Abstract
Breast cancer is the most common cancer type for women in the western world. Despite decades of research, the molecular processes associated with breast cancer progression are still inadequately defined. Here, we focus on the systematic alteration of metabolism by using the state of the art metabolomic profiling techniques to investigate the changes of 157 metabolites during the progression of normal mouse mammary epithelial cells to an isogenic series of mammary tumor cell lines with increasing metastatic potentials. Our results suggest a two-step metabolic progression hypothesis during the acquisition of tumorigenic and metastatic abilities. Metabolite changes accompanying tumor progression are identified in the intracellular and secreted forms in several pathways, including glycolysis, the tricarboxylic acid cycle, the pentose phosphate pathway, fatty acid and nucleotide biosynthesis, and the GSH-dependent antioxidative pathway. These results suggest possible biomarkers of breast cancer progression as well as opportunities of interrupting tumor progression through the targeting of metabolic pathways.
Keywords: Breast Cancer, Metabolism, Metabolomics, Tumor Metabolism, Tumor Metastases
Introduction
Approximately 40,000 American women succumb to breast cancer each year, with metastasis causing the overwhelming majority of these deaths (1). Metastasis is a multistep process, requiring tumor cells to intravasate into the bloodstream, survive in the circulation, adhere to and extravasate from the vascular network in the secondary organ, and finally adapt to a foreign microenvironment (2). Recent functional transcriptomics studies identified genes that play important roles in individual steps of breast cancer metastasis with the MDA-MB-231 xenograft model (3–6) or the 4T1 syngeneic mouse models (7). However, studies with metabolomic approaches to identify key metabolites that characterize metastasis progression are still scarce (8). Metabolic reprogramming was linked to the major hallmarks of cancer, including tissue invasion and metastasis (9). However, its functional role in tumor progression and metastasis remains largely undefined. In a recent study, metabolomic profiling identified increased sarcosine synthesis as a functionally important metabolic alteration during prostate cancer progression (10). Similar efforts to identify important metabolic changes during breast cancer progression hold the potential for providing putative diagnostic and prognostic biomarkers as well as new therapeutics targets. In the current study, we used the 4T1 syngeneic mouse model of breast cancer to systematically identify metabolite changes.
EXPERIMENTAL PROCEDURES
Cell Culture
All cell lines used were cultured in Dulbecco's modified Eagle's medium supplemented with 7.5% dialyzed fetal bovine serum (Thermo Fisher Scientific HyClone).
Quenching of Metabolism and Metabolite Extraction
All cells were grown in 10-cm tissue culture dishes, and the media were replaced 24 and 2 h prior to metabolite extraction. All samples were harvested at subconfluence. Metabolism was quenched, and metabolites were extracted by aspiration of media and immediate addition of 4 ml of 80:20 methanol:water at −80 °C to simultaneously lyse cells and quench metabolism. After 15 min of incubation at −80 °C, cell remnants were scraped from the tissue culture dish and transferred, along with the methanol:water, into a 15-ml conical centrifuge tube. The resulting mixture was centrifuged at 5000 × g for 5 min, and the supernatant was moved to a new tube. The remaining pellet was re-extracted twice more with 500 μl of 80:20 methanol:water at −80 °C, and all the supernatants were combined with the original supernatant. Ten microliters of the extract were injected into each of the liquid chromatography-tandem mass spectrometry (LC-MS/MS)4 separations. Extraction of metabolites in conditioned media was performed by combining 1 volume of 24-h conditioned media with 4 volumes of cold methanol, spinning down, and taking supernatant for analysis. Fresh media unconditioned by cells yet placed in the same incubator for 24 h were used as control.
Targeted Liquid Chromatography-Mass Spectrometry
Two different liquid chromatography (LC) separations were coupled by electrospray ionization (ESI) to triple quadrupole mass spectrometers operating in multiple reaction monitoring mode. The LC method coupled to positive mode ESI was hydrophilic interaction chromatography on an aminopropyl column at basic pH as reported previously (11). The LC method coupled to negative mode ESI was reversed phase chromatography with an amine-based ion pairing agent, with the method used a variant of one reported previously (12), with additional reactions to monitor more metabolites and identical stationary and mobile phases but an altered gradient: t = 0, 0% B; t = 5, 0% B; t = 10, 20% B; t = 20, 20% B; t = 35, 65% B; t = 38, 95% B; t = 42, 95% B, t = 43, 0% B; t = 50, 0% B, where B refers to the methanol-containing mobile phase. LC instrumentation was an LC-20 AD high pressure liquid chromatography system (Shimadzu), autosampler temperature 4 °C, injection volume 10 μl. MS instrumentation was a TSQ Quantum Ultra or a Discovery Max triple-quadrupole mass spectrometer (Thermo Fisher Scientific). Mass spectrometry parameters were as reported previously (11).
