Abstract
The rapid induction of the c-fos gene correlates with phosphorylations of histone H3 and HMGN1 by mitogen- and stress-activated protein kinases. We have used a cell-free system to dissect the mechanism by which MSK1 phosphorylates histone H3 within the c-fos chromatin. Here, we show that the reconstituted c-fos chromatin presents a strong barrier to histone H3 phosphorylation by MSK1; however, the activators (serum response factor, Elk-1, cAMP-response element-binding protein (CREB), and ATF1) bound on their cognate sites recruit MSK1 to phosphorylate histone H3 at Ser-10 within the chromatin. This activator-dependent phosphorylation of histone H3 is enhanced by HMGN1 and occurs preferentially near the promoter region. Among the four activators, CREB plays a predominant role in MSK1-mediated phosphorylation of histone H3, and the phosphorylation of Ser-133 in CREB is essential for this process. Mutational analyses of MSK1 show that its N-terminal inhibition domain is critical for the kinase to phosphorylate chromatin-embedded histone H3 in a CREB-dependent manner, indicating the presence of an intricate regulatory network for MSK1-mediated phosphorylation of histone H3.
Keywords: Chromatin/Modification, Enzymes/Kinase, Histones/Modification, Oncogene/fos, Phosphorylation, Protein/Binding/DNA, Transcription/Enhancers
Introduction
The proto-oncogene c-fos is an immediate-early gene, which is induced by various extracellular stimuli, including growth factors and cellular stresses. Transcription of the c-fos gene is controlled by inducible regulatory elements such as serum-response element, which are bound by SRF2 and Elk-1, and FAP-1 and CRE, which are bound by CREB and ATF1 (1). Upon cell stimulation, the activators become phosphorylated via cascades of protein kinases before they induce c-fos transcription (1, 2). Concomitant with the phosphorylation of the activators, the c-fos gene also undergoes rapid alterations in nucleosomal structure, which coincide with post-translational modifications of chromatin proteins, including acetylations of histones H3 and H4 as well as phosphorylations of histone H3 and non-histone chromatin protein HMGN1 (2). In particular, phosphorylations of serine 10 in histone H3 (H3-S10) and serine 6 in HMGN1 are called the “nucleosomal response” (3) and are likely to play a causal role in the rapid, dramatic induction of c-fos transcription (3, 4).
Originally, phosphorylation of H3-S10 associated with transcriptional induction was observed in immediate-early genes, including c-fos, c-jun, and c-myc (5–7). However, transcription of other types of genes appears to involve H3-S10 phosphorylation as well. In yeast, induction of Gcn5-regulated genes is controlled by H3-S10 phosphorylation (8, 9). In flies, heat shock gene loci display dramatic H3-S10 phosphorylation during heat shock-induced transcription (10). In mammals, the murine Hsp70 gene also undergoes H3-S10 phosphorylation when induced by arsenite (11). Follicle-stimulating hormone, thyroid hormone, and progesterone also induce phosphorylation of H3-S10 to regulate their target genes (12–14). Hence, phosphorylation of H3-S10 may be a histone modification that is more broadly required for transcriptional induction.
It is now firmly established that phosphorylations of histone H3 and HMGN1 are mediated by mitogen- and stress-activated protein kinases (MSK) 1 and 2, two closely related serine/threonine kinases that are activated via both the ERK1/2 and p38 MAPK pathways (15–17). MSK1/2 reside almost exclusively in the nucleus (15, 17), and once activated, they phosphorylate Ser-10 and Ser-28 in histone H3 and Ser-6 in HMGN1 (3, 4) as well as Ser-133 in CREB and Ser-63 in ATF1 (18, 19). Structurally, MSK1/2 are composed of two kinase domains, the N- and C-terminal kinase domains (NTKD and CTKD), which are related to the AGC kinase and calmodulin-dependent protein kinase families, respectively (15, 17). The CTKD is phosphorylated by the upstream MAPKs and then activates the NTKD through multiple phosphorylations within a single mitogen- and stress-activated kinase polypeptide (20).
A puzzling but intriguing aspect of H3-S10 phosphorylation is that it appears to play opposing roles in the regulation of chromatin structure during different phases of the cell cycle. During mitosis or meiosis, Aurora kinases phosphorylate the bulk of H3-S10, which in turn facilitates compaction of chromatin and transcriptional repression (21–23). During interphase, in contrast, extracellular stimuli lead eventually to phosphorylation of H3-S10 by MSK1/2 in a minute fraction of histone H3, in correlation with rapid transcriptional induction of a subset of genes (23, 24). Given the antipodal effects of H3-S10 phosphorylation on transcription during different phases of the cell cycle, the phosphorylation of H3-S10 in both phases must be under strict control.
To dissect the regulatory mechanism of chromatin phosphorylation, we have used a cell-free system reconstituted with purified activators and kinases as well as the recombinant chromatin assembled on the natural c-fos promoter (25). This system allows analyses of the roles for each factor in the regulated phosphorylation of histones within the c-fos chromatin. We show that the chromatin structure is intrinsically inhibitory to MSK1-mediated phosphorylation of histone H3; in contrast, Aurora B phosphorylates histone H3 within the chromatin relatively well, possibly through its direct interactions with the tails of core histones. The phosphorylation of histone H3 by MSK1, however, is restored by the presence of CREB and, to lesser extent, ATF1 but not by the presence of SRF or Elk-1. This CREB-dependent phosphorylation of histone H3 requires the phosphorylation of CREB at Ser-133 and is facilitated by HMGN1, a non-histone protein involved in chromatin decompaction and c-fos induction. Mutational analyses of MSK1 show that it possesses a complex network of intramolecular regulations, which CREB appears to exploit for the regulated phosphorylation of histone H3 embedded within the c-fos chromatin.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies against modified residues in histone H2A and H2B were purchased from Upstate Biotechnology, Inc., and anti-phospho-SRF (Ser-103) was purchased from Cell Signaling. All the other antibodies were purchased from Abcam.
RNA Extraction and Reverse Transcription-PCR
The cDNA, synthesized from the total RNA by a cDNA synthesis kit (TaKaRa), was used for reverse transcription-PCR with a set of primers, 5′-GCTTACTCCAGGGCTGGCGTTGTGAAAGACC-3′ and 5′-AGTCAGCCTCGGGGTAGGTGAAGACGAAGG-3′, to analyze c-fos mRNA levels. The amplified products were analyzed by electrophoresis on an agarose gel.
Plasmids
The mutant c-fos plasmid, pfMC2AT(mt), was constructed from its wild-type counterpart, pfMC2AT (25), using a PCR-based method with the following oligonucleotides to introduce mutations into the serum-response element, FAP-1, and CRE sequences: SRE, 5′-CTTACACTGTATGTCCCAATCGGGACATC-3′ and 5′-GATGTCCCGATTGGGACATACAGTGTAAG-3′; FAP-1, 5′-CCAATCGGGACATCCAACTCAGGCAGG-3′ and 5′-CCTGCCTGAGTTGGATGTCCCGATTGG-3′; and CRE, 5′-GGTTGAGCCCGTAAGGCGTACACTC-3′ and 5′-GAGTGTACGCCTTACGGGCTCAAGG-3′. The cDNA clones and the expression vectors for transcription factors are described (25).
Protein Purification
SRF, Elk-1, CREB, and ATF1 were purified as described (25). Recombinant human histones H2A and H2B or H3 and H4 were co-expressed in Escherichia coli as H2A/H2B dimers or H3/H4 tetramers, respectively. Cells were sonicated in HEG(100) (10 mm Hepes-KOH, pH 7.6, 1 mm EDTA, 10% glycerol, 100 mm NaCl) containing 0.1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 1 mm sodium metabisulfite, and 2 mm benzamidine HCl. HCl was added to the extract to the final concentration of 0.25 m, and the mixture was incubated at −20 °C for 30 min. After centrifugation, the supernatant was neutralized with 0.25 volume of 2 m Tris base and dialyzed against HEGK(100) (10 mm Hepes-KOH, pH 7.6, 1 mm EDTA, 10% glycerol, 10 mm KCl, 100 mm NaCl). Histones were purified further by a HiTrap SP HP column (GE Healthcare) in a linear gradient from 0.1 to 2 m NaCl. Histones were eluted at ∼1.1 m NaCl, and typical yields of H2A/H2B dimer and H3/H4 tetramer were ∼25 and ∼3 mg, respectively, per 1 liter of E. coli culture. Native core histones were purified from suspension culture of HeLa S3 cells. The cells were harvested, washed with phosphate-buffered saline, and homogenized in the lysis buffer (0.1% Triton X-100, 0.25 m sucrose, 10 mm Tris-HCl, pH 7.4, 10 mm sodium butyrate, 4 mm MgCl2) containing 0.1 mm phenylmethylsulfonyl fluoride, and 2 mm benzamidine HCl. After centrifugation, the nuclear pellet was resuspended in the lysis buffer. The nuclei were overlaid onto a 30% sucrose cushion, and after centrifugation, the nuclear pellet was washed twice with the lysis buffer and once with 0.25 mm EDTA, pH 8.0. The nuclear pellet was suspended in HEG(100), and the native core histones were purified further as described for the recombinant histones. Human His-tagged NAP-1 was expressed in insect cells and purified from the cytoplasmic fraction by phenyl-Sepharose and HiTrap Q columns (GE Healthcare). FLAG-tagged human topoisomerase I was expressed in insect cells and purified by SP-Sepharose and hydroxyapatite columns. Drosophila ACF1 and ISWI were co-expressed in insect cells and purified by HiTrap SP, HiTrap Q, and Mono S columns (GE Healthcare). FLAG-tagged p38 MAPK was co-expressed with a constitutively active mutant of MKK6 in E. coli. The activated pp38 was purified by a HiTrap Q column and anti-FLAG M2 affinity gel (Sigma). FLAG-tagged MSK1 and Aurora B were expressed in insect cells and purified with ion-exchange columns and anti-FLAG M2 affinity gel. HMGN1 was expressed in E. coli, and the collected cells were sonicated in 5% perchloric acid. After centrifugation, trichloroacetic acid was added to the supernatant to the final concentration of 25%, and the mixture was incubated at 4 °C for 2 h. After centrifugation, the pellet was resuspended in 0.3 m HCl, and the insoluble proteins were removed by centrifugation. The HMGN1 was then precipitated by acetone, and the pellet was resuspended in BC(100). HMGN1 was further purified by a HiTrap Q column.
