Abstract
Ctf4/AND-1 is a highly conserved gene product required for both DNA replication and the establishment of sister chromatid cohesion. In this report, we examined the mechanism of action of human Ctf4 (hCtf4) in DNA replication both in vitro and in vivo. Our findings show that the purified hCtf4 exists as a dimer and that the hCtf4 SepB domain likely plays a primary role determining the dimeric structure. hCtf4 binds preferentially to DNA template-primer structures, interacts directly with the replicative DNA polymerases (α, δ, and ϵ), and markedly stimulates the polymerase activities of DNA polymerases α and ϵ in vitro. Depletion of hCtf4 in HeLa cells by small interfering RNA resulted in G1/S phase arrest. DNA fiber analysis revealed that cells depleted of hCtf4 exhibited a rate of DNA replication slower than cells treated with control small interfering RNA. These findings suggest that in human cells, hCtf4 plays an essential role in DNA replication and its ability to stimulate the replicative DNA polymerases may contribute to this effect.
Keywords: Cell Cycle, DNA Binding Protein, DNA Polymerase, DNA-Protein Interaction, DNA Replication, Protein Purification, AND-1, Ctf4, Sister Chromatid Cohesin
Introduction
To ensure that DNA replication occurs once during each cell cycle, eukaryotic cells generate replication complexes in a stepwise manner (1). Initially, pre-replication complexes form at replication origins at a point in the G1 phase of the cell cycle when the initiation of replication is prevented due to the absence of cyclin-dependent kinase activity. During the G1-S transition, pre-replication complexes are activated in a manner that simultaneously excludes the production of new pre-replication complexes. This selective discrimination restricts the normal replication cycle so that each region of the chromosome is replicated uniquely during each cell cycle. Activation of the pre-replication complexes at origins during the G1/S transition stage, although poorly understood, depends on the action of two protein kinases, cyclin-dependent kinase and Dbf4-dependent Cdc7 kinase (Cdc7-Dbf4), which are required for the association of the Mcm2–7 complex with other replication proteins to form the replisome progression complex (RPC)2 (2–4). Key RPC components include Cdc45, GINS, and Mcm proteins, which interact to form an active DNA helicase complex at origins that support duplex unwinding as well as the recruitment of DNA polymerases (Pol) and auxiliary factors required for DNA synthesis.
The components of the RPC have been partially characterized in Saccharomyces cerevisiae (5). These elegant studies revealed that the complex contains, in addition to Mcm2–7, Cdc45, and GINS, a variety of proteins involved in activation of the DNA replication checkpoint system (Mrc1, Tof1, and Csm3), Mcm10, previously shown to be essential for the chromatin loading of Cdc45 in Xenopus (2, 3), the histone chaperone FACT (Spt16 and Pob3), which is required for both chromosomal replication and transcription through chromatin (4), and Ctf4. Pol α is also a component of the RPC, although its association with the complex is salt sensitive (6).
Ctf4 (also called AND-1 for acidic nucleoplasmic DNA-binding protein) was initially identified in S. cerevisiae using genetic screens for mutants involved in chromosome transmission fidelity (Ctf4) (7–9). Subsequent experiments documented roles for Ctf4 in cohesion (7) and DNA replication (10). In contrast to S. cerevisiae, Ctf4 is essential for Schizosaccharomyces pombe viability and DNA replication in both Xenopus extracts and human cells (see Refs. 11–13, and this report). In budding yeast, Ctf4 was shown to interact with DNA Pol α and contribute to the stability of the p180 catalytic subunit, and similar observations have been reported for its homologues in fission yeast, Xenopus, and humans (10, 11, 13, 14). Importantly, deletion of ctf4 in budding yeast resulted in the isolation of RPC lacking Pol α (6), suggesting that Ctf4 is required for the recruitment of Pol α to the RPC. Recent studies revealed that Ctf4 interacts with Mcm10, GINS (a key component of the Cdc45-Mcm2–7-GINS (CMG) helicase complex) as well as with Pol α (10, 11, 13). In human cells, in conjunction with RecQL4 and Mcm10, Ctf4 was shown to be essential for the formation of the CMG complex (15).
