Abstract
Lipopolysaccharides (LPS) induce tumor necrosis factor-α (TNF-α) production in cardiomyocytes, which contributes to myocardial depression during sepsis. However, the underlying mechanisms remain not fully understood. This study was undertaken to investigate the contribution of histone deacetylase (HDAC) to TNF-α expression in cardiomyocytes and the signaling mechanism of LPS-induced HDAC activation. Here, we show for the first time that LPS increases HDAC activity and that inhibition of HDAC decreases LPS-stimulated TNF-α expression via the accumulation of NF-κB/p65 at the TNF-α promoter in cardiomyocytes. Using a positive screen, we have further identified HDAC3 as a specific member of the HDAC family able to regulate TNF-α production. Furthermore, our data reveal that LPS-induced HDAC activity is mediated through reactive oxygen species from mitochondria and c-Src signaling. In summary, this study demonstrates a novel signaling mechanism by which LPS via mitochondrial reactive oxygen species/c-Src/HDAC3 pathways mediate TNF-α expression in cardiomyocytes.
Keywords: Cytokines, Histones/Deacetylase, Immunology/Lipopolysaccharide, Signal Transduction, Gene Expression, Cardiomyocytes, Mitochondria, Reactive Oxygen Species
Introduction
Tumor necrosis factor-α (TNF-α),2 a pro-inflammatory cytokine, has been demonstrated to impair cardiac contractile function in intact animals, isolated hearts, and cardiomyocytes (1–3). Lipopolysaccharides (LPS) of Gram-negative bacteria induce TNF-α expression in cardiomyocytes, which may be a major local source of TNF-α in the myocardium during sepsis (4). Blocking TNF-α reduces myocardial depression induced by endotoxemia (1, 4). Thus, modulation of local myocardial TNF-α levels produced by cardiomyocytes may be of therapeutic significance in sepsis-induced myocardial dysfunction. However, regulation of TNF-α expression remains not fully understood in cardiomyocytes.
LPS induces intracellular signaling via a well delineated cascade of events (5). LPS binds to LPS-binding protein, which presents LPS to CD14. Binding of LPS to CD14 activates TLR-4 (Toll-like receptor-4). Activation of TLR-4 results in the recruitment of myeloid differentiation factor 88, Toll/interleukin-1 receptor domain-containing adaptor protein, and interleukin-1 receptor-associated protein kinases, which activates the NF-κB signaling pathway, leading to production of pro-inflammatory factors including TNF-α. However, the signaling pathways downstream of the TLR-4 complex have not been fully defined.
Histone deacetylase (HDAC) is a class of enzymes that remove acetylation from lysine residues within histones, which is important in the regulation of gene expression (6). The HDAC family has 18 members in four classes: class I (HDAC1, -2, -3, and -8), class II (HDAC4, -5, -6, -7, -9, and -10), class III (sirtuin-1,- 2, -3, -4, -5, -6, and -7), and class IV (HDAC11). HDACs have been shown to induce or repress the production of pro-inflammatory cytokines in a range of disease models in mice, including septic shock (7–9). However, the isoform-specific contribution of HDAC remains to be determined. In addition, the role of HDAC in LPS-induced TNF-α expression has never been shown in cardiomyocytes.
The purpose of this study was to investigate the role of HDAC in cardiomyocyte TNF-α expression upon LPS stimulation and to determine the signaling mechanism by which LPS induce HDAC activation in cardiomyocytes. We demonstrate that mitochondrial reactive oxygen species (ROS)/c-Src pathways mediate HDAC3 activation, which promotes TNF-α production in LPS-stimulated cardiomyocytes.
EXPERIMENTAL PROCEDURES
Animals
All experimental protocols were approved by the Animal Use Subcommittee at the University of Western Ontario. Breeding pairs of C57BL/6 mice were purchased from The Jackson Laboratory. A breeding program for mice was implemented at our animal care facilities.
Neonatal Mouse Cardiomyocyte Culture
The neonatal cardiomyocytes were prepared and cultured according to methods we described previously (10).
Reagents
LPS (Salmonella typhosa), rotenone, antimycin A, apicidin, trichostatin A (TSA), and PP2 were purchased from Sigma or Calbiochem. Peptides SS31 and SS20 were synthesized using a published protocol (11) as described previously (12). The class II-specific HDAC inhibitors MC1568 and MC1575 were synthesized as described elsewhere (13, 14).
Adenoviral Infection of Cultured Cardiomyocytes
Cardiomyocytes were infected with adenoviral vectors containing SOD2 (Ad-SOD2) or UCP2 (uncoupling protein 2; Ad-UCP2) (Gene Transfer Vector Core, University of Iowa) or β-galactosidase (Ad-β-galactosidase; Vector Biolabs) as a control at a multiplicity of infection of 100 plaque-forming units/cell. Adenovirus-mediated gene transfer was implemented as described previously (10, 15).
Gene Knockdown Using Small Interfering RNA (siRNA)
To knock down HDAC1, -2, and -3 and c-Src expression, a siRNA against mouse HDAC1, -2, or -3 or c-Src was obtained (Santa Cruz Biotechnology, Santa Cruz, CA), and a scrambled siRNA was employed as a control. Transfection was performed using TransMessenger transfection reagent (Qiagen) according to the manufacturer's instructions as described previously (16).
Analysis of TNF-α mRNA by Real-time Reverse Transcription-PCR
Total RNA was extracted from cardiomyocytes using the TRIzol reagent (Invitrogen) following the manufacturer's instructions. Real-time reverse transcription-PCR for mouse TNF-α and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was performed. The primers for TNF-α and GAPDH were described previously (10).