In Vitro Chromatin Assembly
The template plasmid (0.3 μg) was relaxed in advance with 10 ng of topoisomerase I before the subsequent chromatin assembly reaction. After 0.38 μg of histones and 8.8 μg of NAP-1 were mixed and incubated on ice for 15 min in the assembly buffer (10 mm Hepes-KOH, pH 7.6, 1 mm EDTA, 10% glycerol, 70 mm KCl, 0.3 μg of bovine serum albumin), the relaxed plasmid (0.3 μg), 0.1 μg of ACF1/ISWI, 10 ng of topoisomerase I, and 10 μl of ATP-regenerating system (213 μm creatine phosphate, 21.3 μm ATP, 30 μm MgCl2, 38 μg of creatine kinase) were added to the reactions. When the chromatin was used for the kinase assays in the presence of [γ-32P]ATP, the ATP-regenerating system contained 0.213 μm ATP. Where indicated, 2.5 or 5.0 μg of HMGN1 was also included during the assembly reaction. The reactions were further incubated at 30 °C for 2 h and then used for DNA supercoiling or MNase assays.
In Vitro Chromatin Kinase Assays
The chromatin (equivalent to 150 ng of DNA) was incubated at 30 °C for 1 h with kinases (150 ng of MSK1 plus 6.3 ng of pp38 or 150 ng of Aurora B) and activators (each 0.1 μg) in the kinase buffer (10 mm Hepes-KOH, pH 7.9, 100 mm KCl, 10 mm MgCl2, 5% glycerol, 0.5 mm Na3VO4) containing 10 μm ATP and 10 μCi of [γ-32P]ATP (PerkinElmer Life Sciences). Then, polyethyleneimine (PEI) was added to the mixture to the final concentration of 0.8%, and after incubation on ice for 30 min, the chromatin was precipitated and washed in BC(100) (10 mm Hepes-KOH, pH 7.9, 10% glycerol, and 100 mm KCl). The precipitate was suspended in SDS-PAGE sample buffer, boiled, and separated by SDS-PAGE. The phosphorylated histones were detected by either autoradiography or immunoblotting with an appropriate phosphospecific antibody.
GST Pulldown Assays
GST and GST-SRF, -Elk-1, -CREB, and -ATF1 were expressed in insect cells, and the histone tails fused to the N terminus of GST were expressed in E. coli. The extract from the expressed cells was incubated with glutathione-Sepharose 4B (GE Healthcare) for 1.5 h at 4 °C in the binding buffer (10 mm Tris-HCl, pH 7.3, 1.5 mm MgCl2, 10 mm KCl) and then washed extensively in BC(100). The amount of extract was adjusted so that a roughly equal amount of each GST fusion protein was bound to glutathione-Sepharose 4B. The bound GST fusion proteins were incubated in BC(100) at room temperature at 4 °C with either F:MSK1 or F:Aurora B, which were partially purified by a HiTrap Q column. The bound proteins were eluted in SDS-PAGE sample buffer, resolved by SDS-PAGE, and detected by immunoblotting with anti-FLAG M2 antibody.
In Vivo ChIP Assays
HeLa S3 cells, serum-starved and then treated with 10 μg/ml anisomycin, were fixed with 1% formaldehyde at 37 °C for 5 min. The cells were washed with ice-cold phosphate-buffered saline, resuspended in the lysis buffer (1% SDS, 10 mm EDTA, pH 8.0, 50 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture (Sigma), 1 mm Na3VO4, 50 mm NaF, and 10 mm sodium butyrate), and sonicated. After centrifugation, the supernatant was diluted with 10 volumes of the ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.0, 167 mm NaCl, protease inhibitor mixture (Sigma), 1 mm Na3VO4, 50 mm NaF, and 10 mm sodium butyrate). The supernatant was cleared at 4 °C for 3 h with 20 μl of protein A-agarose (50% slurry) (GE Healthcare) in 0.2 mg/ml sonicated salmon sperm DNA, 0.5 mg/ml bovine serum albumin, 5 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, and 0.025% sodium azide. After centrifugation, an appropriate antibody was added to the supernatant, and the mixture was incubated at 4 °C overnight. Then, 10 μl of protein A-agarose was added, and the mixture was further incubated at 4 °C for 3 h. The protein A-agarose was washed first in Low Salt Buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, pH 8.0, 20 mm Tris-HCl, pH 8.0, and 150 mm NaCl), second in High Salt Buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 500 mm NaCl), third in LiCl Buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, pH 8.0, and 10 mm Tris-HCl, pH 8.0), and finally in TE Buffer (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA). The bound DNA was eluted in 10 mm dithiothreitol, 10% SDS, and 0.1 m NaHCO3 at room temperature. The eluted DNA samples were pooled and heated at 65 °C for at least 6 h in 0.2 m NaCl to reverse the formaldehyde cross-linking. DNA fragments were treated with proteinase K, extracted with phenol/chloroform, and precipitated with ethanol. The obtained DNA was analyzed by real time PCR with a set of primers for the c-fos promoter regions (5′-CGCGAGCAGTTCCCGTCAATCCCT-3′ and 5′-GAATGAGTGTAAACGTCACGGGCTC-3′) and the β-globin coding region (5′-GCTGCTGGTTGTCTACCCTTGGAC-3′ and 5′-CACTGAGGCTGGCAAAGGTGCCCT-3′). PCRs were performed with SYBR Green PCR Master Mix (Applied Biosystems), and the products were analyzed in ABI Prism 7900 sequence detector system (Applied Biosystems).
In Vitro ChIP Assays
In vitro ChIP assays were performed essentially as described for in vivo ChIP assays, except that formaldehyde cross-linking and sonication were not used. Instead, the chromatin was digested with MNase at 37 °C for 10 min, and the digested sample was then diluted with 10 volumes of the ChIP dilution buffer before immunoprecipitation. The recovered DNA samples were analyzed with a set of primers for the c-fos promoter as shown above and for the vector region (5′-GGATTAGCAGAGCGAGGTATGTAGG-3′ and 5′-TTTCGTTCCACTGAGCGTCAGACCC-3′).
RESULTS
Increased Occupancy of MSK1 on the c-fos Promoter upon Induction of c-fos Gene Transcription
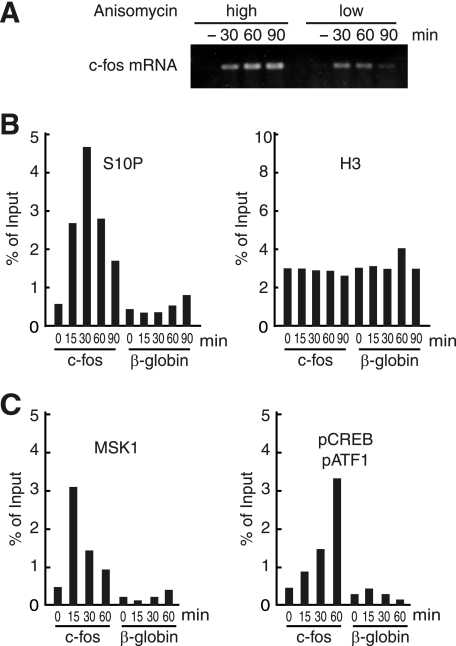
As a first step toward analyzing the mechanisms of histone H3 phosphorylation during transcriptional activation, we chose the induction of c-fos gene transcription by anisomycin as a model system. As reported previously (3, 26), anisomycin treatment (10 μg/ml or 25 ng/ml) of HeLa cells induced c-fos transcription, with higher concentration (10 μg/ml) of anisomycin eliciting a more sustained c-fos transcription (Fig. 1A). Using HeLa cells treated with 10 μg/ml anisomycin, we performed chromatin immunoprecipitation (ChIP) assays to monitor the phosphorylation of serine 10 in histone H3 (H3-S10), recruitment of MSK1, and phosphorylation of CREB/ATF1 on the c-fos promoter. Phosphorylated serine 10 in histone H3 (H3-S10P) appeared within 15 min after anisomycin treatment, peaked at 30 min, and continued to show a severalfold increase until 90 min (Fig. 1B). The occupancy of MSK1 peaked only 15 min after anisomycin treatment, apparently preceding the phosphorylation of H3-S10 (Fig. 1C). Consistent with its role in phosphorylating CREB/ATF1 (18, 19), the occupancy of MSK1 also preceded that of phosphorylated CREB/ATF1 (pCREB/pATF1), which continued to increase until 60 min after anisomycin treatment (Fig. 1C). These results demonstrate that MSK1 is actively recruited to the c-fos promoter before the appearances of pCREB/pATF1 and H3-S10P on the c-fos chromatin.
FIGURE 1.