In this report, we describe the isolation of human Ctf4 and characterization of a number of its biochemical properties. We show that it exists as a homodimer and interacts with primed-template junction DNA as well as with all three replicative polymerases, albeit with different affinities. In vitro, hCtf4 markedly stimulated the polymerase activities of Pol α and Pol ϵ, but marginally affected Pol δ. In vivo studies revealed that depletion of hCtf4 reduced the rate of fork progression and was essential for DNA replication. We also noted that the cellular level of hCtf4 was relatively constant throughout the cell cycle and localized to the nucleus. However, its association with chromosomes was dramatically cell cycle-dependent. During interphase, the period of active replication, hCtf4 was colocalized with chromosomes. This association, however, was not detected when cells progressed through mitosis but re-occurred when cells exited anaphase and entered telophase. Collectively, these findings indicate that hCtf4 plays an important and direct role in replication at the fork, most likely through its interactions with key replication proteins and replicative DNA polymerases.
EXPERIMENTAL PROCEDURES
Plasmids
hCtf4 (GenBank accession number NM_007086) was amplified from a HeLa cDNA library (Clontech) using 5′- and 3′- hCtf4-specific primers containing BamHI and NotI sites, respectively. The hCtf4 cDNA was subcloned into the BamHI and NotI sites of pET28a (Novagen, for bacterial expression), pFastBac1 (Invitrogen, for baculovirus expression of untagged proteins), and pFastBacHtbFlag2 (modified Invitrogen pFastBacHtb, for baculovirus expression of His6-Flag2 fusion proteins). The cDNAs encoding the human Pol α p70 and p58 (primase) subunits were PCR amplified from baculoviruses expressing p70 and p58, and subcloned into pFastBacHtbFlag2. All plasmids were sequenced to verify that mutations were not introduced during PCR and cloning. All baculoviruses were generated according to the manufacturer's instructions.
Purification of hCtf4 from Baculovirus-infected Insect Cells
Monolayers of High Five insect cells (4 × 107 cells/150-cm2 plate) were infected with the recombinant baculovirus expressing hCtf4 at a multiplicity of infection of 10. Infected cells from 40 plates (150 cm2) were grown at 27 °C in Graces medium containing 10% heat-inactivated fetal bovine serum for 48 h and harvested by centrifugation at 600 × g for 10 min at 4 °C. Harvested cells (10 ml) were washed with 100 ml of ice-cold PBS, lysed in 20 ml of hypotonic buffer (50 mm HEPES-NaOH, pH 7.5, 1.5 mm MgCl2, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride/protease inhibitors (2 μg/ml each of aprotinin, leupeptin, and antipain and 0.1 mm benzamidine)) containing 10 mm KCl by Dounce homogenization (20 strokes) and the mixture centrifuged at 2,500 × g for 30 min at 4 °C. The supernatant (cytosolic fraction) was adjusted to 0.42 m NaCl, and the pellet was resuspended in 10 ml of hypotonic buffer containing 0.42 m NaCl. The resuspended pellet and cytosolic fractions were centrifuged at 43,500 × g for 30 min at 4 °C and the supernatants combined (47.5 ml, 275 mg of protein), adjusted to 50 mm NaCl with Q-0 buffer (50 mm HEPES-NaOH, pH 7.5, 1 mm DTT, 0.05% Nonidet P-40, 5% glycerol/protease inhibitors), and applied to a 30-ml Q-Sepharose column (2.5 × 10 cm). Bound proteins were eluted with 300 ml of 0.05 to 0.75 m NaCl linear gradient in Q-0 buffer and the elution of hCtf4 was monitored by Western blotting using hCtf4-specific polyclonal antibodies. Fractions containing hCtf4 (peaking at ∼0.28 m NaCl) were pooled (116 ml, 162 mg of protein), diluted to 0.1 m NaCl with SP-0 buffer (50 mm MES, pH 6.5, 1 mm DTT, 5% glycerol, 0.05% Nonidet P-40/protease inhibitors), and then loaded onto a 20-ml of SP-Sepharose column that was eluted with 200 ml of 0.1 to 0.75 m NaCl linear gradient. Fractions containing hCtf4 (peaking ∼150 mm NaCl) were pooled (27 ml, 27 mg of protein) and dialyzed against 2 liters of storage buffer (50 mm HEPES-NaOH, pH 7.5, 1 mm DTT, 0.05% Nonidet P-40, 25% glycerol, 0.15 m NaCl/protease inhibitor) overnight at 4 °C. The dialyzed preparation (11 ml, 23 mg of protein) was aliquoted and stored at −80 °C.