Measurement of TNF-α Protein
Concentrations of TNF-α protein in culture medium were determined with a mouse TNF-α enzyme-linked immunosorbent assay kit (eBiosciences Inc.) as described in our previous report (10).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay was performed to assess NF-κB/p65 binding to the TNF-α promoter as described previously (17). Briefly, cardiomyocytes were fixed in 1% formaldehyde (to cross-link DNA-proteins) for 10 min at room temperature. Cross-linked chromatin was isolated and sheared by sonication. This process generates chromatin fragments containing DNA ranging in size from 200 to 500 bp. The sheared chromatin was recovered by centrifugation at 12,000 rpm at 4 °C. The chromatin solution was precleared with salmon sperm DNA/protein G-agarose for 2 h at 4 °C. Ten microliters of the chromatin solution were reserved as “input” sample. The remaining chromatin was immunoprecipitated by incubation at 4 °C overnight with antibodies specific to NF-κB/p65 (Santa Cruz Biotechnology). The immunoprecipitated complexes were further incubated for 1 h at 4 °C with protein G-agarose, washed, and then eluted in a buffer containing 1% SDS and 0.1 m NaHCO3. The immunoprecipitated and input sample cross-links were reversed by heating at 65 °C overnight. After proteinase K digestion, DNA was isolated via phenol/chloroform extraction and purified via ethanol precipitation. Real-time PCR was performed to quantify p65 binding to the TNF-α promoter using primers complementary to two sites flanking a ∼300-bp fragment of the murine TNF-α promoter between nucleotides −434 and −722 relative to the transcription start site and encoding κB sites 2 and 2a: 5′-AGTCATACGGATTGGGAGAAATCCTG-3′ (forward) and 5′-AGTTCTTGGAGGAAGTGGCTGAAGGCA-3′ (reverse).
HDAC Activity Assay
Intracellular HDAC activity in cardiomyocytes was determined with the HDAC fluorometric cellular activity assay kit (BIOMOL International) following the manufacturer's instructions. Changes in fluorescence are expressed in arbitrary units.
Intracellular ROS Measurement
The production of ROS was measured by using the ROS-sensitive dye 2,7-dichlorodihydrofluorescein diacetate (Invitrogen) as an indicator. The assay was performed on cultured cardiomyocytes as we described in our recent report (15). The fluorescent product formed was quantified using a spectrofluorometer at 485/525 nm. Changes in fluorescence are expressed in arbitrary units.
Western Blot Analysis
HDAC3, c-Src, phosphorylation of c-Src (Tyr-418), and acetylation of p65 (Lys-310), histone 2A, IκB, and GAPDH were determined by Western blot analysis using the respective specific antibodies.
Statistical Analysis
All data are given as mean ± S.D. Analysis of variance followed by the Newman-Keuls test was performed for multigroup comparisons. A value of p < 0.05 was considered statistically significant.
RESULTS
HDAC3 Activation Contributes to LPS-stimulated TNF-α Expression
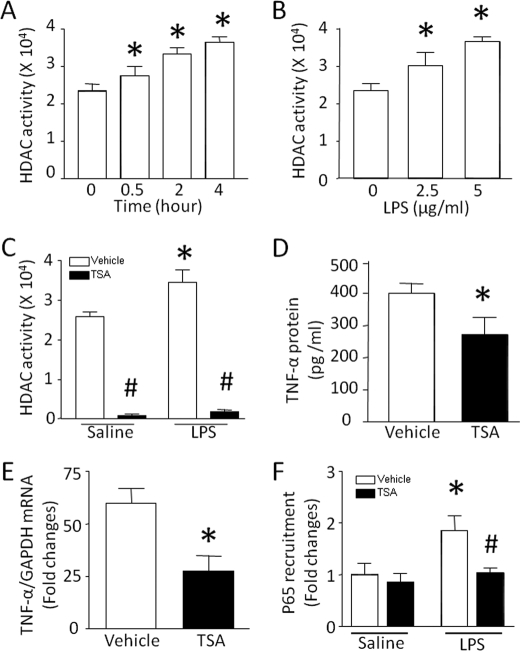
HDAC plays an important role in regulation of gene transcription (6). Interestingly, LPS treatment time- and dose-dependently increased HDAC activity (Fig. 1, A and B). Co-incubation with TSA, a pan-inhibitor for HDAC, abrogated basal and LPS-induced HDAC activity in cardiomyocytes (Fig. 1C). LPS-induced TNF-α production was significantly reduced by TSA (Fig. 1D). TSA also attenuated TNF-α mRNA in LPS-stimulated cardiomyocytes (Fig. 1E), suggesting a putative role for HDAC in the transcriptional control of TNF-α expression. To support the transcriptional regulation by HDAC, we analyzed the recruitment of NF-κB/p65 at the promoter of TNF-α in LPS-stimulated cardiomyocytes because NF-κB is an important transcription factor in control of TNF-α transcription. In line with the down-regulation of TNF-α expression, the accumulation of NF-κB/p65 at the promoter of TNF-α was significantly reduced by TSA in LPS-stimulated cardiomyocytes (Fig. 1F). These data demonstrate that HDAC positively regulates LPS-induced TNF-α expression at least in part through a transcriptional mechanism.
FIGURE 1.
Effects of TSA on HDAC activity and TNF-α expression. A and B, time- and dose-dependent activation of HDAC in response to LPS. A, cardiomyocytes were incubated with LPS (5 μg/ml). At different time points (0, 0.5, 2, and 4 h) after LPS treatment, HDAC activity was determined in cardiomyocytes. B, cardiomyocytes were exposed to different doses of LPS (0, 2.5, and 5 μg/ml). Four hours later, HDAC activity was measured. C–F, effects of TSA. Cardiomyocytes were incubated with LPS or saline in combination with TSA or vehicle for 4 h. TSA treatment significantly inhibited HDAC activity (C), TNF-α protein (D), TNF-α mRNA (E), and the accumulation of NF-κB/p65 at the promoter of TNF-α (F). Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus 0 or vehicle in saline; #, p < 0.05 versus vehicle in LPS.
