (5.46 ± 0.04) |
| WT α1β | 1α:3β | 87 (4.06 ± 0.04) | >300 (<3.5) | ∼300 (∼3.5) |
| 6′ M2 residue mutants | ||||
| α1/α1T6′F | 1 α:1 αT6′F:2 vector | 92 (4.04 ± 0.05) | >300 (<3.5) | >300 (<3.5) |
| α1βF6′T | 1α:3β | 88 (4.06 ± 0.04) | 3.0 (5.52 ± 0.03) | 11 (4.95 ± 0.05) |
| α1βF6′G | 1α:3β | 102 (3.99 ± 0.07) | ∼100 (∼4.0) | ∼100 (∼4.0) |
| α1T6′FβF6′T | 1α:3β | 110 (3.95 ± 0.05) | >300 (<3.5) | >300 (<3.5) |
| 2′ M2 residue mutants | ||||
| α1G2′P | 1α:1 vector | 210 (3.68 ± 0.06) | 4.1 (5.39 ± 0.05) | 1.4 (5.85 ± 0.05) |
| α1βP2′G | 1α:3β | 160 (3.80 ± 0.05) | ∼100 (∼4) | ∼300 (∼3.5) |
| α1βP2′G/F6′T | 1α:3β | 150 (3.82 ± 0.05) | 3.2 (5.49 ± 0.05) | 7.2 (5.14 ± 0.05) |
| 10′ and 13′ M2 residue mutants | ||||
| α1T10′S | 1α:1 vector | NDa,b | ND | ND |
| α1T13′S | 1α:1 vector | NDa | ND | ND |
| α1T10′S/T13′S | 1α:1 vector | NDa | ND | ND |
| α1βS10′T | 1α:3β | 79 (4.10 ± 0.05) | >300 (<3.5) | ∼300 (∼3.5) |
| α1βS13′T | 1α:3β | 91 (4.04 ± 0.05) | >300 (<3.5) | >300 (<3.5) |
| α1βS10′T/S13′T | 1α:3β | 93 (4.03 ± 0.06) | >300 (<3.5) | ∼300 (∼3.5) |
| α1βF6′T/S10′T | 1α:3β | 110 (3.98 ± 0.06) | 2.5 (5.59 ± 0.05) | 7.3 (5.14 ± 0.07) |
| α1βF6′T/S10′T/S13′T | 1α:3β | 110 (3.95 ± 0.05) | 1.2 (5.92 ± 0.05) | 5.2 (5.28 ± 0.04) |
| α1T10′Sβ | 1α:3β | NDa | ND | ND |
| α1T13′Sβ | 1α:3β | NDa | ND | ND |
| α1T10′S/T13′Sβ | 1α:3β | NDa | ND | ND |
| α1T10′SβS10′T | 1α:3β | NDa | ND | ND |
| α1T13′SβS13′T | 1α:3β | NDa | ND | ND |
| α1T10′S/T13′SβS10′T/S13′T | 1α:3β | NDa | ND | ND |
a Significantly elevated basal levels of fluorescence were observed compared to WT GlyR expressing cells.
b ND, not determined.
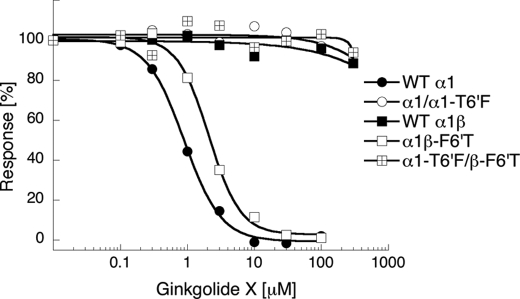
FIGURE 5.
The importance of the 6′ M2 residue for the activity of ginkgolide X at the GlyRs. Concentration-inhibition curves of ginkgolide X at tsA-201 cells transfected with cDNAs for WTα1, WTα1 and α1T6′F, WTα1 and WTβ, WTα1 and βF6′T, or α1T6′F and βF6′T GlyR subunits. The FMP Blue assay was performed as described under “Experimental Procedures” using EC70–EC95 concentrations of glycine as final agonist concentrations in the assays. The figure depicts data from a single representative experiment, and error bars are omitted for reasons of clarity.
The IC50 values obtained for ginkgolide X and picrotoxin at the α1βF6′T GlyR were slightly higher than those at the WT α1 GlyR. Still, it was important to verify that the “α1-like” profiles of the antagonists at α1βF6′T-expressing cells did not simply arise from the mutant β subunit being trapped inside the cells. Hence, we also characterized the functional properties of the antagonists at cells co-expressing α1 and βF6′G subunits. Although the antagonist activities of ginkgolide X and picrotoxin were slightly higher at these cells than at WT α1β-expressing cells, the antagonists displayed 30–100-fold lower antagonist activities at α1βF6′G than at the homomeric α1 GlyR (Table 2). Because there is no reason to expect the ability of the β subunit to reach the cell surface (when co-expressed with α1) to be more compromised by a F6′T than a F6′G mutation, this strongly suggests that the receptor being expressed at the surface of α1βF6′T-expressing cells is indeed the heteromeric receptor.
As for the studies of the effects of the T6′F mutation in α1, only a weak response to glycine application could be measured in α1T6′F-transfected cells in the FMP Blue assay. In previous studies, α1T6′F GlyRs have displayed 4–8-fold lower Imax values for glycine than the WT α1 in electrophysiological recordings (19, 25). Thus, the low signals induced by glycine in α1T6′F-transfected cells may be ascribed to lower sensitivity of the FMP Blue assay compared with the conventional patch-clamp technology. However, in tsA-201 cells co-transfected with WT α1 and α1T6′F subunits (in a 1:1 ratio) a significant glycine-induced response could be measured in the FMP Blue assay. Assuming that the cell surface expression levels of the WT and mutant α1 subunits in these cells are similar and that the ability of the α1 subunit to assemble into pentameric GlyRs is not influenced by the T6′F mutation, the α1:α1T6′F stoichiometry of the majority of the receptor complexes in these cells will be 2:3 or 3:2. Thus, the pseudo-homomeric GlyRs formed in α1/α1T6′F-transfected cells actually are the best representation of the subunit composition of the heteromeric WT α1β GlyR when it comes to the 6′ residue arrangement, although the fixed subunit arrangement of the WT α1β GlyR obviously will not be mirrored in all receptors formed in these cells. The GlyRs in α1/α1T6′F-transfected cells were completely insensitive to both ginkgolide X and picrotoxin at concentrations up to 300 μm (Table 2 and Fig. 5). Finally, co-transfection of α1T6′F and βF6′T subunits also resulted in formation of functional GlyRs (Table 2). The fact that co-expression of the “functionally dead” α1T6′F mutant with βF6′T results in a functional receptor is another indication that the βF6′T mutant is being incorporated into heteromeric α1β complexes expressed on the cell surface (in the presence of an α1 subunit). The α1T6′FβF6′TGlyR was completely insensitive to ginkgolide X and picrotoxin at concentrations up to 300 μm (Fig. 5 and Table 2).