Histone H3 phosphorylation and MSK1 recruitment on the c-fos promoter in response to anisomycin treatment. A, HeLa cells were stimulated by anisomycin with two different concentrations (high, 10 μg/ml; low, 25 ng/ml) after being starved in 0.5% fetal bovine serum for 1.5 days. The c-fos mRNA was amplified by reverse transcription-PCR, followed by electrophoresis on an agarose gel. B, HeLa cells were stimulated by anisomycin (10 μg/ml) for the indicated time and used for ChIP assays with anti-histone H3 and anti-H3-S10P antibodies. The precipitated DNA was quantified by real time PCR using primers for the c-fos gene. As control, the β-globin gene was used. The values were averaged from two independent experiments. C, occupancies of MSK1 and pCREB/pATF1 were determined by ChIP assays using antibodies against MSK1 and pCREB(Ser-133), which also recognizes pATF1(Ser-63). The values were averaged from two independent experiments.
Chromatin Precludes Histone H3 Phosphorylation by MSK1
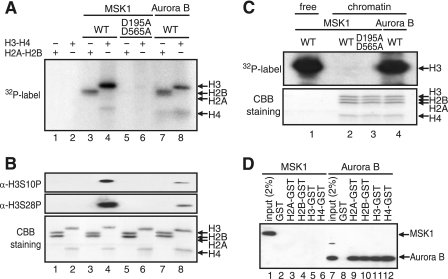
To recapitulate MSK1-mediated phosphorylation of histone H3 within the chromatin in vitro, we first prepared MSK1 and its inactive mutant, D195A/D565A, as well as Aurora B (supplemental Fig. S1). Fig. 2A indicates that MSK1 and Aurora B phosphorylated histones H3 (lanes 4 and 8) as well as H2B (lanes 3 and 7) and, to a lesser extent, histone H4 (lanes 4 and 8). The contribution of contaminating kinases to the observed phosphorylations was ruled out because no phosphorylation was detected when the kinase-deficient MSK1 mutant, D195A/D565A, was used (Fig. 2A, lanes 5 and 6). The immunoblots using anti-S10P and anti-S28P antibodies confirmed that MSK1 and Aurora B phosphorylated both Ser-10 and Ser-28 in histone H3 (lanes 4 and 8).
FIGURE 2.
MSK1 does not phosphorylate histones assembled into chromatin. A, kinase assays contained [γ-32P]ATP and 100 ng of recombinant histones with 100 ng of MSK1 (lanes 3 and 4), D195A/D565A (lanes 5 and 6), or Aurora B (lanes 7 and 8). The position of each histone is indicated on the right. WT, wild type. B, identical kinase assays were performed in the absence of [γ-32P]ATP, and the phosphorylated residues in histone H3 were identified by immunoblotting using anti-H3-S10P and anti-H3-S28P antibodies. CBB, Coomassie Brilliant Blue. C, 150 ng of chromatin assembled on pfMC2AT was phosphorylated in the presence of [γ-32P]ATP with 150 ng of MSK1 (lane 2), D195A/D565A (lane 3), or Aurora B (lane 4). As control, 10 ng of free H3-H4 tetramer were phosphorylated with MSK1 (lane 1). The positions of histone H3 on the autoradiogram (upper panel) and the core histones on the SDS-polyacrylamide gel (lower panel) are indicated on the right. D, N-terminal tails of histones fused to GST were retained on glutathione-Sepharose 4B and incubated with FLAG-tagged MSK1 (lanes 2–6) or Aurora B (lanes 8–12), and the bound proteins were analyzed by immunoblotting using anti-FLAG M2 antibody. The positions of the bound MSK1 or Aurora B are indicated on the right.
Next, chromatin was reconstituted on a c-fos promoter plasmid, pfMC2AT (25), via the ATP-utilizing chromatin assembly and remodeling factor-based chromatin assembly system using recombinant core histones (supplemental Fig. S2, A and B). The recombinant core histones, which were devoid of post-translational modifications such as phosphorylation, acetylation, and methylation (supplemental Fig. S2E), produced a highly regular array of nucleosomes, as opposed to the HeLa cell-derived core histones (supplemental Fig. S2, C and D). Moreover, the regularity of nucleosomes was not affected by the G-less cassette in pfMC2AT. Using the reconstituted c-fos chromatin, we then asked whether MSK1 and Aurora B phosphorylate histone H3 within the context of the chromatin. The assembled c-fos chromatin was incubated with the indicated kinase in the presence of [32P]ATP and precipitated by PEI to remove unincorporated [32P]ATP. The precipitated chromatin was then separated by SDS-PAGE and autoradiographed. As shown in Fig. 2C, MSK1 seemed to phosphorylate histone H3 embedded within the chromatin to a very small extent (upper panel, lane 2). Unlike MSK1, however, Aurora B phosphorylated histone H3 within the chromatin relatively well (Fig. 2C, upper panel, lane 4). To explore a mechanistic basis for their differential sensitivities to the chromatin, we tested the interactions between each kinase and the N-terminal tails of core histones. GST pulldown assays revealed that Aurora B interacted with the N-terminal tails of all four histones (Fig. 2D, lanes 9–12), whereas MSK1 failed to do so under the same condition (Fig. 2D, lanes 3–6). Together, our results show that the chromatin structure presents a strong barrier to the MSK1-mediated phosphorylation of histone H3; however, Aurora B appears to overcome this barrier relatively well, presumably through its direct interactions with the histone tails.
MSK1 Phosphorylates Histone H3 at Ser-10 in an Activator-dependent Manner
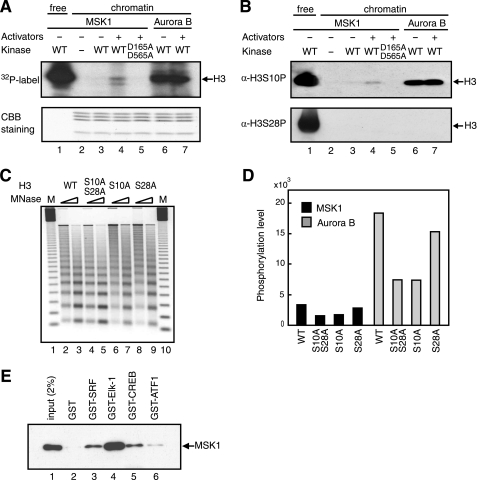
In view of the poor MSK1-mediated phosphorylation of histone H3 within the c-fos chromatin, we wondered whether MSK1 requires factors that assist its interactions with histones in the context of chromatin. Because prime candidates for directing MSK1 to histone H3 within the c-fos chromatin are activators, including SRF, Elk-1, CREB, and ATF1, we examined their effect on histone H3 phosphorylation by MSK1. When the activators were included in the reactions, MSK1 phosphorylated histone H3 within the chromatin noticeably better than in the absence of the activators (Fig. 3A, lanes 2–4). Interestingly, this activator dependence was observed for additional phosphorylation, possibly of histone H2A (Fig. 3A, lane 4), which may represent another physiologically important phosphorylation site (27). The observed phosphorylations were specific to the intrinsic kinase activity of MSK1 because the kinase-deficient MSK1, D195A/D565A, displayed no phosphorylation of histones (Fig. 3A, lane 5). In marked contrast, Aurora B phosphorylated chromatin-embedded histone H3 relatively well regardless of the presence of the activators (Fig. 3A, lanes 6 and 7).
FIGURE 3.
MSK1 phosphorylates serine 10 of histone H3 within chromatin in an activator-dependent manner. A, 150 ng of the c-fos chromatin was incubated in the presence of 150 ng of MSK1 (lanes 3–5) or Aurora B (lanes 6 and 7) and 100 ng of the four activators (lanes 4, 5, and 7) using 10 mm [γ-32P]ATP. D195A/D595A is a kinase-deficient MSK1 (lane 5). 20 ng of free H3/H4 tetramer was phosphorylated by 400 ng of MSK1 as control (lane 1). The gel was stained in Coomassie Brilliant Blue (CBB) (lower panel) and then exposed to an x-ray film (upper panel). WT, wild type. B, same sets of reactions as in A were performed in the absence of [γ-32P]ATP, and the phosphorylated residues were detected by immunoblotting using anti-H3-S10P (upper panel) or anti-H3-S28P (lower panel). C, chromatins assembled with wild-type (lanes 2 and 3) or mutant histone H3 whose serine was replaced with an alanine at both Ser-10 and Ser-28 (lanes 4 and 5), Ser-10 (lanes 6 and 7), or Ser-28 (lanes 8 and 9) were digested with two different concentrations of MNase. Molecular mass marker (M) is a 123-bp DNA ladder (lanes 1 and 10). D, kinase reactions were performed as in A, using the chromatin reconstituted with wild-type or mutant histones in the presence of MSK1 (black bars) or Aurora B (gray bars). The levels of phosphorylation were measured by ImageQuant software (GE Healthcare). E, GST (lane 2) or GST-fused activators (lanes 3–6) bound to glutathione-Sepharose 4B were incubated with FLAG-tagged MSK1, and the bound MSK1 was detected by immunoblotting using anti-FLAG M2 antibody. 2% of input FLAG-tagged MSK1 was loaded as control (lane 1).