Data Collection and Processing
Cells were allowed to grow to subconfluence, at which time the metabolites were methanol-extracted. Four biological replicates prepared over two separate days were used for each cell line. Metabolite extraction and LC-MS/MS analysis were performed. Metabolite signals were normalized to total protein levels within each cell line as well as to the signal in NMuMG from the same experimental day and log2-transformed. Metabolite signals of conditioned media were normalized to the geometric mean for the day the samples were run and log2-transformed.
RESULTS
Intracellular Metabolite Changes Clustering Cell Lines
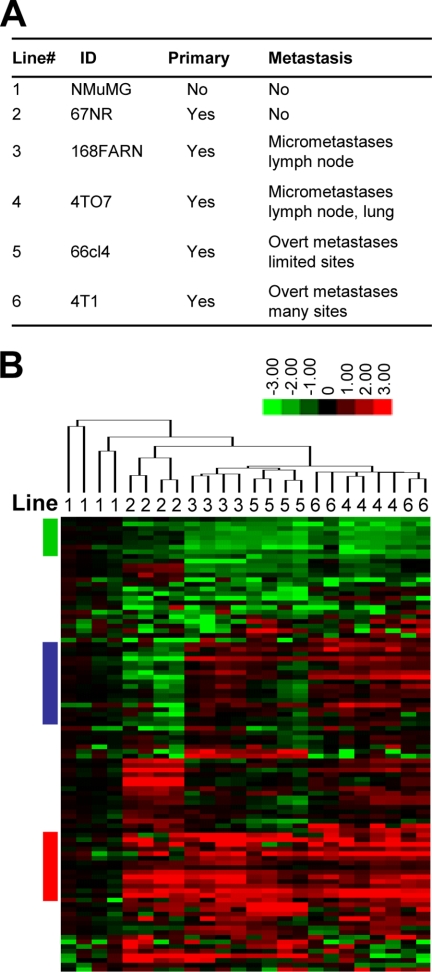
To identify intracellular metabolic changes during tumorigenesis and metastasis, we used LC-MS/MS to profile the levels of 157 different small molecule metabolites (13) in the well characterized 4T1 series of cell lines of a murine mammary cancer model (14). This series includes five isogenic tumorigenic lines (67NR, 168FARN, 4TO7, 66cl4, and 4T1) that originated from one spontaneous tumor in the BALB/cfC3H mouse. Each cell line displays unique tumorigenic and metastatic capabilities (Fig. 1A). We also included a normal murine mammary gland epithelial cell line, NMuMG (15), as the baseline control to observe metabolic changes correlated with tumorigenesis. The normalized intracellular metabolite signals are listed in supplemental Table 1. Unsupervised hierarchical clustering of the normalized metabolite intensities showed clear separation of the non-tumorigenic line NMuMG from other tumorigenic lines (Fig. 1B). In the tumorigenic branch, the non-metastatic line 67NR was separate from the metastatic lines (168FARN, 4TO7, 66cl4, and 4T1). There was no further subgrouping among the four metastatic lines that was consistent with the increased metastatic potential. These clustering results are consistent with the possibility that metabolic reprogramming is required both during the initial transformation process and during the acquisition of metastatic potential. No further metabolic changes were observed correlated with the quantitatively increased metastatic potential, suggesting that acquisition of enhanced metastatic potential did not, at least in these cell lines, require further systematic metabolic reprogramming. Important patterns in the clustering heat map (Fig. 1B, colored bars) are described below.
FIGURE 1.
Metabolomic profiling clustering mouse mammary cells with different tumorigenic potentials. A, six mouse cell lines (represented with numbers throughout the figure) with distinct tumorigenic and metastatic abilities. ID, cell line identification. B, hierarchical clustering of metabolomic profiles of the six cell lines identified subsets of metabolites that correlated with tumorigenicity and metastatic ability. Each cell line has four biological replicates labeled above the heat map with numbers as indicated in A. Red, green, and blue bars indicate three patterns of metabolites examined in Fig. 2, A, B, and D, respectively.