Comparison of Low Energy Conformations of Ginkgolide X, Ginkgolide A, and Picrotoxinin
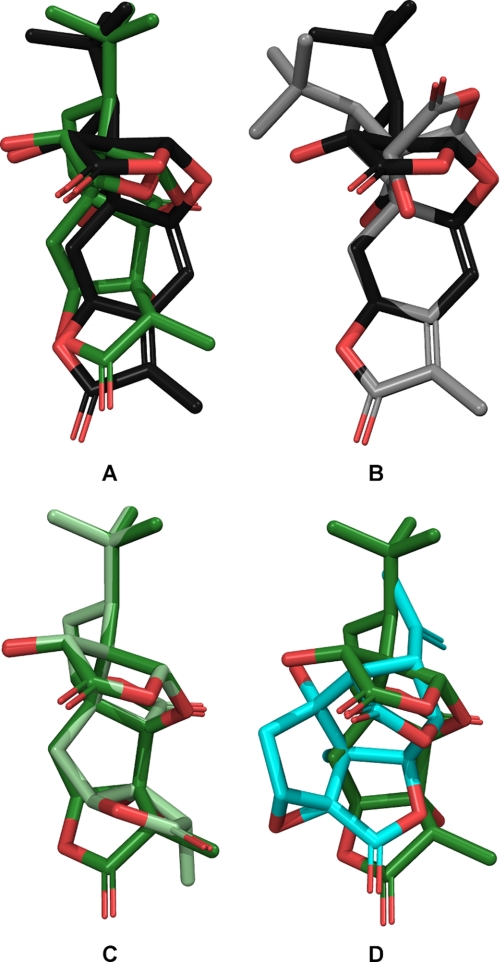
Interestingly, whereas ginkgolide X clearly is a selective antagonist of homomeric over heteromeric GlyRs, ginkgolide A and the other native ginkgolides have been reported to be either non-selective antagonists at the various GlyRs or even to display some preference for the heteromeric subtypes (18, 19, 21, 24). Superimposing ginkgolide A and ginkgolide X in their global energy minima using conserved hetero atoms as fitting points results in a very similar overlay (Fig. 6A). The conformational flexibility of the core ring structures of ginkgolide A and ginkgolide X within a 3 kcal/mol cut-off, considered to be the limit for high affinity binding (48), is shown in Fig. 6, B and C, respectively. For ginkgolide X the only alternative low energy ring conformer (+1.9 kcal/mol) originates from flexibility in the t-butyl end of the molecule, whereas the only alternative ring conformer for ginkgolide A stem from flexibility in the opposite 3-methyldihydrofuran-2-one end of the molecule (+0.8 kcal/mol). In contrast, picrotin and picrotoxinin are completely rigid in their ring frameworks within the 3 kcal/mol cut-off and the only flexibility for these molecules is due to rotation around freely rotatable bonds. The superimposition of picrotoxinin on ginkgolide A is shown in Fig. 6D.
FIGURE 6.
Comparison of low-energy conformations of ginkgolide A, ginkgolide X, and picrotoxinin. A, ginkgolide X (dark gray) superimposed on ginkgolide A (green), both in their lowest energy ring conformations. B, low energy conformations of ginkgolide X. Global minimum (dark gray), low energy conformation (+1.9 kcal/mol) (light gray). C, low energy conformations of ginkgolide A. Global minimum (green), low energy conformation (+0.8 kcal/mol) (pale green). D, picrotoxinin (cyan) superimposed on ginkgolide A (green) in its lowest energy conformation. The figure is prepared with PyMOL 1.2 (39).
Binding Modes of Ginkgolide A, Ginkgolide X, and Picrotoxinin to the α1 GlyR Homology Model
Having identified the 6′ M2 residue as the selectivity determinant in the GlyRs for the actions of ginkgolide X, we set out to elucidate the binding modes of this compound and picrotoxin to the ion channels of the receptors. To investigate the observed differences between the native ginkgolides and ginkgolide X with respect to flexibility and its potential role for their different selectivity profiles at GlyRs we also included ginkgolide A in this study.
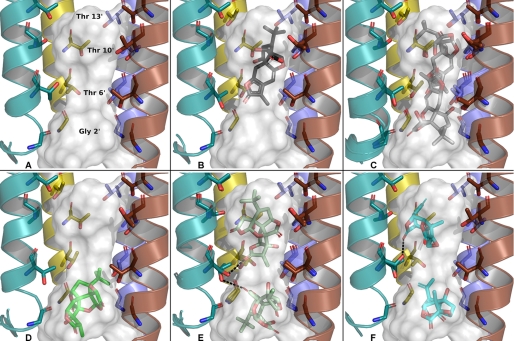
Ginkgolide X, ginkgolide A, and picrotoxinin were docked to the homology model of the transmembrane domain (M1–M4) of the homomeric α1 GlyR using an induced fit protocol that allows protein side chains to adapt to the docked poses of the ligands. Furthermore, to ensure that all relevant conformations of the core ring structures of the ligands were presented to the receptor, both identified low energy ring conformers of ginkgolides A and X were used as input to the docking program. The docking poses were clearly clustered in the two pockets above and below the 6′ residues as illustrated in the homology model (Fig. 7A). Furthermore, poses were clustered according to input conformation, indicating that the docking engine could not handle conformational sampling of the complex ring structures of the ginkgolides.
FIGURE 7.
Top scoring poses of ginkgolide X, ginkgolide A, and picrotoxinin when docked to the α1 GlyR homology model. Transmembrane (TM) helices 1, 3, 4, and one of the TM2 helices are removed for clarity. A, homology model of the transmembrane region of the α1 GlyR, showing the putative binding site in the lower part of the ion channel defined by M2 helices. The shape and size of the cavity is illustrated by the transparent gray surface generated with PASS. The rings of the important 2′, 6′, 10′, and 13′ M2 residues discussed in the text are indicated. B, top scoring pose of the lowest energy ring conformer of ginkgolide X. C, top scoring poses of ginkgolide X in its alternative (+1.9 kcal/mol) conformation in the 6′–13′ and 2′–6′ regions. D, top scoring pose of ginkgolide A in it lowest energy conformation. E, top scoring poses of ginkgolide A in its alternative ring conformation (+0.8 kcal/mol) in the 6′–13′ and 2′–6′ regions (H-bond dashes indicate which T6′ conformation belongs to which pose). F, top scoring poses of picrotoxinin in the 2′–6′ and 6′–13′ region. The figure is prepared with PyMOL 1.2 (39).