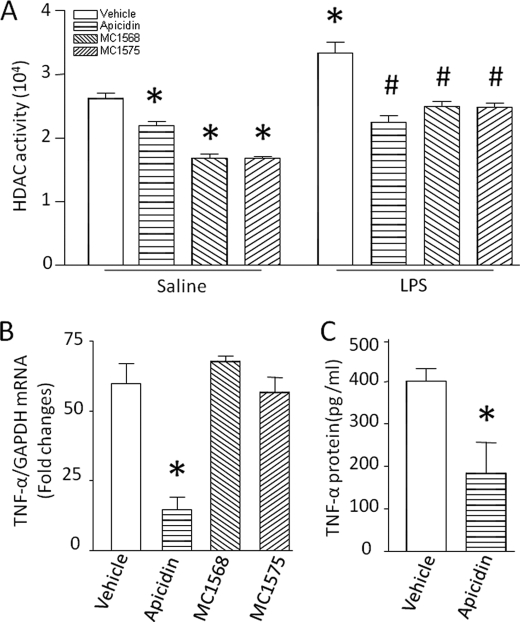
To define the isoform-specific contribution of HDAC, we incubated cardiomyocytes with LPS in the presence of the class I-specific HDAC inhibitor apicidin (5 μm), particularly active against HDAC2 (IC50 = 120 nm) and HDAC3 (IC50 = 43 nm) (18), or the class II-specific HDAC inhibitors MC1575 (20 μm) and MC1568 (20 μm) (19, 20). If LPS-induced HDAC activity resulted from both class I and class II HDAC, the levels of HDAC activity would be similar between basal and LPS-stimulated cardiomyocytes after incubation with their inhibitors. Indeed, treatment with apicidin reduced basal and LPS-stimulated HDAC activity to a similar level, whereas MC1575 or MC1568 reduced only basal but not LPS-stimulated HDAC activity (Fig. 2A), suggesting a putative role of class I HDAC in LPS-induced HDAC activity. Consistently, apicidin inhibited LPS-induced TNF-α mRNA and protein to the same extent as TSA (Fig. 2, B and C). In contrast, the class II-specific HDAC inhibitors MC1575 and MC1568 had no effect on TNF-α expression in LPS-stimulated cardiomyocytes (Fig. 2B). Thus, class I HDAC activation contributes to LPS-induced TNF-α expression.
FIGURE 2.
Role of class I HDAC in TNF-α expression. Cardiomyocytes were incubated with LPS or saline in combination with apicidin, MC1568, MC1575, or vehicle. Four hours later, HDAC activity (A), TNF-α mRNA (B), and TNF-α protein (C) were measured. Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus vehicle in saline or vehicle in LPS; #, p < 0.05 versus vehicle in LPS.
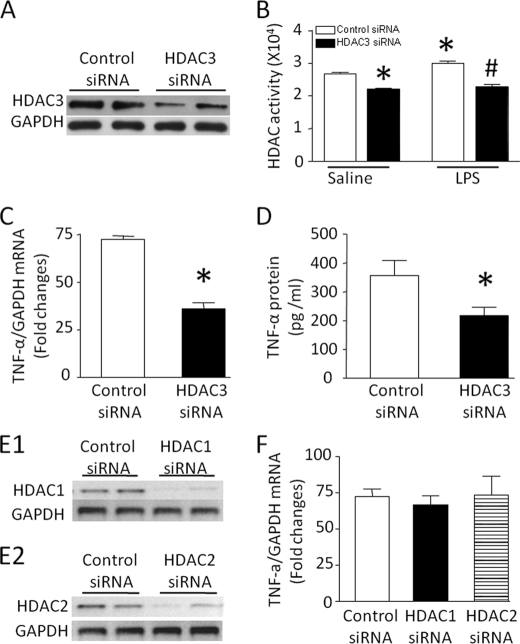
To further confirm and dissect the isoform-specific role of class I HDAC, we employed siRNA to specifically knock down HDAC1, -2, and -3 in cardiomyocytes. Cardiomyocytes were transfected with siRNA against HDAC1, -2, or -3 or a scrambled siRNA as control and then incubated with LPS for 4 h. Transfection with HDAC3 siRNA significantly decreased HDAC3 protein expression (Fig. 3A), suggesting a successful knockdown. Similarly, down-regulation of HDAC3 reduced basal and LPS-stimulated HDAC activity to a similar level (Fig. 3B) and significantly decreased LPS-stimulated TNF-α mRNA and production (Fig. 3, C and D). In contrast, knockdown of HDAC1 or -2 did not alter LPS-induced TNF-α expression (Fig. 3, E and F). These data demonstrate that LPS-induced HDAC activity results from HDAC3 activation, which contributes to TNF-α expression in cardiomyocytes.
FIGURE 3.
Contribution of HDAC3 to TNF-α expression in LPS-stimulated cardiomyocytes. Cardiomyocytes were transfected with siRNA against HDAC1, -2, or -3 or control siRNA for 24 h and then exposed to LPS for 4 h. A–D, effects of HDAC3 siRNA. Transfection of HDAC3 siRNA down-regulated HDAC3 protein (A), decreased HDAC activity (B), and reduced TNF-α mRNA (C) and TNF-α protein (D). E and F, effects of HDAC1 and -2 siRNA. Knockdown of HDAC1 and -2 was confirmed by down-regulation of their mRNA levels (E1 and E2). The TNF-α mRNA levels were not altered by knockdown of HDAC1 or -2 (F). Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus vehicle in saline or control siRNA; #, p < 0.05 versus vehicle in LPS.