To determine whether MSK1 phosphorylates histone H3 at the physiologically relevant residues, Ser-10 and Ser-28, in response to the activators, immunoblot analyses were performed using antibodies that specifically recognize H3-S10P or H3-S28P. As shown in Fig. 3B, MSK1 phosphorylated H3-S10 clearly in the presence of the activators but only marginally in their absence (lanes 3 and 4). This phosphorylation of H3-S10 required the intact kinase domain of MSK1, indicating that the phosphorylation of H3-S10 is specific (Fig. 3B, lane 5). Aurora B phosphorylated H3-S10 to a greater extent than did MSK1, albeit in a completely activator-independent manner (Fig. 3B, lanes 6 and 7). Interestingly, despite their strong activity toward Ser-28 in free histone H3, both kinases did not phosphorylate H3-S28 to detectable levels in the context of chromatin. This predominant phosphorylation of H3-S10 over H3-S28 by MSK1 was further corroborated by the quantitative analyses of 32P-labeled histone H3 on the chromatins reconstituted with histone H3 mutant that harbors serine-to-alanine mutation(s); namely S10A, S28A, or S10A/S28A (Fig. 3, C and D). As quantified in Fig. 3D, the overall level of MSK1-mediated phosphorylation of histone H3 was reduced markedly by the S10A mutation but not by the S28A mutation, indicating that MSK1 phosphorylates histone H3 at Ser-10 but not Ser-28 within the chromatin. Likewise, the levels of phosphorylation of H3-S10A and H3-S10A/S28A were comparative (Fig. 3D), again indicating the absence of Ser-28 phosphorylation. The residual phosphorylation of S10A/S28A also suggests a potential phosphorylation site in histone H3 (Fig. 3D), although its significance remains to be determined. Together, these results demonstrate that the activators on the c-fos chromatin permit MSK1 to phosphorylate histone H3 at Ser-10 but not Ser-28 and that the Ser-10 phosphorylation occurs independently of the phosphorylation status of Ser-28.
Given the role of the activators for MSK1-mediated H3 phosphorylation, we asked whether MSK1 interacts directly with the activators. As shown in Fig. 3E, GST pulldown assays demonstrated that MSK1 interacted with all the four activators with variable affinities. The strongest interaction with MSK1 was observed for Elk-1, followed by SRF and CREB, whereas the interaction of MSK1 with ATF1 was reproducibly the weakest. These results demonstrate that, in contrast to Aurora B, MSK1 phosphorylates histone H3 at Ser-10 by virtue of its interactions with the activators.
HMGN1 Enhances Activator- dependent Histone H3 Phosphorylation by MSK1
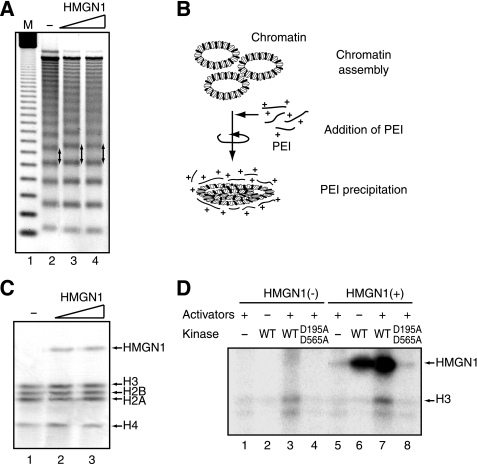
HMGN1 is a nucleosome-binding protein, which decompacts chromatin and is associated with actively transcribed genes (28, 29). Because HMGN1 is phosphorylated by MSK1/2 when c-fos is induced (4), we explored whether HMGN1 has a direct effect on MSK1-mediated histone H3 phosphorylation. Recombinant HMGN1, expressed in and purified from E. coli (supplemental Fig. S3A), served as an efficient substrate for phosphorylation by MSK1 (supplemental Fig. S3B). Consistent with the previous study (30), the purified HMGN1 could be incorporated efficiently into the chromatin and increased the interval between nucleosomes (Fig. 4A, arrows), an effect that is consistent with its role in chromatin decompaction. To obtain additional evidence of HMGN1 incorporation into the chromatin, we precipitated the assembled chromatin with PEI, a positively charged polymer that binds DNA but not histones and HMGN1 (Fig. 4B). Fig. 4C shows that PEI precipitated not only the histones that were assembled into chromatin but HMGN1 as well, further confirming that HMGN1 was indeed incorporated into the assembled chromatin. An SDS-PAGE analysis revealed that the precipitated chromatin contained roughly equivalent moles of HMGN1 and core histones (Fig. 4C), consistent with the incorporation of two HMGN1 molecules per each nucleosome (30, 31).
FIGURE 4.
HMGN1 enhances MSK1-mediated histone H3 phosphorylation within the c-fos chromatin. A, c-fos plasmid, pfMC2AT, was assembled into chromatin in the presence of 0 μg (lane 2), 2.75 μg (lane 3), and 5.5 μg (lane 4) of HMGN1, and then partially digested by MNase. The arrows indicate the increased spacing between nucleosomes by incorporated HMGN1. Lane 1, molecular mass marker (M). B, schematic depiction of chromatin precipitation by PEI. C, chromatin was assembled in the presence of 0 μg (lane 1), 2.75 μg (lane 2), and 5.5 μg (lane 3) of HMGN1 as in A, precipitated by PEI as in B, resolved by SDS-PAGE, and stained with Coomassie Brilliant Blue (CBB). The positions of precipitated proteins are indicated on the right. D, kinase assays were performed with the chromatin assembled in the presence (lanes 5–8) or absence (lanes 1–4) of 5.5 μg of HMGN1. The chromatin was incubated in the presence of [γ-32P]ATP with (lanes 1, 3–5, 7, and 9) or without (lanes 2 and 6) 100 ng each of activators (SRF, Elk-1, CREB, and ATF1). The reactions also contained wild-type (WT) MSK1 (lanes 2, 3, 6, and 7) or D195A/D565A (lanes 4 and 8). The positions of HMGN1 and H3 are indicated on the right.
Next, we examined the effect of the incorporated HMGN1 on the phosphorylation of chromatin. As shown in Fig. 4D, MSK1 phosphorylated histone H3 in response to the activators to a greater extent on the chromatin with HMGN1 than that without HMGN1 (lanes 3 and 7), indicating that HMGN1 facilitates MSK1-mediated phosphorylation of histone H3. Interestingly, MSK1 phosphorylated chromatin-bound HMGN1 quite efficiently regardless of the presence of activators (Fig. 4D, lanes 6 and 7). Nonetheless, there appeared a slight increase in the phosphorylation of HMGN1 in the presence of the activators (Fig. 4D, lanes 6 and 7), suggesting that some residue(s) of HMGN1 may be phosphorylated in an activator-dependent manner as well. Overall, these results indicate that HMGN1 directly enhances MSK1-mediated histone H3 phosphorylation and also suggest a possible role for some phosphorylated residues of HMGN1 in the histone H3 phosphorylation.
Histone H3 Phosphorylation by MSK1 Occurs Preferentially on the Promoter Proximal Region
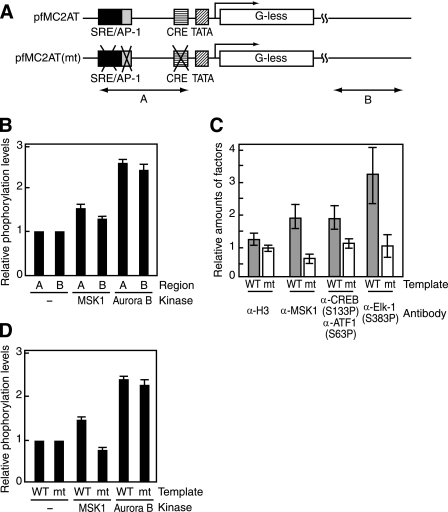
Given the requirement of the activators in MSK1-mediated H3-S10 phosphorylation, we performed in vitro ChIP assays to analyze whether H3-S10P is localized to the c-fos promoter region, where SRF, ATF1, CREB, and ATF1 are present. Following the in vitro chromatin kinase assays, the chromatin was digested by MNase and precipitated by an antibody against either histone H3 or H3-S10P, and then the precipitated DNA was subjected to real time PCR using sets of primers for the promoter region as well as the vector region located opposite from the promoter (A or B in Fig. 5A). As shown in Fig. 5B, MSK1 phosphorylated H3-S10 around the promoter region slightly more efficiently than the vector region. In contrast, Aurora B phosphorylated H3-S10 with little regional preference, albeit at a higher efficiency (Fig. 5B). Although the observed regional specificity for MSK1-mediated H3-S10 phosphorylation was unexpectedly small, this difference may be significant in view of the atomic force microscopic observation that the nucleosomes on the opposite side of a chromatin reconstituted on a typical circular plasmid are not necessarily remote from each other and could often be in close proximity to each other (32).
FIGURE 5.
Preferential phosphorylation of histone H3 near the c-fos promoter region. A, wild-type (pfMC2AT) and mutant (pfMC2AT(m)) c-fos templates used for in vitro ChIP assays and the regions amplified by PCR. Region A overlaps with the c-fos promoter, and region B is within the vector pUC19 and ∼1.5 kb downstream from the transcription start site. B, c-fos chromatin was phosphorylated by MSK1 in the presence of activators or by Aurora B and then analyzed by ChIP assays using anti-H3-S10P. The levels of H3-S10P were normalized by the values obtained with anti-histone H3 antibody. The values obtained without any kinase were set to 1. C, in vitro ChIP assays for the occupancies of MSK1 and phosphorylated activators (CREB, ATF1, and Elk-1). Wild-type (WT) and mutant (mt) c-fos templates were assembled into chromatin, and the kinase reactions were done in the presence of MSK1 and the activators. ChIP assays were performed with antibodies against histone H3, MSK1, pCREB(Ser-133) and pElk-1(Ser-383), using the primers for the region A. The anti-pCREB(Ser-133) antibody also recognizes pATF1(Ser-63). Gray and white bars indicate the relative amounts of histone H3, MSK1, and the activators on the wild-type and mutant templates, respectively. D, wild-type and mutant c-fos templates were assembled into chromatin, and the relative levels of H3-S10 phosphorylation in the presence of MSK1 or Aurora B were determined by ChIP assays using anti-H3-S10P and the primers for region A. The error bars represent S.E.