For ginkgolide X in its lowest energy ring conformer the cluster of poses was situated consistently with the 3-methylfuran-(2)-one ring facing the threonines in the 6′ position. The top scoring pose of ginkgolide X from this cluster of poses is shown in Fig. 7B. The carbonyl group of the 3-methylfuran-(2)-one ring forms a direct hydrogen bond with one of the 6′ threonines and the bulky part of the molecule (the three joined 5-membered rings) is situated at the level of the 10′ threonines, whereas the t-butyl group resides between the 10′ and 13′ M2 residues. Apart from hydrogen bonds directly to the 6′ and 10′ Thr residues a number of water-mediated hydrogen bonds must almost certainly exist. In the alternative ring conformation of ginkgolide X (+1.9 kcal/mol) two different clusters of poses were obtained. Both clusters have the 3-methylfuran-(2)-one ring placed at the level of the 6′ threonines but the two clusters protrude from either the 10′ or the 2′ side as shown in Fig. 7C.
The suggested binding poses for ginkgolide A were also very dependent on the ring conformation. In the lowest energy ring conformation poses in the 2′–6′ region were obtained, and in contrast to ginkgolide X these poses approach the 6′ position with the t-butyl group (Fig. 7D). In the alternative ring conformation (+0.8 kcal/mol) ginkgolide A assumes many different binding poses. The top scoring of these in the 6′ to 13′ region resemble the top scoring pose of ginkgolide X but is situated ∼1.5 Å higher in the ion channel. In the 2′ region poses are perpendicular to the axis through the ion channel with the t-butyl group facing the G2′ residues and with a hydrophilic side of the ligand facing the 6′ residues (Fig. 7E). Interestingly, a number of low scoring poses with an inverted binding mode were also observed (not shown) for both clusters where the hydrophobic t-butyl group is presented to the residues in the 6′ position. For picrotoxinin the highest scoring poses (Fig. 7F, upper pose) were obtained in the 6′–10′ region but a unique binding mode could not be established. Poses in the 2′ region were also obtained (Fig. 7F, lower pose), however, with significantly lower scores as compared with poses in the 6′–10′ region. The scoring values and corresponding docking regions for the different top scoring poses are given in Table 3.
TABLE 3.
Docking scores from ginkgolide A, ginkgolide X, and picrotoxinin to the α1 GlyR homology model
| Ligand | Input conformationa | Glide-XP score | IFD score | Docking region | Fig. |
|---|---|---|---|---|---|
| Ginkgolide X | 0.0 | −8.13 | −959.39 | 6′-10′-13′ | 7B |
| Ginkgolide X | +1.9 | −7.52 | −958.00 | 6′-10′-13′ | 7C (top) |
| Ginkgolide X | +1.9 | −8.17 | −957.84 | 2′-6′ | 7C (bottom) |
| Ginkgolide A | 0.0 | −7.91 | −956.12 | 2′-6′ | 7D |
| Ginkgolide A | +0.8 | −7.65 | −955.83 | 6′-10′-13′ | 7E (top) |
| Ginkgolide A | +0.8 | −6.04 | −954.55 | 2′-6′ | 7E (bottom) |
| Picrotoxinin | 0.0 | −6.16 | −956.79 | 6′-10′ | 7F (top) |
| Picrotoxinin | 0.0 | −5.08 | −955.18 | 2′-6′ | 7F (bottom) |
a Conformational energy (MMFFS94/GBSA) of input conformation.
The Roles of 2′, 10′, and 13′ M2 Residues in Ginkgolide X and Picrotoxin Binding to the GlyR
The 2′ M2 Residue
The 2′ M2 residue is a glycine or an alanine in the four α GlyR subunits, whereas it is a Pro residue in the β subunit (Fig. 4). To probe the role in the 2′ M2 residue in the binding of ginkgolide X and picrotoxin to the GlyR, a P2′G mutation was introduced in the β subunit and the reverse G2′P mutation was introduced in the α1 subunit. Ginkgolide X and picrotoxin both exhibited a slightly increased antagonist potency at the α1βP2′G receptor compared with their activities at WT α1β but they were still ∼100-fold weaker as antagonists at this mutant receptor than at WT α1 (Table 2 and Fig. 8). Introduction of the G2′P mutation in α1 did not lead to major changes in the antagonist potency of ginkgolide X at the receptors, as the IC50 value obtained at cells expressing the homomeric α1G2′P GlyR was similar compared with that at the WT α1 (Table 2 and Fig. 8). The IC50 value for picrotoxin at homomeric α1G2′P was slightly decreased compared with WT α1 (Table 2). This is in agreement with a previous study, although the increase in antagonist potency caused by the mutation was less pronounced in the FMP Blue assay than in the electrophysiological set-up in that study (25).
FIGURE 8.
The importance of the 2′ M2 residue for the activity of ginkgolide X at the GlyRs. Concentration-inhibition curves of ginkgolide X at tsA-201 cells transfected with cDNAs for WT α1, α1G2′P, WT α1 and WT β, WT α1 and βP2′G, or WT α1 and βP2′G/T6′T GlyR subunits. The FMP Blue assay was performed as described under “Experimental Procedures” using EC70–EC95 concentrations of glycine as final agonist concentrations in the assays. The figure depicts data from a single representative experiment, and error bars are omitted for reasons of clarity.
Analogously to the similar IC50 values displayed by ginkgolide X and picrotoxin at tsA-201 cells expressing WT α1 and α1G2′P GlyRs, the antagonist potencies displayed by the two compounds at α1βF6′T and α1βP2′G/F6′T GlyRs did not differ significantly (Table 2 and Fig. 8). Thus, the addition of the P2′G mutation to βF6′T did not appear to further improve the binding of the two compounds to the GlyR, which had been facilitated by the F6′T mutation in the β subunit (Table 2 and Fig. 8).
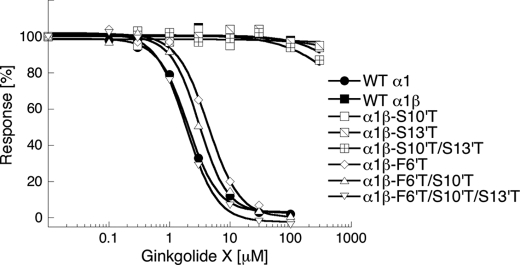
The 10′ and 13′ M2 Residues
The 10′ and 13′ M2 residues are conserved as threonines in all four α GlyR subunits, whereas both corresponding residues in the β subunit are serines (Fig. 4). Introduction of S10′T, S13′T, and S10′T/S13′T mutations into the β subunit did not introduce sensitivity in the α1β GlyR to ginkgolide X or picrotoxin at concentrations up to 300 μm (Table 2 and Fig. 9). Interestingly, both ginkgolide X and picrotoxin displayed slightly reduced IC50 values (2.5- and 2.1-fold, respectively) at α1βF6′T/S10′T/S13′T than at α1βF6′T, exhibiting antagonist potencies at α1βF6′T/S10′T/S13′T not significantly different from those at the WT α1 (Table 2 and Fig. 9).