LPS Induces HDAC Activation via Mitochondrial ROS Production
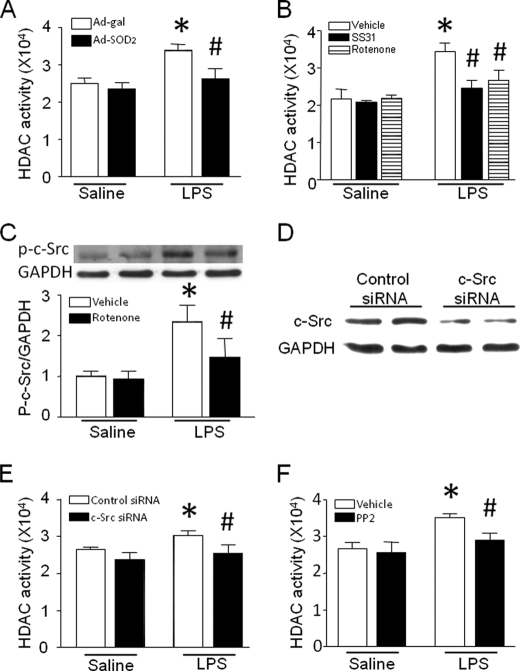
Oxidative stress has been shown to modulate HDAC activation. Mitochondria are important ROS sources in cardiomyocytes. To determine whether ROS from mitochondria contributes to HDAC activation, we analyzed the effect of the mitochondrially targeted antioxidant peptide SS31 (H-d-Arg-dimethyltyrosine-Lys-Phe-NH2) or rotenone on HDAC activity. SS31 specifically quenches mitochondrial ROS (12). The structurally related peptide SS20 (H-Phe-d-Arg-Phe-Lys-NH2), which lacks antioxidant properties, served as a control (12). Furthermore, we examined the effect of SOD2 overexpression on HDAC activation induced by LPS in cardiomyocytes because SOD2 has been shown to specifically neutralize mitochondrial ROS (21). Cardiomyocytes were infected with Ad-SOD2 or Ad-β-galactosidase as a control for 24 h or incubated with SS31 or SS20 (2.5 μm) as a control for 15 min and then exposed to LPS (5 μg/ml) for 4 h. Both infection of Ad-SOD2 and treatment with SS31 or rotenone significantly attenuated LPS-stimulated but not basal HDAC activity in cardiomyocytes (Fig. 4, A and B), suggesting that mitochondrial ROS induce the increase in HDAC activity.
FIGURE 4.
Role of the mitochondrial ROS/c-Src pathway in HDAC activity. A, cardiomyocytes were infected with Ad-SOD2 or Ad-β-galactosidase and followed by LPS. Four hours later, HDAC activity was measured. B, cardiomyocytes were incubated with LPS alone or in combination with SS31, rotenone, or vehicle for 4 h, and HDAC activity was determined. C, cardiomyocytes were incubated with LPS in the presence of rotenone or vehicle for 4 h, and phosphorylated c-Src (Tyr-418) was determined by Western blot analysis. The upper panel is the representative Western blot for c-Src phosphorylation, and the lower panel is the quantification of phosphorylated c-Src relative to GAPDH. D, knockdown of c-Src by siRNA. Cardiomyocytes were transfected with c-Src siRNA or control siRNA for 24 h. A representative Western blot for down-regulation of c-Src protein by siRNA is presented. E and F, inhibition of c-Src by siRNA or pharmacological inhibitor PP2 blocks LPS-induced HDAC activity in cardiomyocytes. Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus Ad-β-galactosidase in saline, vehicle in saline, or control siRNA in saline; #, p < 0.05 versus Ad-β-galactosidase in saline, vehicle in LPS, or control siRNA in LPS.
To identify the link between mitochondrial ROS and HDAC activation, we focused on c-Src because c-Src is ROS-sensitive (22) and has been shown to activate HDAC3 (23, 24). Indeed, LPS treatment increased the levels of c-Src phosphorylation at Tyr-418, indicating c-Src activation, which was inhibited by rotenone (Fig. 4C). This result supports an important role of mitochondrial signals in c-Src activation in LPS-stimulated cardiomyocytes. Furthermore, knockdown of c-Src by siRNA decreased LPS-induced HDAC activity (Fig. 4, D and E). The role of c-Src in HDAC activity was also examined by using a selective Src inhibitor, PP2. Consistently, incubation with PP2 (10 μm) inhibited LPS-induced HDAC activity in cardiomyocytes (Fig. 4F). These results demonstrate that mitochondrial ROS induces HDAC activity in a c-Src-dependent manner in LPS-stimulated cardiomyocytes.
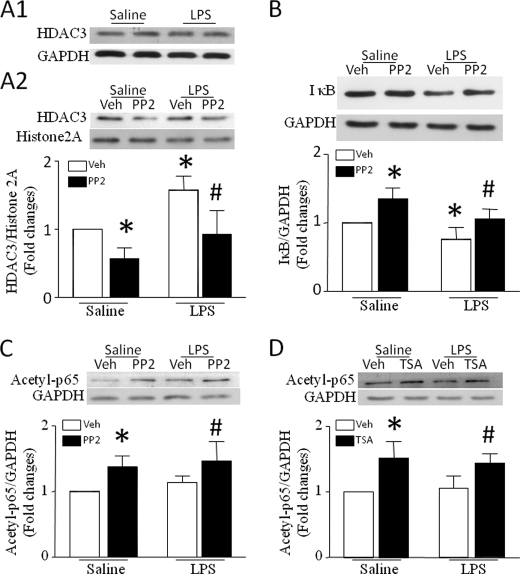
To explore the potential signaling mechanisms of c-Src-mediated HDAC3 activity, we first measured the protein levels of HDAC3 in nuclei. Although HDAC3 levels in whole cell lysates were not altered in response to LPS, LPS treatment significantly elevated nuclear HDAC3, which was prevented by inhibition of c-Src with PP2 (Fig. 5A). This result suggests that c-Src activation increases the nuclear translocation of HDAC3. It has been shown that IκB degradation promotes translocation of HDAC3 from the cytoplasm to the nucleus (25). In this regard, we also determined the levels of IκB protein in cardiomyocytes. LPS reduced the levels of IκB in cardiomyocytes. However, inhibition of c-Src with PP2 preserved IκB in response to LPS (Fig. 5B). Thus, c-Src activation may induce the nuclear translocation of HDAC3 through IκB degradation in LPS-stimulated cardiomyocytes.
FIGURE 5.
Role of c-Src in nuclear translocation of HDAC3 and IκB degradation. A and B, cardiomyocytes were incubated with saline or LPS in the presence of PP2 or vehicle (Veh). Four hours later, nuclei were isolated from cardiomyocytes. HDAC3 protein was determined in isolated nuclei and whole cell lysates by Western blot analysis. Histone 2A (A2) and GAPDH (A1) were measured as load controls for nuclear and whole cell HDAC3, respectively. The protein levels of IκB were also measured by Western blot analysis. Whole cell HDAC3 is shown in A1. Upper panels are the representative Western blots from three to five different experiments for nuclear HDAC3 (A2) and IκB protein (B), and lower panels are the quantification of the ratios of HDAC3 to histone 2A (A2) and IκB to GAPDH (B). C and D, role of c-Src and HDAC3 in p65 acetylation. Cardiomyocytes were incubated with saline or LPS in the presence of PP2, TSA, or vehicle for 4 h. Western blotting was performed to analyze the effect of PP2 (C) and TSA (D) on acetylated p65. Upper panels are the representative Western blots from three to five different experiments for acetylated p65, and lower panels are the quantification of the ratio of acetylated p65 to GAPDH (C and D). Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus vehicle in saline; #, p < 0.05 vehicle in LPS.
