Abstract
Inorganic polyphosphate (poly P) is a polymer made from as few as 10 to several hundred phosphate molecules linked by phosphoanhydride bonds similar to ATP. Poly P is ubiquitous in all mammalian organisms, where it plays multiple physiological roles. The metabolism of poly P in mammalian organisms is not well understood. We have examined the mechanism of poly P production and the role of this polymer in cell energy metabolism. Poly P levels in mitochondria and intact cells were estimated using a fluorescent molecular probe, 4′,6-diamidino-2-phenylindole. Poly P levels were dependent on the metabolic state of the mitochondria. Poly P levels were increased by substrates of respiration and in turn reduced by mitochondrial inhibitor (rotenone) or an uncoupler (carbonyl cyanide p-trifluoromethoxyphenylhydrazone). Oligomycin, an inhibitor of mitochondrial ATP-synthase, blocked the production of poly P. Enzymatic depletion of poly P from cells significantly altered the rate of ATP metabolism. We propose the existence of a feedback mechanism where poly P production and cell energy metabolism regulate each other.
Keywords: Bioenergetics, Bioenergetics/ATP Synthase, Metabolism/Energy, Methods/Fluorescence, Methods/Microscopic Imaging, Organisms/Mammal, Subcellular Organelles/Mitochondria, Polyphosphate
Introduction
Inorganic polyphosphate (poly P)2 is found in all living organisms ranging from bacteria to mammals (1). Poly P performs multiple physiological functions, which are distinct and dependent on the type of organism and the subcellular localization of the polymer. In microorganisms, poly P primarily plays a role in transcription. Additionally, poly P serves as an energy store (2) and as a reserve pool of inorganic phosphates (3). However, in mammalian organisms, poly P plays predominantly a regulatory role (4) and has been implicated in the regulation of enzyme activity in cancer cells (5), stimulation of blood coagulation (6), regulation of mitochondrial ion transport (7), and regulation of respiratory chain activity (8).
Although a specific enzyme(s) responsible for poly P production in mammals is currently not known (1), poly P synthesis has been detected in intact mammalian cells. Lysis of mammalian cells leads to loss of poly P synthesis activity, suggesting that poly P synthesis in mammalian cells is likely an energy-dependent process linked to membrane transport and integrity (1, 9). Taking into account that the membrane potential generated at the mitochondrial inner membrane is a major energy source for cellular metabolism, we hypothesized that mitochondria may be the likely source of poly P production in mammalian cells.
Poly P is found in mammalian cells at significantly lower levels when compared with microorganisms (9); therefore, it is very difficult to adapt poly P measurement methods developed for bacterial studies for the study of mammalian cells. Recently we developed a protocol, which we optimized for suitability for measuring low amounts of poly P using the fluorescent probe 4′,6-diamidino-2-phenylindole (DAPI) (10). In our previous study we used this method to confirm poly P hydrolyzing activity of yeast polyphosphatase expressed in mitochondria of mammalian cultured cells (8). Here we take advantage of this tool to examine real time changes in poly P amounts in mammalian cultured cells, as well as in isolated mitochondria. The aim of this study was to establish key pathways for poly P metabolism in mammalian cells and elucidate how production of mitochondrial poly P is related to energy metabolism and consumption. Another study attempted to assess poly P changes in mammalian mitochondria using isolated rat liver mitochondria in the early 1960's, but lead to inconclusive results, presumably due to insufficient sensitivity and reliability of assay methods available at that time (11, 12). Thus, to our knowledge this is the first study that addresses the issue of poly P metabolism in mammalian mitochondria.
EXPERIMENTAL PROCEDURES
Cell Culture and Mitochondria Isolation
Isolated cortical astrocytes were prepared as previously described (13). Cerebra taken from 2–5-day-old Sprague-Dawley rats (UCL breeding colony) or 4-day-old mice were used. The cerebra were chopped and triturated until homogenous, passed through a 297-μm mesh, and trypsinized (50,000 units ml−1 porcine pancreas, Sigma) with 336 units ml−1 DNase I (bovine pancreas, Sigma), and 1.033 units ml−1 collagenase (Sigma) at 37 °C for 15 min. After the addition of fetal bovine serum (10% of final volume) and filtering through 140-μm mesh, the tissue was centrifuged through 0.4 m sucrose (400 g, 10 min), and the resulting pellet transferred to minimal essential medium, which was supplemented with 5% fetal calf serum, 2 mm glutamine, and 1 mm malate in tissue culture flasks pre-coated with 0.01% poly-d-lysine. The cells reached confluency at 12–14 days in vitro and were harvested and reseeded onto glass coverslips precoated with 0.01% poly-d-lysine for fluorescence measurements and used at 2–4 days.
Mitochondria were isolated by differential centrifugation as previously described (7). Liver from one Sprague-Dawley rat was homogenized using Teflon-glass homogenizer and resuspended in 50 ml of isolation buffer, which contained 70 mm sucrose, 230 mm mannitol, 1 mm EGTA, and 5 mm Hepes-KOH, pH 7.4. Unbroken cells were centrifuged at 600 × g for 15 min. The supernatant was collected and spun at 8500 × g for 20 min. The resulting pellet was re-suspended in 10 ml of the isolation buffer without EGTA. Functional activity of mitochondrial preparations were tested by measuring the oxygen consumption following activation of the respiratory chain by addition of succinate (5 mm) in the presence of rotenone (5 μm) in a buffer solution containing 150 mm KCl, 20 mm Hepes-KOH, pH 7.0, using a Clark oxygen electrode (Rank Brothers).