FIGURE 9.
The importance of the 10′ and 13′ M2 residues for the activity of ginkgolide X at the GlyRs. Concentration-inhibition curves of ginkgolide X at tsA-201 cells transfected with cDNA for WT α1 or with the cDNA for WT α1 together with cDNAs for WT β, βS10′T, βS13′T, βS10′T/S13′T, βF6′T, βF6′T/S10′T, or βF6′T/S10′T/S13′T GlyR subunits. The FMP Blue assay was performed as described under “Experimental Procedures” using EC70–EC95 concentrations of glycine as final agonist concentrations in the assays. The figure depicts data from a single representative experiment, and error bars are omitted for reasons of clarity.
All of the mutant GlyRs mentioned above (i.e. all receptors composed of α1 and/or β subunits where one or both subunits were mutated in the 2′ and/or 6′ M2 positions and receptors composed of WT α1 and mutant β subunits with mutations in the 10′ and/or 13′ M2 positions) displayed “WT-like GlyR” profiles when challenged with glycine in the FMP Blue assay. This means that the basal levels of fluorescence recorded from wells with cells transfected with all these mutant receptors were similar to the basal fluorescence levels measured in wells with similar numbers of WT α1- or WT α1β-transfected cells. Furthermore, a concentration-dependent increase in fluorescent intensity was observed upon application of increasing concentrations of glycine to the cells expressing these mutant GlyRs, with glycine displaying similar EC50 values at the mutant GlyRs as at the WT GlyRs (Table 2). In contrast to the functional properties of these mutants, substitutions of the Thr residues in the 10′ and/or 13′ M2 positions of the α1 subunit with Ser residues had dramatic effects on the observed basal fluorescence levels and glycine-induced responses in the FMP Blue assay. The basal levels of fluorescence recorded from cells expressing homomeric α1T10′S, α1T13′S, and α1T10′S/T13′S GlyRs were significantly higher than from cells expressing the WT α1 (Table 2 and exemplified in supplemental Fig. S3). Furthermore, application of glycine in assay concentrations up to 3 mm at the cells only gave rise to small, if any, increases in fluorescence intensity (supplemental Fig. S3). Elevated basal levels of fluorescence and negligible responses to glycine application were also recorded in cells coexpressing α1T10′S, α1T13′S, or α1T10′S/T13′S subunits together with the WT β subunit (data not shown) and in cells transfected with the cDNAs encoding for the α1T10′SβS10′T, α1T13′SβS13′T, or α1T10′S/T13′SβS10′T/S13′T combinations (Table 2 and exemplified in supplemental Fig. S3). Interestingly, the elevated basal levels of fluorescence in these cells could be reduced concentration-dependently by applications of ginkgolide X, picrotoxin, or the competitive GlyR antagonist strychnine (data not shown). Thus, Thr to Ser mutations in these positions of the α1 subunit appear to change the distribution of the FMP Blue dye across the cell membrane during incubation with the dye, thus giving rise to higher basal levels of fluorescence in cells expressing these mutants than in those expressing WT GlyRs.
The Importance of Stoichiometry and Arrangement of the 6′ M2 Residues for Ginkgolide X and Picrotoxin Antagonism of the Anionic Cys-loop Receptor
The heteromeric GABAAR with its 2α/2β/1γ stoichiometry and fixed β:α:β:α:γ subunit arrangement constitute a good model system to probe the functional consequences of incorporation of subunits with different 6′ M2 residues into the anionic Cys-loop receptor for the activity of picrotoxin and ginkgolide X.
T6′F mutations were introduced in the α5 and γ2s subunits, and the sensitivities of the mutant α5β2γ2s GABAARs incorporating these subunits toward ginkgolide X and picrotoxin were characterized in the FMP Blue assay. GABA displayed 10- and 6-fold higher EC50 values at the α5T6′Fβ2γ2s and α5β2γ2sT6′F, respectively, than at the WT α5β2γ2s receptor (Table 4). Furthermore, the intensities of the fluorescence signals elicited by GABA application on the mutant receptor-expressing cells were significantly lower than from WT receptor-expressing cells (data not shown). Both picrotoxin and ginkgolide X were inactive as antagonists of α5T6′Fβ2γ2s and α5β2γ2sT6′F GABAARs at concentrations up to 300 μm (Table 4).
TABLE 4.
Functional characteristics of ginkgolide X and picrotoxin at wild type and mutant α5β2γ2s GABAARs transiently expressed in tsA-201 cells in the FMP Blue assay
For the antagonist experiments, EC70–EC95 concentrations of GABA at the respective receptors were used as agonist.
| Receptor | GABA | Ginkgolide X | Picrotoxin |
|---|---|---|---|
| EC50 (pEC50 ± S.E.) | IC50 (pIC50 ± S.E.) | IC50 (pIC50 ± S.E.) | |
| WT α5β2γ2s | 1.4 (5.85 ± 0.04) | 2.4 (5.62 ± 0.05) | 3.4 (5.46 ± 0.04) |
| WT α5β2 | 1.7 (5.77 ± 0.05) | 3.4 (5.47 ± 0.05) | 2.9 (5.54 ± 0.04) |
| α5T6′Aβ2γ2s | 3.2 (5.49 ± 0.04) | >300 (< 3.5) | >300 (<3.5) |
| α5T6′Sβ2γ2s | 7.2 (5.14 ± 0.05) | ∼300 (∼3.5) | ∼300 (∼3.5) |
| α5T6′Fβ2γ2s | 15 (4.83 ± 0.07) | >300 (<3.5) | >300 (<3.5) |
| α5β2γ2sT6′A | 3.5 (5.45 ± 0.04) | ∼100 (∼4.0) | ∼100 (∼4.0) |
| α5β2γ2sT6′S | 2.6 (5.59 ± 0.05) | ∼100 (∼4.0) | ∼100 (∼4.0) |
| α5β2γ2sT6′F | 8.7 (5.06 ± 0.07) | >300 (<3.5) | >300 (<3.5) |
To investigate the effects of more subtle variations in the side chains of the 6′ residues, Ala and Ser residues were introduced in this position of the α5 and γ2s subunits. The T6′A and T6′S mutations also resulted in pronounced impairment of the antagonist activities of ginkgolide X and picrotoxin, although inhibition of the signaling through some of these mutant α5β2γ2s receptors were observed when high micromolar concentrations of the two antagonists were used (Table 4).
DISCUSSION
Due to limited amounts of ginkgolide X available for this study, the pharmacological properties of ginkgolide X at WT and mutant Cys-loop receptors was characterized using the fluorescence-based FMP Blue assay. The pharmacological profiles exhibited by GlyRs, GABAARs, and other Cys-loop receptors in this assay have been found to be very representative of those obtained from electrophysiological recordings (26, 34, 45–47). Nevertheless, the assay still represents an indirect and somewhat coarse measurement of Cys-loop receptor signaling compared with conventional electrophysiology, and thus it is not possible to extract the same level of information about the finer aspects of receptor signaling, such as activation and deactivation/desensitization kinetics, and cooperativity, from this assay. Furthermore, one should be cautious not to overinterpretate observed trends and minor differences and only base conclusions on substantial differences in the obtained data.
