Abstract
During immunoglobulin heavy chain (Igh) V(D)J recombination, D to J precedes V to DJ recombination in an ordered manner, controlled by differential chromatin accessibility of the V and DJ regions and essential for correct antibody assembly. However, with the exception of the intronic enhancer Eμ, which regulates D to J recombination, cis-acting regulatory elements have not been identified. We have assembled the sequence of a strategically located 96-kb V-D intergenic region in the mouse Igh and analyzed its activity during lymphocyte development. We show that Eμ-dependent D antisense transcription, proposed to open chromatin before D to J recombination, extends into the V-D region for more than 30 kb in B cells before, during, and after V(D)J recombination and in T cells but terminates 40 kb from the first V gene. Thus, subsequent V antisense transcription before V to DJ recombination is actively prevented and must be independently activated. To find cis-acting elements that regulate this differential chromatin opening, we identified six DNase I-hypersensitive sites (HSs) in the V-D region. One conserved HS upstream of the first D gene locally regulates D genes. Two further conserved HSs near the D region mark a sharp decrease in antisense transcription, and both HSs bind CTCF in vivo. Further, they both possess enhancer-blocking activity in vivo. Thus, we propose that they are enhancer-blocking insulators preventing Eμ-dependent chromatin opening extending into the V region. Thus, they are the first elements identified that may control ordered V(D)J recombination and correct assembly of antibody genes.
Keywords: Antisense RNA, Chromatin, DNase, Gene Regulation, Transcription Regulation, Immunoglobulin, Insulator, V(D)J Recombination
Introduction
V(D)J recombination of the multigene antigen receptor loci is essential for the generation of a diverse antigen receptor repertoire. Recombination is strictly regulated, occurring only in lymphocytes due to restricted expression of the recombination activating gene enzymes, RAG1 and RAG2, therein. Further, T cell receptors only recombine in T cells, B cell receptors only recombine in B cells, and the loci only recombine at specific stages in lymphocyte differentiation. In B cells, the Igh recombines before the Ig light chains. Finally some antigen receptor loci (e.g. the Igh) have two ordered recombination events. A D gene first recombines with a J gene on both alleles, followed by recombination of a V gene to the DJ recombined segment. Once a productive VDJ rearrangement has been generated, further V to DJ recombination is prevented on the second allele, a process termed allelic exclusion, which in B cells ensures that each B cell expresses a monoclonal IgH (1).
Ordered recombination is crucial for antigen receptor integrity, but key questions remain: how is recombination order achieved, and how is it regulated? Numerous studies have suggested that order is achieved through alterations in the chromatin conformation of individual gene domains at sequential stages of lymphocyte development (2). In the mouse Igh locus, the D-J-C region acquires histone post-translational modifications characteristic of open chromatin before the V region (3, 4). Non-coding RNA transcripts, including Iμ, generated from Eμ, located 3′ of the J genes (5), and μo, transcribed from the promoter of the most 3′ D gene, DQ52, occur on germ line alleles (6). Following D to J recombination, non-coding transcripts are generated from the V genes (7, 8). Furthermore, extensive antisense intergenic transcription occurs throughout the D and J domains before D to J, and then throughout the V domain before V to DJ recombination (9, 10). Nuclear positioning may also play a role in ordered V(D)J recombination. The Igh locus is tethered at the nuclear periphery via the V region in non-B cells (11, 12). Relocation toward euchromatic regions occurs preferentially from the DJC end, favoring D to J recombination. Furthermore, locus compaction through DNA looping is required for distal V gene recombination (13, 14). Several transcription factors, including Pax5 (13), YY1 (15), and Ikaros (16), play a role in looping, and in their absence, only the D-proximal V genes recombine. Following productive V(D)J recombination and cell surface expression of an IgH polypeptide, several of the above processes are reversed to silence V to DJ recombination of the second allele by allelic exclusion. Both Igh V regions decontract, V region germ line transcription is lost, and the second Igh allele is recruited to pericentric heterochromatin via the D-distal V genes (1). In contrast, both DJC regions remain transcriptionally active (9, 17). Thus, there is differential chromatin regulation of both activation and inactivation of the DJC versus V regions of the Igh locus.
With the exception of the intronic enhancer Eμ, the regulatory elements that control ordered recombination and allelic exclusion have not been identified. Eμ is required for efficient D to J recombination (18, 19). It acts in part by activation of antisense intergenic transcription, which is abrogated in the DJ region by Eμ deletion (10). It is unclear whether Eμ is required for V to DJ recombination. However, the V region is transcribed in its absence (10, 18, 19), suggesting that additional elements that activate the V region are present in the Igh locus. The only other element identified in the V-D-J region, the PDQ52 promoter/enhancer, is unlikely to play a role because its deletion does not affect germ line V gene transcription (19) or V to DJ recombination (20). Furthermore, the large V region (2.5 Mb), contains 195 V genes (500 bp) separated by intergenic sequences of 10–20 kb (21). Active histone modifications and germ line transcription associated with V gene promoters are very localized (22), suggesting that they are insufficient to activate the entire V region. To date, the only candidate element implicated in V to DJ recombination is a pro-B cell-specific DNase I-hypersensitive site (HS)5 5′ of the V region (23). However, preliminary studies suggest that it may repress V to DJ recombination.
We have previously assembled the V and D region sequences of the C57BL/6 mouse Igh locus (10, 21), revealing that they are separated by 96 kb of DNA sequence. Here we test the hypothesis that this uncharacterized region contains cis-acting regulatory elements, strategically positioned to influence ordered V(D)J recombination. Such elements may act as insulators, either to prevent heterochromatin spreading from the V to the D region in pro-B cells undergoing D to J recombination or to prevent enhancer-mediated activating processes spreading from the D to the V region. Alternatively, the region may contain enhancers that activate the V region.
Here we have characterized the mouse Igh V-D intergenic region to determine its activity during lymphocyte development and to identify putative regulatory elements therein. We show here that antisense transcription extends 30 kb upstream from the D region in B and T cells. We identify six novel DNase I HSs and investigate their roles by determining their lineage specificity and by identifying key interacting factors and functions in vivo. Two HSs interact with CTCF and have enhancer-blocking activity in B cells. Our results suggest that the V-D intergenic region contains a V region-activating element and insulator elements that separate the V and D regions into distinct chromatin domains.
EXPERIMENTAL PROCEDURES
Mice, Cell Lines, and Cell Sorting
Rag1−/− mice (24), back-crossed to C57BL10, and C57BL6/J wild type mice were maintained in the Babraham Institute Small Animal Barrier Unit, and all animal work was performed under project license PPL 80/2084, in compliance with Home Office guidelines. CD19-positive cells from Rag1−/− bone marrow were sorted using anti-CD19 MACsTM magnetic beads (Miltenyi Biotech; purity >80% CD19+). Fractions representing sequential stages of bone marrow B cell development (25) (Hardy fractions A, B/C, and C′) were sorted on a FACSAria as follows: Fraction A, B220+CD43+CD19−; fraction B, B220+CD19+CD43+BP1−; fraction C, small B220+CD19+CD43+BP1+; fraction C′, large B220+CD19+CD43+BP1+. The antibodies used (BD Pharmingen) were as follows: CyChrome-labeled anti-B220 (RA3–6B2), allophycocyanin-labeled anti-CD19 (1D3), phycoerythrin-labeled anti-BP1, and fluorescein isothiocyanate-labeled CD43 (S7). The Rag2−/− pro-B cell line (26), the TK-1 and BW5147 thymoma cell lines, and the RAW264 macrophage line were maintained in RPMI 1640 with Glutamax and 10% (v/v) fetal bovine serum, 50 μm 2-mercaptoethanol.