To further validate the role of the activators in recruiting MSK1 to the promoter region, we used a c-fos template with mutated serum-response element, FAP-1, and CRE that do not bind the activators. ChIP assays showed that the mutant template did not bind the activators (ATF1/CREB and Elk-1) and failed to recruit MSK1 to the template (Fig. 5C). Moreover, MSK1 did not phosphorylate H3-S10 on the mutant c-fos template even in the presence of the activators, indicating that the activators must be bound to their cognate sites to elicit MSK1-mediated H3-S10 phosphorylation (Fig. 5D). In contrast, Aurora B phosphorylated H3-S10 on both wild-type and mutant c-fos templates to comparable levels, indicating that the phosphorylation of H3-S10 by Aurora B is not influenced by the activators (Fig. 5D). Together, these experiments indicate that the activators bound on their cognate sites recruit MSK1 to the promoter and allow preferential phosphorylation of histone H3 at Ser-10 near the promoter.
Phosphorylation of Ser-133 in CREB Is Essential for the Phosphorylation of H3-S10
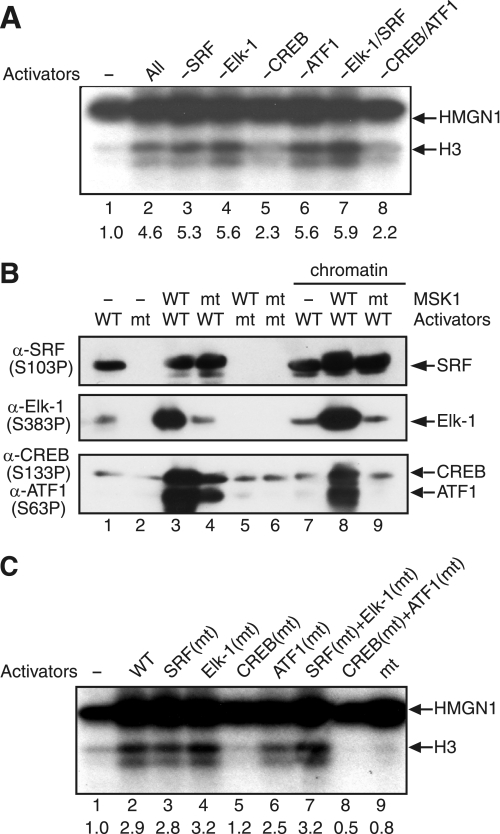
Having demonstrated the role of activators in directing MSK1 to phosphorylate H3-S10, we determined which of the four activators are essential for this process. Fig. 6A shows that only the omission of CREB reduced the MSK1-mediated H3-S10 phosphorylation significantly, although it reduced HMGN1 phosphorylation only marginally (lane 5). ATF1 also had a small effect in the MSK1-mediated H3-S10 phosphorylation because the simultaneous omission of CREB and ATF1 reduced the phosphorylation more than the omission of CREB alone (Fig. 6A, lanes 5 and 8). In contrast, MSK1 phosphorylated H3-S10 efficiently regardless of the presence of SRF and Elk-1 (Fig. 6A, lanes 3, 4, and 7). These results indicate that CREB is the predominant transcription factor that directs the MSK1-mediated histone H3 phosphorylation.
FIGURE 6.
MSK1-mediated chromatin phosphorylation requires phosphorylation of Ser-133 in CREB. A, c-fos chromatin was phosphorylated by MSK1 in the presence of various combinations of activators. The reactions contained the following: no activator (lane 1), SRF, Elk-1, CREB, and ATF1 (lane 2); Elk-1, CREB, and ATF1 (lane 3); SRF, CREB, and ATF1 (lane 4); SRF, Elk-1, and ATF1 (lane 5); SRF, Elk-1, and CREB (lane 6); CREB and ATF1 (lane 7); and SRF and Elk-1 (lane 8). The positions of histone H3 and HMGN1 are indicated on the right. B, wild-type (WT) or mutant (mt) activators were analyzed by immunoblotting using the indicated phosphospecific antibodies. The control reactions (lanes 1–6) contained either wild-type or mutant MSK1 and activators in the absence of chromatin, whereas the reactions in lanes 7–9 were performed in the presence of the c-fos chromatin. C, phosphorylation-defective mutants of SRF (lane 3), Elk-1 (lane 4), CREB (lane 5), ATF1 (lane 6), SRF and Elk-1 (lane 7), and CREB and ATF1 (lane 8) were used in place of the corresponding wild-type activator(s). Control reactions contained no activator (lane 1), wild-type activators (lane 2), and mutant activators (lane 9). The positions of histone H3 and HMGN1 are indicated on the right.
Because the activators become phosphorylated together with the phosphorylation of H3-S10 during c-fos induction in vivo (Fig. 1C), we asked whether the phosphorylations of the activators are functionally related to that of histone H3. Although the activators obtained from insect cells were partially phosphorylated, the purified activators could be further phosphorylated by MSK1 in vitro. For instance, partially phosphorylated Ser-383 in Elk-1, Ser-133 in CREB, and Ser-63 in ATF1 could be phosphorylated further by MSK1 in vitro in the absence (Fig. 6B, lanes 1–6) or presence (Fig. 6B, lanes 7–9) of the c-fos chromatin. To test the functional roles of these and other phosphorylations, we mutated the following amino acid residues that became phosphorylated upon cell stimulation into an alanine, namely Ser-77, Ser-79, Ser-83, Ser-85, and Ser-103 in SRF (33, 34); Thr-363, Thr-368, Ser-383, and Ser-389 in Elk-1 (35); Ser-133 in CREB (36); and Ser-63 in ATF1 (37). When the phosphorylation-defective activators were used in place of their respective wild-type counterparts, S133A strongly inhibited the phosphorylation of histone H3 (Fig. 6C, lane 5), and S63A reduced the phosphorylation slightly. Notably, S133A also eliminated the low background phosphorylation observed when the reactions contained MSK1 but not the activators (Fig. 6C, lanes 1 and 5), indicating that S133A may inhibit the kinase activity of MSK1 in a dominant negative manner. Consistent with the dispensability of SRF and Elk-1 (Fig. 6A), phosphorylation-defective SRF and Elk-1 also did not affect the histone H3 phosphorylation at all (Fig. 6C, lanes 3, 4, and 7). Taken together, these experiments make it highly likely that the phosphorylation of CREB at Ser-133 and, to a lesser extent, ATF1 at Ser-63 serves as a regulatory cue for MSK1-mediated H3-S10 phosphorylation.
MSK1 Is Subject to Both Positive and Negative Autoregulation for Phosphorylating Chromatin-embedded Histone H3
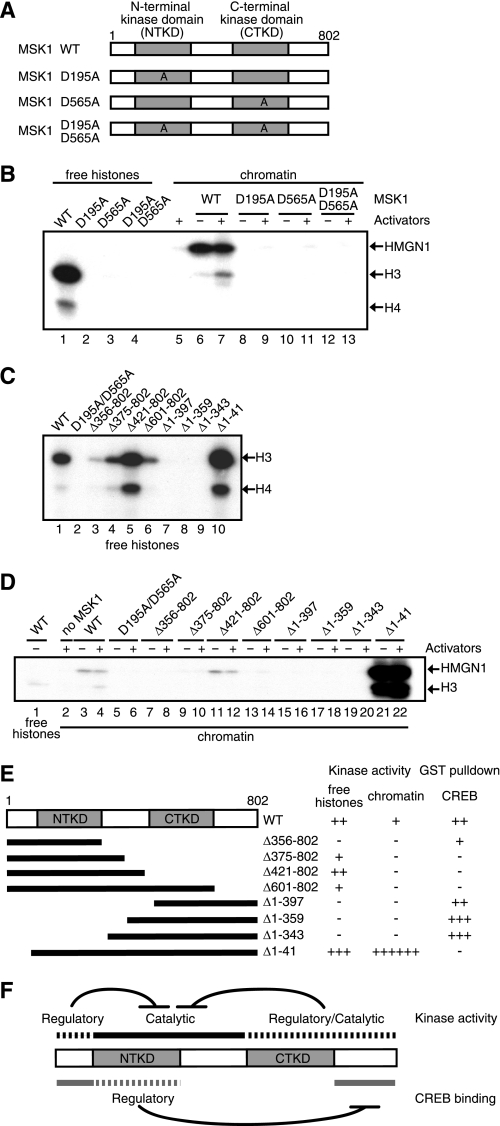
MSK1 is one of unique kinases that possess two kinase domains termed N- and C-terminal kinase domain (NTKD and CTKD) (15, 17). Because MSK1 appears to phosphorylate HMGN1 and histone H3 concomitantly during c-fos induction (4), we hypothesized that each kinase domain may be dedicated to phosphorylate separate substrates. We also wondered whether one of the kinase domains may be dispensable for phosphorylating free histones but not for chromatin-embedded histones. To test these hypotheses, we constructed three MSK1 mutants, each of which lacks N-terminal kinase activity (D195A), C-terminal kinase activity (D565A), or both kinase activities (D195A/D565A) (Fig. 7A). Similar to D195A/D565A, both D195A and D565A lost the kinase activity toward free and chromatin-bound histones alike (Fig. 7B). Likewise, D195A and D565A completely lost its kinase activity toward chromatin-bound HMGN1 (Fig. 7B). These results indicate that the both N- and C-terminal kinase activities are necessary for phosphorylating different substrates and for both free and chromatin-embedded histones.