The 6′ M2 Residue Is the Principal Molecular Determinant of Ginkgolide X Activity at Anionic Cys-loop Receptors
The antagonist activities displayed by ginkgolide X at α1βF6′T and α1/α1T6′F GlyRs compared with WT α1 and WT α1β GlyRs, respectively, unequivocally identify the 6′ M2 residue as the principal molecular determinant of the selectivity profile of ginkgolide X (Table 2 and Fig. 5). This observation is in concordance with the importance of the residue for the activity of native ginkgolides and for the subtype selectivity of picrotoxin at the GlyRs (19, 20, 25, 27, 28). Furthermore, the crucial role of the residue for ginkgolide X activity is underlined by the similar antagonist potencies displayed by the compound at the heteromeric GABAARs. The T6′ residue present in all α GlyR subunits is conserved in the vast majority of GABAAR subunits (Fig. 4), albeit not all (49). Thus it is not surprising that ginkgolide X displays similar IC50 values at homomeric GlyRs and the GABAARs tested in this study (Table 1). An interesting incongruity from this observation is the significantly lower antagonist potency of ginkgolide X at the homomeric ρ1 GABAAR. Not only does the ρ1 subunit contain a T6′ M2 residue, it also has threonines in the 10′ and 13′ positions (see below). This suggests that the 6′ M2 residue may not be the only molecular determinant of ginkgolide X activity at the anionic Cys-loop receptor.
The α7 nAChR and 5-HT3AR, which both possess a uniform 6′ M2 ring of threonines, are also insensitive to ginkgolide X. However, this is not necessarily surprising, because the ability of ginkgolide X to access a site in the cytoplasmic end of the ion pore could differ significantly from cationic to anionic channels. Furthermore, the 2′, 10′, and 13′ M2 residues in the anionic Cys-loop receptors are not conserved in the α7 and 5-HT3A subunits, and thus the contributions of these residues to a putative binding site for the ginkgolide differ in the cationic receptors.
Importance of Stoichiometry and Arrangement of 6′ M2 Residues in the Anionic Cys-loop Receptor for Ginkgolide X and Picrotoxin Activity
The arrangement of residues in the 6′ M2 rings of the homomeric GlyRs and the GABAARs is T:T.T:T:T, whereas it is believed to be T:F:T:F:F in the heteromeric GlyR (50). To investigate the role of the individual 6′ M2 residues in the anionic Cys-loop receptor for ginkgolide X and picrotoxin binding in greater detail we used the α5β2γ2s GABAAR as a model receptor. Interpretations of data outlined below are made under the assumptions that the five 6′ M2 residues in the pentamer contribute equally to the binding site, and that the subunit composition of the GABAAR is not altered by 6′ residue mutations in the respective subunits.
The inactivity of ginkgolide X and picrotoxin at WT α1β GlyR (T:F:T:F:F) and α1T6′FβF6′T GlyR (F:T:F:T:T) is mirrored by the inactivity of the compounds at α5T6′Fβ2γ2s (T:F:T:F:T) and α5T6′Fβ2γ2sT6′F GABAARs (T:F:T:F:F). Furthermore, the insensitivity of the α5β2γ2sT6′F GABAAR (T:T:T:T:F) to ginkgolide X and picrotoxin demonstrates that the presence of a single F6′ residue in the pentamer is enough to completely disrupt the binding of both antagonists (Table 3). In the case of picrotoxin, this observation is in concordance with a previous study (51).
Introduction of the more conservative T6′A and T6′S mutations in the α5 and γ2s subunits also profoundly impair the ability of ginkgolide X and picrotoxin to inhibit α5β2γ2s signaling, although receptors containing A6′ or S6′ subunits are not as insensitive to the antagonists as F6′ subunit-containing receptors (Table 3). The importance of a uniform 6′ M2 ring consisting solely of threonines for proper binding of ginkgolide X and picrotoxin is likely to apply for the GlyRs as well, a hypothesis supported by the significantly decreased antagonist activities of the two compounds at the α1βF6′G GlyR mutant. Thus, although the most obvious explanation for the insensitivity of the heteromeric GlyR to the two antagonists is the bulk introduced into the ion pore by the F6′ residues in the β subunits, the shear fact that the heteromeric receptor presents a non-uniform 6′ ring to the antagonists may also be a contributing factor.
Molecular Basis for Ginkgolide X, Ginkgolide A, and Picrotoxin Binding to the GlyR
The involvement of the 6′ M2 residue in the GlyR in the binding of the native ginkgolides is well established (19, 20). However, considering the equipotency or even preference for heteromeric over homomeric GlyRs displayed by these compounds (18, 19, 21, 24), it is evident that they must bind differently to the GlyR ion pore from picrotoxin and ginkgolide X.
To rationalize the different functional properties of picrotoxinin, ginkgolide X, and ginkgolide A, we docked low energy conformations of the compounds into a homology model of the α1 GlyR. Interestingly, the differences in conformational flexibility between ginkgolide X and ginkgolide A may offer an explanation for the different GlyR activity profiles of the compounds. In their lowest energy conformations there is a striking resemblance between the two compounds. However, whereas ginkgolide X is flexible in the t-butyl end of the molecule, ginkgolide A is flexible in the other end. It is noteworthy, that when adopting the more caved alternative (+0.8 kcal/mol) ring conformation, the tip of ginkgolide A turns hydrophobic (Fig. 6C). In contrast, the tip of ginkgolide X remains the carbonyl oxygen of the 3-methylfuran-(2)-one ring within all energetically accessible conformations. Furthermore, the difference in energy of 1.9 and 0.8 kcal/mol between the respective low energy conformations of ginkgolide X and ginkgolide A indicates that the latter can easily adopt the alternative conformation, whereas for ginkgolide X it would result in a ∼25-fold decrease in binding affinity.
When docked to the α1 GlyR homology model in their lowest energy conformations, ginkgolide X docks exclusively in the 6′–13′ region (Fig. 7B), whereas ginkgolide A docks in the 2′–6′ region (Fig. 7D). In the suggested binding mode for ginkgolide X, the 3-methylfuran-(2)-one ring is situated at the level of the T6′ residues forming a direct hydrogen bond to one of the threonines. It is obvious that this binding mode would be impossible in the heteromeric α1β GlyR with its T:F:T:F:F 6′ M2 ring arrangement due to the steric bulk of the phenylalanines (Fig. 7B). The same argument applies to the alternative binding modes of ginkgolide X (Fig. 7C). The suggested binding modes for the alternative conformation of ginkgolide A reside either above or below the 6′ residues and would thus be less affected by a T6′F mutation in this position or by the mixed T/F 6′ M2 ring in the WT α1β GlyR (Fig. 7E). Finally, picrotoxinin has a completely different and more compact shape than both ginkgolide X and ginkgolide A. Considering the high number of different docking poses obtained for picrotoxinin in the homology model we will refrain from further interpretations regarding this compound (Fig. 7F). It is obvious that the crude nature of the homology model applied here prompts for extremely careful interpretation of the results. Taking receptor flexibility and solvation of the ion channel into account is necessary to get a more detailed picture of the binding modes of the compounds.