Bioinformatic Analysis
BLAST searches of the National Center for Biotechnology Information, Baylor College of Medicine, and Ensembl data bases identified bacterial artificial chromosome sequences from the C57BL6/J Mus musculus and Rattus norvegicus Igh V-D intergenic region. The sequence of the mouse region was established through bacterial artificial chromosome assembly using Sequencher (Gene Codes): RP23-109B20, RP24-275L15, RP23-404D8, and RP23-270B12 (the last two cover the Igh D region) (27). These bacterial artificial chromosomes provide at least 2-fold coverage except in four regions of 12,735, 1087, 528, and 430 bp, which have single strand coverage. Sequence analysis was performed using Nucleotide Identity X, provided by the Human Genome Mapping Project (21) and RepeatMasker (available on the World Wide Web). Large sequences were compared using VISTA global alignment (available on the World Wide Web), Pipmaker (available on the World Wide Web), or the Artemis Comparison Tool (ACT, version 3) local alignment programs with the default settings. Small sequences (<9 kbp) were compared by ClustalW. Long interspersed nucleotide elements (LINEs) were characterized with the L1Base data base (available on the World Wide Web).
Real-time and Strand-specific RT-PCR
RNA was purified using the RNeasy kit (Qiagen). DNA was removed using the RQ1 DNase I kit (Promega). The RNA was repurified using the RNeasy kit and RNA cleanup protocol (Qiagen). 1 μg of RNA was reverse transcribed using 100 ng of random hexamers (Amersham Biosciences) and Superscript III (Invitrogen) at 50 °C for 1 h. Samples were analyzed by real-time PCR using an ABI PRISM 7000 sequence detection system and SYBR Green Fluorogenic dye (Applied Biosytems). Thermocycling conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60–64 °C (primer-dependent) for 1 min. Relative quantification was performed with standard curves of serial dilutions of genomic DNA. Samples were normalized to a normalization factor calculated for each sample from four stably expressed housekeeping genes (β2m (β2-microglobulin), Tbp (TATA box-binding protein), hprt1 (hypoxanthine phosphoribosyltransferase 1), and sdha (succinate dehydrogenase complex, subunit A)), using the geNORM method (28). The normalization factor was set to 1 arbitrary unit. Alternatively, samples were normalized to β-actin and then expressed relative to transcription of DFL16.1 in thymus. For strand-specific RT-PCR, RNA was reverse transcribed with sequence-specific primers. For detection of the 3′Adam6 gene, a two-round PCR approach was used (15 cycles and then a 1:20 volume diluted in a second PCR with nested primers, 35 cycles). Primers are detailed in supplemental Table 1.
DNase I Hypersensitivity Assays
DNase I hypersensitivity assays were performed based on previous experimental procedures (29). 2 × 106 nuclei were treated with DNase (0.05–1.00 units) (Roche Applied Science) at 37 °C for 3 min. Following the addition of 300 mm NaCl, 10 mm Tris-HCl, pH 8.0, 0.5% (w/v) SDS, 5 mm EDTA, and proteinase K (250 μg/ml), the DNA was purified by phenol/chloroform/isoamyl alcohol extraction. DNase I-treated DNA (5–15 μg) was digested to completion with restriction endonucleases. For fine mapping of HSs, an extra control sample was digested with the appropriate restriction endonucleases. For Southern blotting, DNA was fractionated through 0.8% (w/v) agarose gels, transferred to nylon membranes (Hybond N+, GE Healthcare), and hybridized at 65 °C for 14–18 h in modified Church and Gilbert buffer (50 mm Na2HPO4, pH 7.5, 7% (w/v) SDS, 10 mm EDTA) with DNA probes labeled with [α-32P]dCTP using the Radprime DNA labeling kit (Invitrogen). Probes were generated by PCR (primers detailed in supplemental Table 2) and cloned into pGEM-Teasy (Promega). After hybridization, membranes were analyzed using phosphor imager (Fujifilm) or BioMax MS-1 film (Eastman Kodak Co.).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed on Rag1−/− CD19+ cells essentially as described previously (30), with the following modifications. The sample was sonicated with a Diagenode Bioruptor (high power, 10 cycles of 30 s on/off). Dynal Protein A beads (Invitrogen) were incubated with 5 mg of CTCF antibody (Upstate) and 5 mg of rabbit anti-goat nonspecific control antibody (Sigma). Bound fractions were diluted 1:10, and input was diluted to 10 ng/ml, and 1 ml of each was used in triplicate and compared with a genomic DNA standard to normalize for different primer efficiencies. Results were compared with input to calculate -fold enrichment. Relative enrichment of MTA was set to 1 for comparison between experiments.
Enhancer-blocking Assay
The backbone plasmid, pNI, generously provided by Gary Felsenfeld, contains a neomycin resistance gene linked to the human γ-globin promoter and the hypersensitive site 2 enhancer (mHS2) from the murine β-globin LCR, with an intervening AscI site for cloning putative enhancer-blocking elements. The 1.2-kb full-length chicken β-globin HS4 insulator protects the construct from position effect variegation. DNA fragments comprising HS4, HS4 paralogue, HS5, and HS6 were PCR-amplified from Pro B cell genomic DNA with primers containing AscI linkers (supplemental Table 1) and cloned into pTeasy. After sequence verification, the inserts were cloned into the AscI site of pNI in both orientations to create pNI-HSF (forward) and pNI-HSR (reverse). The forward direction reflects the endogenous orientation in the Igh locus, pointing toward DFL16 and the intronic enhancer. A 250-bp fragment comprising the core chicken β-globin 5′HS4 insulator was cloned into pNI to create the positive control insulator pNI-cINS. pNI-cINS was partially digested with AscI, and a second copy of the 250-bp insulator was cloned in tandem to generate pNI-2cINS. K562 cells (106) were transfected with the HS constructs (2 μg of linearized DNA) by Amaxa nucleofection, with Nucleofector Kit V, optimized for K562 cells, according to the manufacturer's instructions. The cells were transferred to 2 ml of Iscove's modified Dulbecco's medium with 10% fetal bovine serum and incubated overnight at 37 °C, 5% CO2. The next day, 1 ml of cell suspension was mixed with 29 ml of Iscove's modified Dulbecco's medium with 10% fetal bovine serum, 750 mg/ml active G418 (Invitrogen) plus 3.5 ml of 3% cell culture agar (Sigma), poured into a 140-mm tissue culture dish, and incubated at 37 °C in 5% CO2. The number of G418-resistant colonies was counted after 2–3 weeks.
RESULTS
Bioinformatic Characterization of the Mouse Igh V-D Intergenic Region
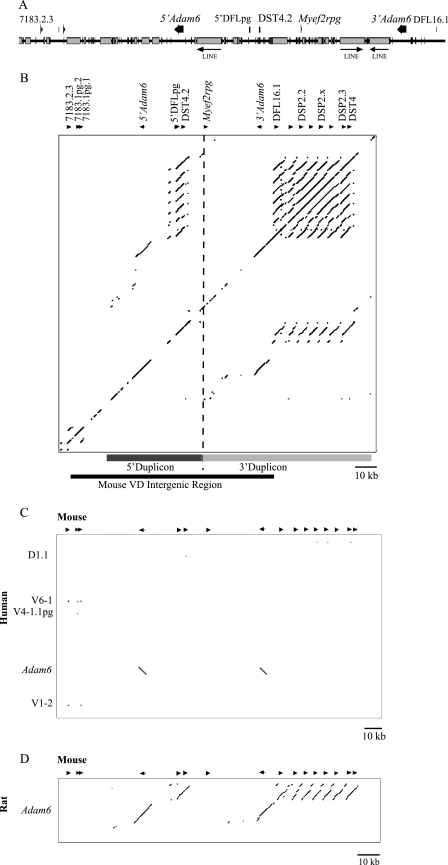
Our recent assembly of the C57BL6/J mouse Igh locus V region revealed a 90kb sequence between the most 3′ V gene, 7183.1.1pg and the most 5′ D gene, DFL16.1. Because the first two V genes (7183.1.1pg and Q52.1.2pg) are pseudogenes, we have included these in an extended region of 96,314 bp for analysis, which extends from 3′ of the first functional V gene, VH7183.2.3 (also previously named 81X) to the first D gene, DFL16.1. The V-D intergenic region contains a high proportion (56%) of interspersed repeats, similar to the Igh V region (52.4%) (21), including three 6-kb full-length LINE-1 repeats (Fig. 1A). Nucleotide Identity X analysis shows that it contains one previously reported DH gene (DST4.2), which has never undergone D to J recombination (27), and Myef2rpg (myelin basic expression factor 2 repressor pseudogene) (GenBankTM accession number XM_621300). Most notably, the V-D region encodes two genes, Adam6a and Adam6b (a distintegrin and metalloproteinase domain 6a and -b) (31). Adam6 belongs to a large protein family involved in cell adhesion (32). The Adam6 genes are here renamed 5′Adam6 and 3′Adam6, respectively, to denote their position with respect to the V genes. They are both oriented in the opposite direction to the V and D genes. These genes showed 99.8 and 94.2% nucleotide identity, respectively, to the mouse Adam6 cDNA sequence (GenBankTM accession AY158689). Two Adam6 copies suggested that this region might have been duplicated. This was further confirmed by local alignment of the sequence against a repeat-masked copy of itself (Fig. 1B), which showed that the duplicated sequences included part of the DH region, the Adam6 gene, and some upstream intergenic sequence. The DH-like region contains the DST4.2 gene and a DH pseudogene, here named 5′DFLpg because it has 72.5% identity to the DFL16.1 gene.