Inhibition of c-Src and HDAC3 Promotes p65 Acetylation
Acetylation of p65 has been suggested to attenuate NF-κB transcriptional activity (26). We therefore investigated the role of c-Src and HDAC in acetylation of p65 in LPS-induced cardiomyocytes. As shown in Fig. 5 (C and D), both c-Src inhibitor PP2 and HDAC inhibitor TSA significantly increased the levels of acetylated p65. This result suggests that c-Src/HDAC signaling deacetylates p65, presumably promoting NF-κB transcriptional activity in cardiomyocytes.
Selective Inhibition of Mitochondrial ROS Reduces TNF-α Biosynthesis in Response to LPS
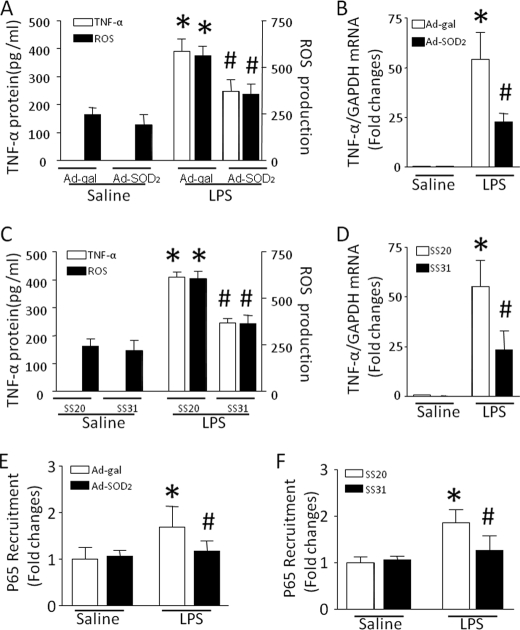
To investigate the functional link between mitochondrial ROS and TNF-α biosynthesis, we analyzed the effect of SS31 and SOD2 on TNF-α production in LPS-stimulated cardiomyocytes. Incubation with SS31 or overexpression of SOD2 significantly reduced TNF-α production in response to LPS, which was mirrored by a reduction in ROS production (Fig. 6, A and C). Mitochondrially targeted antioxidants also down-regulated TNF-α mRNA accumulation (Fig. 6, B and D) and the accumulation of NF-κB/p65 at the promoter of TNF-α (Fig. 6, E and F). These results demonstrate that selectively blocking mitochondrial ROS down-regulates TNF-α expression in LPS-stimulated cardiomyocytes.
FIGURE 6.
Effects of mitochondrially targeted antioxidants on TNF-α expression, NF-κB/p65 accumulation, and ROS production. Cardiomyocytes were infected with Ad-SOD2 or Ad-β-galactosidase for 24 h and then exposed to LPS for 4 h. In a separate experiment, cardiomyocytes were incubated with SS31 or SS20 for 15 min, followed by LPS for 4 h. A and C, TNF-α protein in culture medium and ROS production in cardiomyocytes were measured. B and D, TNF-α mRNA was quantified by real-time reverse transcription-PCR. E and F, chromatin immunoprecipitation assay was performed to determine the accumulation of NF-κB/p65 at the TNF-α promoter. Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus Ad-β-galactosidase in saline or SS20 in saline; #, p < 0.05 versus Ad-β-galactosidase in LPS or SS20 in LPS.
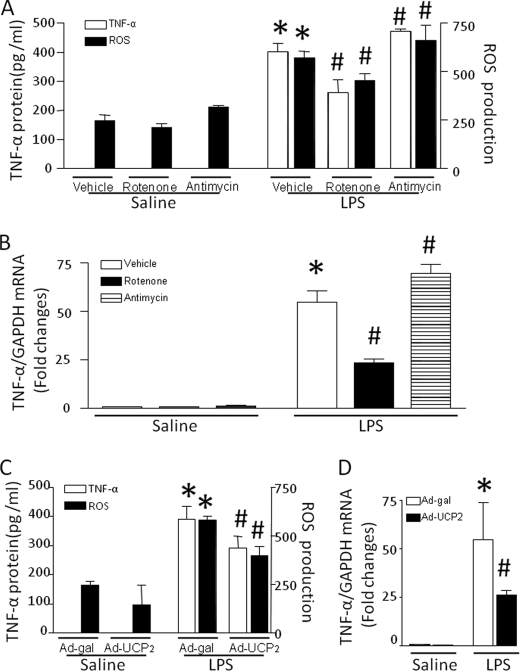
To identify the source of mitochondrial ROS in regulation of TNF-α expression, we analyzed the ability of cardiomyocytes to produce TNF-α in culture conditions affecting mitochondrial ROS generation. Because mitochondrial respiratory chain complexes I and III are the major sites for superoxide generation (27), we incubated cardiomyocytes with LPS alone or in combination with rotenone or antimycin A, inhibitors for mitochondrial respiratory chain complex I or III, respectively. Co-incubation with rotenone attenuated whereas antimycin A slightly enhanced the capacity to produce TNF-α in response to LPS, which paralleled the changes in ROS production (Fig. 7, A and B). This result suggests that ROS from mitochondrial complex I contribute to TNF-α expression. To further confirm the contribution of mitochondrial ROS, we overexpressed UCP2 in cardiomyocytes by infection with Ad-UCP2 and then incubated these cells with LPS for 4 h. UCP2 has been shown to limit mitochondrial ROS production by decreasing the mitochondrial membrane potential (28, 29). Consistent with these previous reports, overexpression of UCP2 reduced ROS production induced by LPS (Fig. 7C). Similarly, TNF-α mRNA and protein were significantly decreased in Ad-UCP2- compared with Ad-β-galactosidase-infected cardiomyocytes (Fig. 7, C and D). Thus, these data further support the fact that mitochondrial ROS promote TNF-α expression in response to LPS.
