Abstract
MicroRNAs (miRs) have been reported to play a critical role in muscle differentiation and function. The purpose of this study is to determine the role of miRs during smooth muscle cell (SMC) differentiation from embryonic stem cells (ESCs). MicroRNA profiling showed that miR-10a expression is steadily increased during in vitro differentiation of mouse ESCs into SMCs. Loss-of-function approaches using miR-10a inhibitors uncovered that miR-10a is a critical mediator for SMC lineage determination in our retinoic acid-induced ESC/SMC differentiation system. In addition, we have documented for the first time that histone deacetylase 4 is a novel target of miR-10a and mediates miR-10a function during ESC/SMC differentiation. To determine the molecular mechanism through which retinoic acid induced miR-10a expression, a consensus NF-κB element was identified in the miR-10a gene promoter by bioinformatics analysis, and chromatin immunoprecipitation assay confirmed that NF-κB could bind to this element. Finally, inhibition of NF-κB nuclear translocation repressed miR-10a expression and decreased SMC differentiation from ESCs. Our data demonstrate for the first time that miR-10a is a novel regulator in SMC differentiation from ESCs. These studies suggest that miR-10a may play important roles in vascular biology and have implications for the diagnosis and treatment of vascular diseases.
Keywords: Cell Differentiation, Embryonic Stem Cell, Gene Expression, MicroRNA, Smooth Muscle
Introduction
Embryonic stem cells (ESCs)7 have been widely used as an effective tool to study the molecular mechanisms governing cell differentiation (1, 2). The reason why ESCs are such a valuable tool is that each stage of early development is recapitulated in the process of ESC differentiation to a specific cell lineage (3, 4). ESCs have been shown to be able to differentiate into smooth muscle cells (SMCs) through embryoid body formation under various differentiation conditions with combinations of molecular modifications (5–10). We have recently developed a highly efficient and reproducible differentiation protocol, independent of embryoid body formation or additional molecular modifications, to generate relatively pure and functional SMC populations by treating ESCs with a high concentration of retinoid acid (RA) (5, 7). This ESC/SMC differentiation system has provided a valuable platform to explore molecular mechanisms of SMC differentiation in vitro, which can be instructive to in vivo disease models related to SMC differentiation.
SMCs express a number of smooth muscle-specific genes, referred to as SM markers, which include SM α-actin (SMA), SM22α, calponin, and SM myosin heavy chain (11). SMCs can exhibit a spectrum of phenotypes ranging from the more differentiated “contractile” state in which high levels of SMC differentiation markers are expressed to the less differentiated “synthetic” state in which SMC markers are down-regulated (11–13). Alterations in SMC differentiation state play a major role in the development of a number of cardiovascular diseases such as atherosclerosis, restenosis, hypertension, and aneurysm (14). Therefore, a better understanding of the molecular mechanisms that controls SMC differentiation is very important to help develop new ways to prevent and treat these diseases. SMC differentiation has been documented to be tightly regulated through a complex and synergistic combination of 1) DNA binding transcription factors, 2) accessory cofactors for the DNA-binding proteins, and 3) DNA/histone modifications present within promoter chromatin. This multilayered orchestra works to coordinate regulation of the multiple transcriptional programs that underlie differentiation and plasticity of the SMC phenotype (15). Recent works have identified a new layer of post-transcriptional regulation involving miR-mediated mechanisms, which in turn are involved in the control of SMC differentiation players (16). Specifically, several miRs, including miR-21 (17, 18), 143 and 145 (19–22), and 221 and 222 (23, 24), have been shown to be involved in vascular SMC development and related-disease progression. Here, we have documented for the first time that miR-10a is a novel mediator of SMC differentiation from ESCs.
EXPERIMENTAL PROCEDURES
SMC in Vitro Differentiation System
We have previously reported the ESC/SMC in vitro differentiation system (7). Briefly, mouse ESCs were dissociated into individual cells and resuspended and plated on 6-well, gelatin-coated plastic Petri dishes (Falcon) at a density of ∼3 × 104/cm2. Cells were then incubated at 37 °C with 5% of CO2 in a differentiation medium. The differentiation medium consisted of Dulbecco's modified Eagle's medium/F-12 supplemented with 10% of fetal calf serum, 1 mmol/liter l-glutamine, 0.1 mmol/liter nonessential amino acids, penicillin/streptomycin (Invitrogen), 0.1 mmol/liter β-mercaptoethanol, and added 10−5 mol/liter RA (Sigma). The cells were cultured for 8–10 days with a daily change of fresh differentiation medium. A404 cells (kindly provided by Dr. Gary K. Owens at the University of Virginia) were induced to differentiate into SMCs following the previous report, with minor modification (25). Briefly, A404 cells were treated with 1 μm RA for 4 days.
RNA Extraction and Quantitative Real-time (qRT)-PCR
Total RNA from cultured cells was extracted by using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized from 5 μg of total RNA with Superscript III first-strand synthesis system (Invitrogen). cDNA samples were subjected to qRT-PCR amplification using custom primers on an Bio-Rad MyIQ detection system (Bio-Rad) according to the protocol provided by the manufacturer. Quantitative RT-PCR custom primers are described in supplemental Table 1. 18 S RNA served as an internal standard.