Measurements of Fluorescence
DAPI fluorescence was measured using a DeltaRAM V fluorimeter (Photon Technology International, Lawrenceville, NJ). All recordings for isolated mitochondria were done in a plastic cuvette with 2 ml of buffer solution containing 150 mm KCl, 20 mm Hepes-KOH, pH 7.0. A silicone stirrer was used to continuously mix the recording solution during fluorescence measurements. DAPI stock solution was prepared in water to the final concentration of 20 mm DAPI. Stock solutions of poly P standards (sodium salt with poly P content of 60% (provided by Dr. T. Shiba Regenetiss, Inc., Japan)) were prepared in recording buffer to final concentrations of 0.5 mg/ml or 50 μg/ml. DAPI and poly P additions were made directly to the cuvette containing the recording buffer. For all results presented here, the total amount of poly P added to the cuvette is reported. For fluorescence spectra, 1 μg of poly P corresponds to the 6 μm expressed in orthophosphates. In our control experiments, similar fluorescent changes were detected using mitochondria amounts ranging from 0.1 to 0.8 mg of protein/ml, which suggests that the observed effects are not caused by changes in fluorescence due to spectroscopic artifacts of added mitochondria. For the DAPI experiments presented in this study, mitochondria were added to the recording solution at a concentration defined by a protein content of 0.5 mg/ml. Alternatively, spectra were collected with a Fluorolog-3 with a dual excitation monochrometer to alleviate scattering artifacts (Jobin-Yvon Horriba, NJ).
For imaging of poly P, [Mg2+]c and Δψm cells were loaded for 30 min at room temperature with 0.5 μm DAPI (Molecular Probes) or MagFura-2 AM (with 0.005% Pluronic) in a Hepes-buffered salt solution containing (in mm units): 156 NaCl, 3 KCl, 2 MgSO4, 1.25 KH2PO4, 2 CaCl2, 10 glucose, and 10 HEPES, pH adjusted to 7.35, with NaOH. For measurement of Δψm, 20 nm tetramethylrhodamine methylester (Molecular Probes) was added into the cultures 40 min prior to the experiment.
Fluorescence measurements of MagFura were obtained using an Olympus epifluorescence inverted microscope with a ×20 fluorite objective. Excitation light from a xenon arc lamp was selected using a monochromator at 340 and 380 nm (Cairn Research, Faversham, UK). Emitted light passed through a long-pass filter to a cooled CCD camera (Retiga, QImaging, Canada) and digitized to a 12-bit resolution (Digital Pixel Ltd., UK). All imaging data were collected and analyzed using software from Andor (Belfast, UK). Cells were protected from phototoxicity by interposing a shutter in the light path to limit exposure between acquisition of successive images.
Confocal images were obtained using a Zeiss 510 CLSM equipped with a META detection system and a ×40 oil immersion objective. A 543-nm laser line and a 560-nm long pass filter were used to obtain tetramethylrhodamine methylester measurements. DAPI emission spectra were measured using 405 nm laser for excitation. The specific DAPI-poly P emission signal was measured above 550 nm, using the Zeiss “META” system and presented as arbitrary units. Illumination intensity was kept to a minimum (at 0.1% of laser output) to avoid phototoxicity and the pinhole was set to give an optical slice of ∼2 μm. All data presented were obtained from at least 5 coverslips and from 2 to 3 different cell preparations. Ca2+ uptake by isolated canine cardiac sarcoplasmic reticulum vesicles was monitored using Fura-2 as previously described (14). 100 μg of sarcoplasmic reticulum vesicles were continuously stirred in a cuvette with 2 ml of uptake buffer containing: 100 mm KCl, 4 mm MgCl2, 10 mm oxalate, 20 mm Hepes, pH 7.0, in the presence of 2.9 μm Fura-2. Uptake was initiated by the addition of 1.5 mm NaATP, 1.5 mm creatine phosphate, and 0.4 units/ml of creatine phosphokinase. Fura-2 fluorescence signals (alternating 340/380 nm excitation and 510 nm emission) were measured at 1-s intervals.
Luminescence Measurements
Cell luminescence was measured in a home built luminometer. Cells (400,000–600,000 per coverslip) were constantly perfused with a modified Krebs-Ringer buffer containing: 125 mm NaCl, 5 mm KCl, 1 mm Na3PO4, 1 mm MgSO4, 1 mm CaCl2, 20 μm luciferin, and 20 mm Hepes, pH 7.4, at 37 °C, supplemented with either 5.5 mm glucose or 0.1 mm pyruvate, 1 mm lactate. Under these conditions, the light output of a coverslip of transfected cells was in the range of 500 counts/s for each measurement versus a background lower than 5 counts/s. Luminescence was entirely dependent on the presence of luciferin and proportional to the perfused luciferin concentration ∼50 μm.
Extraction and Assay of Poly P
Acid-soluble and salt-soluble poly P fractions were extracted at 4 °C with 0.5 n HClO4 and a saturated solution of NaClO4 in 1 n HClO4, respectively. The remaining biomass was treated with 0.5 n HClO4 for 30 min at 90 °C, and the level of acid-insoluble poly P fraction was measured by the amount of released Pi (15). Nucleotides were removed from the acid-soluble fraction by adsorption to Norit A charcoal (16). The level of poly P in the acid-soluble and salt-soluble poly P fractions was calculated as a difference in the Pi amount before and after hydrolysis of samples in the presence of 1 n HCl for 10 min at 100 °C (labile phosphorus). The DE81 filter paper treatment was used in parallel to estimate the amount of poly P (17). Protein was determined as described in Ref. 18 using bovine serum albumin as a standard.
Poly P Electrophoresis
Extracted poly P was subjected to electrophoresis in a 20% polyacrylamide gel in the presence of 7 m urea. The gel was stained with toluidine blue (9). Poly P with chain lengths of P14, 60, and 130 (Regenetiss Inc., Japan) were used as standards.
Statistical Analysis
Statistical analysis and exponential curve fitting were performed using Origin 8 software (Microcal Software Inc., Northampton, MA). Results were expressed as mean ± S.E.
Curve Fitting
Curve fitting was performed using Sigma Plot software with the following functions: y = a × x + b and y = Y0 + a × exp(−b × x) for analysis of reaction kinetics, and y = a × exp(−0.5 ×((x-X0)/b)∧2) for spectral analysis.