FIGURE 7.
Roles of the mitochondrial electron transport chain in TNF-α expression and ROS production. Cardiomyocytes were infected with Ad-UCP2 or Ad-β-galactosidase for 24 h and then exposed to LPS for 4 h or incubated with LPS in combination with rotenone, antimycin A, or vehicle for 4 h. A and C, TNF-α protein in culture medium and ROS production in cardiomyocytes were measured. B and D, TNF-α mRNA was quantified by real-time reverse transcription-PCR. Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus Ad-β-galactosidase in saline or vehicle in saline; #, p < 0.05 versus Ad-β-galactosidase in LPS or vehicle in LPS.
c-Src Signaling Induces LPS-stimulated TNF-α Expression
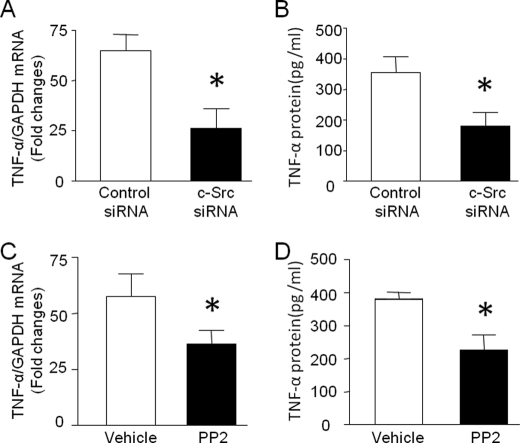
Having shown that mitochondrial ROS promoted TNF-α expression and induced HDAC activation via c-Src signaling, we hypothesized that c-Src activation mediated TNF-α biosynthesis in LPS-stimulated cardiomyocytes. In support of this hypothesis, knockdown of c-Src by siRNA or inhibition of c-Src by PP2 significantly reduced TNF-α mRNA and production in response to LPS (Fig. 8, A–D).
FIGURE 8.
Role of c-Src in TNF-α expression during LPS stimulation. A and B, cardiomyocytes were transfected with c-Src siRNA or control siRNA for 24 h and then exposed to LPS for 4 h. TNF-α mRNA in cardiomyocytes (A) and TNF-α protein in culture medium (B) were measured. C and D, cardiomyocytes were incubated with LPS in combination with PP2 or vehicle for 4 h, and TNF-α mRNA (C) and TNF-α protein (D) were quantified. Data are mean ± S.D. (n = 3–5). *, p < 0.05 versus vehicle in LPS or control siRNA in LPS.
DISCUSSION
The major findings of this study are that LPS induces HDAC3 activation via mitochondrial ROS and c-Src signaling and that inhibition of HDAC3 suppresses TNF-α expression, suggesting an important role of HDAC3 in regulation of LPS-induced TNF-α expression. Selective inhibition of mitochondrial ROS or c-Src attenuates TNF-α expression in cardiomyocytes during LPS stimulation. Thus, our study demonstrates a novel signaling mechanism by which LPS induces TNF-α expression in cardiomyocytes.
Role of HDAC3 in LPS-induced TNF-α Expression
HDAC activation is an important mechanism in regulation of gene transcription. However, it has never been shown whether HDAC plays a role in LPS-induced TNF-α expression in cardiomyocytes. Our study aimed to investigate the role of HDAC in LPS-induced TNF-α expression in cardiomyocytes. Using cultured cardiomyocytes, we demonstrated the following. First, LPS increases HDAC activity. This up-regulation of HDAC activity is abrogated by a pan-HDAC inhibitor (TSA), a class I-specific HDAC inhibitor (apicidin), or knockdown of HDAC3, but not by class II HDAC inhibitors, suggesting that the increase in HDAC activity results from HDAC3 activation in LPS-stimulated cardiomyocytes. Second, TSA, apicidin, or knockdown of HDAC3 inhibits TNF-α expression to the same extent. Third, inhibition of class II HDAC or knockdown of HDAC1 and -2 does not alter LPS-stimulated TNF-α expression. These results provide definite evidence to support the view that HDAC3 activation induces TNF-α expression in LPS-stimulated cardiomyocytes. This is apparently different from a recent report that demonstrated that HDAC3 activation repressed TNF-α expression in LPS-stimulated mononuclear phagocytes (30). It is currently unknown what causes this discrepancy, but it may be due to different cell types used in different studies. Indeed, HDAC inhibitors have been shown to potentiate TNF-α expression in microglial cells (31) but to suppress TNF-α expression in cultured human peripheral blood mononuclear cells and macrophages in response to LPS (32, 33). Thus, the role of HDAC3 in regulation of TNF-α expression may be cell type-specific.
The molecular mechanisms by which HDAC3 induces TNF-α expression remain to be determined in LPS-stimulated cardiomyocytes. Our study shows that inhibition of HDAC activity reduces the accumulation of NF-κB/p65 at the promoter of TNF-α during LPS stimulation, suggesting that NF-κB may be a potential downstream pathway of the HDAC3 signaling in regulation of TNF-α expression. Our data further suggest that HDAC3 may promote NF-κB transcriptional activity through direct deacetylation of p65 because acetylation of p65 plays a key role in IκBα-mediated attenuation of NF-κB transcriptional activity (26). However, this study could not exclude other mechanisms such as modulation of histone acetylation and other signaling pathways upstream of NF-κB (23).
Contribution of Mitochondrial ROS to TNF-α Expression during LPS Stimulation
ROS production by mitochondria has been shown to play an important role in cardiac pathophysiology (34). In this study, we have demonstrated that mitochondrial ROS from complex I promote LPS-stimulated TNF-α expression in cardiomyocytes. This conclusion is supported by the following lines of evidence. First, overexpression of SOD2 or incubation with SS31 reduces LPS-induced ROS and TNF-α production. SOD2 and SS31 per se may also inhibit cytosolic ROS, which have been suggested to contribute to TNF-α expression in LPS-stimulated cardiomyocytes (35, 36). However, SOD2 and SS31 are predominantly present in mitochondria (12, 21), and thus, the possible inhibitory effect of SOD2 and SS31 on cytosolic ROS is negligible in this study. Second, overexpression of UCP2 inhibits ROS and TNF-α production in LPS-stimulated cardiomyocytes. Third, rotenone but not antimycin A down-regulates ROS and TNF-α production induced by LPS, supporting an important role of mitochondrial respiratory chain complex I.