TaqMan miR Assay for Mature miR Expression Analysis
To measure mature miR expression, total RNA, including mature miRs, was extracted from cultured cells by using the microRNeasy mini kit (Qiagen) according to the manufacturer's instructions. Primers specific for miRs and U6 (Applied Biosystems) were used, and each mature miR expression was quantified using TaqMan microRNA assays (Applied Biosystems) following the manufacturer's protocol. In brief, TaqMan microRNA assays include two steps; that is, stem-loop RT followed by real-time PCR. For each 15-μl RT reaction, 10 ng of purified total RNA, 1× RT primer, 1× RT buffer, 1 mm dNTPs, 0.2 units/μl RNase inhibitor, and 3.33 units/μl MultiScribe reverse transcriptase from the TaqMan microRNA reverse transcription kit were added and incubated for 30 min at 16 °C followed by 30 min of incubation at 42 °C. qRT-PCR was performed using a standard protocol on a Bio-Rad MyIQ System (Bio-Rad). All TaqMan microRNA assays were performed in triplicate. Total RNA input was normalized based on the cycle threshold values of the U6 assay as an endogenous control. The -fold change was calculated based on delta cycle threshold between endogenous U6 control and miR-10a.
Fluorescence-activated Cell Sorting Analysis
Fluorescence-activated cell sorting analysis was performed as previously reported (26). Briefly, cell pellets were fixed, permeabilized using a BD Biosciences cytofix/cytopermTM kit, and incubated overnight with anti-SMA antibody (Millipore). Mouse IgG2a served as isotypic control (Dakocytomation), whereas rat vascular SMCs served as positive controls. Goat anti-mouse IgG Alexa Fluor 488 was used as the secondary antibody (Molecular Probes). Finally, fluorescence was analyzed using the FACSCaliburTM system (BD Biosciences) following the user's guide.
Construction of Reporter Plasmids and Reporter Assays
To generate the miR-10a-luciferase reporter plasmid, an ∼900-bp fragment (−868 to +110 bp) containing the NF-κB putative binding site was amplified from C57/B6L mouse genomic DNA and then cloned into KpnI and HindIII sites of the pGL4.10 [Luc2] vector (Promega). The following primers were used for cloning the miR-10a promoter: ggaggggtaccagaatcccattttggcca (forward primer) and ggaggaagcttgcggagtgtttatgtcaact (reverse primer). For the construction of the p65mut-miR-10a luciferase plasmid, the NF-κB binding site was mutated from the miR-10a- luciferase construct using QuikChange XL mutagenesis (Stratagene) following the manufacturer's instructions. Primers used were as follows, resulting in the mutation of the putative NF-κB element (capitalized is the partial mutated region from the predicted miR-10a promoter): NF-κB-mF (forward primer, cttcttctcaggttgtaaacggaattTTTAgtcattaaatatggg) and NF-κB-mR (reverse primer, cccatatttaatgacTAAAaattccgtttacaacctgagaagaag). A DNA fragment containing the HDAC4 3′-UTR sequence with a putative miR-10a binding site was cloned into the SacI and HindIII restriction sites of the pMIR-REPORTTM vector (Promega) to generate the HDAC4–3′-UTR reporter plasmid. Primers used to amplify the DNA fragment containing the HDAC4 3′-UTR from cDNA were HDAC4-F (forward primer, atactcagaagggacatcaag) and HDAC4-F (reverse primer, ccaaggcggccagaagagc). Site-directed mutagenesis against the putative miR-10a binding site located in the HDAC4–3′-UTR was generated by standard PCR to generate the HDAC4-mut-3′-UTR reporter plasmid. Primers used for site-directed mutagenesis are as follows: HDAC4-mF (forward primer, ctgaatttggtggcatttacagCCGCgatgggaactgggtcaagc) and HDAC4-mR (reverse primer, gcttgacccagttcccatcGCGGctgtaaatgccaccaaattcag). The final sequences of all constructed plasmids were confirmed by DNA sequencing. Constructed plasmids (0.2 μg of plasmid/well of 24-well plates) together with Renilla luciferase control reporter vector (pRL-TK) were transfected into HEK 293 cells using Lipofectamine 2000 (Invitrogen) for 4 h. Luciferase activity was measured 48 h after transfection using the Dual-Luciferase assay kit (Promega) with a TD-20/20 luminometer (Turner Designs). Individual luciferase activity was normalized to the responding TK promoter-Renilla-luciferase activity.
Western Blot Analysis
Whole cell lysate samples were extracted from cells using the mammalian protein extraction reagent M-Per (Promega) supplemented with a protease inhibitor mixture (Roche Applied Science). Cytoplasmic and nuclear fractions were obtained using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) following the manufacturer's instructions. Antibodies against p65 (Santa Cruz Biotechnology, 1:1000), SMA (Millipore, 1:3000), SM-myosin heavy chain (SMMHC, BTI, 1:2000), glyceraldehyde-3-phosphate dehydrogenase (Santa Cruz Biotechnology, 1:1000), lamin A/C (Santa Cruz Biotechnology, 1:1000), and HDAC4 (Abcam, 1:500) were used for testing individual protein expression. Immuno-activity and band density was visualized by the enhanced chemiluminescence detection system (Amersham Biosciences) or Odyssey system (LI-COR Biosciences, Lincoln, NE) according to the manufacturer's instructions.
Expression Plasmids, miR Inhibitors, and Transfection
A complex of plasmids or oligonucleotides was transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. miR-10a inhibitor and inhibitor negative control were purchased from Dharmacon. These inhibitors are RNA oligonucleotides with novel secondary structure designed to inhibit the function of endogenous miR (27). HDAC4, p65, Sp1, and SMAD3 expression plasmids were purchased from Addgene. PathDetect NF-κB Cis-Reporter Plasmid was purchased from Stratagene (NF-κB binding site luciferase).