RESULTS
Measurement of Poly P Changes in Live Mammalian Cells
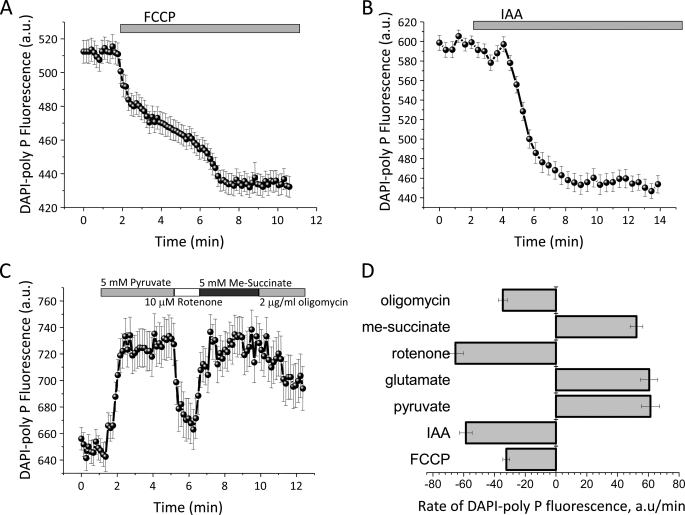
To investigate the role of poly P in cell energy metabolism, we utilized the modified the DAPI fluorescence measurement of poly P, and monitored the changes in poly P levels in response to activators and inhibitors of cellular metabolism. Application of 1 μm FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone, a mitochondrial uncoupler that dissipates mitochondrial membrane potential) to the astrocytes induced a decrease of 58.7 ± 4.1% in DAPI-poly P fluorescence in the mitochondrial area (n = 47; p < 0.001; Fig. 1A). FCCP also reduced the DAPI fluorescence signal in the cytosolic area (data not shown), suggesting an actual decrease in the amount of poly P by the protonophore, rather than a redistribution of poly P from the mitochondria to other cellular compartments. FCCP caused a complete depolarization of the mitochondrial membrane, which resulted in induction of the maximal rate of respiration and inhibited production of ATP by oxidative phosphorylation. Furthermore, FCCP stimulated ATP consumption by the mitochondrial F1F0-ATPase and the release of Ca2+ from the mitochondria into the cytosol (19). The observed decrease in the amount of poly P after FCCP addition suggests that the poly P metabolism in astrocytes is linked to the ability of the cells to produce ATP and/or the possibility that the poly P concentration is linked to the ability of the mitochondria to maintain their membrane potential.
FIGURE 1.
Inhibition of the activation of mitochondrial metabolism changes the level of DAPI-poly P in the cells. Mitochondrial uncoupler, FCCP (1 μm, A), and an inhibitor of glycolysis, IAA (20 μm, B), decrease poly P-DAPI fluorescence in mitochondrial and cytosolic areas of cortical astrocytes. C, effect of substrates for respiratory complexes I (pyruvate, 5 mm) and II (Me-succinate, 5 mm), inhibitor of complex I (rotenone, 5 μm) and the F1F0-ATP synthatase inhibitor (oligomycin, 2 μg/ml) on the level of poly P of cortical astrocytes. D, the rate of changes in the poly P level in response to mitochondrial substrates and inhibitor and inhibitors of glycolysis and oxidative phosphorylation in cortical astrocytes. a.u., arbitrary units.
The addition of 20 μm iodoacetic acid (IAA), an inhibitor of glycolysis, significantly reduced DAPI-poly P fluorescence in both mitochondrial (by 43.5 ± 3.7%; Fig. 1 B) and cytosolic (by 42.4 ± 2.7%; data not shown) areas of cortical astrocytes (n = 56; p < 0.05). Because glycolysis provides substrates for mitochondrial respiratory complexes; it is possible that the level of poly P in these cells depends on the activity of the respiratory chain. Application of 5 mm glutamate or pyruvate, substrates for complex I, increased fluorescence of DAPI-poly P by 23.1 ± 1.7%, n = 47 (21.5 ± 0.9%, for pyruvate). Subsequent inhibition of complex I with 10 μm rotenone decreased the DAPI-poly P signal by 34.6 ± 3.2% as compared with control. Activation of respiration with addition of 5 mm methyl succinate (Me-succinate), a membrane permeable analogue of succinate and a substrate for complex II, increased the poly P level by 27.4 ± 1.9% (Fig. 1C). The observed effects of FCCP, IAA, mitochondrial substrates, and rotenone support the idea that the rate of production and consumption of poly P depends on the activity of the oxidative phosphorylation machinery.
We also observed that the rate of changes in DAPI-poly P fluorescence was dependent on mitochondrial substrates, inhibitors of mitochondrial respiration, oxidative phosphorylation, and glycolysis. We should note here that fluorescent measurements do not allow us to quantitatively estimate the amount of poly P present in mitochondria. However, in vitro experiments indicate that DAPI fluorescence responds linearly to the amount of poly P present (10). Thus, the kinetics of DAPI fluorescence change reflect the kinetics of relative poly P change and can be used for calculations of rates of poly P flux in levels. The rate of changes in DAPI-poly P fluorescence under application of substrates for complex I was higher (61.4 ± 5.9 arbitrary units/min for pyruvate; 60.6 ± 5.6 arbitrary units/min for glutamate comparing to 51.2 ± 4.1 arbitrary units/min for Me-succinate; Fig. 1D), although this difference is not considered significant. Note that IAA and rotenone induced a faster decrease of the poly P level in comparison to FCCP and oligomycin (Fig. 1D).