His-FLAG-tagged fusions of full-length hCtf4 and various hCtf4 derivatives (as described below) were expressed in Sf9 and purified as described previously for the isolation of Ctf18-RFC (16). Following glycerol gradient centrifugation, 12, 17, 18, 20, 0.35, and 0.50 mg of His6-Flag2-hCTF4 (full-length), WD, SepB, HMG, WD-SepB, and SepB-HMG fragments were obtained from 0.8 liter of Sf9 cells, respectively. Factors contributing to the low yield of the WD-SepB and SepB-HMG fragments have not been explored.
Purification of Pol α-Primase Complexes
High Five insect cells (1.6 × 109 cells) were infected with baculoviruses expressing the four subunits of Pol α (p180 and p70) and the primase (His6-Flag2-p58 and p48) complex each at a multiplicity of infection of 3 and grown for 48 h at 27 °C. Cells (19 ml) isolated from 40 plates (150 cm2) were washed with ice-cold PBS and lysed in 36 ml of hypotonic buffer (20 mm HEPES-NaOH, pH 7.5, 5 mm KCl, 1.5 mm MgCl2, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride/protease inhibitors) by Dounce homogenization (20 strokes). The NaCl concentration of the mixture was adjusted to 0.2 m NaCl and centrifuged at 43,000 × g for 45 min at 4 °C. The supernatant (21 ml, 336 mg of protein) was dialyzed against 3 liters of dialysis buffer (25 mm HEPES-NaOH, pH 7.5, 1 mm DTT, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 0.01% Nonidet P-40, 0.1 mm phenylmethylsulfonyl fluoride, 25 mm NaCl/protease inhibitors) overnight at 4 °C. The dialyzed material (14 ml, 210 mg of protein) was loaded onto a 10-ml Q-Sepharose column (1.5 × 10 cm) pre-equilibrated with dialysis buffer. After washing with 10 ml of dialysis buffer, bound proteins were eluted with 44 ml of Q-elution buffer (25 mm HEPES-NaOH, pH 7.5, 1 mm DTT, 1 mm EDTA, 10% glycerol, 0.01% Nonidet P-40, 0.3 m NaCl/protease inhibitors). FLAG-M2-agarose beads (Sigma, 1 ml) were added to the Q fraction (44 ml, 60 mg of protein) and the mixture gently rocked overnight at 4 °C. Collected beads were washed three times with 10 ml of FLAG buffer (25 mm HEPES-NaOH, pH 7.5, 10% glycerol, 0.3 m NaCl, 1 mm DTT, 0.01% Nonidet P-40, 1 mm EDTA/protease inhibitors), and bound proteins were eluted three times consecutively with 0.5 ml of FLAG buffer supplemented with 1 mg/ml of Flag3 peptide. The peptide-eluted fractions were pooled (1.2 ml, 0.7 mg of protein) and dialyzed against 2 liters of polymerase storage buffer (20 mm Tris-HCl, pH 7/25% glycerol, 0.1 mm EDTA, 0.01% Nonidet P-40, 0.1 mm DTT, 0.1 m NaCl) overnight at 4 °C. The dialyzed material (0.67 ml, 0.75 mg of protein) was aliquoted and stored in −80 °C. The two-subunit Pol α (p180 and His6Flag2-p70) complex was expressed and purified in a similar manner except that infections were carried out with 1.5 liters of Sf9 cells (2.3 × 106 cells/ml). Primase (His6Flag2-p58 and p48) complex was expressed and purified from 1.5 liters of Sf9 cells (2.3 × 106 cells/ml) as described previously for the isolation of Ctf18-RFC (16). Preparations of the four-subunit Pol α and primase complexes were further purified by glycerol gradient centrifugation that yielded stoichiometric complexes of these materials. Primase activity of the four-subunit Pol α-primase and two-subunit primase complexes varied between 5 and 7000 nmol/mg of protein/h; similar activities were observed for the Pol α activity of the four- and two-subunit complexes.