FIGURE 7.
Multiple domains within MSK1 are required for histone H3 phosphorylation. A, D195A, D565A, or D195A/D565A has an alanine in place of an aspartate at residue(s) 195, 565, or 195 and 565, respectively. WT, wild type. B, histone H3-H4 tetramers (lanes 1–4) or reconstituted chromatin (lanes 5–13) were phosphorylated by wild-type (lanes 1, 6, and 7) or mutant MSK1 (lanes 2–4 and 8–13). Mitogen- and stress-activated kinase (MSK) was omitted from lane 5. In the chromatin kinase assays, the reactions were performed in the presence (+) or absence (−) of activators (SRF, Elk-1, CREB, and ATF1). Phosphorylated HMGN1 and histones were detected by autoradiography. The positions of HMGN1 and histone H3 and H4 are indicated on the right. C, histone H3-H4 tetramers were phosphorylated by the deletion mutants of MSK1 as depicted in E. The positions of histone H3 and H4 are indicated on the right. D, chromatin kinase assays were performed with wild-type (lanes 3 and 4) or mutant MSK1 (lanes 5–22) in the presence or absence of activators. As controls, free histone H3-H4 tetramers were phosphorylated by wild-type MSK1 (lane 1). MSK1 was omitted from lane 2. E, schematic depiction of MSK1 deletion mutants and the summarized results of C and D as well as those of GST pulldown assays between the MSK1 mutants and CREB. No phosphorylation or interaction is indicated by −, and the approximate levels of phosphorylation or interaction are indicated by the increasing number of +. F, multiple intramolecular regulations control the kinase activity of MSK1 and its interactions with CREB.
Previous studies on MSKs or their related RSKs revealed that their activities are regulated by a complex relay of phosphorylations (15, 17, 38–41). Such an elaborate activation mechanism implies that the phosphorylation of histone H3 within the chromatin may be under a similarly intricate regulation as well. To explore this, we constructed a series of MSK1 deletion mutants (Fig. 7E) and examined their activities for phosphorylating free and chromatin-bound histones (Fig. 7, C and D). Deletion of the C-terminal region up to the CTKD (Δ601–802) slightly diminished the kinase activity; however, complete removal of the CTKD (Δ421–802) recovered a kinase activity, which elicited spurious phosphorylation of histone H4 (Fig. 7C). These experiments show that the NTKD is the kinase activity that directly phosphorylates free histones, that the CTKD regulates the activity and, to some extent, the substrate specificity of the NTKD, and that the NTKD is constitutively repressed by the CTKD unless the latter is activated. Further removal of the linker region (Δ356–804) virtually eliminated the kinase activity (Fig. 7C), consistent with the essential role of the linker region for the NTKD activity (20, 42). Deletion of the NTKD, on the other hand, completely abolished the kinase activity (Δ1–343), consistent with the notion that the NTKD is the kinase domain of the whole MSK1 protein (Fig. 7C). Of special note in these analyses is Δ1–41, which lacks only 41 N-terminal amino acid residues but exhibited an activity much higher than wild-type MSK1, with the ability to promiscuously phosphorylate histone H4 (Fig. 7C). These experiments revealed that, in addition to the CTKD (residues 431–600), the N-terminal 41 residues and the region between residues 601 and 802 play regulatory roles as well.
We then tested the above MSK1 mutants for their ability to phosphorylate histone H3 and HMGN1 in the context of the c-fos chromatin. As is evident from Fig. 7D, none of the deletion mutants phosphorylated histone H3 in an activator-dependent manner, indicating that almost all regions of MSK1 are required for the proper phosphorylation of histone H3 within the c-fos chromatin. Interestingly, Δ421–804 could phosphorylate HMGN1, but not histone H3, within the chromatin regardless of the presence of the activators (Fig. 7D, lanes 11 and 12). Moreover, Δ1–41 phosphorylated histone H3 extremely efficiently regardless of the presence of the activators (Fig. 7D, lanes 21 and 22), as if the chromatin structure presented no barrier whatsoever to this particular mutant, an activity that is reminiscent of Aurora B. Furthermore, Δ1–41 phosphorylated HMGN1 much better than did wild-type MSK1 (Fig. 7D, lanes 21 and 22), indicating that this mutant has completely lost the regulatory mechanism when phosphorylating substrates in the context of chromatin.
To further understand the complex regulatory mechanism, we tested the interactions between CREB and the deletion mutants of MSK1 (Fig. 7E). GST pulldown experiments with a series of MSK1 deletion mutants revealed that CREB interaction domains were located in the residues 1–41 and 601–802. In addition, gradual N-terminal deletion of MSK1 revealed that the C-terminal CREB interaction domain (residues 601–802) was under regulation by two regions between residues 42 and 344 and between residues 1 and 42; the CREB-interaction domain (residues 601–802) was strongly repressed by the regions between 42 and 344, which in turn was counteracted by the N-terminal 41 amino acids. The presence of two CREB interaction regions (residues 1–42 and 601–802) is consistent with the fact that MSK1 requires the N- and C-terminal regions that are located outside the canonical kinase domains to phosphorylate histone H3 within the chromatin in response to activators (Fig. 7D).
In sum, in addition to the well known regulation of the NTKD by the CTKD, MSK1 possesses a network of both positive and negative autoregulations with regard to the overall kinase activity and CREB interactions (Fig. 7F). These autoregulatory networks within MSK1 seem to be exploited by CREB via protein-protein interactions to regulate histone H3 phosphorylation within chromatin.
DISCUSSION
In this study, we demonstrate that CREB and its phosphorylation at Ser-133 play a pivotal role in MSK1-mediated histone H3 phosphorylation, which is augmented by HMGN1. Earlier studies in vivo strongly implicated the role of CREB and HMGN1 in regulating the MSK1-mediated phosphorylation of histone H3 at the promoters of immediate-early genes (4, 19, 20, 24, 43–45). In particular, Msk1/Msk2 double knock-out cells show that the kinases are essential for the stress-induced phosphorylations of CREB and ATF1 and that lack of both kinases results in a 50% reduction in c-fos mRNA induction (19). Moreover, in these cells histone H3 and HMGN1 phosphorylations were severely diminished or abolished (4). Likewise, Hmgn1 knock-out cells show that HMGN1 modulates histone H3 phosphorylation upon anisomycin treatment (44). Our biochemical analyses not only corroborate these in vivo studies but also provide molecular insights into the regulatory mechanisms that underlie histone H3 phosphorylation by MSK1.
MSK1 Recruitment to the c-fos Promoter
MSK1, like its closely related MSK2, possesses two nuclear localization signals within its CTKD and is localized almost exclusively within the nucleus (15). Despite its nuclear localization, however, it is implausible that MSK1 is continuously associated with chromatin. Indeed, the occupancy of MSK1 on the endogenous c-fos promoter increases dramatically upon c-fos gene induction but promptly decreases thereafter (Fig. 1C) (46); moreover, MSK1 does not interact stably with the N-terminal tails of core histones in our GST pulldown assays (Fig. 2D). In view of these observations, MSK1 may be actively recruited to, and perhaps removed from, the c-fos chromatin in vivo. Active recruitment of kinases has been documented for other kinases as well (47), including the Hog1 kinase, which is a yeast MAPK involved in osmotic stress response (48). Such kinases are now considered to be a transient but active component of the transcriptional machinery (49) and to regulate a broad range of transcriptional processes that occur on chromatin (50, 51). For instance, the Hog1 kinase, once recruited to the promoter, facilitates subsequent recruitment of RNAPII (51), the Rpd3 histone deacetylase complex (52) and the RSC chromatin remodeling complex (53).
The recruitment of MSK1 and the subsequent phosphorylation of H3-S10 on the c-fos chromatin requires CREB and, to a lesser extent, ATF1 but surprisingly not SRF and Elk-1 (Fig. 6). Relevant to this result, a study on Msk1/2 double knock-out cells reported that anisomycin-induced transcription is markedly reduced in CRE-dependent immediate-early genes such as c-fos but not in serum-response element-dependent immediate-early genes such as Egr-1 (19). Moreover, transcriptional induction of Nur77, which is mainly mediated via CREB bound to the two AP-1-like elements on the Nur77 promoter, depends strongly upon the activation of MSK1/2 (54). Thus, these results argue strongly for an intimate functional relationship between MSK1/2 and CREB/ATF1. An important question then concerns how MSK1 selectively utilizes CREB and ATF1 as a platform for phosphorylating histone H3 within the chromatin. The simplest notion is that only CREB and ATF1 interact specifically with MSK1 and recruit it to the chromatin and that the MSK1 recruitment is sufficient for subsequent histone H3 phosphorylation. This notion, however, seems unlikely because MSK1 interacts with SRF and Elk-1 even better than CREB and ATF1 (Fig. 3E), and Elk-1 does recruit MSK1 to the c-fos and Egr-1 promoters in vitro and in vivo (46). Instead, in view of the complex functional and physical interplays between CREB and MSK1 (Fig. 7), it is more plausible that only a certain type of activators interact with MSK1 in a manner that is conducive to MSK1-mediated histone H3 phosphorylation. Because anisomycin-stimulated cells show a punctate distribution of H3-S10P within the nucleus (5) and a mere fraction of histone H3 becomes phosphorylated in these cells (7, 55), activators that assist MSK1 to phosphorylate H3-S10 may include a fairly small number of activators.