FIGURE 1.
The mouse Igh V-D intergenic region has undergone a tandem duplication. A, sequence map of the V-D intergenic region showing the position and orientation of genes (arrows) and repeat sequences (gray boxes) identified. Predicted full-length LINE sequences are labeled. Note in particular the antisense orientation of the Adam6 genes with respect to the orientation of V and D genes. For clarity, locus positions of the Adam6 genes will refer to their geographical position with respect to gene V7183.2.3. B, dot plot showing local alignment of the mouse Igh locus V-D intergenic region against a repeat-masked copy of the same sequence. Arrows, drawn to scale, above the dot plot, show the gene positions and orientations. The 5′ duplicon is indicated by a dark gray line, the 3′ duplicon is shown by a light gray line, and the boundary between them is shown by a dashed line. The black line indicates the V-D intergenic region. C and D, dot plots showing local alignment of the repeat-masked mouse V-D intergenic sequence on the x axis to the human V-D (C) and the partial rat V-D intergenic sequence (D) on the y axis. Genes in the mouse sequence are shown as in B, and genes in the human and rat sequence are labeled.
Sequence Conservation of the V-D Region
To initially assess whether the Igh locus V-D intergenic region has a regulatory function, conserved non-coding sequences were sought. The human Igh sequence (33) contains a single Adam6 gene just upstream of the most 3′ V gene and a neighboring V pseudogene. We therefore analyzed an extended sequence surrounding the human V-D intergenic region, including the Adam6 gene. Alignment to the repeat-masked mouse V-D intergenic sequence revealed nucleotide conservation of the flanking immunoglobulin genes and the Adam6 genes but not of non-coding sequences (Fig. 1C). Only one Adam6 gene in the human Igh indicates that the mouse sequence duplication is not conserved. Identification and alignment of a partial sequence of the rat (R. norvegicus) V-D intergenic region (RNOR03303655), containing a DH gene, an Adam6 gene, and 14,252 bp of upstream sequence, to the repeat-masked mouse V-D intergenic region sequence showed several conserved non-coding sequences (Fig. 1D). These included 600 bp positioned downstream of both mouse Adam6 genes, with respect to the locus (77% nucleotide identity), and a second 500-bp sequence (76% identity), between the sequence downstream of the 3′Adam6 gene and the DFL16 gene. Due to the absence of complete sequence, it is not yet known whether the rat V-D region contains a sequence duplication.
Antisense Transcription Continues Upstream of the Igh D throughout B Cell Development but Terminates 40 kb from the V Region
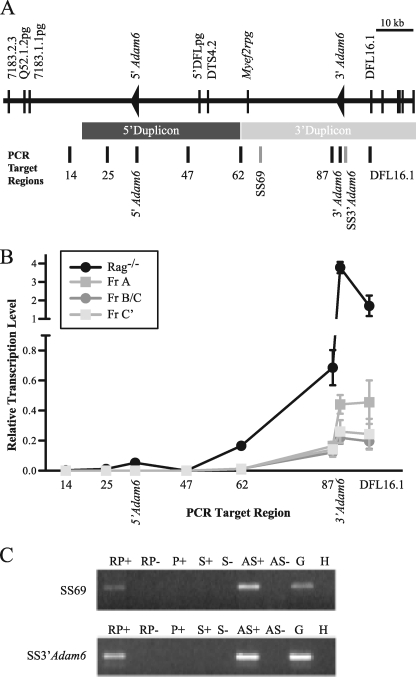
We have recently shown that antisense intergenic transcription, initiating 5′ of and dependent upon the Igh intronic enhancer, occurs throughout the >60-kb Igh D and J region prior to D to J recombination (10). We have proposed that it remodels chromatin to facilitate D to J recombination. Importantly, this transcription is distinct from antisense intergenic transcription in the V region, which occurs at the next B cell developmental stage before V to DJ recombination. This raised a number of possibilities. First, transcription may be actively inhibited from progressing upstream of the D region, either permanently or until after D to J recombination. Alternatively, the V-D intergenic region may also be transcribed before D to J recombination due to the continued transit of the RNA polymerase complex. In the latter case, either transcription inefficiency, coupled with the large sequence distance, may passively prevent it from extending all the way to the V region, or it may be actively blocked immediately adjacent to the upstream V region. To distinguish between these possibilities, random-primed quantitative real-time RT-PCR was performed across the region (Fig. 2A), in ex vivo Rag1−/− bone marrow CD19+B cells. Recombination does not occur in these cells, and both the DJ and V regions are in a chromatin state poised for V(D)J recombination (3, 10). RNA samples from ex vivo bone marrow B cell fractions A, B/C, and C′ and lymphocyte cell lines representing sequential activation states of the Igh locus were also included to analyze developmental patterns of transcription. Fractions A, B/C, and C′ have the Igh locus in germ line/DJ, DJ/VDJ, and VDJ (on one or both alleles), respectively. The Adam6 genes were included in this analysis to distinguish whether they are actively transcribed, which might suggest protein-coding function, or passively transcribed along with other V-D intergenic sequences. Analysis of ex vivo Rag−/− pro-B cells and fractions A, B/C, and C′ is shown in Fig. 2B. All samples showed transcription across a large part of the V-D region, highest at the DFL16.1 gene, and decreasing toward the VH region, suggesting that transcription of the V-D intergenic region represents the continuation of DH antisense transcription. Strand-specific RT-PCR (Fig. 2C) demonstrated that transcription occurs in the antisense direction, with respect to the orientation of the V, D, and J genes, further supporting a continuation of antisense transcription from the D region. Transcription exhibited a biphasic pattern, decreasing most acutely between DFL16.1 and the 87-kb site upstream of the 3′Adam6 gene, followed by more gradual reduction until transcription was virtually undetectable at the 5′Adam6 gene. Thus, transcription terminated at least 41 kb from the first V gene, 7183.1.1pg. This is consistent with the expression of the DH and VH antisense transcripts at sequential developmental stages (9, 10) and suggests that the V-D intergenic region or elements therein prevent DH antisense transcription from continuing into the VH region.
FIGURE 2.
DH region antisense transcription decreases through the V-D intergenic region. A, schematic representation of the Igh V-D intergenic region illustrating the regions analyzed for transcription by real-time and strand-specific RT-PCR. The numbers below the PCR target regions indicate distance from the 7183.2.3 gene. B, graph depicting relative transcription levels analyzed by real-time RT-PCR in Rag−/− pro-B and wild type fraction A, B/C, and C′ cells. Transcription levels were compared with that of the geNORM housekeeping gene normalization factor for each individual cell type. This value was arbitrarily set to 1. Distance of PCR amplicons from 7183.2.3 is drawn to scale. C, representative examples of PCR products generated by strand-specific RT-PCR. RT reactions were carried out with random hexamers (RP), no primer (P), antisense primer to detect sense transcription (S), or sense primer to detect antisense transcription (AS). RT reactions were performed with (+) and without (−) reverse transcriptase. Genomic DNA (G) and water (W) were included as controls.