MicroRNA Target Prediction
Target genes for miR-10a were predicted using the free software PicTar and TargetScan 4.2 with the cutoff set at p values less than 0.05.
Chromatin Immunoprecipitation (ChIP) Assay
Mouse ESCs were treated with 10 μm RA. After 6 h, cellular proteins were cross-linked, and a ChIP assay was performed by using the EZ-ChIP assay kit (Millipore). The size of the sonicated DNA fragments used for immunoprecipitation was between 500 and 1000 bp in length as determined by ethidium bromide gel electrophoresis. Purified chromatin was immunoprecipitated using anti-p65 antibody (Santa Cruz Biotechnologies). Eluted DNA fragments were purified to serve as templates for the PCR amplification. The input fraction corresponded to 2% of the chromatin solution before immunoprecipitation. IgG control serves as the negative control to the p65 antibody. By using rVista software to analyze conserved regions on the pre-mmu-miR-10a promoter, we identified a potential NF-κB binding site, which is located in the −638 to −627-bp region upstream of the transcription start site. The primers used to amplify the area containing this NF-κB binding site are: forward primer (5′-gctgcaacttttatggttcc-3′) and reverse primer 5′-gggagaggtgtctgggtgtg-3′), resulting in a 225-bp fragment.
Statistical Analysis
Data were analyzed for statistical significance by the Student-Newman-Keuls test using SYSTAT software (SYSTAT Inc.) with values of p < 0.05 considered to be significant. All experiments were independently repeated at least three times.
RESULTS
miR-10a Is Up-regulated during SMC Differentiation from Mouse ESCs
Recently, we have established the ESC/SMC in vitro differentiation system (7). To determine whether miRs regulate RA-induced ESC/SMC differentiation, we profiled miR expression levels during ESC/SMC differentiation at days 0, 1, and 3 using a TaqMan miR assay. Among 226 commercially available miR primer sets profiled in our assay, the expression levels of 11 miRs were increased, and 7 miRs decreased by more than 3-fold (supplemental Table S2). Interestingly, miR-10a was among the most strongly up-regulated miRs during RA-induced ESC/SMC differentiation (supplemental Table S2). The TaqMan miR assay showed that miR-10a is up-regulated during RA-induced SMC differentiation by ∼20-fold at day 3 and continued to increase during later stages of differentiation (supplemental Fig. S1). DMSO-treated cells also showed moderate up-regulation of miR-10a (5-fold by day 6); however, it was not as remarkable or sustained as the up-regulation observed in the RA-treated cells. The up-regulation observed in the DMSO-treated cells may result from a low percentage of spontaneously differentiated SMC population.
Inhibition of miR-10a Impairs SMC Differentiation from Mouse ESCs
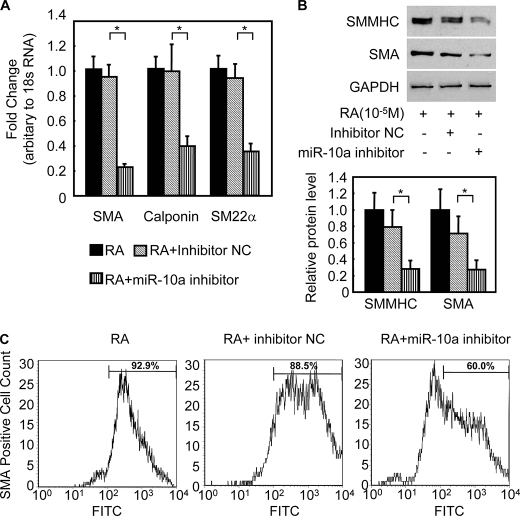
To examine the potential role of miR-10a during RA-induced ESC/SMC differentiation, a miR-10a inhibitor was transfected into ESCs. After 24 h of the transfection, ESCs were treated with RA for 3, 6, and 8 days and subjected to analysis of SMC markers by qRT-PCR for mRNA levels and Western blots for protein levels. The miR-10a inhibitor effectively reduced the expression of miR-10a to less than 20% that of control cells transfected with an inhibitor negative control (supplemental Fig. S2A). Intriguingly, when mature miR-10a levels were repressed, SMC marker expression during RA-induced ESC/SMC differentiation dramatically declined (Fig. 1, A and B) and differentiation efficiency to SMCs reduced, as evidenced by a decreased percentage of SMA-positive cell population in the derived cells (61.73 ± 3.38% in miR-10a inhibitor group versus 87.45 ± 2.45% in miR negative control, p < 0.05, Fig. 1C). In addition, no effect on other cell lineage-specific marker expression was observed during miR-10a repression in the context of this ESC/SMC differentiation system by day 6 (supplemental Fig. S2B). Interestingly, miR-10a expression was also up-regulated upon the RA treatment in the A404-SMC differentiation system (supplemental Fig. S3A). Furthermore, blockage of miR-10a expression with miR-10a inhibitor impaired SMC differentiation in this A404 system, as evidenced by down-regulation of SMC-specific markers (supplemental Fig. S3, B and C). Taken together, these results suggest that miR-10a is important for the RA-induced SMC differentiation process, probably by targeting specific genes involved in the SMC phenotype.