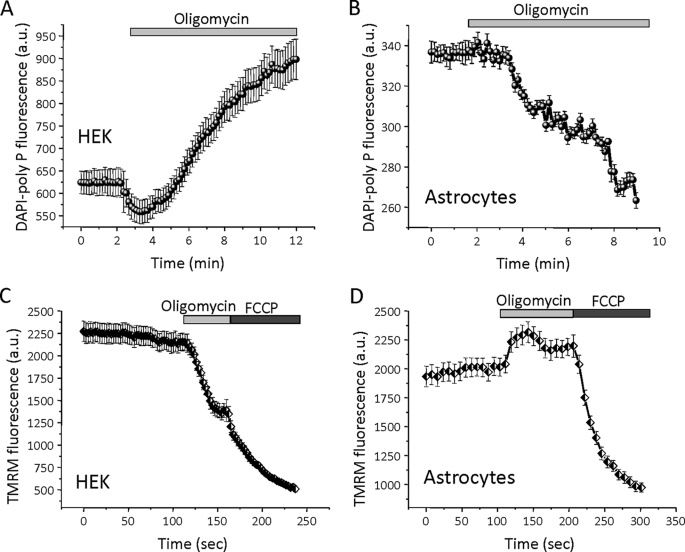
The effect of the F1F0-ATP synthetase inhibitor oligomycin on the level of poly P was dependent on cell type. Application of oligomycin (2 μg/ml) to HEK cells induced a large increase in DAPI-poly P fluorescence (n = 44; Fig. 2A). Interestingly, when the same inhibitor was added to primary astrocytes, the DAPI-poly P signal decreased by 54.6 ± 3.6% (n = 71, p < 0.001; Fig. 2B; see also Fig. 1C). We further investigated the differences seen between these two types of cells in response to the addition of oligomycin by measuring the effects of oligomycin on mitochondrial membrane potential (Δψm) using the potentially sensitive dye tetramethylrhodamine methylester. Addition of oligomycin to cortical astrocytes resulted in “classical” hyperpolarization of the mitochondrial membrane (n = 39, Fig. 2D). On the other hand, oligomycin addition to HEK cells induces depolarization of the mitochondrial membrane (n = 41; Fig. 2C). In cells with normal oxidative phosphorylation, the Δψm is maintained by the proton pumping activity of the respiratory chain. However, if respiration is impaired, hydrolysis of ATP by the F1F0-ATPase (complex V) may take over, pumping protons across the inner membrane and thus maintaining Δψm (20). Our data suggest that in HEK cells, oligomycin inhibits ATP consumption, which leads to an increase in the poly P level. Conversely, in cortical astrocytes, oligomycin inhibits ATP production, which results in a decrease in the level of poly P. These observations further support the notion that the level of poly P in mammalian cells is directly dependent on the level of ATP.
FIGURE 2.
Oligomycin causes the opposite effect on DAPI-poly P fluorescence in HEK cells and astrocytes. Inhibitor of the F1F0-ATP synthase, oligomycin (2 μg/ml), increases the level of poly P in HEK cells (A) and decrease DAPI-poly P fluorescence in cortical astrocytes (B). Note that oligomycin-induced mitochondrial depolarization in HEK cells (mitochondrial membrane potential was measured as tetramethylrhodamine methylester fluorescence, C), but hyperpolarized mitochondria in cortical astrocytes (D). Uncoupler FCCP (1 μm) indicates complete mitochondrial depolarization.
Calculation of Rate Constants for Poly P Hydrolysis
To obtain further insight into the kinetics of poly P metabolism, we attempted to measure rate constants of chemical reactions responsible for these changes. Taking into account that current work deals with the whole organelle in an intact cell rather than purified enzyme(s), it is impossible to tell at this level how many enzymes are involved in the process, and which substrates are potentially used for the synthesis. For this reason, results of rate constant measurements should be taken with care, as we cannot tell how many reactions are potentially involved in the process and the order of these reactions. Despite these difficulties, we were able to find significant differences in the kinetics of poly P hydrolysis depending on the applied conditions of study. In our analysis on the kinetics of poly P hydrolysis we tested two possibilities: 1) zero order reaction, which will take place if the poly P polymer binds to the enzyme and is not released until the whole molecule is hydrolyzed, in this case enzymatic cleavage of poly P will be the rate-limiting factor and decease of fluorescence should follow zero order linear kinetics; 2) a first order reaction that would take place if poly P is used as a substrate for some other reaction and becomes bound and released after cleavage of a single orthophosphate. We have found that kinetics of poly P hydrolysis induced by addition of FCCP (Fig. 1A) or IAA (Fig. 1B) follows a first order reaction and can be well fitted with a single exponential, but not with a linear function. With this model reaction rate constants were 0.25 ± 0.04 min−1 for FCCP and 0.66 ± 0.04 min−1 for IAA. To the contrary, application of oligomycin stimulated poly P hydrolysis with kinetics, which were best fit with linear rather than exponential function. This suggested zero order kinetics of this reaction with rate constants of 12 ± 1 min−1 for experiments shown at Fig. 1C and 9.9 ± 0.4 min−1 for the experiment shown at Fig. 2B.
Overall, these results allow us to suggest that the mechanism of poly P hydrolysis in the presence of the F1F0-ATPase inhibitor is fundamentally different. These results support the idea of direct involvement of F1F0-ATPase in the process of poly P metabolism. Additionally, they contribute significantly to the notion that poly P hydrolysis under conditions of the respiratory chain are uncoupled or inhibited.
Measurement of Poly P in Isolated Mitochondria
To ensure that our observations were not simply due to changes in DAPI fluorescence in response to changes in ATP levels, we determined how the presence of ATP as well as ADP might affect the fluorescence of DAPI-poly P. The DAPI fluorescence spectrum was measured in vitro using a fluorimeter in the presence of ATP and poly P. For these experiments, the excitation wavelength was set at 405 nm, which corresponds to the excitation wavelength used in the confocal fluorescence experiments.