It is unclear why Rag−/− pro-B had 10-fold greater transcription levels than fractions A, B/C, and C′, but this finding agrees with our previous studies of V region antisense transcription, which was also higher in Rag−/− pro-B cells versus wild type pro-B cells (9). Rag−/− pro-B cells are blocked at the stage when antisense transcription is occurring and may continue to transcribe rather than progress to the next developmental stage. At the DFL16 gene, Rag−/− pro-B cells had 2–4-fold higher transcription levels than the geNORM normalization control, whereas the fractions had 2–5-fold lower than the control. Nevertheless, this is a high frequency for non-coding transcripts, which are often 100-fold lower than coding transcripts. The patterns of transcription were similar in fractions A, B/C, and C′. However, notably, the quantity of transcripts in fraction A was roughly twice that of fractions B/C and C′. All alleles in fraction A are germ line or DJ recombined and thus retain the V-D region, whereas only 25–50% of those in the other fractions do due to ongoing V(D)J recombination. This suggests that the pattern and rate of transcription is constant on individual alleles throughout early B cell development. Because in fraction C′, in particular, the remaining DJ allele retaining the V-D region is silenced by allelic exclusion, this suggests that V-D transcription must continue to be actively blocked to prevent V to DJ recombination of the second allele.
Because wild type fractions have heterogeneous Igh locus configurations, we sought to determine the transcription patterns of individual locus configurations, by employing cell lines with clonal Igh locus configurations. BW5147 thymoma cells (34) represent a silent Igh locus because it is unrearranged and expresses negligible levels of Iμ and μ0 transcripts (data not shown). TK-1 thymoma cells (35) transcribe Iμ and μ0 but do not undergo DH to JH recombination (data not shown) and thus contain an Igh locus actively poised before D to J recombination. Ex vivo wild type thymus cells express Iμ, μ0, and DH antisense transcripts (10) and undergo D to J but not V to DJ recombination; thus, the Igh locus is poised before V to DJ recombination (36). Transcription of the V-D intergenic region was undetectable in the BW5147 line, indicating that V-D is not transcriptionally active in a silent Igh locus. Transcription was detected in all other cell lines, with a transcriptionally active DJ region (supplemental Fig. 1). The pattern in thymus and the Rag−/− cell line was identical to fractions A, B/C, and C′. In TK-1 cells, transcription was sustained at similar levels to a distance of 30 kb upstream of DFL16, decreasing more sharply thereafter between the 62 and 47 kb sites to reach basal levels similar to those of the other cell lines at 5′Adam6.
Transcription levels did not increase significantly at the Adam6 genes, suggesting that the Adam6 promoters are inactive in lymphocytes and that transcription from the 3′Adam6 gene in particular was due to transcriptional read-through from the D region rather than active messenger RNA production. This suggests that the protein products of these genes are not expressed in lymphocytes. This hypothesis was further supported by RT-PCR analysis of nuclear and cytoplasmic RNA, which demonstrated that Adam6 transcripts are restricted to the nucleus in lymphocytes (supplemental Fig. 2). In contrast, high levels of cytoplasmic Adam6 transcripts were detected in testis cells, a predicted site of ADAM6 protein expression.
Identification of DNase I-hypersensitive Sites in the Mouse Igh V-D Intergenic Region
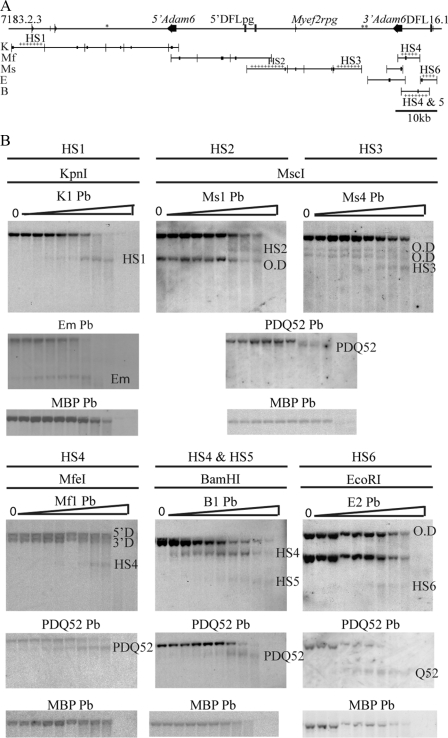
To determine whether the mouse Igh V-D intergenic region contains regulatory elements, DNase I hypersensitivity assays were performed. These assays detect increased accessibility of chromatin structure, usually caused by trans-acting factor interactions with DNA, and are used to detect cis-acting regulatory elements (37). The entire 96-kb V-D region was analyzed by Southern blotting, using the cell lines described in supplemental Fig. 1, since these had identical transcription patterns to fractions A, B/C, and C′, had more homogeneous Igh locus configurations, and provided large cell numbers. Initial studies on the Rag2−/− cell line, using 18 restriction fragments and sequence-specific probes, are detailed in Fig. 3A and supplemental Table 2. 17 of the 18 Southern blots gave an uncut restriction fragment (parental band) of the expected size, validating the assembled V-D sequence. We detected six DNase I HSs (Fig. 3B), indicated by subfragments generated upon increasing DNase I digestion. Their positioning was complicated by the V-D intergenic region duplication, but careful use of different sizes of cut fragments enabled accurate HS positioning. In particular, a full-length LINE between 5′Adam6 and 5′DFLpg provides a significant gap in the duplication, allowing HS4 to be unambiguously localized to the 3′ duplicon. The HSs were localized to the restriction fragments shown in Fig. 3, which range in size from 5180 to 9666 bp. Specificity and sensitivity were validated by probing for known DNase I HSs associated with Eμ and PDQ52 (3, 6), proximally located in the Igh locus. The neuron-specific MBP (myelin basic protein) promoter was used as negative control. The DNase I HSs were further validated using a second set of restriction digest/probe combinations, and their positions were further determined by fine mapping relative to proximal restriction endonuclease sites (supplemental Fig. 3). This resolved the HS sizes to ∼1 kb (supplemental Table 3). HS1 resides upstream of a PvuII site in the VH7183.2.3 gene and thus probably represents the VH7183.2.3 promoter. HS2 is located within a partial LINE1 sequence downstream of the DST4.2 gene, and HS3 is located within a full-length LINE1 upstream of the 3′Adam6 gene. HS4, HS5, and HS6 are located between the 3′Adam6 gene and the DFL16.1 gene, with HS6 residing only 0.4–1.3 kb upstream of DFL16.1. The fine mapping also revealed that HS4 and HS5 are composed of multiple subfragments. Bioinformatic analysis showed that HS6 is part of the tandem repeat that comprises the DFL/DSP D gene array (supplemental Fig. 4). Homologous sequences to HS6 are therefore present upstream of each DFL and DSP gene. The HS5 sequence occurred only once within the mouse Igh locus, whereas the HS4 sequence was also present in the V-D 5′ duplicon, although DNase I hypersensitivity was only detected in the 3′ duplicon. Notably, HS4 and HS5 correspond to the 600- and 500-bp non-coding regions of greatest homology between the rat and mouse V-D intergenic sequences (Fig. 1). Identification of the six novel DNase I HSs suggests that the V-D intergenic region may contain regulatory regions.
FIGURE 3.