In Vitro Interactions of hCtf4 with Pol α, δ, and ϵ
Reaction mixtures (100 μl) containing 2.5 pmol of His6-Flag2-tagged Pol α/primase (or δ or ϵ) and varying amounts of hCtf4 (1.9 to 15 pmol) were incubated in binding buffer (50 mm HEPES-NaOH, pH 7.5, 0.05% Nonidet P-40, 100 mm NaCl, 1 mm DTT, 0.1 mg/ml of BSA/protease inhibitors) overnight at 4 °C. FLAG-tagged Pol-hCtf4 complexes were immunoprecipitated by adding 15 μl of FLAG-M2 antibody-agarose to 50-μl aliquots of the reaction mixture. FLAG peptide (1 mg/ml) was added to the remaining 50 μl of reaction mixture, which was then also treated with 15 μl of FLAG-M2 antibody-agarose (representing a negative control). Reaction mixtures containing agarose beads were mixed gently at 4 °C for 30 min, washed three times with 0.5 ml of binding buffer, and bound proteins eluted with 20 μl of 1× SDS loading buffer were subjected to 10% SDS-PAGE. hCtf4 and subunits of various polymerases were detected by Western blotting with specific antibodies, as indicated in the figure legends.
Oligonucleotide Primer-Templates
The 94-mer hairpin C oligonucleotide (5′-(C)36 TTT CCC TGT GCC CTT CGT ATA CGA TGG GTT TTT CCC ATC GTA TAC GAA GGG CAC AGG G-3′), the 89-mer TG40 repeat ((5′-(TG)19 TTGG TTG GCC GAT CAA GTG CCC AGT CAC GAC GTT GTA AAA CGA GCC CGA GT-3′), and the 50-mer primer (5′-CAC TCG GGC TCG TTT TAC AAC GTC GTG ACT GGG CAC TTG ATC GGC CAA CC-3′) were synthesized by International Technology and purified by 6% SDS-PAGE. The 50-mer was annealed to the 89-mer TG40 repeat template strand by mixing the two oligomers in a 2:1 primer:template molar ratio in 0.1 m NaCl, 10 mm Tris-HCl, pH 7.5, and 1 mm EDTA and the mixture was heated to 95 °C for 5 min followed by slow cooling to room temperature.
DNA Binding Assay
Reaction mixtures (15 μl) containing 50 fmol of 5′-32P-labeled 94-mer hairpin C oligonucleotide (possessing the TTT(C)36 5′-single stranded region) (2000 cpm/fmol) and varying amounts of hCtf4, as described in figure legends, were incubated in DNA binding buffer (50 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, 1 mm DTT, 10% glycerol, 50 mm NaCl) for 20 min at 30 °C. hCtf4-DNA complexes formed were separated from free DNA by electrophoresis through a 5% native PAGE in 0.5× Tris borate-EDTA at 16 V/cm. Following electrophoresis, gels were dried and labeled bands were visualized and quantified by phosphorimaging. The oligonucleotides used in the DNA binding competition assays were as described in the legend to Fig. 2.
FIGURE 2.