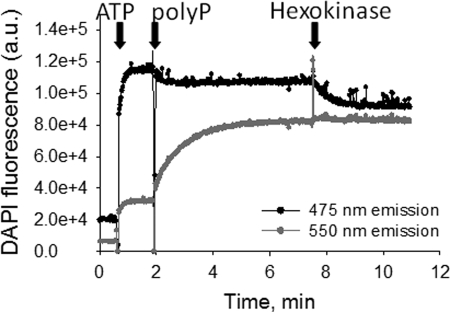
The intensity of DAPI fluorescence can be modulated by a variety of compounds, but only DAPI bound to poly P demonstrates a specific shift in both the excitation and emission spectra (21). This property of DAPI is determined by the unique, high density distribution of negative charges found only in poly P. In our experimental conditions, changes in the DAPI fluorescence induced by non-poly P compounds are most prominent at 475 nm, whereas fluorescence changes induced by poly P binding to DAPI are further red shifted and thus best detectable at 550 nm. As shown in Fig. 3, addition of ATP (1.25 mm) increased the intensity at both 475 and 550 nm fluorescence emissions. However, subsequent addition of 1 μg (6 μm) of poly P induces a large increase in the 550-nm fluorescence intensity, and a small decrease in the 475-nm fluorescence, consistent with a poly P-induced shift of DAPI fluorescence. Addition of hexokinase, which converts ATP to ADP, decreased the fluorescence intensity at 475 nm, whereas the signal at 550 nm remained virtually unchanged, indicating that the fluorescence intensity at 550 nm is independent of the ratio between ATP and ADP. These experiments demonstrate that the fluorescence intensity changes at 475 nm provide reliable estimates of the fluorescence change induced by nonspecific DAPI interactions, whereas fluorescence changes at 550 nm reflect a specific change in DAPI fluorescence due to poly P interaction with DAPI.
FIGURE 3.
DAPI fluorescence in the presence of purified ATP and poly P. Fluorescence was excited at 405 nm and emission was monitored simultaneously at 475 and 550 nm. Arrows indicate points of addition of 6 μm poly P and 1.25 mm ATP. The change in DAPI fluorescence in response to ATP addition is larger when emission is measured 475 nm as compared with 550 nm. Subsequent addition of poly P substantially increased the 550-nm emission fluorescence while reducing the 475-nm emission fluorescence, consistent with a shift in DAPI-poly P emission spectrum. Addition of hexokinase to convert all the ATP in the cuvette to ADP reduced the fluorescence emission at 475 nm without any measurable effect on the 550 nm emission.
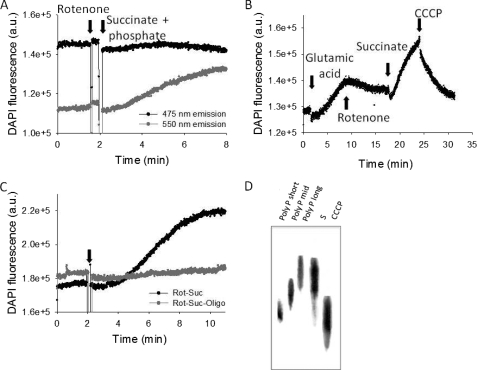
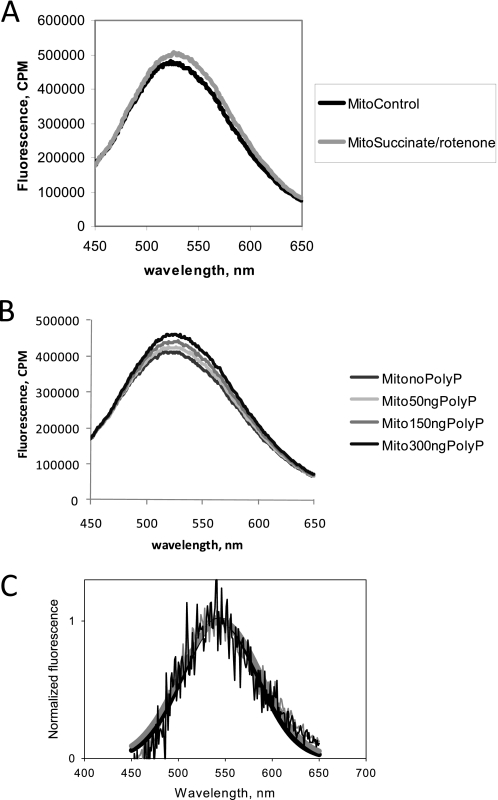
We further confirmed the results we obtained in intact cells using isolated mitochondria and monitoring the DAPI fluorescence simultaneously at both 475 and 550 nm with excitation at 415 nm. Stimulation of mitochondrial respiration with substrates for respiratory complex II (succinate, 5 mm) + phosphate (1 mm) in the presence of rotenone (5 μm) significantly increased poly P-dependent DAPI fluorescence (550 nm) with no changes in the 475-nm emission fluorescence (Fig. 4A; n = 5). These results confirmed the dependence of poly P synthesis on activity of the mitochondrial respiration.
FIGURE 4.
Changes in DAPI fluorescence in isolated rat liver mitochondria in response to activation and inhibition of the respiratory chain. A, the emission fluorescence of DAPI was monitored at 475 and 550 nm after addition of succinate (5 mm) and phosphate (1 mm) in the presence of rotenone. The increase in emission fluorescence at 550 nm with no change in 475 nm confirmed that the measured changes in DAPI fluorescence were due to direct changes in poly P levels. B, changes in DAPI fluorescence measured at 550 nm emission in response to substrates (glutamate, 5 mm, and succinate, 5 mm); inhibitor (rotenone, 5 μm), and uncoupler (CCCP, 1 μm) of the respiratory chain. C, the presence of oligomycin (2 μg/ml) to block mitochondrial ATP synthase prevented the increase in DAPI fluorescence (gray trace) observed with activation of mitochondrial respiration after addition of rotenone (5 μm) and succinate (5 mm, black trace). D, change in poly P length in mitochondria treated with succinate (5 mm) in the absence (S) and presence of CCCP. Average length of poly P standards is as follows: short, 14; middle, 60; long, 130. a.u., arbitrary units.