The Roles of the 2′, 10′, and 13′ M2 Residues for Ginkgolide X, Ginkgolide A, and Picrotoxin Binding to the GlyR
Despite the crude nature of the α1 GlyR homology model and the resulting limitations to the information extractable from the docking of ligands into it, the differential binding modes of gingkolide A and ginkgolide X in the model prompted us to perform a mutagenesis study to evaluate the contributions of the pore lining 2′, 10′, and 13′ M2 residues to the binding of ginkgolide X.
The 2′ M2 residue does not appear to be important for the binding of ginkgolide X to the GlyR. The ginkgolide displays a slightly increased IC50 value at α1G2′P than at WT α1 (2.9-fold). Furthermore, it displays a weak but significant antagonist activity at the α1βP2′G in high micromolar concentrations, an activity not observed at the WT α1β (see Table 2 and Fig. 8). However, none of these differences are substantial enough to indicate a direct involvement of the α1 G2′ residue in the binding of ginkgolide X. In contrast, ginkgolide A has displayed a 30-fold increased IC50 value at the α1G2′P GlyR compared with WT α1 and a >30-fold increased IC50 value at α1G2′Pβ compared with WT α1β, strongly indicating a direct involvement of the 2′ residue in binding of this ginkgolide (19). In contrast to the G2′P mutation, G2′A and G2′S mutations in the α1 GlyR have been found not to alter the antagonist activities of native ginkgolides significantly (20). Finally, the antagonist activity of picrotoxin has been reported not to be significantly different at WT α1β and α1βP2′G GlyRs, whereas picrotoxin displays 3–6-fold lower IC50 values at α1G2′P GlyR than at WT α1 (19, 25), findings in good agreement with the observations made for picrotoxin in this study.
Investigation of the involvement of 10′ and 13′ M2 residues in GlyR binding of ginkgolide X and picrotoxin was complicated by the dramatically increased basal responses measured in tsA-201 cells expressing the α1T10′S, α1T13′S, or α1T10′S/T13′S subunits in the FMP Blue assay (supplemental Fig. S3). With the exception of the β GlyR subunit, the 10′ and 13′ M2 residues are conserved as threonines throughout the GABAA and GlyR subunits, suggestive of a role of these residues for proper anionic Cys-loop receptor function (Fig. 4). A likely explanation for this observation is that the mutations have rendered the receptors constitutively active or hypersensitive to trace concentrations of glycine in the assay buffer. This hypothesis is supported by numerous previous studies demonstrating the importance of the 10′ and 13′ M2 residues for proper gating of both cationic and anionic Cys-loop receptors (52–59).
The compositions of the 10′ and/or 13′ M2 rings in homomeric α1T10′S, α1T13′S, and α1T10′S/T13′S GlyRs and in heteromeric α1T10′Sβ, α1T13′Sβ, and α1T10′S/T13′Sβ GlyRs are S:S:S:S:S, whereas they are T:T:T:T:T and T:S:T:S:S in the WT α1 and WT α1β GlyRs, respectively. Interestingly, coexpression of α1T10′S, α1T13′S, or α1T10′S/T13′S with the corresponding βS10′T, βS13′T, and βS10′T/S13′T mutants (forming receptors with S:T:S:T:T rings) also resulted in elevated basal levels of fluorescence (supplemental Fig. S3). Thus, it does not seem to be the total number of serines in the 10′ and 13′ M2 rings of the GlyR pentamer but rather the identity of the 10′ and 13′ residues in α1 that determine the basal activity levels. This is not necessarily surprising, considering that α1 and β subunits have been shown to have asymmetric contributions to heteromeric GlyR signalling, and similar observations have been made for the α and β subunits contributions to GABAAR signaling (56, 60). Although constitutive activity or agonist hypersensitivity of α1T10′S, α1T13′S, and α1T10′S/T13′S containing GlyRs seems to be a reasonable explanation for the observed elevated basal levels of fluorescence in the FMP Blue assay, this observation clearly needs to be investigated and verified in a more sophisticated assay before any solid conclusions can be drawn. Hence, we decided not to include these mutants in our study of ginkgolide X and picrotoxin.
Introduction of the reverse S10′T, S13′T, or S10′T/S13′T mutations into the β subunit did not result in α1β receptors more sensitive to either ginkgolide X or picrotoxin than WT α1β, at least not at the concentrations tested in this study (see Table 2 and Fig. 9). Clearly, the Ser to Thr substitutions are not enough to facilitate binding of the compounds as long as the β subunit in the heteromeric receptor contains the F6′ residue. Interestingly, however, both ginkgolide X and picrotoxin display similar antagonist potencies at α1βF6′T/S10′T/S13′T and WT α1. It is tempting to speculate that the presence of five Thr residues in the 10′ and 13′ positions in the α1βF6′T/S10′T/S13′T pentamer has added further to the binding affinities of picrotoxin and ginkgolide X already facilitated by the F6′T mutation in the β subunit, and that the increased hydrophobic character of Thr compared with Ser in these positions stabilizes the binding of the two antagonists. However, because the observed changes in IC50 values are fairly small, they should be considered trends rather than significant changes.
In conclusion, we propose that the structural differences between gingkolide X and the native ginkgolide give rise to different binding modes of the compounds to the GlyR channel, and that this in turn results in entirely different pharmacological profiles. The binding mode of ginkgolide A to the 2′–6′ region of the α1 GlyR ion channel observed for the top scoring pose of the compound in the docking experiments in this study is in good agreement with the binding mode previously proposed for the native ginkgolides and supported by extensive mutagenesis work (Fig. 7D) (19, 20). Although it seems to be possible for the alternative ring conformation of ginkgolide A to dock in the 6′–13′ region as well (see Fig. 7E), it is slightly less favorable from an energy perspective. If the compound is capable of binding to both sites in the channel, the 2′–6′ binding mode is likely to be favored. However, this preference could possibly shift in the α1β GlyR as the tip of ginkgolide A, due to conformational flexibility in the 3-methyldihydrofuran-2-one end of the molecule, turns hydrophobic in the alternative (+0.8 kcal/mol) ring conformer and thus could be stabilized by hydrophobic interactions with the F6′ residues in the heteromeric receptor.