hCtf4 binds preferentially to DNAs containing primer-template junctions. A, diagram of labeled and unlabeled DNAs used in DNA binding experiments. B, binding of hCtf4 to DNA. Varying levels of hCtf4 were incubated with the 5′-32P-labeled 94-mer 3′-recessed hairpin C oligonucleotide as described under “Experimental Procedures.” Labeled bands, corresponding to free DNA and DNA complexed to hCtf4, are indicated. C, influence of unlabeled DNAs on the binding of the hCtf4 to 5′-32P-labeled 94-mer 3′-recessed hairpin C DNA. Reaction mixtures, as described under “Experimental Procedures,” contained 2.5 pmol of hCtf4 and 50 fmol of radiolabeled 94-mer hairpin C DNA. Competition reactions were carried out in the presence of the indicated unlabeled DNAs including: 3′-recessed 94-mer hairpin C (3′R Hairpin C), 5′-recessed 94-mer hairpin C possessing the identical 94-mer-hairpin C sequence but with opposite phosphodiester bond polarity (5′R Hairpin C), a double-stranded 94-mer hairpin C (ds Hairpin C), and single-stranded 39-mer dC oligonucleotide (dC39). D, binding of the hCtf4 derivatives to 3′R hairpin C. Increasing levels of purified truncated proteins (WD, SepB, HMG, WD + SepB, and SepB + HMG) or full-length (FL) hCtf4 were incubated with 150 fmol of 5′-32P-labeled 3′R hairpin C in DNA binding buffer (50 mm Tris-Cl, pH 7.5, 0.5 mm EDTA, 1 mm DTT, 10% glycerol) for 30 min at 25 °C. After incubation, 10 μl of the reaction mixture was applied onto an alkali-washed nitrocellulose membrane that was subsequently washed with 3 ml of ice-cold wash buffer (50 mm Tris-Cl, pH 7.5, 0.5 mm EDTA, 1 mm DTT) on a vacuum manifold. The membranes were dried and the radioactivity retained on the filters quantified by scintillation counting.
DNA Replication Assays
DNA polymerase activity was measured using either the primed 89-mer TG40 repeat or singly primed M13. Reaction mixtures with the oligonucleotide primer-template (20 μl) contained 40 mm HEPES-NaOH, pH 7.5, 100 μg/ml of BSA, 0.75 mm DTT, 10 mm magnesium acetate, 17.5 μm [α-32P]dATP (∼10,000 cpm/pmol), 50 μm dCTP, 5 pmol of the 89-mer TG40 repeat oligonucleotide hybridized to the 50-mer primer and other additions, as indicated in the legends. Reactions were incubated for 30 min at 37 °C and halted with 5 μl of stop solution (10% SDS, 0.5 m EDTA). DNA synthesis was measured by adsorption to DEAE-cellulose paper and/or visualized by autoradiography following denaturing 6 m urea, 10% PAGE.
Reaction mixtures (15 μl) using singly primed M13 contained 40 mm HEPES/NaOH, pH 7.5, 1 mm DTT, 100 μg/ml of BSA, 30 μm [α-32P]dATP (10–20,000 cpm/pmol), 100 μm each of dTTP, dGTP, and dCTP, 10 mm magnesium acetate, 10 fmol of singly primed M13 mp18, and the two-subunit Pol α (p180-p70) complex, as indicated. Reactions were incubated for 45 min at 37 °C and halted with 5 μl of stop solution. Incorporation was monitored by DEAE-cellulose paper adsorption and alkaline-agarose (1.1%) gel electrophoresis followed by autoradiography.
RNA Interference Protocol
HeLa cells (ATCC) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. Duplex siRNA (small interfering RNA) (21 bp) with 3′-dU overhangs targeting hCtf4, and a random sequence (AAT TCT CCG AAC GTG TCA CGT), which acted as a negative control, were synthesized by Dharmacon. The nucleotide sequence targeted by the hCtf4 siRNA was GAU CAG ACA UGU GCU AUU A. For transfections, HeLa cells were seeded at 60% confluency (2 × 106 cells) in 6-well plates and grown until ∼90% confluent. Transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs were introduced into cells at a final concentration of 50 nm. Cells were analyzed by flow cytometry, indirect immunofluorescence, and DNA fiber analyses 48 h post-transfection.
Flow Cytometry
To measure DNA synthesis during S-phase, cells were incubated with 10 μm BrdUrd for 90 min, harvested, washed once with PBS, and fixed with 70% ethanol for 2 h in the dark. After washing with PBS buffer (containing 0.5% BSA), DNA was denatured with 2 m HCl for 20 min, cells were washed with PBS buffer and then incubated in 0.1 m sodium borate for 2 min. Cells were then incubated for 30 min in the dark with 10 μl of BD Pharmingen (catalogue number 556028) fluorescein isothiocyanate-conjugated BrdUrd antibody diluted with 90 μl of PBS containing 0.5% bovine serum albumin and 0.5% Tween 20. After several washes, DNA was stained with propidium iodide (10 μg/ml) in PBS containing 100 μg/ml of RNase A for 15 min. Fluorescence-activated cell sorter and data analyses were carried out with a FACS Calibur Cytometer and CellQuest software, respectively.