The relative contribution of mitochondrial complexes and respiration in general to poly P homeostasis can be estimated by the effects of added substrates or the presence of respiratory chain inhibitors on DAPI fluorescence. Application of 5 mm glutamate, a substrate for complex I, to mitochondria increased the level of DAPI-poly P by 12 ± 4% (n = 5, p < 0.01; Fig. 4B). Addition of rotenone (5 μm) decreased DAPI fluorescence but, interestingly, not to basal levels. Subsequent addition of substrate for complex II (succinate, 5 mm) not only restored, but also increased the level of poly P in the mitochondria (26 ± 10%, n = 7; p < 0.001). Addition of a mitochondrial uncoupler, CCCP (1 μm), reduced DAPI-poly P fluorescence by 13 ± 4% (n = 4; p < 0.01) but, again, not to basal levels (Fig. 4B). The experiments described in this section were done in the presence of phosphate and absence of ADP. It is established that under these conditions the mitochondrial respiratory chain is active, but its activity is not coupled to ATP production (22, 23). In our control experiments conducted in the absence of ADP, addition of succinate induced a 2.5 ± 1-fold (n = 3) increase of the rate of respiration. This rate was 34 ± 8% (n = 3) of the maximal rate of mitochondrial respiration achieved in the presence of CCCP. Thus, the level of poly P in mitochondria is dependent on the activity of mitochondrial respiration and not the production of ATP. These results suggest that mitochondria do not use ATP as a substrate for poly P synthesis.
Fig. 4C shows the application of rotenone and succinate to mitochondria in the presence of oligomycin. Under these conditions the effect of substrates of respiration on DAPI fluorescence in mitochondria was completely abolished. This suggests that in mammalian mitochondria poly P production is closely related to the activity of the oligomycin-dependent F1F0-ATP synthase.
To confirm that DAPI fluorescence changes reflect changes in poly P amount, rather than its complex formation with other molecules, we compared the total amount of poly P in succinate-stimulated mitochondria in the presence and absence of CCCP using a biochemical assay. It is important to note that it was not possible to do similar experiments using tissue culture cells due to the requirement of large amounts of material and sample differences arising from mitochondrial isolation from cell culture. For these experiments poly P was extracted from large preparations (50 mg/protein) of pretreated mitochondria and their amounts were measured using a biochemical assay as described under “Experimental Procedures.” We have found that in the presence of CCCP the level of poly P in mitochondria was reduced to 35 ± 5 from 75 ± 6 pmol/mg in the absence of CCCP. Interestingly, addition of CCCP not only decreased the total concentration of poly P, but also induced a significant decrease in the poly P chain length (Fig. 4D).
Finally, we further confirmed that fluorescent changes seen in our experiments reflect changes in DAPI/poly P fluorescence by analyzing the full spectrum of mitochondrial fluorescence before and after stimulation with succinate in the presence of rotenone. As can be seen from Fig. 5A, stimulation of mitochondria results in an increase of fluorescence in the red edge, but not on the blue side of the spectrum. The shape of the spectra was analyzed by performing Gaussian fits. We have found that upon stimulation, both fluorescence intensity, and maximum wavelength, changed. Fluorescence intensity increased from 110,200 ± 200 to 119,000 ± 200 counts/min, whereas the peak of the spectrum shifted toward the longer wavelengths ranging from 511.8 ± 0.1 to 515.6 ± 0.1 nm. A similar effect was observed when low (50 to 300 ng) amounts of synthetic poly P were added to unstimulated mitochondria (Fig. 5B). The difference in fluorescence spectra obtained by subtraction of stimulated mitochondria spectrum from control mitochondria spectrum, and then compared with the spectrum obtained by subtraction of mitochondrial spectrum in the presence and absence of added synthetic poly P was virtually indistinguishable. Fig. 5C shows the overlay of these two normalized spectra along with Gaussian fits, in turn illustrating the similarity in both cases. In the case of stimulated mitochondria, the emission maximum was at 544.9 ± 0.6 nm compared with 544.3 ± 0.9 nm in the case of added synthetic poly P. We should note that the results indicated some difference in spectral widths of 43.4 ± 0.7 versus 39.7 ± 0.9, which might be explained by the difference in microenvironment of poly P (intra- versus extramitochondrial). Thus, supporting the idea that the full spectrum represents fluorescence from both nonspecific DAPI binding, and DAPI/poly P fluorescence, changes caused by stimulation of mitochondria can still be observed in the red edge that corresponds to changes in poly P concentration.
FIGURE 5.
Fluorescence spectrum of DAPI in the presence of mitochondria. A, emission spectrum of DAPI was first collected after addition of rat liver mitochondria and again from the same sample following stimulation by 5 mm succinate in the presence of 5 μm rotenone. B, emission spectrum of DAPI was first collected after addition of mitochondria and again following the addition of various amounts of synthetic poly P. C, normalized fluorescence along with results of Gaussian fits obtained by subtraction of spectrum shown on panel A of stimulated mitochondria minus control mitochondria (gray lines) and B, mitochondria in the presence of 150 ng of poly P minus control mitochondria (black lines). Samples were excited at 415 nm wavelength.
Production and Consumption of Poly P and ATP
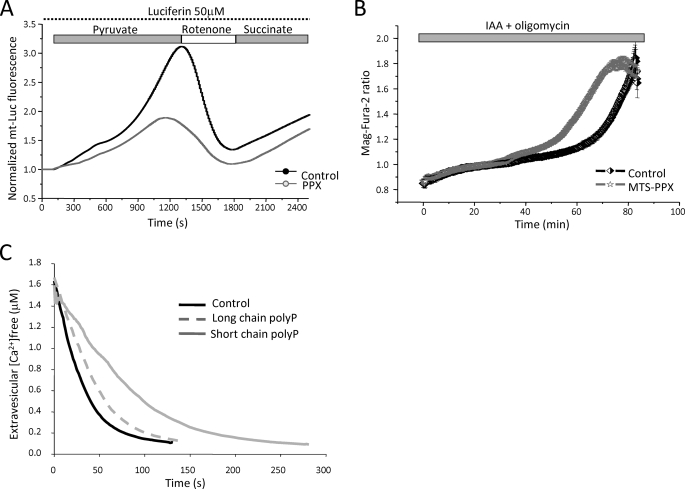
To date the enzymes responsible for poly P synthesis and consumption have not been identified in mammalian mitochondria. Previously we demonstrated that the level of poly P in mitochondria of intact cells can be reduced to a minimum level by overexpression of the mitochondrion-targeted yeast polyphosphatase (MTS-GFP-PPX) (8). To further investigate the role of poly P in the regulation of ATP production, we co-transfected MTS-GFP-PPX and mitochondrion-targeted luciferase in C2C12 cells. We found that the rate of ATP production in cells with mitochondrial PPX in response to substrates for complexes I and II (Fig. 6A) is slower when compared with the control. This is consistent with alteration of the redox index and Δψm in MTS-GFP-PPX cells (8), confirming the role of poly P in oxidative phosphorylation.