This study further shows that blocking mitochondrial ROS decreases the recruitment of NF-κB/p65 at the promoter of TNF-α during LPS stimulation, suggesting that mitochondrial ROS-promoted TNF-α expression may be related to the NF-κB/p65-dependent pathway because NF-κB activation is an important mechanism in mediating TNF-α expression in response to LPS. Mitochondrial ROS-induced NF-κB activation has also been suggested in cancer cells under hypoxia (37). Thus, NF-κB may represent an important mechanism that mediates the cross-talk of mitochondria to nuclei.
It is worthwhile to mention that selective inhibition of mitochondrial ROS attenuates but does not abrogate cellular ROS production and TNF-α expression induced by LPS, suggesting that additional ROS-related pathways may be involved in regulation of TNF-α expression. Indeed, NAD(P)H oxidase is another important source of ROS in cardiomyocytes, which has been demonstrated to contribute to LPS-stimulated TNF-α expression in cardiomyocytes (36). Both mitochondria and NAD(P)H oxidase have been suggested to cross-talk to each other in producing ROS (38, 39). However, whether such cross-talk is operative in regulation of ROS production and consequent TNF-α expression in cardiomyocytes during LPS stimulation merits future studies.
Mitochondrial ROS in HDAC3 Activation during LPS Stimulation
Another important finding of this study is that blocking mitochondrial ROS inhibits HDAC3 activation in LPS-stimulated cardiomyocytes. This suggests that mitochondrial ROS are important signals in HDAC3 activation in response to LPS. However, the involved signaling pathways remain not fully understood. It has been shown that mitochondrial ROS induce c-Src activation during hypoxia (22, 37). In agreement with those previous reports, this study demonstrates that LPS induces c-Src activation, which is blocked by selective inhibition of mitochondrial ROS in cardiomyocytes. Furthermore, blocking c-Src inhibits LPS-induced TNF-α expression. Thus, mitochondrial ROS-promoted TNF-α is mediated through c-Src activation. A recent study has suggested that c-Src signaling may directly activate HDAC3 and increase the nuclear translocation of HDAC3 (24). c-Src can also induce IκB phosphorylation and degradation, which promote localization of HDAC3 from the cytoplasm to the nucleus (25). In this regard, our study shows that blocking c-Src signaling either by knockdown of c-Src or by using the pharmacological inhibitor PP2 abrogates the increase in HDAC3 activity and the protein levels of nuclear HDAC3 in cardiomyocytes during LPS stimulation. These results support the role of c-Src in HDAC3 activation. Thus, it is likely that mitochondrial ROS activate c-Src, which may be present in mitochondria (40). c-Src then induces HDAC3 nuclear translocation and activation, leading to TNF-α expression in cardiomyocytes during LPS stimulation.
In summary, this study demonstrates for the first time that activation of HDAC3 and mitochondrial ROS promote LPS-stimulated TNF-α expression in cardiomyocytes. Moreover, mitochondrial ROS induces HDAC3 activation through c-Src signaling in LPS-stimulated cardiomyocytes. These findings provide a novel pathway in which LPS via mitochondrial ROS/c-Src/HDAC3 pathways produces TNF-α expression in cardiomyocytes.
This work was supported by Grant-in-aid T6717 (to T. P.) from the Heart and Stroke Foundation of Ontario.
- TNF-α
- tumor necrosis factor-α
- LPS
- lipopolysaccharide/s
- HDAC
- histone deacetylase
- ROS
- reactive oxygen species
- Ad
- adenovirus
- siRNA
- small interfering RNA
- TSA
- trichostatin A
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Grandel U., Fink L., Blum A., Heep M., Buerke M., Kraemer H. J., Mayer K., Bohle R. M., Seeger W., Grimminger F., Sibelius U. (2000) Circulation 102, 2758–2764 [DOI] [PubMed] [Google Scholar]
- 2.Bozkurt B., Kribbs S. B., Clubb F. J., Jr., Michael L. H., Didenko V. V., Hornsby P. J., Seta Y., Oral H., Spinale F. G., Mann D. L. (1998) Circulation 97, 1382–1391 [DOI] [PubMed] [Google Scholar]
- 3.Oral H., Dorn G. W., 2nd, Mann D. L. (1997) J. Biol. Chem. 272, 4836–4842 [DOI] [PubMed] [Google Scholar]
- 4.Peng T., Lu X., Lei M., Moe G. W., Feng Q. (2003) Cardiovasc. Res. 59, 893–900 [DOI] [PubMed] [Google Scholar]
- 5.Pålsson-McDermott E. M., O'Neill L. A. (2004) Immunology 113, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode K. A., Schroder K., Hume D. A., Ravasi T., Heeg K., Sweet M. J., Dalpke A. H. (2007) Immunology 122, 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leoni F., Zaliani A., Bertolini G., Porro G., Pagani P., Pozzi P., Donà G., Fossati G., Sozzani S., Azam T., Bufler P., Fantuzzi G., Goncharov I., Kim S. H., Pomerantz B. J., Reznikov L. L., Siegmund B., Dinarello C. A., Mascagni P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glauben R., Batra A., Fedke I., Zeitz M., Lehr H. A., Leoni F., Mascagni P., Fantuzzi G., Dinarello C. A., Siegmund B. (2006) J. Immunol. 176, 5015–5022 [DOI] [PubMed] [Google Scholar]