In contrast to ginkgolide A, ginkgolide X appears to target the 6′–13′ region of the ion pore of the GlyR. This proposal is based primarily on the results of the docking experiments, where the lowest energy conformation as well as many of the alternative conformations of the ginkgolide targets this region, although an alternative conformation of the compound is capable of binding to the 2′–6′ region (Fig. 7, B and C). Just as for ginkgolide A, we cannot exclude that ginkgolide X potentially could bind to both sites. However, based on the docking results and the considerable energy penalty (1.9 kcal/mol) for ginkgolide X to exist in its alternative low energy conformation, the 6′–13′ region must be considered the primary site. The mutagenesis analysis of the contributions of the 2′, 10′, and 13′ M2 residues to ginkgolide X activity cannot be claimed to provide substantial support for any of the two binding modes, because the changes observed for all of these mutants compared with WT GlyRs are relatively subtle.
Conclusion
In the present study a novel naturally occurring ginkgolide analog, ginkgolide X, has been found to exhibit a unique functional profile at anionic Cys-loop receptors. The ginkgolide is only the second compound published to date that displays complete functional selectivity for homomeric over heteromeric GlyRs, picrotoxin being the other. Like picrotoxin, ginkgolide X also inhibits signaling through various heteromeric GABAAR subtypes. However, in contrast to its relative potent pan-activity at these receptors, the ginkgolide is a much weaker antagonist of the homomeric ρ1 GABAAR. Finally, ginkgolide X has been found to be inactive at selected nAChR subtypes and at the 5-HT3AR at concentrations up to 300 μm in the FMP Blue assay (Table 1). This functional profile makes the ginkgolide a potential valuable pharmacological tool in studies of native GlyRs and further underlines the potential of the ginkgolide molecule as lead structure for the development of ligands with interesting properties at anionic Cys-loop receptors.
The 6′ M2 residue in the GlyR subunit has been demonstrated to be the key molecular determinant of the selectivity profile of ginkgolide X at these receptors. Furthermore, the presence of a 6′ M2 ring consisting exclusively of Thr residues seems to be crucial for the binding of ginkgolide X and picrotoxin to anionic Cys-loop receptors, something that clearly is not a requirement for proper binding of the native ginkgolides. Based on conformational analysis and docking of low-energy conformations of ginkgolide A and ginkgolide X into a homology model of the α1 GlyR, we propose that their different functional profiles may arise from a significant difference in the flexibility of the two molecules. Although both compounds may adopt alternative low energy ring conformers, the flexibility resides in different ends of the molecules, and this may facilitate different binding modes of the ginkgolides to the GlyR, where ginkgolide A appears to target the 2′–6′ region and ginkgolide X the 6′–13′ region in the ion pore.
Supplementary Material
Acknowledgments
We thank Dr. H. Jaggy and Dr. S. S. Chatterjee (both from Dr. Willmar Schwabe Research Laboratories) for isolating, identifying, and supplying pure samples of ginkgolide X. Dr. Whiting and Merck, Sharp and Dohme are thanked for the generous gifts of the human GABAAR cDNAs, and Drs. Weiss, Schofield, Betz, Lindstrom, and Millar are thanked for their gifts of various other cDNAs. We thank Drs. Kellar, Xiao, Stitzel, and Egebjerg for their gifts of cell lines.
This work was supported by The Lundbeck Foundation, The Carlsberg Foundation, Direktør Ib Henriksen Foundation, and the Danish Medical Research Council. Part of this work was presented at the Society of Neuroscience conference, November 15–19, 2008, in Washington D.C.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
S. S. Chatterjee, personal communication.
- GABA
- γ-aminobutyric acid
- ACh
- acetylcholine
- GABAAR
- γ-aminobutyric acid type A receptor
- FMP
- FLIPR Membrane Potential
- GlyR
- glycine receptor
- nAChR
- nicotinic acetylcholine receptor
- WT
- wild type
- 5-HT3
- 5-hydroxytryptamine type 3.
REFERENCES
- 1.Kirsch J. (2006) Cell Tissue Res. 326, 535–540 [DOI] [PubMed] [Google Scholar]
- 2.Mody I., Pearce R. A. (2004) Trends Neurosci. 27, 569–575 [DOI] [PubMed] [Google Scholar]
- 3.Whiting P. J. (2003) Drug Discov. Today 8, 445–450 [DOI] [PubMed] [Google Scholar]
- 4.Lynch J. W. (2004) Physiol. Rev. 84, 1051–1095 [DOI] [PubMed] [Google Scholar]
- 5.Betz H., Laube B. (2006) J. Neurochem. 97, 1600–1610 [DOI] [PubMed] [Google Scholar]
- 6.Sieghart W., Sperk G. (2002) Curr. Top. Med. Chem. 2, 795–816 [DOI] [PubMed] [Google Scholar]
- 7.Taly A., Corringer P. J., Guedin D., Lestage P., Changeux J. P. (2009) Nat. Rev. Drug Discov. 8, 733–750 [DOI] [PubMed] [Google Scholar]
- 8.Jensen A. A., Frølund B., Liljefors T., Krogsgaard-Larsen P. (2005) J. Med. Chem. 48, 4705–4745 [DOI] [PubMed] [Google Scholar]
- 9.Barnes N. M., Hales T. G., Lummis S. C., Peters J. A. (2009) Neuropharmacology 56, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiting P. J. (2003) Curr. Opin. Drug Discov. Devel. 6, 648–657 [PubMed] [Google Scholar]
- 11.Nemeroff C. B. (2003) Psychopharmacol. Bull. 37, 133–146 [PubMed] [Google Scholar]
- 12.Da Settimo F., Taliani S., Trincavelli M. L., Montali M., Martini C. (2007) Curr. Med. Chem. 14, 2680–2701 [DOI] [PubMed] [Google Scholar]
- 13.Orser B. A., Canning K. J., Macdonald J. F. (2002) Curr. Opin. Anaesthesiol. 15, 427–433 [DOI] [PubMed] [Google Scholar]
- 14.Korpi E. R., Sinkkonen S. T. (2006) Pharmacol. Ther. 109, 12–32 [DOI] [PubMed] [Google Scholar]