FIGURE 6.
Role of poly P in ATP metabolism. A shows representative normalized traces of C2C12 cells expressing mitochondrially targeted luciferase (mt-Luc) and mitochondrially targeted GFP (mt-GFP) as control (black trace) or MTS-PPX (gray trace) perfused with 50 μm luciferin and challenged in sequence with 5 mm pyruvate, 10 μm rotenone, and 5 mm Me-succinate (n = 3 different transfections). B, measurements of [Mg2+]i, which increase as ATP is hydrolyzed (see text) in control and MTS-PPX C2C12 cells in response to inhibitors of ATP production, IAA (20 μm) and oligomycin (2 μg/ml). C, effect of poly P on the sarcoplasmic-endoplasmic reticulum Ca2+-ATPase activity estimated as the rate of Ca2+ uptake into isolated canine cardiac sarcoplasmic reticulum vesicles.
Because Mg2+ is released from MgATP upon hydrolysis (24), we measured cellular free [Mg2+]c as a measure of ATP consumption using a Mg2+-sensitive fluorescent probe, MagFura-2. Application of inhibitors of glycolysis (IAA, 20 μm) and F1F0-ATP synthase (oligomycin, 2 mg/ml) completely block ATP production in cells, which eventually leads to an increase in MagFura-2 fluorescence due to ATP depletion and consequent Mg2+ release. In addition to binding Mg2+, MagFura-2 is also used as a low affinity Ca2+ indicator. Thus, the energy capacity of the cell can be estimated using MagFura-2 by measuring the time between the application of inhibitors of glycolysis, ATP-synthase, and energetic collapse when cells depleted all the ATP needed for maintenance of Ca2+ homeostasis (25). As shown in Fig. 6B, there was no difference in the rate of ATP consumption between the control and MTS-PPX C2C12 cells (compare 0.021 ± 0.001 MagFura-2 ratio/min in control C2C12 cells, n = 99 to 0.022 ± 0.001 in MTS-GFP-PPX, n = 67). However, addition of 2 μg/ml of oligomycin + 5 mm IAA induced cell collapse at a faster rate in C2C12 cells with no poly P in mitochondria compared with control (48.4 ± 3.2 min compared with 67.2 ± 5.1 min in control, p < 0.05). These results suggest that poly P may be used by the cells as a source of energy, prolonging the time required for complete energetic collapse.
In light of these findings, we explored the ability of sarcoplasmic-endoplasmic reticulum Ca2+-ATPase to use poly P as an alternate source of energy to ATP by measuring the rate of Ca2+ uptake into isolated sarcoplasmic reticulum vesicles. Sarcoplasmic-endoplasmic reticulum Ca2+-ATPase did not demonstrate any measurable activity in the absence of ATP and the presence of up to 1 mm of either short (poly P-14) or long (poly P-130) chain poly P. Moreover, short chain poly P and to a lesser degree long chain poly P, decreased the maximal rate of Ca2+ uptake from 0.16 μmol/min mg in control to 0.06 μmol/min mg in the presence of short chain poly P and 0.12 μmol/min mg in the presence of long chain poly P (Fig. 6C). These results demonstrate that poly P cannot be used directly by sarcoplasmic-endoplasmic reticulum Ca2+-ATPase as an energy source, but do not rule out the possibility that other enzymes in the cell could use poly P to meet the energy demand of the cell.
DISCUSSION
The results presented in this study demonstrate that the level of poly P in eukaryotic cells is very dynamic and rapidly changes in response to various inhibitors and activators of mitochondrial respiration. In agreement with previous reports (9) we propose that poly P is constantly produced and consumed by mammalian cells. A high rate of turnover of poly P in eukaryotic cells is consistent with a regulatory role of poly P in cell physiology. We have found that production of poly P is directly linked to mitochondrial respiration and oxidative phosphorylation. Inhibition of poly P synthesis by oligomycin and reduction of the poly P levels in response to mitochondrial inhibitors suggests that the production of this polymer might occur through a mechanism that involves the F1F0-ATP synthase using a proton gradient by mechanisms similar to ATP synthesis. Considering the opposing effects of oligomycin in cells with F1F0-ATPase working in reverse mode (Fig. 2), we propose that there are at least two enzymes that produce poly P in mammalian cells. Inhibition of glycolysis by IAA also decreased the level of poly P in the cells, although this observation may not be due to a direct effect of glycolysis on production of poly P. This could be related to the interaction of glycolysis and mitochondrial respiration.
Recently it was demonstrated that activity of the F1F0-ATPase can be regulated by cyclophilin D, a key protein involved in mitochondrial permeability transition pore regulation (26). Our previous reports indicate that poly P plays an essential role in activation and/or formation of the mitochondrial permeability transition pore (7, 8). We suggest that a link exists between F1F0-ATPase regulation of poly P metabolism and mitochondrial permeability transition pore activation.