Identification of DNase I-hypersensitive sites in the V-D intergenic region. A, schematic representation of the mouse V-D intergenic region showing restriction fragments and positions of probes used in Southern blots to map DNase I hypersensitivity. Enzymes were as follows: ApaI (A), BamHI (B), BglI (BgI), BglII (BgII), BstEII (BsE), BstXI (BsX), EcoRI (E), KpnI (K), MfeI (Mf), MscI (Ms), PstI (P), PvuII (Pv), SapI (Sa), SpeI (S), XbaI (X), and XcmI (Xc). The positions of the probes (Pb) are indicated by black rectangles on the restriction fragment they were used to detect. The asterisks indicate regions that could not be assayed for DNase I HSs, and crosses indicate restriction fragments in which DNase I HSs were detected. B, Southern blots of DNA from Rag2−/− cell line in which DNase I HS sites were identified. Nuclei were treated with 0–1 unit of DNase/2 × 106 nuclei, indicated by triangles. The HSs are named by their position from the V proximal end of the region. The detection of more than one parental band by the cross-hybridization of the probe to the other duplicon (O.D) in the V-D intergenic region is indicated. Southern blots were reprobed with either the PDQ52 probe or Eμ probe as a positive control, followed by reprobing with the Mbp probe as a negative control.
Lineage Specificity of DNase I-hypersensitive Sites
The lineage specificity of DNase I HSs 2, 3, 4, and 5 was determined in primary cells and cell lines to gain further understanding of their functions. Analysis of Rag1−/− CD19+ and CD19− ex vivo bone marrow cells (Fig. 4) confirmed that the DNase I HSs are present in non-transformed primary cells and further defined the lineage specificity of these sites because the CD19+ cell population is composed of B cells, whereas the CD19− cell population is composed mainly of myeloid cells (38). The β2-microglobulin promoter was used as an additional positive control because Eμ and PDQ52 are not DNase I-hypersensitive in non-lymphoid cells. HS2 and HS3 may be restricted to B cells because the Rag2−/− pro-B cell line was the only cell line in which they were detected (Table 1). For HS3, this observation was substantiated by the finding that HS3 was present in Rag1−/− CD19+ cells but absent from CD19− cells (Fig. 4). However, HS2 could not be detected in either Rag1−/− cell population, suggesting that this site is unique to the Rag2−/− cell line and represents either a strain-specific difference or an artifact. HS4 may be active in all hematopoietic cell lineages because it was detected in all of the cell lines and at equivalent levels in both of the Rag1−/− cell populations. HS5 was detected only in the B and T cell lines and at a greatly reduced level in the Rag1−/− CD19− compared with the Rag1−/− CD19+ population (Fig. 4), suggesting that it is limited to the lymphocyte lineage. Fine mapping of HS4 and HS5 again showed multiple subfragments (data not shown).
FIGURE 4.
Lineage specificity of DNaseI HSs. Southern blots of DNA from Rag1−/− CD19+ and Rag1−/− CD19− bone marrow cells treated with DNase I are shown. Rag1−/− Southern blots with BamHI digested DNA show nuclei treated with 0.0, 0.3, 0.5, 0.6, 0.8, and 1.0 unit of DNase, indicated by triangles. Rag1−/− Southern blots with ApaI-digested DNA show nuclei treated with 0.0, 0.3, 0.4, 0.5, 0.6, and 0.8 units of DNase, indicated by triangles. Southern blots are labeled by the DNase I HS they were designed to detect and in parentheses the restriction enzymes and probes (Pb) used. Analysis of the β2-microglobulin promoter, and myelin basic protein promoter was carried out on all Southern blots, but only representative results from the BamHI Southern blot are shown. Cross-hybridization of the probe to the other duplicon within the V-D region is labeled by the other duplicon (O.D).
TABLE 1.
Lineage specificity of DNase I hypersensitive sites in cell lines
| HS | Rag−/− Pro-B | TK-1 T | BW5146 T | RAW264 macrophage |
|---|---|---|---|---|
| HS2 | + | − | − | − |
| HS3 | + | − | − | − |
| HS4 | + | + | + | + |
| HS5 | + | + | + | − |
HS4 and HS5 Have Functional CTCF Binding Sites
Because HS4 and HS5 sequences are highly conserved and are upstream of local regulation of D genes, we hypothesized that these sites might function as insulators. Insulators can exhibit barrier (boundary) function, which prevents spreading of histone modifications (e.g. those associated with heterochromatin) across the insulator, and/or enhancer-blocking function, which protects promoters from the activity of enhancers or silencers, and almost invariably requires CTCF binding, which has been proposed to isolate chromatin domains by facilitating looping out of DNA (39, 40). A computational search using the CTCF consensus binding site common to the well studied β-globin 5′HS4 boundary element and imprinted H19 promoter and X chromosome imprinting center (CCGCNNGGNGGCAG) (41), allowing two mismatches, revealed consensus CTCF binding sites in both HS4 and HS5 (CACCAAGGGGGAAG and CACAAGAGGGCAG), respectively. We next determined whether these putative sites were functional in vivo by performing CTCF chromatin immunoprecipitation in Rag1−/− CD19+ BM. Unique primers and stringent PCR conditions were used to amplify only the 3′HS4 sequence and not its homologous DNase-insensitive counterpart in the 5′ duplicon. A sequence from the Igh 3′ regulatory region hypersensitive site 7 (Igh3′RR-HS7) was used as a positive control because it contains multiple active CTCF sites in pro-B cells (42). Probes specific for MTA1 (metastasis-associated protein 1), downstream of the Igh3′RR-HS7, and IL5 (the interleukin-5 gene) were negative controls. As expected, Igh3′RR-HS7 was greatly enriched in the CTCF-bound fraction (110-fold), compared with the adjacent negative control, MTA1 (Fig. 5). Both HS4 and HS5 were also greatly enriched in the CTCF-bound chromatin fraction (55- and 80-fold respectively), despite each having only a single putative CTCF site, demonstrating that both HSs bind CTCF with high frequency in pro-B cells.
FIGURE 5.
CTCF binding to HS4 and HS5 by chromatin immunoprecipitation. The bar chart depicts results of chromatin immunoprecipitation with an anti-CTCF antibody, followed by real-time PCR analyses in Rag1−/− CD19+ BM cells. Nonspecific binding using a rabbit control antibody was minimal and subtracted before plotting. Results were compared with the input fraction to calculate -fold enrichment. Relative enrichment of the negative control, MTA, was set to 1 for comparison between experiments. For each primer pair, the bars depict a representative biological sample, in which experiments were performed twice in triplicate. Two independent Rag1−/− CD19+ BM samples were immunoprecipitated with anti-CTCF and analyzed in this manner with similar results.
HS4 and HS5 Are Enhancer-blocking Elements
CTCF confers the enhancer-blocking activity observed in vertebrate insulator elements. Therefore, we asked if HS4 and HS5 served as classical enhancer-blocking elements in a standard cellular assay, in which intervening enhancer-blocking elements prevent the murine β-globin HS2 enhancer from activating a neomycin-resistant gene, thereby preventing formation of neomycin-resistant colonies in soft agar (43). The constructs are depicted in Fig. 6, left. HS4 and HS5 were cloned in both orientations, and the HS4 5′ paralogous sequence and HS6 were included as controls. The classical 250-bp core insulator element (cINS) from the chicken β-globin HS4 reduced colony numbers to 60% of the control (Fig. 6, right). Inclusion of a second copy of cINS (pNI-2×cINS) reduced colony numbers to 30% of control, compared with the single copy cINS, indicating that enhancer-blocking activity was proportional to copy number. HS4-F had enhancer-blocking activity similar to that of cINS. When HS4 was cloned in the reverse orientation (HS4-R), the enhancer-blocking activity was reduced, indicating that it is orientation-dependent, as is often observed with CTCF sites. Some enhancer-blocking activity, albeit less than HS4, was detected with HS4-para, the upstream sequence paralogous to HS4, which is not DNase I-hypersensitive in vivo and does not contain a consensus CTCF site. This sequence is not in its normal silent chromatin context in this enhancer-blocking assay; thus, it exhibits activity that may not occur in vivo. Notably, its lack of a CTCF binding site indicates first that the full enhancer-blocking activity of HS4 is dependent on CTCF and, second, that it may also depend on additional factors. HS5 exhibited stronger enhancer binding activity than the control cINS insulator. This activity was ablated in HS5 cloned in reverse orientation. This underlines the orientation specificity of the CTCF site but in this case also suggests that HS5 insulator activity also depends on another factor that is highly orientation-dependent. HS6 had insignificant enhancer-blocking activity, supporting our hypothesis that it serves as a D gene promoter. Notably, because these assays were performed in human erythroleukemia K562 cells, the observed enhancer-blocking activity of HS4 and HS5 does not require a lymphoid-specific factor.