FIGURE 1.
miR-10a regulates SMC differentiation from ESCs. A and B, miR-10a inhibitor represses SMC differentiation. 50 nm miR-10a inhibitor or negative control was transfected into ESCs. Twenty-four hours after transfection, the cells were treated with 10 μm RA for 6 days and harvested for analysis of SMC markers expression, including SMA and SM myosin heavy chain (SMMHC), as evidenced by qRT-PCR (A) and Western blotting (B). NC, negative control. *, p < 0.05. 18 S RNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as internal controls for qRT-PCR and Western blot, respectively. C, miR-10a inhibitor reduces SMC differentiation efficiency. Mouse ESCs were transfected with either miR-10a inhibitor or miR inhibitor negative control, treated with RA for 6 days, and subjected to fluorescence-activated cell sorting analysis. The proportion of SMA-positive cells is shown in the histograms, representing RA alone (left panel), RA + inhibitor negative control (central panel), and RA + miR-10a inhibitor (right panel). n = 4. Normal IgG2a served as the fluorescence-activated cell sorting analysis control. FITC, fluorescein isothiocyanate.
miR-10a Mediates SMC Differentiation through Targeting on HDAC4
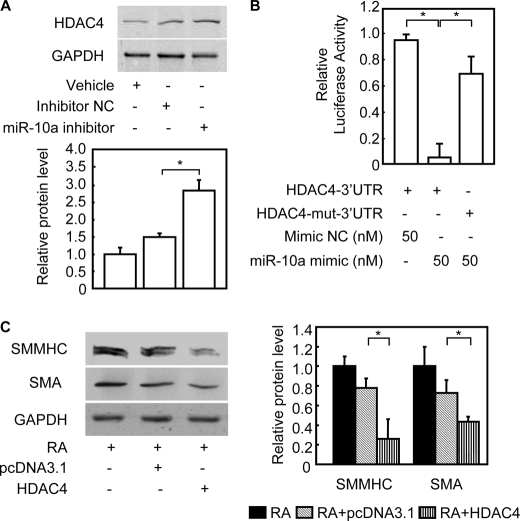
To better understand the miR-10a-dependent molecular mechanism regulating SMC differentiation, we performed a bioinformatics search for targets that might mediate this effect. Remarkably, one of the targets consistently predicted by two different algorithms, TargetScan 4.0 (28) and PicTar (29), was HDAC4. The predicted hybridization site was to a seed sequence in the HDAC4 3′-UTR that is evolutionarily conserved among vertebrate species (supplemental Fig. S4A). This result suggests that miR-10a may act to suppress HDAC4 and its activity during SMC differentiation. Secondary structure analysis also showed a favorable minimum free energy (−29.1 kcal/mol) in the formation of the miR-10a; HDAC4 3′-UTR duplex stem-loop (supplemental Fig. S4B). Consistent with this prediction, our results uncovered an inverse relationship between miR-10a expression and HDAC4 protein levels (supplemental Fig. S4C) during ESC/SMC differentiation without a significant change of HDAC4 mRNA levels (supplemental Fig. S4D). Furthermore, down-regulation of HDAC4 protein level was rescued when mature miR-10a expression was blocked by miR-10a inhibitor in this ESC/SMC differentiation system (Fig. 2A), without changes to HDAC4 mRNA levels (data not shown). This result is consistent with the functional prediction that miR-10a binding to the HDAC4 3′-UTR would lead to HDAC4 translational repression.
FIGURE 2.
HDAC4 is a miR-10a target during SMC differentiation from ESCs. Mouse ESCs were transfected with miR-10a mimic or mimic negative control (NC) and treated with RA for 6 days. A, cells were harvested at day 6, and cell lysate was used for the Western blot with an anti-HDAC4 antibody, or an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was used as the internal control. Representative Western blots are shown. B, either wild type or mutant HDAC4–3′-UTR reporter constructs were cotransfected into HEK 293 cells together with miR-10a mimic or mimic negative control (NC), as indicated. Forty-eight hours after transfection, cells were lysed, and luciferase activity was determined. Individual luciferase activity was normalized to the responding TK promoter-Renilla-luciferase activity. Relative luciferase activities were expressed as the mean ± S.D. Data shown are representative samples from at least three independent experiments, each done in triplicate. *, p < 0.05. C, representative Western blots in mouse ESCs were transfected with HDAC4. Mouse ESCs were transfected with human HDAC4-FLAG and subsequently induced to differentiate into SMCs with RA. The derived cells were subject to Western blot analysis. The expression of SMC markers was detected in extracts from cells differentiated for 6 days. Glyceraldehyde-3-phosphate dehydrogenase served as the internal control for Western blot. The right panel shows the quantitative analysis of SMC specific markers from Western blots. *, p < 0.05. SMMHC, SM-myosin heavy chain.
We next tested whether HDAC4 was a target for repression by miR-10a by cloning a fragment of the HDAC4 3′-UTR, containing either wild type or a mutant (mut) miR-10a binding site, downstream from the firefly luciferase gene in the pMIR- ReportTM vector (supplemental Fig. S4A). Co-transfections of miR-10a mimic with the HDAC4–3′-UTR reporter resulted in the repression of luciferase activity when compared with the mimic negative control (p < 0.05) (Fig. 2B). On the other hand, the HDAC4-mut-3′-UTR reporter was not repressed by miR-10a mimic. Neither mutation of the putative miR-10a complementary seed sequence nor the miR mimic negative control repressed luciferase activity, indicating that the repression was specific for miR-10a binding.