Metabolites Associated with Transformation
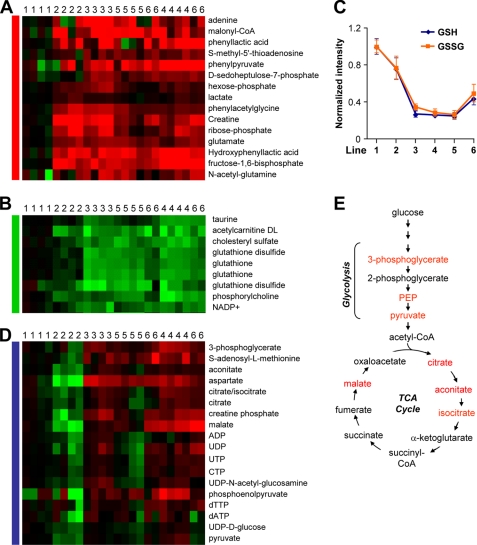
When compared with NMuMG, all five tumorigenic lines contained a set of metabolites that were significantly more abundant (Fig. 2A). Several intermediates in glycolysis increased in tumorigenic lines, including hexose phosphate (i.e. glucose-6-phosphate and fructose-6-phosphate) and fructose-1,6-bisphosphate, consistent with the “Warburg effect” of increased glycolysis rate in tumor cells (16). Additionally, ribose phosphate was significantly higher in these cells when compared with NMuMG (Fig. 2A). Ribose phosphate is a product of the pentose phosphate pathway, which also produces NADPH. Ribose is necessary for nucleic acid biosynthesis, and NADPH is the major reductant in fatty acid and steroid synthesis. The building block of fatty acids, malonyl-CoA, was also significantly more abundant in tumorigenic lines (Fig. 2A). This result was consistent with the finding of active fatty acid synthesis in tumor tissues (17). The Warburg effect includes increased lactate production. However, we noticed that lactate was not universally higher in tumorigenic cells (Fig. 2A). This may reflect non-universality of the Warburg effect (18), in vitro culture condition (19), or metabolic plasticity in response to hypoxia (20).
FIGURE 2.
Characteristic metabolic changes during transformation and metastasis. A, metabolites increased in transformed cells. B, metabolites decreased in transformed and metastatic cells. C, GSH and GSSG levels in six cell lines. Data represent average ± S.D. on log2 scale. D, metabolites increased in cells with metastatic potential. E, simplified schematic of glycolysis and tricarboxylic acid cycle (TCA cycle) showing in red the metabolites that are elevated in metastatic cell lines. Six cell lines were labeled with numbers in the same scheme as in Fig. 1. PEP, phosphoenolpyruvate.
Cellular transformation was also characterized by decreased levels of a set of metabolites, which were lower in five tumorigenic lines than NMuMG (Fig. 2B). Most notably, the levels of GSH and the oxidized form of glutathione (GSSG) were decreased in tumorigenic lines (Fig. 2C). GSH and GSSG are crucial regulators of the cellular reduction/oxidation (redox) state, with GSH protecting cells from oxidative stress (21). Thus, the lower GSH levels in transformed cells suggest that reduced antioxidant activity may sometimes occur early in oncogenesis, perhaps predisposing cells to reactive oxygen species-induced mutations necessary for full-fledged oncogenesis. Continued GSH/GSSG depletion may also sometimes contribute to the acquisition of general metastatic ability as there was a further decrease of glutathiones in the metastatic cell lines when compared with 67NR (Fig. 2C). The free energy of the antioxidant reactions is dependent upon the ratio [GSH]2/[GSSG]. Despite the large change in the combined GSH/GSSG pool size, the ratio of the two metabolites does not change significantly. Thus, the free energy of the reaction is dependent on the level of GSH, and the decrease in GSH levels is mirrored by a decrease in the free energy of the antioxidant reactions, further lowering the ability of the cells to cope with redox stress. To further elucidate the role of the glutathione redox system in tumorigenesis and metastasis, pharmacological inhibitors may be used. For example, treatment of NMuMG with methionine sulfoximine, an inhibitor of γ-glutamylcysteine synthetase (22), could be used to determine whether GSH depletion might facilitate the transformation of the cells. Treatment of the tumorigenic cells with GSHest, a permeable compound that can increase the intracellular content of GSH (23), might help restore the redox status and decrease tumorigenesis and metastatic abilities.