- 15.Breitinger H. G., Becker C. M. (2002) ChemBioChem 3, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 16.Strømgaard K., Nakanishi K. (2004) Angew. Chem. Int. Ed. Engl. 43, 1640–1658 [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi K. (2005) Bioorg. Med. Chem. 13, 4987–5000 [DOI] [PubMed] [Google Scholar]
- 18.Kondratskaya E. L., Betz H., Krishtal O. A., Laube B. (2005) Neuropharmacology 49, 945–951 [DOI] [PubMed] [Google Scholar]
- 19.Hawthorne R., Cromer B. A., Ng H. L., Parker M. W., Lynch J. W. (2006) J. Neurochem. 98, 395–407 [DOI] [PubMed] [Google Scholar]
- 20.Heads J. A., Hawthorne R. L., Lynagh T., Lynch J. W. (2008) J. Neurochem. 105, 1418–1427 [DOI] [PubMed] [Google Scholar]
- 21.Kondratskaya E. L., Fisyunov A. I., Chatterjee S. S., Krishtal O. A. (2004) Brain Res. Bull. 63, 309–314 [DOI] [PubMed] [Google Scholar]
- 22.Ivic L., Sands T. T., Fishkin N., Nakanishi K., Kriegstein A. R., Strømgaard K. (2003) J. Biol. Chem. 278, 49279–49285 [DOI] [PubMed] [Google Scholar]
- 23.Huang S. H., Duke R. K., Chebib M., Sasaki K., Wada K., Johnston G. A. (2004) Eur. J. Pharmacol. 494, 131–138 [DOI] [PubMed] [Google Scholar]
- 24.Jensen A. A., Begum N., Vogensen S. B., Knapp K. M., Gundertofte K., Dzyuba S. V., Ishii H., Nakanishi K., Kristiansen U., Strømgaard K. (2007) J. Med. Chem. 50, 1610–1617 [DOI] [PubMed] [Google Scholar]
- 25.Shan Q., Haddrill J. L., Lynch J. W. (2001) J. Neurochem. 76, 1109–1120 [DOI] [PubMed] [Google Scholar]
- 26.Jensen A. A. (2005) Eur. J. Pharmacol. 521, 39–42 [DOI] [PubMed] [Google Scholar]
- 27.Pribilla I., Takagi T., Langosch D., Bormann J., Betz H. (1992) EMBO J. 11, 4305–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z., Cromer B. A., Harvey R. J., Parker M. W., Lynch J. W. (2007) J. Neurochem. 103, 580–589 [DOI] [PubMed] [Google Scholar]
- 29.Jaracz S., Nakanishi K., Jensen A. A., Strømgaard K. (2004) Chemistry 10, 1507–1518 [DOI] [PubMed] [Google Scholar]
- 30.Chatterjee S. S., Klessing K., Jaggy H., Seederup E., Squires R. F. (2002) Naunyn-Schmiedebergs Arch. Pharmacol. 365, Suppl. 1, R79 [Google Scholar]
- 31.Xiao Y., Meyer E. L., Thompson J. M., Surin A., Wroblewski J., Kellar K. J. (1998) Mol. Pharmacol. 54, 322–333 [DOI] [PubMed] [Google Scholar]
- 32.Karadsheh M. S., Shah M. S., Tang X., Macdonald R. L., Stitzel J. A. (2004) J. Neurochem. 91, 1138–1150 [DOI] [PubMed] [Google Scholar]
- 33.Jensen A. A., Zlotos D. P., Liljefors T. (2007) J. Med. Chem. 50, 4616–4629 [DOI] [PubMed] [Google Scholar]
- 34.Jensen A. A., Kristiansen U. (2004) Biochem. Pharmacol. 67, 1789–1799 [DOI] [PubMed] [Google Scholar]
- 35.SchrödingerLLC (2008) Impact, Version 5.0, Schrödinger, LLC, New York [Google Scholar]
- 36.SchrödingerLLC (2008) MacroModel, version 9.6, Schrödinger, LLC, New York [Google Scholar]
- 37.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unwin N. (2005) J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 39.DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.2, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 40.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 41.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 42.SchrödingerLLC (2008) Maestro, version 8.5, Schrödinger, LLC, New York [Google Scholar]
- 43.Brady G. P., Jr., Stouten P. F. (2000) J. Comput. Aided Mol. Des. 14, 383–401 [DOI] [PubMed] [Google Scholar]
- 44.SchrödingerLLC (2008) Schrödinger Suite 2008 Induced Fit Docking Protocol, Schrödinger, LLC, New York [Google Scholar]
- 45.Fitch R. W., Xiao Y., Kellar K. J., Daly J. W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4909–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joesch C., Guevarra E., Parel S. P., Bergner A., Zbinden P., Konrad D., Albrecht H. (2008) J. Biomol. Screen. 13, 218–228 [DOI] [PubMed] [Google Scholar]
- 47.Price K. L., Lummis S. C. (2005) J. Neurosci. Methods 149, 172–177 [DOI] [PubMed] [Google Scholar]
- 48.Boström J., Norrby P. O., Liljefors T. (1998) J. Comput. Aided Mol. Des. 12, 383–396 [DOI] [PubMed] [Google Scholar]
- 49.Sinkkonen S. T., Hanna M. C., Kirkness E. F., Korpi E. R. (2000) J. Neurosci. 20, 3588–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grudzinska J., Schemm R., Haeger S., Nicke A., Schmalzing G., Betz H., Laube B. (2005) Neuron 45, 727–739 [DOI] [PubMed] [Google Scholar]
- 51.Gurley D., Amin J., Ross P. C., Weiss D. S., White G. (1995) Receptors Channels 3, 13–20 [PubMed] [Google Scholar]
- 52.Devillers-Thiéry A., Galzi J. L., Bertrand S., Changeux J. P., Bertrand D. (1992) Neuroreport 3, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 53.Dang H., England P. M., Farivar S. S., Dougherty D. A., Lester H. A. (2000) Mol. Pharmacol. 57, 1114–1122 [PubMed] [Google Scholar]
- 54.Wenningmann I., Barann M., Vidal A. M., Dilger J. P. (2001) Mol. Pharmacol. 60, 584–594 [PubMed] [Google Scholar]
- 55.Tierney M. L., Birnir B., Cromer B., Howitt S. M., Gage P. W., Cox G. B. (1998) Receptors Channels 5, 113–124 [PubMed] [Google Scholar]
- 56.Dalziel J. E., Birnir B., Everitt A. B., Tierney M. L., Cox G. B., Gage P. W. (1999) Eur. J. Pharmacol. 370, 345–348 [DOI] [PubMed] [Google Scholar]
- 57.Francis M. M., Vazquez R. W., Papke R. L., Oswald R. E. (2000) Mol. Pharmacol. 58, 109–119 [DOI] [PubMed] [Google Scholar]
- 58.Chen L., Durkin K. A., Casida J. E. (2006) J. Biol. Chem. 281, 38871–38878 [DOI] [PubMed] [Google Scholar]
- 59.Goren E. N., Reeves D. C., Akabas M. H. (2004) J. Biol. Chem. 279, 11198–11205 [DOI] [PubMed] [Google Scholar]
- 60.Shan Q., Nevin S. T., Haddrill J. L., Lynch J. W. (2003) J. Neurochem. 86, 498–507 [DOI] [PubMed] [Google Scholar]
- 61.SchrödingerLLC (2005) Prime, version 1.7, Schrödinger, LLC, New York [Google Scholar]
- 62.UniProt Consortium (2008) Nucleic Acids Res. 36, D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.SchrödingerLLC (2008) Glide, version 5.0, Schrödinger LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.