Although HDAC4 has been shown to mediate platelet-derived growth factor-BB pro-proliferation of SMCs (30), the function of HDAC4 on ESC/SMC differentiation is unclear. To further explore the function of HDAC4 on SMC differentiation in this differentiation system, the down-regulated HDAC4 expression was rescued by transient transfection of a HDAC4 expression plasmid in mouse ESCs, and subsequently, transfected cells were induced to differentiate with RA. As shown in Fig. 2C and supplemental Fig. S5, A–C, the expression of SMC-specific markers, including SM myosin heavy chain and SMA on both mRNA and protein levels, were significantly reduced after HDAC4 overexpression. Taken together, these results indicate that miR-10a represses the expression of HDAC4 at the post-transcriptional level during ESC/SMC differentiation, thus preventing its HDAC4-antimyogenic effects.
NF-κB Transcriptionally Regulates miR-10a Expression during RA-induced SMC Differentiation
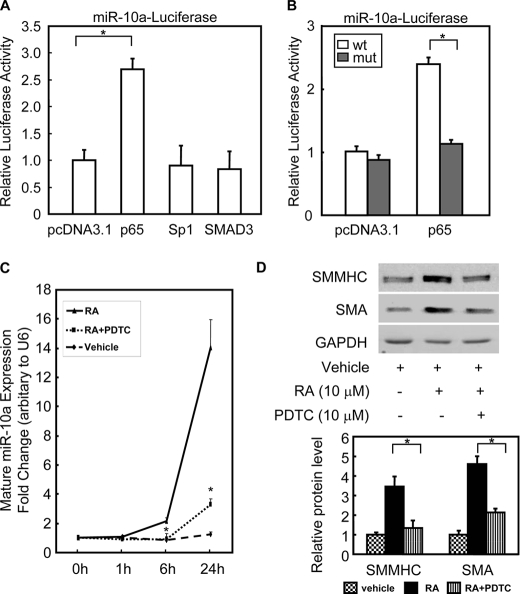
Although the effects of miRs on the regulation of their targets have been extensively studied, the transcriptional regulation of miRs themselves is only beginning to be elucidated. In this study, we were particularly interested in the transcriptional regulation of the miR-10a gene. We used rVISTA (31) to find potential transcription factor binding sites within ∼1 kb upstream region of the miR-10a gene, which is conserved among species (supplemental Fig. S6A). One NF-κB consensus binding site was identified in −638 to −627 bp upstream of the miR-10a gene (supplemental Fig. S6A). This 1-kb miR-10a promoter also contains several conserved cis-elements for other transcription factors, including Sp1 and SMAD3. Among these transcription factors, p65 was shown to be able to transactivate the miR-10a promoter (Fig. 3A).
FIGURE 3.
NF-κB transcriptionally regulates miR-10a expression. A, HEK 293 cells were cotransfected with miR-10a-luciferase reporter plasmid and expression plasmids encoding pdDNA3.1, p65, Sp1, or Smad3. Luciferase activity was determined 48 h after transfection. Individual luciferase activity was normalized to the responding TK promoter-Renilla-luciferase activity. Relative luciferase activities were expressed as the mean ± S.D. Data shown are representative samples from at least three independent experiments, each done in triplicate. *, p < 0.05. B, either the miR-10a-luciferase (wt) or the p65mut-miR-10a-luciferase (mut) reporter plasmid was cotransfected with pcDNA3.1 or p65 expression plasmid, as indicated. Luciferase activity was determined 48 h post-transfection. Individual luciferase activity was normalized to the responding TK promoter-Renilla-luciferase activity. Relative luciferase activities were expressed as the mean ± S.D. Data shown are representative samples from at least three independent experiments, each done in triplicate. *, p < 0.05. C and D, mouse ESCs were treated with p65 inhibitor pyrrolinedithiocarbamate (PDTC, 10 μm) for 30 min and then treated with RA to induce SMC differentiation. The expression of mature miR-10a levels was measured by TaqMan miR assay at different time points (C), and the expression of SMC markers was determined by Western blotting after 6 days of RA treatment (D). n = 3. *, p < 0.05. SMMHC, SM-myosin heavy chain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To test whether p65 could bind to the predicted NF-κB binding site in the miR-10a promoter, either the wild type or the mutated p65 binding site miR-10a-luciferase reporter plasmid was individually transfected into HEK 293 cells together with the p65 expression plasmid. As shown in Fig. 3B, p65 was able to enhance miR-10a-luciferase reporter activity. However, when the p65 binding site was mutated, p65 could no longer activate the expression of the miR-10a-luciferase reporter. In addition, the mature miR-10a expression in HEK 293 was significantly increased by overexpression of p65 in a separate experiment (data no shown). Moreover, p65 nuclear translocation inhibitor, pyrrolinedithiocarbamate, reduced miR-10a expression levels and negatively regulated SMC differentiation marker expression, as shown in Fig. 3, C and D.
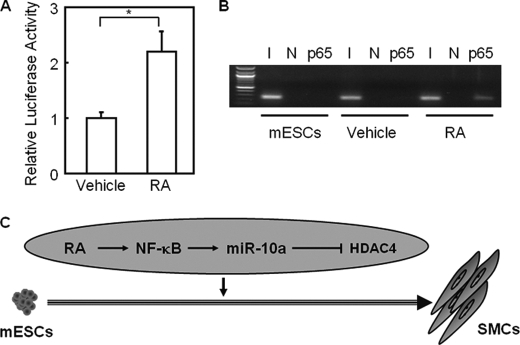
To determine whether NF-κB binding sites are a direct target of RA signaling, we used a plasmid containing the NF-κB binding element followed by the luciferase reporter gene (referred to as NF-κB binding site luciferase). As shown in Fig. 4A, NF-κB binding site luciferase reporter activity was increased by RA treatment in HEK 293 cells as determined by luciferase reporter assay (p < 0.05). Furthermore, the expression of a list of NF-κB target genes was up-regulated in ESCs in response to RA treatment (supplemental Fig. S6B). To determine whether NF-κB binds to the miR-10a promoter in response to RA, a ChIP assay was performed with extracts prepared from mouse ESCs treated with RA for 6 h. Results indicated that RA treatment can induce NF-κB binding to its consensus element in the miR-10a promoter (Fig. 4B). In summary, RA can promote nuclear translocation of NF-κB, which then binds to the miR-10a promoter and enhances miR-10a expression, consequently modulating SMC differentiation.