Metabolites Associated with Metastatic Ability
Metastatic ability- related metabolites were recognized as those differentially represented in the metastatic cell lines (168FARN, 4TO7, 66cl4, and 4T1) when compared with both NMuMG and 67NR. A subset of investigated metabolites showed such a pattern; they are significantly more abundant in the metastatic lines than NMuMG and 67NR (Fig. 2D). The majority of these metabolites fall into two groups: the tricarboxylic acid cycle or nucleotides. In addition, three of the four metabolites in the last steps of glycolysis were enriched in metastatic cells, 3-phosphoglycerate, phosphoenolpyruvate (PEP), and pyruvate, suggesting differences in lower glycolysis when compared with NMuMG and 67NR (Fig. 2E). Although the exact mechanism is not clear, these findings in lower glycolysis may relate to the pivotal role of specific pyruvate kinase isozymes in oncogenesis (24). Moving down from pyruvate, several tricarboxylic acid cycle (TCA cycle) intermediates were enriched in the metastatic lines, including aconitate, citrate, isocitrate, and malate (Fig. 2E). The Warburg effects suggest that tumor cells prefer aerobic glycolysis to the tricarboxylic acid cycle for producing ATP and reductants. Our observation that tricarboxylic acid intermediates are exclusively up-regulated in metastatic cell lines suggests that invasive cancer cells are using the tricarboxylic acid cycle differently than the non-metastatic cells. It is unclear, however, whether these differences are associated with increased tricarboxylic acid cycle flux, and if so, whether this flux is driven primarily by glucose or glutamine. Fluxomic analysis is well suited for answering these questions in future studies (25).
The changes of nucleotide species follow an interesting pattern. Levels are lower in the non-metastatic tumor cells (67NR) than in the non-transformed cells, perhaps due to enhanced nucleotide consumption to feed growth and DNA replication in the transformed cells. The metastatic tumor cells, however, have increased nucleotide levels, reflecting altered nucleotide turnover.
Secreted Metabolites Associated with Metastatic Potential
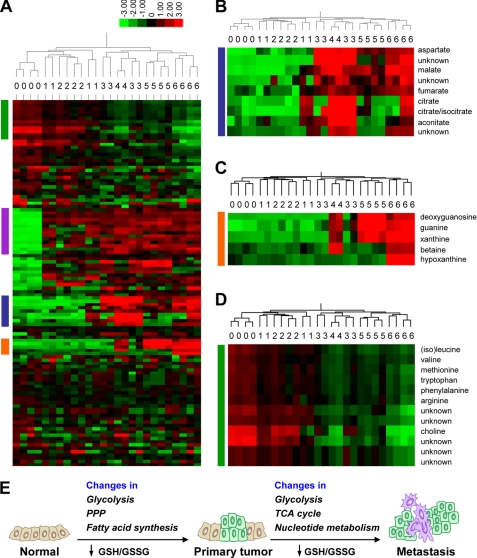
To identify any changes of extracellular metabolites, growth media conditioned by cells for 24 h were profiled (supplemental Table 2). Largely consistent with the intracellular metabolite clustering, the extracellular metabolites also clustered metastatic lines (168FARN, 4TO7, 66cl4, and 4T1) in one branch, separated from the branch containing NMuMG and 67NR as well as the unconditioned media control (Fig. 3A). This result suggested quantitative differences in extracellular metabolites between non-metastatic cells and metastatic cells, yet the influence of transformation was not revealed, possibly due to the limited number of metabolites profiled. One challenge in interpreting the results is to distinguish which metabolites were secreted by tumor cells and which were left over from the consumption of the media. Because the growth media were Dulbecco's modified Eagle's medium (DMEM) with 7.5% dialyzed fetal bovine serum, all the small molecule compounds should derive from DMEM (amino acids, vitamins, glucose, and inorganic salts). Among the metabolites not included in the DMEM components, i.e. secreted from cells, two interesting patterns were noted (Fig. 3A, blue and orange bars). In the cluster of metabolites highlighted by the blue bar, several metabolites were more abundant in the metastatic lines (Fig. 3B). In particular, malate and fumarate were consistently higher in all four metastatic lines, whereas citrate/isocitrate and aconitate were higher in at least three of the lines. These metabolites all belong to the tricarboxylic acid cycle, consistent with the increased levels of these compounds as the intracellular form in the metastatic cell lines (Fig. 2, D and E). Another interesting pattern, highlighted by the orange bar, was the higher level of several nucleotide metabolism players, including guanine, xanthine, hypoxanthine, and deoxyguanosine, in the more metastatic cell lines 4TO7, 66cl4, and 4T1 when compared with NMuMG, 67NR, and the weakly metastatic 168FARN (Fig. 3C). Again, this trend was consistent with the finding of changes of intracellular nucleotide species (Fig. 2D). Overall, the data reinforce the conclusion from the analysis of intracellular metabolites; tricarboxylic acid cycle and nucleotide metabolism alterations accompany the acquisition of metastatic potential in the cell line series. Several amino acids showed significantly less abundance in the metastatic cell conditioned media (Fig. 3A, green bar, and Fig. 3D), presumably due to more consumption of amino acids by the more aggressive cells; thus, they were not considered as changes of secreted metabolites. Another group of metabolites showed a similarly higher level in all cell lines when compared with the unconditioned media (Fig. 3A, purple bar) and were not further scrutinized.