Immunofluorescence Staining
HeLa cells (1.5 × 105 cells), grown in a LAB-TEK chamber slide (Nalge), were incubated in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum at 37 °C for 24 h and then transfected with either hCtf4 or random siRNA. Cells were then incubated with 10 μm BrdUrd for 90 min, fixed in 4% paraformaldehyde for 20 min at room temperature, washed once in cold PBS, and then permeabilized with 0.3% Triton X-100 in PBS for 10 min. After incubation in blocking buffer (3% BSA and 0.3% Triton X-100 in PBS) for 30 min, cells were incubated with rabbit antibodies to hCtf4 (1:100 in blocking buffer) for 60 min at room temperature. Cells were washed 3 times with PBS and then incubated with 1:300 Alexa Fluor 594-conjugated goat anti-rabbit antibody (Invitrogen) in blocking buffer for 60 min. After 3 washes with PBS, cells were treated with 2 m HCl for 10 min at room temperature, washed once with cold PBS, and incubated with 1:500 anti-mouse monoclonal BrdUrd antibody (BD Biosciences) in blocking buffer. After two washes with PBS, cells were incubated for 3 min with 1:350 Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen) in blocking buffer for 30 min. For DNA staining, cells were incubated with 0.2 μg/ml of DAPI.
DNA Fiber Analyses
After siRNA transfection, cells were doubly labeled by incubating with 50 μm IdU for 20 min followed by incubation with 50 μm CldU for 20 min. DNA spreads were prepared as described previously (17) with some modifications. Briefly, 2 aliquots of cells, resuspended in ice-cold PBS at ∼1 × 106 cells/ml, were spotted onto a microscope slide and then overlaid with 15 μl of spreading buffer (0.5% SDS in 200 mm Tris-HCl, pH 7.4, 50 mm EDTA). After 6 min, the slides were tilted 15 degrees to allow lysates to slowly move down the slide, and the resulting DNA spreads were air dried, fixed in 3:1 mixture of methanol/acetic acid for 2 min at −20 °C, and stored at 4 °C overnight. The slides were then treated with 2.5 m HCl for 30 min, washed 3 times in PBS, incubated in blocking buffer (1% BSA in PBS containing 0.05% Tween 20) for 1 h followed by 1 h at room temperature with 1:500 rat anti-BrdUrd antibody (to detect CldU) (abCAM) plus 1:100 mouse anti-BrdUrd (to detect IdU) (BD Biosciences) diluted in blocking buffer. After rinsing 3 times with PBS, the slides were incubated for 30 min in 1:350 Alexa Fluor 594-conjugated goat anti-rat antibody and 1:350 Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen). Slides were rinsed three times with PBS, air dried, and mounted in Prolong Gold antifade reagent (Invitrogen). Microscopy was carried out using an Olympus AX70 microscope and the collected images processed using MetaMorph software.
RESULTS
Isolation of hCtf4
hCtf4 is a 126-kDa protein containing 1129 amino acids that includes three conserved domains: WD repeats (aa 1–427), SepB (aa 428–850), and HMG (aa 851–1139) (Fig. 1A). To characterize the properties of hCtf4, we expressed the full-length and truncated forms that contain various domains of hCtf4 in baculovirus-infected insect cells and purified them to >90% homogeneity (Fig. 1B). The hydrodynamic properties of hCtf4 and its truncated derivatives were assessed by size exclusion chromatography (Fig. 1C and data not shown) and glycerol gradient sedimentation (Fig. 1D and data not shown), and the results summarized in Fig. 1E. Based on the Monty-Siegel equation, which includes both the sedimentation value and the Stokes radius (18), the apparent molecular mass of the full-length hCtf4 was 262 kDa, approximately twice the predicted value derived from the known peptide sequence. These findings indicate that hCtf4 appears to be a dimer, similar to that reported for the Xenopus homologue (19). The domain responsible for dimerization of hCtf4 mapped to the SepB region. In keeping with this notion, the apparent molecular weights of hCtf4 and derivatives, including the SepB domain alone, truncated derivates WD + SepB and SepB + HMG, were approximately twice their calculated molecular weights (based on amino acid content). Although the apparent molecular weight for the HMG domain alone was ∼1.5-fold larger than its calculated mass, we classified it as a monomer. As shown in Fig. 1B, both the WD and HMG domains migrate aberrantly during SDS-PAGE analysis. WD is a highly acidic protein that could account for its slow migration but the reasons for the altered mobility of the HMG domain are unclear. Consistent with these observations, both the WD + SepB and SepB + HMG derivatives ran larger than their expected sizes. Based on these findings, we conclude that the SepB domain contributes importantly to the dimeric structure of hCtf4.