FIGURE 4.
RA enhances p65 binding to its target gene promoters during SMC differentiation from ESCs. A, HEK 293 cells were transfected with NF-κB-binding site-luciferase reporter plasmid and treated with either 10 μm RA or vehicle for 24 h. Luciferase activity was determined and normalized to the responding TK promoter-Renilla-luciferase activity. Relative luciferase activities were expressed as the mean ± S.D. Data shown are representative samples from at least three independent experiments, each done in triplicate. *, p < 0.05. B, mouse ESCs (mESCs) were treated with either vehicle (DMSO) or RA for 6 h, and ChIP was performed with primers specific for a putative p65 binding site on the miR-10a promoter. Undifferentiated mouse ESCs were used as a control. Total inputs are indicated. I, input; N, negative control. C, a model depicts the role of the RA-p65-miR-10a-HDAC4 regulatory pathway in SMC differentiation from ESCs. In this in vitro differentiation system, RA induces binding of p65 to the miR-10a promoter, leading to an increase in miR-10a expression levels. Enhanced miR-10a expression suppresses HDAC4 expression, therefore relieving its repression on SMC markers and driving differentiation into SMC lineage.
DISCUSSION
In this study we investigated the function of miRs during SMC differentiation from ESCs. The results shown here revealed that RA triggered NF-κB nuclear translocation, which in turn leads to up-regulation of NF-κB target genes including miR-10a. Furthermore, up-regulated miR-10a enhanced SMC differentiation via repression of HDAC4, which is a negative regulator of SMC differentiation. Thus, our data have demonstrated that miR-10a is a novel regulator in SMC differentiation from ESCs.
The miR-10a gene and mature miR-10a sequence are highly evolutionarily conserved. The function of miR-10a has been indicated in apoptosis (32), protein synthesis (33), embryo development and differentiation (34, 35), hematopoietic stem cell mobilization (36), inflammation (37), and various tumorigenesis including hematopoietic (38, 39), liver (40), and urinary (41). Although miR-10a has been shown to be highly expressed in rat artery and slightly down-regulated by day 7 after angioplasty (18), there is no further literature available dissecting the intrinsic relationship between miR-10a and SMC differentiation. Here, we provide novel evidence showing that miR-10a is induced during SMC differentiation from both ESCs and A404, whereas the miR-10a inhibitor dramatically repressed SMC differentiation. These results indicate that miR-10a expression can modulate SMC differentiation. Interestingly, miR-10a mimic per se cannot drive SMC differentiation in the absence of RA treatment (data no shown), which implies that miR-10a up-regulation is elegantly integrated in harmony with other regulatory mechanisms operating during RA-induced SMC differentiation. As previously reported (35, 42–44), there is a list of miRs that show the same dynamic change as miR-10a during the differentiation of ESCs. More studies are needed to unravel the function of these miRs during this differentiation process.
We also showed that the molecular mechanism by which miR-10a function to modulate SMC differentiation is by targeting HDAC4. Here we showed an inverse relationship in the expression pattern between miR-10a and HDAC4 during ESC/SMC differentiation. Luciferase assays further demonstrated that the mature miR-10a was able to bind to the ACAGGGU seed sequence in the HDAC4 3′-UTR. Furthermore, inhibition of miR-10a expression resulted in up-regulation of HDAC4 protein levels. Remarkably, HDAC4 was consistently predicted by two algorithms, TargetScan (28) and PicTar (29), as a target of miR-10a. HDACs are part of a vast family of enzymes that have critical roles in diverse biological processes, largely through transcriptional repression (45). Results of several previous studies have shown HDAC4 negative function in terms of SMC differentiation. For example, the HDAC inhibitor trichostatin A has been shown to increase the activity of the SM22α gene in 10T1/2 embryonic fibroblasts (46). Furthermore, studies have shown that HDAC4 can be recruited to enhance the platelet-derived growth factor-BB-mediated repression of SM α-actin gene expression (30) through the activation of ERK1/2 (extracellular signal-regulated kinase 1/2) pathway (47). In addition, HDAC4 has been shown to interact with serum response factor (48) and its coactivator myocardin (49).
Here we showed that overexpression of HDAC4 inhibited ESC/SMC differentiation, as shown by the down-regulated expression of SMC markers. However, the detailed mechanism through which HDAC4 is able to regulate SMC differentiation in this ESC/SMC differentiation system remains to be determined. The data shown here indicate that miR-10a mediates its positive effects in the modulation of SMC differentiation by targeting HDAC4. However, it is difficult to know whether the effects of miR-10a on SMC differentiation is primarily mediated through HDAC4 or whether HDAC4 could serve simultaneously as a target for miR-10a and other co-regulatory miRs. It should be noted that overexpression or inhibition of miR-10a could have also affected other cell factors in the differentiating cultures. Further studies are needed to identify and characterize additional miR-10a target genes.