FIGURE 3.
Characterization of metabolite changes in conditioned media. A, hierarchical clustering of metabolomic profiles of the conditioned media identified subsets of metabolites that correlated with metastatic ability. Each cell line has four biological replicates (except 4TO7 with two samples) labeled above the heat map with numbers as indicated in Fig. 1A. Number 0 represents the unconditioned media. Blue, orange, green, and purple bars indicate patterns further examined. B, metabolites increased in most of the metastatic cells. C, metabolites increased in more metastatic cells when compared with the non-tumorigenic NMuMG and the non-metastatic 67NR (represented by 1 and 2) as well as the weakly metastatic 168FARN (represented by 3). D, metabolites with lower abundance in conditioned media of cells with metastatic potential. E, schematic of the two-step metabolic progression hypothesis. PPP, pentose phosphate pathway.
DISCUSSION
Malignant transformation of normal epithelial cells and metastatic ability acquisition have usually been studied with the aim to identify genes and proteins that play tumor-promoting or suppressive roles (26–28). The pace of finding such molecules has been tremendously accelerated with the development of transcriptomics and proteomics to look for transcripts and proteins with altered abundance during malignancy. Another aspect of molecular changes, altered metabolism, was less explored, although the phenomenon of aerobic glycolysis was one of the first major discoveries in cancer research (16). Fortunately, the situation is improving because of the recent technical advances in global analysis of metabolites using mass spectrometry or high resolution 1H nuclear magnetic resonance spectroscopy (8). High throughput metabolomic analysis allows simultaneous quantification of hundreds of metabolites belonging to a diverse array of metabolic pathways in a panel of cell lines or tissues. As illustrated in this study, 157 metabolites were profiled in six cell lines with progressively increased tumorigenicity and metastatic ability. The analysis of intracellular metabolites clustered the lines into three categories: normal, tumorigenic (but non-metastatic), and metastatic in general. Results from the analysis favor a two-step metabolic progression hypothesis during mammary tumor progression (Fig. 3E). The first step accompanies the acquisition of tumorigenicity and includes altered glycolysis, pentose phosphate pathway (PPP), and fatty acid synthesis, as well as decreased GSH/GSSG redox pool; the second step is correlated with the gain of the general metastatic ability and includes further changes in glycolysis and tricarboxylic acid cycle (TCA cycle), further depletion of the glutathione species, and increased nucleotides. No further metabolite alterations correlated with stepwise increase of metastasis potential in the four metastatic lines were resolved in the analysis. This model suggests that the fine regulation of the ability to colonize distant organs in breast cancer may not require further dramatic biochemical reprogramming and may instead rely more on alterations in gene expression regulation and cellular behaviors, although we cannot rule out the potential importance of metabolomic changes in metastatic lesions in vivo in different target organs. Our analysis of extracellular metabolites identified increased abundance of tricarboxylic acid cycle components as well as nucleotide metabolism intermediates, similar to the intracellular results. Our findings agree with a recent study profiling a more limited set of metabolites in the MCF10 model of mammary carcinoma (19). Both studies find evidence for increased pentose phosphate pathway, tricarboxylic acid cycle, and fatty acid biosynthetic activity in transformed and/or metastatic cells (19). Further efforts should investigate the universality of our findings with other in vitro and in vivo preclinical models as well as with human samples. Confirmed altered metabolic pathways may open new therapeutic avenues for treating malignant breast cancer. Several secreted metabolites accompanying the increased metastatic potential (malate, fumarate, deoxyguanosine, guanine, xanthine, and hypoxanthine) should be tested for their value as diagnostic and prognostic biomarkers of malignant breast cancer in future studies.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health R01 Grant R01CA134519 (to Y. K.) and R21 Grant R21CA128620 (to J. R.). This work was also supported by the Brewster Foundation and a Department of Defense Era of Hope Scholar Award BC051647.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- ESI