FIGURE 6.
HS4 and HS5 are enhancer blockers. Left, the enhancer-blocking test constructs are depicted, with open rectangles representing the full-length chicken HS4 insulator and the neomycin resistance gene, the latter with an arrow to represent promoter position. The open oval represents the β-globin HS2 enhancer. The AscI restriction enzyme site in pNI is replaced by rounded rectangles representing putative insulator elements in forward (F) or reverse (R) orientation. The filled ovals depict copies of the chicken core HS4 insulator. All constructs derive from pNI. Right, the number of neomycin-resistant colonies, reflecting the enhancer-blocking activity of each construct, was normalized to the backbone vector pNI, which lacked any putative enhancer-blocking elements, set to a value of 1. The data presented are the mean ± S.D. of three independent enhancer-blocking experiments, each with duplicate transfections.
DISCUSSION
We and others (9, 10, 44) have previously identified extensive antisense non-coding transcription in the Igh D and V regions, before D to J and V to DJ recombination, respectively. We have proposed that this transcription opens up chromatin to enable V(D)J recombination. This model is supported by studies showing that intergenic transcription is required for V to J recombination in the Tcrα locus (45). Igh D antisense transcripts (Eμ-dependent), and V region antisense transcripts (Eμ-independent) rarely occur simultaneously on the same Igh allele (10), suggesting that the V region is activated by a separate enhancer and/or they are actively maintained in separate chromatin domains. Here we show that antisense transcription from the D region continues into the V-D region in Igh loci poised for VDJ recombination but terminates 40 kb from the V region. This demonstrates that transcription in the D and V regions are separate events and that the later appearance of V antisense transcription (10) is not due simply to later transcription read-through. The V-D region in the human Igh locus is noticeably shorter (20.2 kb) (33). However, in the mouse Igh, most of the transcription was lost within 5 kb of the DFL16 gene, suggesting that similar regulation may nonetheless occur in the human locus, despite its smaller size. Importantly, our finding of antisense transcription in the V-D region before, during, and after V(D)J recombination refutes the possibility that this region only becomes transcriptionally active after D to J recombination, thereby ruling out an exclusive role in activating the V region. Together, these data support an alternative role in actively maintaining the DJ region in a separate chromatin domain to the inactive V region during D to J recombination. Notably, continued V-D transcription and termination on the DJ recombined allele after VDJ recombination of the first allele indicates that mechanisms and elements that ablate this transcription persist as part of the allelic exclusion mechanism that prevents further V to DJ recombination on the second allele. This is the first site-specific transcription checkpoint identified in the Igh locus and supports the model that non-coding RNA transcription plays a functional role that must be tightly regulated.
Does Adam6 Have a Function in B Lymphocytes?
We also identified two Adam6 genes within the sequence. ADAM6 proteins participate in cell adhesion, by interaction with αβ integrins, and activating membrane-bound cytokines (32). We hypothesized that they might participate in stromal cell-pro-B cell adhesion, which is essential for pro-B cell development but is lost once pro-B cells undergo V to DJ recombination, with consequent loss of V-D and Adam6 sequence. However, these genes do not appear to generate cytoplasmic protein-coding transcripts in B cells (supplemental Fig. 2) but rather appear to be transcribed only as a consequence of the antisense transcription proceeding through this region (Fig. 2 and supplemental Fig. 1). The analogous V-D region in the Tcrβ locus contains a trypsinogen gene, which is expressed in pancreas but not in T cells (46). These findings eliminate the possibility that ADAM6 plays a role in stromal cell-pro-B cell adhesion.
HS6 Is a Putative D Gene Promoter
HS6 is the most D-proximal HS site, 0.4–1.3 kb upstream of the DFL16.1 gene. Active histone modifications, including histone H3 lysine 9 acetylation and histone H3 lysine 4 dimethylation, have been reported here in pro-B cells, both in vitro (4) and in vivo (47). We propose that HS6 is a D gene promoter because the HS6 sequence is repeated upstream of all DSP genes, and weak HSs could be detected from those sequences also (data not shown). Second, HS6 overlaps at least 500 bp of a DSP promoter (48). Third, it has been suggested that there is a bidirectional promoter between 0.7 and 1.2 kb upstream of DFL16.1 (44). This coincides with our positioning of HS6 and supports our finding of antisense transcription upstream of DFL16.1 prior to D to J recombination. Fourth, we have shown here that HS6 has negligible enhancer-blocking activity, ruling it out as an insulator. Notably, HS6 corresponds to a site in which a V gene and its regulatory sequences were inserted in vivo (500 bp upstream of DFL16.1) (47). The V gene circumvented the normal constraints of ordered recombination. It was proposed that this was because it was now part of the DJ region and that order is normally maintained because the V region is in a different chromatin domain, perhaps separated by a boundary element upstream of the V gene knock-in site (47). Our studies support this model in which insulator elements in the V-D region are upstream of this knock-in site (discussed below). However, our findings reveal that not only was the V gene placed adjacent to the DFL16.1 gene, it was also placed in a much more open chromatin context (i.e. a HS site, compared both with the V region and with adjacent D genes) (3, 49). Either or both contexts could give it a significant recombination advantage that could explain the promiscuous recombination reported. Thus, to validate the model above, it will be important to knock in a V gene close to DFL16.1 but not at a HS site and ideally up- and downstream of HS4/HS5 to show insulator effects.
HS4 and HS5 Are Enhancer-blocking Insulators
Substantial evidence of differential histone modification and germ line transcription of the V and D regions during B cell development supports a model in which insulator elements in the V-D region regulate ordered Igh recombination by actively separating these regions (3, 4, 10). In particular, before D to J recombination, the D region has active histone modifications, whereas the V region has repressive histone H3 lysine 9 methylation marks (50). Because Eμ is a potent enhancer of DJ region antisense transcription and D to J recombination (10, 18, 19) and enhancers can act over several hundred kb (51), enhancer blocking may be required to prevent Eμ from activating V genes. We propose that HS4 and HS5, 3 and 5 kb upstream of the DFL16.1 gene, perform this function. First, they are either active in all hematopoietic cells (HS4) or restricted to the lymphocyte lineage (HS5) (Figs. 3 and 4 and Table 1), the appropriate developmental stages to insulate the V region from the Eμ-induced activation of the D region, particularly in T lymphocytes (36). Alternatively, they may have boundary function to protect the active DJ region from heterochromatin spreading from the V region (50). Second, active histone marks, including histone H3 lysine 9 acetylation, peak 2 kb upstream of DFL16.1 (1 kb downstream of HS5) (44), and histone H3 lysine 4 dimethylation peaks over DFL16.1 in pro-B cells and has been proposed to mark a chromatin boundary (4). Conversely, repressive histone H3 lysine 9 dimethylation only appears 6–10 kb upstream of DFL16.1 (i.e. immediately upstream of HS4) (44). Thus, HS4 and HS5 are strategically placed at the interface between two opposing histone modifications, a characteristic feature of insulators (39). Third, we have identified a 71-bp putative scaffold/matrix attachment region element 300 bp upstream of HS4, with 13 predicted SATB (special AT-rich sequence-binding protein) binding sites (52). Thus, HS4 may be associated with the nuclear matrix through SATB1/SATB2 interactions, which could stabilize interaction with other cis-acting elements. Further, scaffold/matrix attachment regions can function as boundary elements (53), which may contribute to HS4 and HS5 insulator function. Fourth, HS4 was detected in all of the cell lines (TK-1, AKR/Cum; BW5147, AKR/J; RAW264, BALB/c; Rag1−/−, C57/BL6/MF1), and HS5 was detected in the C57BL/6 and AKR strains, indicating that these elements are conserved between mouse strains, in stark contrast to Igh genic regions, where restriction fragment length polymorphisms are extremely common. Moreover, HS4 and HS5 comprise the region of greatest non-coding homology between the rat and mouse, with 77 and 76% identity, respectively, to corresponding rat V-D intergenic sequences. This compares with 91% homology between mouse and rat Eμ enhancer and 82% at the HS1/2 enhancer (54, 55). Using the rVista program (available on the World Wide Web), we have identified conserved binding sites for Pax5 in both HS4 and HS5. Pax5 binds V genes and recruits the RAG complex (56) and is required for Igh DNA looping (13), but it is unknown whether it participates directly. It will be interesting to determine whether it binds to HS4 and HS5 and whether this contributes to relocation of V genes proximal to DJ genes. Additionally, HS4 contains a conserved binding site for Stat3 (57), and HS5 contains a conserved binding motif for PU.1 (58), both factors involved in B lineage commitment. Together, these data suggest that these elements are under greater evolutionary pressure than the Igh genic regions, supporting a conserved functional role in V(D)J recombination.