Additional requirement for MSK1-mediated H3-S10 phosphorylation on the c-fos chromatin is that CREB must bind DNA and be phosphorylated at Ser-133 (Figs. 5 and 6). Although more rigorous analyses are required to determine the exact temporal order of CREB and H3-S10 phosphorylations, it would be mechanistically logical to assume that CREB becomes phosphorylated at Ser-133 before it facilitates the phosphorylation of H3-S10. Given that the phosphorylation of Ser-133 induces an alteration in the structure of CRE-bound CREB but not that of free CREB (56), this structurally altered pCREB bound to the CRE may be an essential platform whereby mitogen- and stress-activated kinase phosphorylates H3-S10 on the c-fos chromatin.
Finally, although both H3-S10 and H3-S28 are phosphorylated upon cell stimulation by MSK1/2 (4), recent analyses revealed that each phosphorylation occurs in spatially segregated locations within the nucleus (45, 57). In our kinase assays using the c-fos chromatin, MSK1 does not phosphorylate H3-S28 to a detectable level, although MSK1 phosphorylates free histone H3 at both Ser-10 and Ser-28 (Figs. 2 and 3). We suspect that the Ser-28 phosphorylation within the chromatin may require additional modification(s) of histone H3 or some activators other than CREB and ATF1. Such modifications or activators, however, remain to be identified.
Chromatin Structure and Histone H3 Phosphorylation
Chromatin imposes a strong barrier to the reactions that occur on DNA and also impedes accessibility to histones within nucleosomes. Even the N-terminal tails of histones, which protrude from the nucleosome, are not necessarily accessible. For example, a histone acetyltransferase Gcn5 can acetylate the N-terminal tails of free histones but not those of histones within chromatin (58, 59) unless it is complexed as the SAGA complex (59). Likewise, MSK1 phosphorylates the tails of free histones efficiently, yet its phosphorylation of histone tails within chromatin is far less efficient (Fig. 2).
This intrinsic nucleosomal barrier appears to be modulated to some extent by HMGN1, an architectural chromatin protein that binds to nucleosomes and decompacts chromatin (29, 60, 61). Although Lim et al. (44) reported that HMGN1 diminishes the MSK1-mediated phosphorylation of histone H3 within a nucleosome core particle, our assays show that HMGN1 enhances the MSK1-mediated histone H3 phosphorylation in an activator-dependent manner on a reconstituted nucleosome array, a more physiological context in which HMGN1 interacts with nucleosomes in vivo. Nevertheless, both in vitro studies agree in that HMGN1 has a certain degree of influence on the phosphorylation of histone H3 by MSK1. Together with the concurrent appearance of HMGN1 and H3-S10 phosphorylations upon cell stimulation (7), the regulation of histone H3 phosphorylation may involve HMGN1 and, possibly, its phosphorylation by MSK1.
HMGN1 consists of multiple domains, including the nucleosomal binding domain and chromatin unfolding domain (60, 61). Because the chromatin unfolding domain is located near, and probably interacts with, the residues 20–50 of histone H3 (62), this domain may directly shift the position of the histone H3 tail in a manner that would facilitate H3-S10 phosphorylation by MSK1. Moreover, MSK1 phosphorylates HMGN1 at Ser-20 and Ser-24, two serine residues that are critical for the interaction of HMGN1 with a nucleosome (44). Because the phosphorylations of Ser-20 and Ser-24 have a marked impact on the HMGN1-nucleosome interaction (44, 63), they may provide an additional regulatory mechanism to the H3-S10 phosphorylation by MSK1.
Intra- and Intermolecular Regulations of MSK1-mediated Histone H3 Phosphorylation
MSK1 possesses two kinase domains, termed the NTKD and CTKD, which are connected by a linker region (15, 17). Consistent with the regulatory role for the CTKD, the isolated CTKD does not phosphorylate free or chromatin-embedded histone H3, further confirming its role only as the regulator of the NTKD even in the chromatin-based assays (Fig. 7). Because the activity of the NTKD can be repressed in a dominant negative manner by the kinase-deficient CTKD, the CTKD functions as a positive as well as negative regulator of the NTKD. In addition to the regulation by the CTKD, this study revealed another major regulation by the region between residues 1 and 41, which we term the N-terminal inhibition domain (NID), located immediately upstream of the NTKD. The importance of the NID is 2-fold. First, this region represses the overall activity of MSK1, an effect most dramatically observed when chromatin-embedded histone H3 is used as a substrate. Second, the NID is essential for CREB-dependent activation of MSK1 in the context of chromatin. Indeed, despite a region of a mere 41 residues, removal of the NID is sufficient to convert MSK1 into a kinase that phosphorylates chromatin-embedded histone H3 indiscriminately, in a manner akin to Aurora B.
This regulatory role of the NID is attested by a recent structural study on the NTKD of MSK1 (64). Smith et al. (64) reported that the N-terminal region of MSK1 forms a three-stranded β-sheet, consisting of the β1L0-strand (residues 26–32), the βB-strand (residues 84–91), and the β9-strand (residues 198–202) and that this structure is unique to MSK1 among the AGC family members. Because the three-stranded β-sheet keeps the ATP-binding site sterically blocked, it serves as an auto-inhibitory structure that holds MSK1 in an inactive conformation, a mechanism that is entirely in agreement with our biochemical results (Fig. 7). Moreover, the study predicts that when the NTKD of MSK1 is activated, the βB-strand would change from the β-sheet into an α-helix before allowing the entry of ATP into the catalytic site. Because the β1L0-strand (residues 26–32) overlaps the NID (residues 1–41), it is tempting to hypothesize that the interaction between the NID and CREB would displace the β1L0-strand from the βB-strand. This would convert the βB-strand from the β-strand into an α-helix, a structural transition that is predicted to evoke activation of MSK1.
Finally, we note that various regulatory roles are played by the N-terminal extensions in protein kinase B, protein kinase C-related kinase, and protein kinase C, all of which belong to the AGC family kinases that resemble the NTKD of MSK1. For example, protein kinase B becomes anchored to plasma membrane through the interaction with phosphoinositides via the pleckstrin homology domain in its N terminus (65–68). Similarly, protein kinase C interacts with and is activated by diacylglycerol via the C1 domain located in its N terminus before the kinase is recruited to plasma membrane (69–71). Protein kinase C-related kinase becomes activated upon its interaction with Rho A via the HR1 and HR2 motifs located in its N terminus (72). Thus, despite their structural diversity, the N-terminally extended residues in MSK1, protein kinase B, protein kinase C, and protein kinase C-related kinase, and perhaps other members of the AGC family as well, may be generally involved in regulating the location and/or activity of each kinase.
Supplementary Material
Acknowledgments
We are grateful to Dario R. Alessi of the human MSK1 cDNA and James T. Kadonaga for the baculoviruses for Drosophila ATP-utilizing chromatin assembly and remodeling factor. We also thank laboratory members for discussion and Michiyo Takeuchi for technical support.
This work was supported in part by grants-in-aid for scientific research (to M. S., T. N., A. F., and K. H.) and special coordination funds for promoting science and technology (to A. F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- SRF
- serum response factor
- NID
- N-terminal inhibition domain
- H3-S10
- serine 10 in histone H3
- NTKD
- N-terminal kinase domain
- CTKD
- C-terminal kinase domain
- ChIP
- chromatin immunoprecipitation
- PEI
- polyethyleneimine
- CRE
- cAMP-response element
- CREB
- cAMP-response element-binding protein
- GST
- glutathione S-transferase
- MNase
- micrococcal nuclease.