- electrospray ionization
- DMEM
- Dulbecco's modified Eagle's medium.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Murray T., Thun M. J. (2008) CA Cancer J. Clin. 58, 71–96 [DOI] [PubMed] [Google Scholar]
- 2.Lu X., Kang Y. (2007) J Mammary Gland Biol. Neoplasia. 12, 153–162 [DOI] [PubMed] [Google Scholar]
- 3.Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 4.Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu X., Wang Q., Hu G., Van Poznak C., Fleisher M., Reiss M., Massagué J., Kang Y. (2009) Genes Dev. 23, 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Y. (2005) Expert Rev. Mol. Diagn. 5, 385–395 [DOI] [PubMed] [Google Scholar]
- 7.Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R. A. (2004) Cell 117, 927–939 [DOI] [PubMed] [Google Scholar]
- 8.Griffin J. L., Shockcor J. P. (2004) Nat. Rev. Cancer 4, 551–561 [DOI] [PubMed] [Google Scholar]
- 9.Kroemer G., Pouyssegur J. (2008) Cancer Cell 13, 472–482 [DOI] [PubMed] [Google Scholar]
- 10.Sreekumar A., Poisson L. M., Rajendiran T. M., Khan A. P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R. J., Li Y., Nyati M. K., Ahsan A., Kalyana-Sundaram S., Han B., Cao X., Byun J., Omenn G. S., Ghosh D., Pennathur S., Alexander D. C., Berger A., Shuster J. R., Wei J. T., Varambally S., Beecher C., Chinnaiyan A. M. (2009) Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11.Bajad S. U., Lu W., Kimball E. H., Yuan J., Peterson C., Rabinowitz J. D. (2006) J. Chromatogr. A 1125, 76–88 [DOI] [PubMed] [Google Scholar]
- 12.Luo B., Groenke K., Takors R., Wandrey C., Oldiges M. (2007) J. Chromatogr. A 1147, 153–164 [DOI] [PubMed] [Google Scholar]
- 13.Munger J., Bajad S. U., Coller H. A., Shenk T., Rabinowitz J. D. (2006) PLoS Pathogens 2, e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aslakson C. J., Miller F. R. (1992) Cancer Res. 52, 1399–1405 [PubMed] [Google Scholar]
- 15.Owens R. B., Smith H. S., Hackett A. J. (1974) J. Natl. Cancer Inst. 53, 261–269 [DOI] [PubMed] [Google Scholar]
- 16.Warburg O., Wind F., Negelein E. (1927) J. Gen. Physiol. 8, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhajda F. P. (2000) Nutrition 16, 202–208 [DOI] [PubMed] [Google Scholar]
- 18.Zu X. L., Guppy M. (2004) Biochem. Biophys. Res. Commun. 313, 459–465 [DOI] [PubMed] [Google Scholar]
- 19.Richardson A. D., Yang C., Osterman A., Smith J. W. (2008) Breast Cancer Res. Treat. 110, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonveaux P., Végran F., Schroeder T., Wergin M. C., Verrax J., Rabbani Z. N., De Saedeleer C. J., Kennedy K. M., Diepart C., Jordan B. F., Kelley M. J., Gallez B., Wahl M. L., Feron O., Dewhirst M. W. (2008) J. Clin. Invest. 118, 3930–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filomeni G., Rotilio G., Ciriolo M. R. (2002) Biochem. Pharmacol. 64, 1057–1064 [DOI] [PubMed] [Google Scholar]
- 22.Griffith O. W., Meister A. (1979) J. Biol. Chem. 254, 7558–7560 [PubMed] [Google Scholar]
- 23.Filomeni G., Rotilio G., Ciriolo M. R. (2003) FASEB J. 17, 64–66 [DOI] [PubMed] [Google Scholar]
- 24.Christofk H. R., Vander Heiden M. G., Wu N., Asara J. M., Cantley L. C. (2008) Nature 452, 181–186 [DOI] [PubMed] [Google Scholar]
- 25.Munger J., Bennett B. D., Parikh A., Feng X. J., McArdle J., Rabitz H. A., Shenk T., Rabinowitz J. D. (2008) Nat. Biotechnol. 26, 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn W. C., Weinberg R. A. (2002) Nat. Rev. Cancer 2, 331–341 [DOI] [PubMed] [Google Scholar]
- 27.Gupta G. P., Massagué J. (2006) Cell 127, 679–695 [DOI] [PubMed] [Google Scholar]
- 28.Kang Y. (2006) Breast Dis. 26, 129–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.