- 10.Peng T., Lu X., Lei M., Feng Q. (2003) J. Biol. Chem. 278, 8099–8105 [DOI] [PubMed] [Google Scholar]
- 11.Schiller P. W., Nguyen T. M., Berezowska I., Dupuis S., Weltrowska G., Chung N. N., Lemieux C. (2000) Eur. J. Med. Chem. 35, 895–901 [DOI] [PubMed] [Google Scholar]
- 12.Zhao K., Zhao G. M., Wu D., Soong Y., Birk A. V., Schiller P. W., Szeto H. H. (2004) J. Biol. Chem. 279, 34682–34690 [DOI] [PubMed] [Google Scholar]
- 13.Mai A., Massa S., Pezzi R., Simeoni S., Rotili D., Nebbioso A., Scognamiglio A., Altucci L., Loidl P., Brosch G. (2005) J. Med. Chem. 48, 3344–3353 [DOI] [PubMed] [Google Scholar]
- 14.Mai A., Massa S., Pezzi R., Rotili D., Loidl P., Brosch G. (2003) J. Med. Chem. 46, 4826–4829 [DOI] [PubMed] [Google Scholar]
- 15.Li Y., Arnold J. M., Pampillo M., Babwah A. V., Peng T. (2009) Free Radic. Biol. Med. 46, 51–61 [DOI] [PubMed] [Google Scholar]
- 16.Peng T., Lu X., Feng Q. (2005) FASEB J. 19, 293–295 [DOI] [PubMed] [Google Scholar]
- 17.Hall G., Singh I. S., Hester L., Hasday J. D., Rogers T. B. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H2103–H2111 [DOI] [PubMed] [Google Scholar]
- 18.Khan N., Jeffers M., Kumar S., Hackett C., Boldog F., Khramtsov N., Qian X., Mills E., Berghs S. C., Carey N., Finn P. W., Collins L. S., Tumber A., Ritchie J. W., Jensen P. B., Lichenstein H. S., Sehested M. (2008) Biochem. J. 409, 581–589 [DOI] [PubMed] [Google Scholar]
- 19.Duong V., Bret C., Altucci L., Mai A., Duraffourd C., Loubersac J., Harmand P. O., Bonnet S., Valente S., Maudelonde T., Cavailles V., Boulle N. (2008) Mol. Cancer Res. 6, 1908–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nebbioso A., Manzo F., Miceli M., Conte M., Manente L., Baldi A., De Luca A., Rotili D., Valente S., Mai A., Usiello A., Gronemeyer H., Altucci L. (2009) EMBO Rep. 10, 776–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X., Zheng S., Metreveli N. S., Epstein P. N. (2006) Diabetes 55, 798–805 [DOI] [PubMed] [Google Scholar]
- 22.Sato H., Sato M., Kanai H., Uchiyama T., Iso T., Ohyama Y., Sakamoto H., Tamura J., Nagai R., Kurabayashi M. (2005) Cardiovasc. Res. 67, 714–722 [DOI] [PubMed] [Google Scholar]
- 23.Matteucci E., Ridolfi E., Maroni P., Bendinelli P., Desiderio M. A. (2007) Mol. Cancer Res. 5, 833–845 [DOI] [PubMed] [Google Scholar]
- 24.Longworth M. S., Laimins L. A. (2006) Oncogene 25, 4495–4500 [DOI] [PubMed] [Google Scholar]
- 25.Gao Z., He Q., Peng B., Chiao P. J., Ye J. (2006) J. Biol. Chem. 281, 4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiernan R., Brès V., Ng R. W., Coudart M. P., El Messaoudi S., Sardet C., Jin D. Y., Emiliani S., Benkirane M. (2003) J. Biol. Chem. 278, 2758–2766 [DOI] [PubMed] [Google Scholar]
- 27.Turrens J. F. (1997) Biosci. Rep. 17, 3–8 [DOI] [PubMed] [Google Scholar]
- 28.Teshima Y., Akao M., Jones S. P., Marbán E. (2003) Circ. Res. 93, 192–200 [DOI] [PubMed] [Google Scholar]
- 29.Arsenijevic D., Onuma H., Pecqueur C., Raimbault S., Manning B. S., Miroux B., Couplan E., Alves-Guerra M. C., Goubern M., Surwit R., Bouillaud F., Richard D., Collins S., Ricquier D. (2000) Nat. Genet. 26, 435–439 [DOI] [PubMed] [Google Scholar]
- 30.Mahlknecht U., Will J., Varin A., Hoelzer D., Herbein G. (2004) J. Immunol. 173, 3979–3990 [DOI] [PubMed] [Google Scholar]
- 31.Suuronen T., Huuskonen J., Pihlaja R., Kyrylenko S., Salminen A. (2003) J. Neurochem. 87, 407–416 [DOI] [PubMed] [Google Scholar]
- 32.Choi Y., Park S. K., Kim H. M., Kang J. S., Yoon Y. D., Han S. B., Han J. W., Yang J. S., Han G. (2008) Exp. Mol. Med. 40, 574–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S. B., Lee J. K. (2009) Arch. Pharm. Res. 32, 613–624 [DOI] [PubMed] [Google Scholar]
- 34.Di Lisa F., Kaludercic N., Carpi A., Menabò R., Giorgio M. (2009) Basic Res. Cardiol. 104, 131–139 [DOI] [PubMed] [Google Scholar]
- 35.Shang F., Zhao L., Zheng Q., Wang J., Xu Z., Liang W., Liu H., Liu S., Zhang L. (2006) Biochem. Biophys. Res. Commun. 351, 947–952 [DOI] [PubMed] [Google Scholar]
- 36.Peng T., Lu X., Feng Q. (2005) Circulation 111, 1637–1644 [DOI] [PubMed] [Google Scholar]
- 37.Lluis J. M., Buricchi F., Chiarugi P., Morales A., Fernandez-Checa J. C. (2007) Cancer Res. 67, 7368–7377 [DOI] [PubMed] [Google Scholar]
- 38.Wosniak J., Jr., Santos C. X., Kowaltowski A. J., Laurindo F. R. (2009) Antioxid. Redox Signal. 11, 1265–1278 [DOI] [PubMed] [Google Scholar]
- 39.Doughan A. K., Harrison D. G., Dikalov S. I. (2008) Circ. Res. 102, 488–496 [DOI] [PubMed] [Google Scholar]
- 40.Tibaldi E., Brunati A. M., Massimino M. L., Stringaro A., Colone M., Agostinelli E., Arancia G., Toninello A. (2008) J. Cell. Biochem. 104, 840–849 [DOI] [PubMed] [Google Scholar]