Although miR biology has been greatly advanced in the last few years, much of the effort has focused on exploring the targets of miRs rather than understanding the regulation of miR genes themselves. To determine cis regulatory elements in volved in the up-regulation of miR-10a, we have documented for the first time that NF-κB can bind to a consensus site on the miR-10a promoter region and transactivate miR-10a expression. The NF-κB family of transcription factors is essential in a broad range of physiological processes, including immunity, inflammation, development, and differentiation (50, 51). Recent data suggests that NF-κB plays a role in proliferation, migration, and differentiation of stem cells (52). Previous studies have shown that the NF-κB pathway is up-regulated upon withdrawal of LIF and that impaired NF-κB signaling delays loss of pluripotency markers (53). The NF-κB activity has also been shown to be inhibited in undifferentiated ES cells by Nanog (one of critical transcription factors to maintain stem cell pluripotency) and up-regulated during differentiation to function as a positive differentiation stimulus (53, 54), which is consistent with the finding that the activation of NF-κB may contribute to changes in cell adhesion and cytokine production during ESC differentiation (55). Here, our data have showed that RA can induce the expression of NF-κB target genes including miR-10a in ESCs. Inhibition of NF-κB nuclear translocation blocked miR-10a up-regulation and impaired SMC differentiation as indicated by reduced expression of SMC markers. Taken together, these results shed further light on the role of NF-κB in ESC/SMC differentiation.
In summary, the results presented here show for the first time the involvement of NF-κB-miR-10a-HDAC4 in a regulatory signaling pathway (Fig. 4C) relevant in positively modulating SMC differentiation from ESCs. These findings not only provide new insights into the interplay between transcription factors and miRs during ESC/SMC differentiation but also have implications for the diagnosis and treatment of cardiovascular diseases.
Supplementary Material
Acknowledgment
We greatly appreciate the gift of A404 cells from Dr. Gary K. Owens at University of Virginia.
This work was supported, in whole or in part, by National Institutes of Health Grants HL092421, HL068878, and HL89544 (to Y. E. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- ESC
- embryonic stem cell
- miR
- microRNA
- SMC
- smooth muscle (SM) cell
- RA
- retinoid acid
- HDAC4
- histone deacetylase 4
- ChIP
- chromatin immunoprecipitation
- SMA
- SM α-actin
- TK
- thymidine kinase
- qRT-PCR
- quantitative real-time-PCR
- UTR
- untranslated region.
REFERENCES
- 1.Murry C. E., Keller G. (2008) Cell 132, 661–680 [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R., Young R. (2008) Cell 132, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley A., Evans M., Kaufman M. H., Robertson E. (1984) Nature 309, 255–256 [DOI] [PubMed] [Google Scholar]
- 4.Doetschman T. C., Eistetter H., Katz M., Schmidt W., Kemler R. (1985) J. Embryol. Exp. Morphol. 87, 27–45 [PubMed] [Google Scholar]
- 5.Huang H., Zhao X., Chen L., Xu C., Yao X., Lu Y., Dai L., Zhang M. (2006) Biochem. Biophys. Res. Commun. 351, 321–327 [DOI] [PubMed] [Google Scholar]
- 6.Sinha S., Hoofnagle M. H., Owens G. K. (2009) Methods Mol. Biol. 482, 345–367 [DOI] [PubMed] [Google Scholar]
- 7.Xie C., Huang H., Wei S., Song L. S., Zhang J., Ritchie R. P., Chen L., Zhang M., Chen Y. E. (2009) Stem Cells Dev. 18, 741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drab M., Haller H., Bychkov R., Erdmann B., Lindschau C., Haase H., Morano I., Luft F. C., Wobus A. M. (1997) FASEB J. 11, 905–915 [DOI] [PubMed] [Google Scholar]
- 9.Levenberg S., Golub J. S., Amit M., Itskovitz-Eldor J., Langer R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4391–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Q., Zeng L., Zhang Z., Hu Y., Xu Q. (2007) Am. J. Physiol. Cell Physiol. 292, C342–C352 [DOI] [PubMed] [Google Scholar]
- 11.Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 12.Sartore S., Chiavegato A., Faggin E., Franch R., Puato M., Ausoni S., Pauletto P. (2001) Circ. Res. 89, 1111–1121 [DOI] [PubMed] [Google Scholar]
- 13.Walsh K., Takahashi A. (2001) Z. Kardiol. 90, 12–16 [DOI] [PubMed] [Google Scholar]
- 14.Hirschi K. K., Lai L., Belaguli N. S., Dean D. A., Schwartz R. J., Zimmer W. E. (2002) J. Biol. Chem. 277, 6287–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald O. G., Owens G. K. (2007) Circ. Res. 100, 1428–1441 [DOI] [PubMed] [Google Scholar]
- 16.Neppl R. L., Wang D. Z. (2009) Cell Stem Cell 5, 130–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) Circ. Res. 100, 1579–1588 [DOI] [PubMed] [Google Scholar]