REFERENCES
- 1.Price M. A., Hill C., Treisman R. (1996) Philos. Trans. R Soc. Lond. B Biol. Sci. 351, 551–559 [DOI] [PubMed] [Google Scholar]
- 2.Thomson S., Mahadevan L. C., Clayton A. L. (1999) Semin. Cell Dev. Biol. 10, 205–214 [DOI] [PubMed] [Google Scholar]
- 3.Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M. J., Mahadevan L. C. (1999) EMBO J. 18, 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. (2003) EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chadee D. N., Hendzel M. J., Tylipski C. P., Allis C. D., Bazett-Jones D. P., Wright J. A., Davie J. R. (1999) J. Biol. Chem. 274, 24914–24920 [DOI] [PubMed] [Google Scholar]
- 6.Cheung P., Tanner K. G., Cheung W. L., Sassone-Corsi P., Denu J. M., Allis C. D. (2000) Mol. Cell 5, 905–915 [DOI] [PubMed] [Google Scholar]
- 7.Clayton A. L., Rose S., Barratt M. J., Mahadevan L. C. (2000) EMBO J. 19, 3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo W. S., Trievel R. C., Rojas J. R., Duggan L., Hsu J. Y., Allis C. D., Marmorstein R., Berger S. L. (2000) Mol. Cell 5, 917–926 [DOI] [PubMed] [Google Scholar]
- 9.Lo W. S., Duggan L., Emre N. C., Belotserkovskya R., Lane W. S., Shiekhattar R., Berger S. L. (2001) Science 293, 1142–1146 [DOI] [PubMed] [Google Scholar]
- 10.Nowak S. J., Corces V. G. (2000) Genes Dev. 14, 3003–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson S., Hollis A., Hazzalin C. A., Mahadevan L. C. (2004) Mol. Cell 15, 585–594 [DOI] [PubMed] [Google Scholar]
- 12.DeManno D. A., Cottom J. E., Kline M. P., Peters C. A., Maizels E. T., Hunzicker-Dunn M. (1999) Mol. Endocrinol. 13, 91–105 [DOI] [PubMed] [Google Scholar]
- 13.Vicent G. P., Ballaré C., Nacht A. S., Clausell J., Subtil-Rodríguez A., Quiles I., Jordan A., Beato M. (2006) Mol. Cell 24, 367–381 [DOI] [PubMed] [Google Scholar]
- 14.Li J., Lin Q., Yoon H. G., Huang Z. Q., Strahl B. D., Allis C. D., Wong J. (2002) Mol. Cell. Biol. 22, 5688–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deak M., Clifton A. D., Lucocq L. M., Alessi D. R. (1998) EMBO J. 17, 4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.New L., Zhao M., Li Y., Bassett W. W., Feng Y., Ludwig S., Padova F. D., Gram H., Han J. (1999) J. Biol. Chem. 274, 1026–1032 [DOI] [PubMed] [Google Scholar]
- 17.Pierrat B., Correia J. S., Mary J. L., Tomás-Zuber M., Lesslauer W. (1998) J. Biol. Chem. 273, 29661–29671 [DOI] [PubMed] [Google Scholar]
- 18.Arthur J. S., Cohen P. (2000) FEBS Lett. 482, 44–48 [DOI] [PubMed] [Google Scholar]
- 19.Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. (2002) Mol. Cell. Biol. 22, 2871–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arthur J. S. (2008) Front. Biosci. 13, 5866–5879 [DOI] [PubMed] [Google Scholar]
- 21.Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F., Lin R., Smith M. M., Allis C. D. (2000) Cell 102, 279–291 [DOI] [PubMed] [Google Scholar]
- 22.Giet R., Glover D. M. (2001) J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prigent C., Dimitrov S. (2003) J. Cell Sci. 116, 3677–3685 [DOI] [PubMed] [Google Scholar]
- 24.Nowak S. J., Corces V. G. (2004) Trends Genet. 20, 214–220 [DOI] [PubMed] [Google Scholar]
- 25.Fukuda A., Nakadai T., Shimada M., Hisatake K. (2009) J. Biol. Chem. 284, 23472–23480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards D. R., Mahadevan L. C. (1992) EMBO J. 11, 2415–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Griffin K., Mondal N., Parvin J. D. (2004) J. Biol. Chem. 279, 21866–21872 [DOI] [PubMed] [Google Scholar]
- 28.Gerlitz G., Hock R., Ueda T., Bustin M. (2009) Biochem. Cell Biol. 87, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bustin M. (2001) Trends Biochem. Sci. 26, 431–437 [DOI] [PubMed] [Google Scholar]
- 30.Paranjape S. M., Krumm A., Kadonaga J. T. (1995) Genes Dev. 9, 1978–1991 [DOI] [PubMed] [Google Scholar]
- 31.Postnikov Y. V., Trieschmann L., Rickers A., Bustin M. (1995) J. Mol. Biol. 252, 423–432 [DOI] [PubMed] [Google Scholar]
- 32.An W., Palhan V. B., Karymov M. A., Leuba S. H., Roeder R. G. (2002) Mol. Cell 9, 811–821 [DOI] [PubMed] [Google Scholar]
- 33.Rivera V. M., Miranti C. K., Misra R. P., Ginty D. D., Chen R. H., Blenis J., Greenberg M. E. (1993) Mol. Cell. Biol. 13, 6260–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manak J. R., Prywes R. (1991) Mol. Cell. Biol. 11, 3652–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marais R., Wynne J., Treisman R. (1993) Cell 73, 381–393 [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez G. A., Montminy M. R. (1989) Cell 59, 675–680 [DOI] [PubMed] [Google Scholar]
- 37.Gupta P., Prywes R. (2002) J. Biol. Chem. 277, 50550–50556 [DOI] [PubMed] [Google Scholar]
- 38.McCoy C. E., Campbell D. G., Deak M., Bloomberg G. B., Arthur J. S. (2005) Biochem. J. 387, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy C. E., Macdonald A., Morrice N. A., Campbell D. G., Deak M., Toth R., McIlrath J., Arthur J. S. (2007) Biochem. J. 402, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomás-Zuber M., Mary J. L., Lesslauer W. (2000) J. Biol. Chem. 275, 23549–23558 [DOI] [PubMed] [Google Scholar]
- 41.Tomás-Zuber M., Mary J. L., Lamour F., Bur D., Lesslauer W. (2001) J. Biol. Chem. 276, 5892–5899 [DOI] [PubMed] [Google Scholar]
- 42.Vermeulen L., Berghe W. V., Beck I. M., De Bosscher K., Haegeman G. (2009) Trends Biochem. Sci. 34, 311–318 [DOI] [PubMed] [Google Scholar]
- 43.Clayton A. L., Mahadevan L. C. (2003) FEBS Lett. 546, 51–58 [DOI] [PubMed] [Google Scholar]
- 44.Lim J. H., Catez F., Birger Y., West K. L., Prymakowska-Bosak M., Postnikov Y. V., Bustin M. (2004) Mol. Cell 15, 573–584 [DOI] [PubMed] [Google Scholar]
- 45.Dunn K. L., Davie J. R. (2005) Oncogene 24, 3492–3502 [DOI] [PubMed] [Google Scholar]
- 46.Zhang H. M., Li L., Papadopoulou N., Hodgson G., Evans E., Galbraith M., Dear M., Vougier S., Saxton J., Shaw P. E. (2008) Nucleic Acids Res. 36, 2594–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokholok D. K., Zeitlinger J., Hannett N. M., Reynolds D. B., Young R. A. (2006) Science 313, 533–536 [DOI] [PubMed] [Google Scholar]
- 48.Alepuz P. M., Jovanovic A., Reiser V., Ammerer G. (2001) Mol. Cell 7, 767–777 [DOI] [PubMed] [Google Scholar]
- 49.Edmunds J. W., Mahadevan L. C. (2006) Science 313, 449–451 [DOI] [PubMed] [Google Scholar]
- 50.Proft M., Struhl K. (2002) Mol. Cell 9, 1307–1317 [DOI] [PubMed] [Google Scholar]
- 51.Alepuz P. M., de Nadal E., Zapater M., Ammerer G., Posas F. (2003) EMBO J. 22, 2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., Posas F. (2004) Nature 427, 370–374 [DOI] [PubMed] [Google Scholar]
- 53.Mas G., de Nadal E., Dechant R., Rodríguez de la Concepción M. L., Logie C., Jimeno-González S., Chávez S., Ammerer G., Posas F. (2009) EMBO J. 28, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darragh J., Soloaga A., Beardmore V. A., Wingate A. D., Wiggin G. R., Peggie M., Arthur J. S. (2005) Biochem. J. 390, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barratt M. J., Hazzalin C. A., Cano E., Mahadevan L. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4781–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma N., Lopez D. I., Nyborg J. K. (2007) J. Biol. Chem. 282, 19872–19883 [DOI] [PubMed] [Google Scholar]
- 57.Dyson M. H., Thomson S., Inagaki M., Goto H., Arthur S. J., Nightingale K., Iborra F. J., Mahadevan L. C. (2005) J. Cell Sci. 118, 2247–2259 [DOI] [PubMed] [Google Scholar]
- 58.Yang X. J., Ogryzko V. V., Nishikawa J., Howard B. H., Nakatani Y. (1996) Nature 382, 319–324 [DOI] [PubMed] [Google Scholar]
- 59.Grant P. A., Duggan L., Côté J., Roberts S. M., Brownell J. E., Candau R., Ohba R., Owen-Hughes T., Allis C. D., Winston F., Berger S. L., Workman J. L. (1997) Genes Dev. 11, 1640–1650 [DOI] [PubMed] [Google Scholar]
- 60.Trieschmann L., Alfonso P. J., Crippa M. P., Wolffe A. P., Bustin M. (1995) EMBO J. 14, 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding H. F., Bustin M., Hansen U. (1997) Mol. Cell. Biol. 17, 5843–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trieschmann L., Martin B., Bustin M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5468–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prymakowska-Bosak M., Misteli T., Herrera J. E., Shirakawa H., Birger Y., Garfield S., Bustin M. (2001) Mol. Cell. Biol. 21, 5169–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith K. J., Carter P. S., Bridges A., Horrocks P., Lewis C., Pettman G., Clarke A., Brown M., Hughes J., Wilkinson M., Bax B., Reith A. (2004) Structure 12, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 65.James S. R., Downes C. P., Gigg R., Grove S. J., Holmes A. B., Alessi D. R. (1996) Biochem. J. 315, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andjelkoviæ M., Alessi D. R., Meier R., Fernandez A., Lamb N. J., Frech M., Cron P., Cohen P., Lucocq J. M., Hemmings B. A. (1997) J. Biol. Chem. 272, 31515–31524 [DOI] [PubMed] [Google Scholar]
- 67.Frech M., Andjelkovic M., Ingley E., Reddy K. K., Falck J. R., Hemmings B. A. (1997) J. Biol. Chem. 272, 8474–8481 [DOI] [PubMed] [Google Scholar]
- 68.Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., Tsichlis P. (1998) Oncogene 17, 313–325 [DOI] [PubMed] [Google Scholar]
- 69.Ono Y., Fujii T., Igarashi K., Kuno T., Tanaka C., Kikkawa U., Nishizuka Y. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 4868–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quest A. F., Bardes E. S., Bell R. M. (1994) J. Biol. Chem. 269, 2961–2970 [PubMed] [Google Scholar]
- 71.Kazanietz M. G., Wang S., Milne G. W., Lewin N. E., Liu H. L., Blumberg P. M. (1995) J. Biol. Chem. 270, 21852–21859 [DOI] [PubMed] [Google Scholar]
- 72.Flynn P., Mellor H., Palmer R., Panayotou G., Parker P. J. (1998) J. Biol. Chem. 273, 2698–2705 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.