Most notably, both HS4 and HS5 contain functional CTCF binding sites, characteristic of enhancer-blocking insulators, in Rag−/− pro-B cells in vivo (Fig. 5). These sites were also recently identified in a CTCF chromatin immunoprecipitation-chip microarray analysis in Rag−/− pro-B cells (59). Furthermore, we show here that HS4 and HS5 have substantial CTCF-dependent enhancer-blocking activity in vivo (Fig. 6). CTCF binding can generate DNA loops that sequester promoters and enhancers in distinct chromatin domains (40), which might block activating signals originating from the Eμ enhancer. In support of this model, we have also found a sharp loss of antisense transcription immediately upstream of HS4 (Fig. 2).
A recent study using DNA-FISH and three-dimensional modeling proposed that a DNA sequence close to HS4/HS5 is sequestered adjacent to the 3′ regulatory region by DNA looping in uncommitted prepro-B cells, along with Eμ. Both relocate proximal to the V region in Rag−/− pro-B cells poised for V(D)J recombination (60). We propose that HS4, active in all hematopoietic progenitors, keeps the V-D and DJ regions separated from the V region in non-B cells by interaction with CTCF sites in the 3′ regulatory region. Subsequently, lymphocyte-specific activation of HS5 and CTCF binding to this site may then redirect the V-D and DJ regions toward the V region in pro-B cells. Here HS4 and HS5 may synergize to provide stronger insulator activity when ordered D-J versus V-DJ recombination is most critical. After D to J recombination, it remains unclear where the V region binds proximal to the DJ domain. Association with elements in the V-D region is an attractive possibility. CTCF also mediates long range intrachromosomal interactions, by formation of DNA loops (61). Furthermore, the Igh V region contains multiple functional CTCF binding sites (59). HS4 and HS5 may recruit distal V region CTCF sites to form DNA loops proximal to DJ recombined genes.
Our studies suggest that similar insulators may be present in other antigen receptor loci. Notably, targeting of a V gene into the V-D intergenic region of the Tcrβ locus, 7 kb upstream of DβJβ, failed to increase its recombination frequency (62), whereas removal of the entire V-D region did (63). In the former study, it was concluded that the flanking sequences controlled V recombination frequency, independent of its location. We suggest alternatively that the 7-kb region contains an insulator that prevents spreading of active chromatin from DβJβ to the targeted V gene, effectively maintaining the V gene in its “normal” separate V region context. This position and putative function are analogous to HS4 and HS5 in the Igh V-D region. Accordingly, we also predict that insertion of a V gene upstream of HS4 and HS5, instead of at HS6 (47), would not alter recombination frequency.
HS3 Is a Putative B Cell-specific V Region Enhancer
Regulatory elements that activate the Igh V region have not been identified. We propose that HS3 may play this role. Importantly, it is the only HS restricted to the B cell lineage, where V region activation occurs. Taken together with its location upstream of the HS4/HS5 insulators, this suggests that it may be a stage-specific enhancer of V region activation. However, it is situated within a full-length LINE, albeit it is retrotransposition-inactive. Nevertheless, such LINEs can be transcribed and retrotransposed by retrotransposition-active FLI-L1s (64). Furthermore, many LINEs have tissue-specific cis-regulatory function (65). Thus, location of HS3 within a LINE does not preclude an enhancer role in V(D)J recombination. Alternatively, HS3 may be involved in Igh locus compaction because LINEs are enriched in scaffold/matrix attachment regions (65), which can form DNA loops (14, 53), and it is active in B cells, where looping of V genes to the DJ region occurs.
In summary, we have characterized the 96-kb V-D intergenic sequence in the mouse Igh locus, strategically placed to regulate V(D)J recombination and allelic exclusion. We have shown that it is transcribed in a manner that may regulate separation of the V and D chromatin domains. It contains several HS sites. Two of these are enhancer-blocking insulators, which we propose prevent activation of the V region before D to J recombination has occurred. These studies identify novel regulatory elements and provide new insight into how ordered V(D)J recombination may be regulated. They set the stage for testing this model by functional characterization of these putative regulatory elements by gene targeting in vivo.
Supplementary Material
Acknowledgments
We thank G. Morgan and A. Davis for fluorescence-activated cell sorting, P. Fraser and D. Bolland for critical reading of the manuscript, and A. West for advice on insulator assays.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–4.
- HS
- hypersensitive site
- RT
- reverse transcription
- CTCF
- CCCTC-binding factor
- LINE
- long interspersed nucleotide element.