The enzyme responsible for poly P synthesis in mammalian cells remains elusive. A DNA data base search shows no obvious homology between any of the bacterial poly P kinases and the mammalian genome, although it has been suggested that a homologous kinase might still exist in mammals (27), and mammals have DdPPK2 homologs. On the other hand, it is possible that mammals lack a dedicated poly P kinase, rather, poly P production may occur in these cells as a byproduct of multiple enzymes complexes. Supporting this idea, the plasma membrane Ca2+-ATPase in human erythrocytes, in addition to transporting Ca2+, can also be involved as a polyphosphate kinase, i.e. it exhibits ATP-polyphosphate transferase and polyphosphate-ADP transferase activities (28). Interestingly, bacteria with double knock-out poly P kinases still retain the ability to produce significant levels of poly P (1), further emphasizing the notion that poly P may be produced by a variety of enzymes. In conclusion, we have shown that: 1) mammalian mitochondria poly P production is directly linked to their energetic state; 2) poly P synthesis is closely linked to the activity of ATP synthase but does not require ATP as a substrate; and 3) the level of poly P regulates the level of cellular ATP.
Acknowledgments
We thank Dr. M. Kargacin and Dr. G. Kargacin for the use of the fluorimeter. We thank Dr. T. Shiba for providing poly P standards. We also thank Samantha Herauf for help proofing this manuscript.
This work was supported by a Clinical Research and Development Committee Research Funding grant and the Heart and Stroke Foundation of Alberta.
- poly P
- polyphosphate
- DAPI
- 4′,6-diamidino-2-phenylindole
- IAA
- iodoacetic acid
- Δψm
- mitochondrial membrane potential
- MTS-PPX
- mitochondrion-targeted yeast polyphosphatase
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- GFP
- green fluorescent protein
- Me
- methyl
- CCCP
- carbonyl cyanide p-chlorophenylhydrazone.
REFERENCES
- 1.Kornberg A., Rao N. N., Ault-Riché D. (1999) Annu. Rev. Biochem. 68, 89–125 [DOI] [PubMed] [Google Scholar]
- 2.Beauvoit B., Rigoulet M., Raffard G., Canioni P., Guérin B. (1991) Biochemistry 30, 11212–11220 [DOI] [PubMed] [Google Scholar]
- 3.Akiyama M., Crooke E., Kornberg A. (1993) J. Biol. Chem. 268, 633–639 [PubMed] [Google Scholar]
- 4.Kulaev I., Vagabov V., Kulakovskaya T. (1999) J. Biosci. Bioeng. 88, 111–129 [DOI] [PubMed] [Google Scholar]
- 5.Wang L., Fraley C. D., Faridi J., Kornberg A., Roth R. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11249–11254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith S. A., Morrissey J. H. (2008) Blood 112, 2810–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlov E., Zakharian E., Bladen C., Diao C. T., Grimbly C., Reusch R. N., French R. J. (2005) Biophys. J. 88, 2614–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abramov A. Y., Fraley C., Diao C. T., Winkfein R., Colicos M. A., Duchen M. R., French R. J., Pavlov E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18091–18096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumble K. D., Kornberg A. (1995) J. Biol. Chem. 270, 5818–5822 [DOI] [PubMed] [Google Scholar]
- 10.Aschar-Sobbi R., Abramov A. Y., Diao C., Kargacin M. E., Kargacin G. J., French R. J., Pavlov E. (2008) J. Fluoresc. 18, 859–866 [DOI] [PubMed] [Google Scholar]
- 11.Lynn W. S., Brown R. H. (1963) Biochem. Biophys. Res. Commun. 11, 367–371 [DOI] [PubMed] [Google Scholar]
- 12.Harold F. M. (1966) Bacteriol. Rev. 30, 772–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramov A. Y., Jacobson J., Wientjes F., Hothersall J., Canevari L., Duchen M. R. (2005) J. Neurosci. 25, 9176–9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beca S., Aschar-Sobbi R., Ponjevic D., Winkfein R. J., Kargacin M. E., Kargacin G. J. (2009) Arch. Biochem. Biophys. 490, 110–117 [DOI] [PubMed] [Google Scholar]
- 15.Vagabov V. M., Trilisenko L. V., Kulaev I. S. (2000) Biochemistry 65, 349–354 [PubMed] [Google Scholar]
- 16.Ault-Riché D., Fraley C. D., Tzeng C. M., Kornberg A. (1998) J. Bacteriol. 180, 1841–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao N. N., Liu S., Kornberg A. (1998) J. Bacteriol. 180, 2186–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 19.Nicholls D. G. (1974) Eur. J. Biochem. 50, 305–315 [DOI] [PubMed] [Google Scholar]
- 20.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M. R., Abramov A. Y., Tinker A., Duchen M. R. (2008) Cell Metab. 8, 13–25 [DOI] [PubMed] [Google Scholar]
- 21.Tijssen J. P., Beekes H. W., Van Steveninck J. (1982) Biochim. Biophys. Acta 721, 394–398 [DOI] [PubMed] [Google Scholar]
- 22.Chance B., Williams G. R. (1955) J. Biol. Chem. 217, 383–393 [PubMed] [Google Scholar]
- 23.Chance B., Williams G. R. (1955) J. Biol. Chem. 217, 429–438 [PubMed] [Google Scholar]
- 24.Silverman H. S., Di Lisa F., Hui R. C., Miyata H., Sollott S. J., Hanford R. G., Lakatta E. G., Stern M. D. (1994) Am. J. Physiol. Cell Physiol. 266, C222–C233 [DOI] [PubMed] [Google Scholar]
- 25.Abramov A. Y., Scorziello A., Duchen M. R. (2007) J. Neurosci. 27, 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giorgio V., Bisetto E., Soriano M. E., Dabbeni-Sala F., Basso E., Petronilli V., Forte M. A., Bernardi P., Lippe G. (2009) J. Biol. Chem. 284, 33982–33988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooley P., Whitehead M. P., Brown M. R. (2008) Trends Biochem. Sci. 33, 577–582 [DOI] [PubMed] [Google Scholar]
- 28.Reusch R. N., Huang R., Kosk-Kosicka D. (1997) FEBS Lett. 412, 592–596 [DOI] [PubMed] [Google Scholar]