- 19.Boettger T., Beetz N., Kostin S., Schneider J., Krüger M., Hein L., Braun T. (2009) J. Clin. Invest. 119, 2634–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Y., Liu X., Yang J., Lin Y., Xu D. Z., Lu Q., Deitch E. A., Huo Y., Delphin E. S., Zhang C. (2009) Circ. Res. 105, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin M., Small E. M., Sutherland L. B., Qi X., McAnally J., Plato C. F., Richardson J. A., Bassel-Duby R., Olson E. N. (2009) Genes Dev. 23, 2166–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2009) J. Biol. Chem. 284, 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Cheng Y., Zhang S., Lin Y., Yang J., Zhang C. (2009) Circ. Res. 104, 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manabe I., Owens G. K. (2001) Circ. Res. 88, 1127–1134 [DOI] [PubMed] [Google Scholar]
- 26.Xie C. Q., Zhang J., Villacorta L., Cui T., Huang H., Chen Y. E. (2007) Arterioscler. Thromb. Vasc. Biol. 27, e311–312 [DOI] [PubMed] [Google Scholar]
- 27.Ruby J. G., Jan C., Player C., Axtell M. J., Lee W., Nusbaum C., Ge H., Bartel D. P. (2006) Cell 127, 1193–1207 [DOI] [PubMed] [Google Scholar]
- 28.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 29.Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T., Gan Q., Shang Y., Owens G. K. (2007) Am. J. Physiol. Cell Physiol. 292, C886–C895 [DOI] [PubMed] [Google Scholar]
- 31.Loots G. G., Ovcharenko I. (2004) Nucleic Acids Res. 32, W217–W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ovcharenko D., Kelnar K., Johnson C., Leng N., Brown D. (2007) Cancer Res. 67, 10782–10788 [DOI] [PubMed] [Google Scholar]
- 33.Ørom U. A., Nielsen F. C., Lund A. H. (2008) Mol. Cell 30, 460–471 [DOI] [PubMed] [Google Scholar]
- 34.Woltering J. M., Durston A. J. (2008) PLoS ONE 3, e1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzur G., Levy A., Meiri E., Barad O., Spector Y., Bentwich Z., Mizrahi L., Katzenellenbogen M., Ben-Shushan E., Reubinoff B. E., Galun E. (2008) PLoS ONE 3, e3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin P., Wang E., Ren J., Childs R., Shin J. W., Khuu H., Marincola F. M., Stroncek D. F. (2008) J. Transl. Med. 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai G., Ambalavanar R., Wei D., Dessem D. (2007) Mol. Pain 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garzon R., Volinia S., Liu C. G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., Flomenberg N., Marcucci G., Calin G. A., Kornblau S. M., Kantarjian H., Bloomfield C. D., Andreeff M., Croce C. M. (2008) Blood 111, 3183–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agirre X., Jiménez-Velasco A., San José-Enériz E., Garate L., Bandrés E., Cordeu L., Aparicio O., Saez B., Navarro G., Vilas-Zornoza A., Pérez-Roger I., García-Foncillas J., Torres A., Heiniger A., Calasanz M. J., Fortes P., Román-Gómez J., Prósper F. (2008) Mol. Cancer Res. 6, 1830–1840 [DOI] [PubMed] [Google Scholar]
- 40.Varnholt H., Drebber U., Schulze F., Wedemeyer I., Schirmacher P., Dienes H. P., Odenthal M. (2008) Hepatology 47, 1223–1232 [DOI] [PubMed] [Google Scholar]
- 41.Veerla S., Lindgren D., Kvist A., Frigyesi A., Staaf J., Persson H., Liedberg F., Chebil G., Gudjonsson S., Borg A., Månsson W., Rovira C., Höglund M. (2009) Int. J. Cancer 124, 2236–2242 [DOI] [PubMed] [Google Scholar]
- 42.Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., Guenther M. G., Johnston W. K., Wernig M., Newman J., Calabrese J. M., Dennis L. M., Volkert T. L., Gupta S., Love J., Hannett N., Sharp P. A., Bartel D. P., Jaenisch R., Young R. A. (2008) Cell 134, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C., Ridzon D., Lee C. T., Blake J., Sun Y., Strauss W. M. (2007) Mamm. Genome 18, 316–327 [DOI] [PubMed] [Google Scholar]
- 44.Gu P., Reid J. G., Gao X., Shaw C. A., Creighton C., Tran P. L., Zhou X., Drabek R. B., Steffen D. L., Hoang D. M., Weiss M. K., Naghavi A. O., El-daye J., Khan M. F., Legge G. B., Wheeler D. A., Gibbs R. A., Miller J. N., Cooney A. J., Gunaratne P. H. (2008) PLoS One 3, e2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haberland M., Montgomery R. L., Olson E. N. (2009) Nat. Rev. Genet 10, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu P., Li L. (2002) Circ. Res. 90, 858–865 [DOI] [PubMed] [Google Scholar]
- 47.Zhou X., Richon V. M., Rifkind R. A., Marks P. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis F. J., Gupta M., Camoretti-Mercado B., Schwartz R. J., Gupta M. P. (2003) J. Biol. Chem. 278, 20047–20058 [DOI] [PubMed] [Google Scholar]
- 49.Cao D., Wang Z., Zhang C. L., Oh J., Xing W., Li S., Richardson J. A., Wang D. Z., Olson E. N. (2005) Mol. Cell. Biol. 25, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 51.Perkins N. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 49–62 [DOI] [PubMed] [Google Scholar]
- 52.Widera D., Mikenberg I., Kaltschmidt B., Kaltschmidt C. (2006) Int. J. Dev. Neurosci. 24, 91–102 [DOI] [PubMed] [Google Scholar]
- 53.Magness S. T., Jijon H., Van Houten Fisher N., Sharpless N. E., Brenner D. A., Jobin C. (2004) J. Immunol. 173, 1561–1570 [DOI] [PubMed] [Google Scholar]
- 54.Torres J., Watt F. M. (2008) Nat. Cell Biol. 10, 194–201 [DOI] [PubMed] [Google Scholar]
- 55.Narayanan R., Higgins K. A., Perez J. R., Coleman T. A., Rosen C. A. (1993) Mol. Cell. Biol. 13, 3802–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.