REFERENCES
- 1.Corcoran A. E. (2005) Semin. Immunol. 17, 141–154 [DOI] [PubMed] [Google Scholar]
- 2.Jung D., Giallourakis C., Mostoslavsky R., Alt F. W. (2006) Annu. Rev. Immunol. 24, 541–570 [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury D., Sen R. (2001) EMBO J. 20, 6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morshead K. B., Ciccone D. N., Taverna S. D., Allis C. D., Oettinger M. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11577–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lennon G. G., Perry R. P. (1985) Nature 318, 475–478 [DOI] [PubMed] [Google Scholar]
- 6.Thompson A., Timmers E., Schuurman R. K., Hendriks R. W. (1995) Eur. J. Immunol. 25, 257–261 [DOI] [PubMed] [Google Scholar]
- 7.Yancopoulos G. D., Alt F. W. (1985) Cell 40, 271–2812578321 [Google Scholar]
- 8.Corcoran A. E., Riddell A., Krooshoop D., Venkitaraman A. R. (1998) Nature 391, 904–907 [DOI] [PubMed] [Google Scholar]
- 9.Bolland D. J., Wood A. L., Johnston C. M., Bunting S. F., Morgan G., Chakalova L., Fraser P. J., Corcoran A. E. (2004) Nat. Immunol. 5, 630–637 [DOI] [PubMed] [Google Scholar]
- 10.Bolland D. J., Wood A. L., Afshar R., Featherstone K., Oltz E. M., Corcoran A. E. (2007) Mol. Cell. Biol. 27, 5523–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosak S. T., Skok J. A., Medina K. L., Riblet R., Le Beau M. M., Fisher A. G., Singh H. (2002) Science 296, 158–162 [DOI] [PubMed] [Google Scholar]
- 12.Yang Q., Riblet R., Schildkraut C. L. (2005) Mol. Cell. Biol. 25, 6021–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M. (2004) Genes Dev. 18, 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayegh C., Jhunjhunwala S., Riblet R., Murre C. (2005) Genes Dev. 19, 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H., Schmidt-Supprian M., Shi Y., Hobeika E., Barteneva N., Jumaa H., Pelanda R., Reth M., Skok J., Rajewsky K., Shi Y. (2007) Genes Dev. 21, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynaud D., Demarco I. A., Reddy K. L., Schjerven H., Bertolino E., Chen Z., Smale S. T., Winandy S., Singh H. (2008) Nat. Immunol. 9, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly J., Licence S., Nanou A., Morgan G., Mårtensson I. L. (2007) EMBO J. 26, 4273–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlot T., Alt F. W., Bassing C. H., Suh H., Pinaud E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14362–14367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afshar R., Pierce S., Bolland D. J., Corcoran A., Oltz E. M. (2006) J. Immunol. 176, 2439–2447 [DOI] [PubMed] [Google Scholar]
- 20.Nitschke L., Kestler J., Tallone T., Pelkonen S., Pelkonen J. (2001) J. Immunol. 166, 2540–2552 [DOI] [PubMed] [Google Scholar]
- 21.Johnston C. M., Wood A. L., Bolland D. J., Corcoran A. E. (2006) J. Immunol. 176, 4221–4234 [DOI] [PubMed] [Google Scholar]
- 22.Johnson K., Angelin-Duclos C., Park S., Calame K. L. (2003) Mol. Cell. Biol. 23, 2438–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawlitzky I., Angeles C. V., Siegel A. M., Stanton M. L., Riblet R., Brodeur P. H. (2006) J. Immunol. 176, 6839–6851 [DOI] [PubMed] [Google Scholar]
- 24.Spanopoulou E., Roman C. A., Corcoran L. M., Schlissel M. S., Silver D. P., Nemazee D., Nussenzweig M. C., Shinton S. A., Hardy R. R., Baltimore D. (1994) Genes Dev. 8, 1030–1042 [DOI] [PubMed] [Google Scholar]
- 25.Li Y. S., Hayakawa K., Hardy R. R. (1993) J. Exp. Med. 178, 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirch S. A., Rathbun G. A., Oettinger M. A. (1998) EMBO J. 17, 4881–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J. (2004) Immunogenetics 56, 399–404 [DOI] [PubMed] [Google Scholar]
- 28.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Genome Biol. 3, RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forrester W. C., Epner E., Driscoll M. C., Enver T., Brice M., Papayannopoulou T., Groudine M. (1990) Genes Dev. 4, 1637–1649 [DOI] [PubMed] [Google Scholar]
- 30.Mitchell J. A., Fraser P. (2008) Genes Dev. 22, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi I., Oh J., Cho B. N., Ahnn J., Jung Y. K., Han Kim D., Cho C. (2004) Genomics 83, 636–646 [DOI] [PubMed] [Google Scholar]
- 32.Huovila A. P., Turner A. J., Pelto-Huikko M., Kärkkäinen I., Ortiz R. M. (2005) Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 33.Matsuda F., Ishii K., Bourvagnet P., Kuma K., Hayashida H., Miyata T., Honjo T. (1998) J. Exp. Med. 188, 2151–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ralph P. (1973) J. Immunol. 110, 1470–1475 [PubMed] [Google Scholar]
- 35.Butcher E. C., Scollay R. G., Weissman I. L. (1980) Eur. J. Immunol. 10, 556–561 [DOI] [PubMed] [Google Scholar]
- 36.Born W., White J., Kappler J., Marrack P. (1988) J. Immunol. 140, 3228–3232 [PubMed] [Google Scholar]
- 37.Gross D. S., Garrard W. T. (1988) Annu. Rev. Biochem. 57, 159–197 [DOI] [PubMed] [Google Scholar]
- 38.Rolink A., ten Boekel E., Melchers F., Fearon D. T., Krop I., Andersson J. (1996) J. Exp. Med. 183, 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaszner M., Felsenfeld G. (2006) Nat. Rev. Genet. 7, 703–713 [DOI] [PubMed] [Google Scholar]
- 40.Wallace J. A., Felsenfeld G. (2007) Curr. Opin. Genet. Dev. 17, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao W., Huynh K. D., Spencer R. J., Davidow L. S., Lee J. T. (2002) Science 295, 345–347 [DOI] [PubMed] [Google Scholar]
- 42.Garrett F. E., Emelyanov A. V., Sepulveda M. A., Flanagan P., Volpi S., Li F., Loukinov D., Eckhardt L. A., Lobanenkov V. V., Birshtein B. K. (2005) Mol. Cell. Biol. 25, 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung J. H., Whiteley M., Felsenfeld G. (1993) Cell 74, 505–514 [DOI] [PubMed] [Google Scholar]
- 44.Chakraborty T., Chowdhury D., Keyes A., Jani A., Subrahmanyam R., Ivanova I., Sen R. (2007) Mol. Cell 27, 842–850 [DOI] [PubMed] [Google Scholar]
- 45.Abarrategui I., Krangel M. S. (2006) Nat. Immunol. 7, 1109–1115 [DOI] [PubMed] [Google Scholar]
- 46.Tripathi R., Jackson A., Krangel M. S. (2002) J. Immunol. 168, 2316–2324 [DOI] [PubMed] [Google Scholar]
- 47.Bates J. G., Cado D., Nolla H., Schlissel M. S. (2007) J. Exp. Med. 204, 3247–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alessandrini A., Desiderio S. V. (1991) Mol. Cell. Biol. 11, 2096–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maës J., Chappaz S., Cavelier P., O'Neill L., Turner B., Rougeon F., Goodhardt M. (2006) J. Immunol. 176, 5409–5417 [DOI] [PubMed] [Google Scholar]
- 50.Johnson K., Pflugh D. L., Yu D., Hesslein D. G., Lin K. I., Bothwell A. L., Thomas-Tikhonenko A., Schatz D. G., Calame K. (2004) Nat. Immunol. 5, 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawwari A., Krangel M. S. (2005) J. Exp. Med. 202, 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagomi K., Kohwi Y., Dickinson L. A., Kohwi-Shigematsu T. (1994) Mol. Cell. Biol. 14, 1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heng H. H., Goetze S., Ye C. J., Liu G., Stevens J. B., Bremer S. W., Wykes S. M., Bode J., Krawetz S. A. (2004) J. Cell Sci. 117, 999–1008 [DOI] [PubMed] [Google Scholar]
- 54.Banerji J., Olson L., Schaffner W. (1983) Cell 33, 729–740 [DOI] [PubMed] [Google Scholar]
- 55.Lieberson R., Giannini S. L., Birshtein B. K., Eckhardt L. A. (1991) Nucleic Acids Res. 19, 933–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., Espinoza C. R., Yu Z., Stephan R., He T., Williams G. S., Burrows P. D., Hagman J., Feeney A. J., Cooper M. D. (2006) Nat. Immunol. 7, 616–624 [DOI] [PubMed] [Google Scholar]
- 57.Chou W. C., Levy D. E., Lee C. K. (2006) Blood 108, 3005–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott E. W., Simon M. C., Anastasi J., Singh H. (1994) Science 265, 1573–1577 [DOI] [PubMed] [Google Scholar]
- 59.Degner S. C., Wong T. P., Jankevicius G., Feeney A. J. (2009) J. Immunol. 182, 44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jhunjhunwala S., van Zelm M. C., Peak M. M., Cutchin S., Riblet R., van Dongen J. J., Grosveld F. G., Knoch T. A., Murre C. (2008) Cell 133, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams A., Flavell R. A. (2008) J. Exp. Med. 205, 747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sieh P., Chen J. (2001) J. Immunol. 167, 2121–2129 [DOI] [PubMed] [Google Scholar]
- 63.Senoo M., Wang L., Suzuki D., Takeda N., Shinkai Y., Habu S. (2003) J. Immunol. 171, 829–835 [DOI] [PubMed] [Google Scholar]
- 64.Wei W., Gilbert N., Ooi S. L., Lawler J. F., Ostertag E. M., Kazazian H. H., Boeke J. D., Moran J. V. (2001) Mol. Cell. Biol. 21, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jordan I. K., Rogozin I. B., Glazko G. V., Koonin E. V. (2003) Trends Genet. 19, 68–72 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.