FIGURE 1.
Purity and properties of purified hCtf4 and its derivatives. A, structure and domains of full-length (FL) hCtf4. See text for details. B, SDS-PAGE analysis of hCtf4 derivatives. Various hCtf4 derivatives (0.5 μg), isolated as described under “Experimental Procedures,” were subjected to SDS-10% PAGE followed by Coomassie staining. The specific proteins loaded (indicated above each lane) were as follows: lane 1, full-length hCtf4; lane 2, WD domain; lane 3, SepB domain; lane 4, HMG domain; lane 5, WD + SepB; lane 6, SepB + HMG domain. C, elution profile of hCtf4 from a Superose 6 column. hCtf4 (6 μg) was loaded onto a Superose 6 PC 3.2/30 column equilibrated with 25 mm Tris-HCl, pH 7.5, 10% glycerol, 0.2 m NaCl, and 1 mm EDTA. The column was developed at a rate of 30 μl/min at 4 °C and fractions (50 μl) collected. Aliquots (20 μl) were then subjected to SDS-PAGE and Coomassie staining. D, glycerol gradient sedimentation of hCtf4. SP-Sepharose-isolated hCtf4 (70 μg) was sedimented through a 5-ml 15–40% glycerol gradient for 18 h at 250,000 × g at 4 °C. Fractions were subjected to 10% SDS-PAGE and Coomassie staining. E, characterization of the hydrodynamic properties of hCtf4 and its derivatives. The hydrodynamic properties of each isolated protein were, including Stokes radii, s values, apparent molecular weight, and predicted molecular weight of hCtf4 derivatives. The results presented were derived as previously described (27).
hCtf4 Binds Preferentially to DNA Primer-Template Structures
Because previous findings revealed that Ctf4 was associated with chromatin and various replisome proteins, we examined its ability to bind to DNA directly using a mobility shift assay. A 5′-32P-labeled 94-mer hairpin C DNA (3′-recessed Hairpin C), containing a 39-nucleotide single-stranded (ss) region, a 24-bp duplex region, and a 4-nucleotide single strand loop, was used for DNA binding studies (Fig. 2A). The results indicated that hCtf4 bound to the 3′-recessed hairpin C DNA (Fig. 2B) with an apparent association constant of 1.5 × 107 m−1. Near quantitative binding of the labeled DNA required ∼50–100-fold molar excess of hCtf4. For comparison, the RPA binding association constant to ssDNA is reported to be ∼1010 m−1 (20). We noted that formation of the hCtf4-DNA complex was salt sensitive. The experiment described in Fig. 2B was carried out in the presence of 50 mm NaCl. The level of complex formed was reduced 50% at a higher NaCl concentration (200 mm) (data not shown).
The binding of hCtf4 to different DNA structures was examined (Fig. 2C). For this purpose, hCtf4 binding to the 5′-32P-labeled 94-mer 3′-recessed hairpin C DNA was carried out in the presence of increasing competitor levels of unlabeled ss- (39-nucleotide), ds-hairpin (lacking the ssDNA region), the 5′-recessed 94-mer hairpin C DNA (containing the identical sequence as the 3′-recessed 94-mer but with opposite phosphodiester polarity) and the 3′-recessed 94-mer hairpin C DNAs (see Fig. 2A for structures of competitors). As shown, hCtf4 binding to the labeled 3′-recessed 94-mer hairpin C DNA was competed effectively by unlabeled 5′- and 3′-recessed hairpin C DNAs but not by the ss- or dsDNAs. In the absence of competitor, hCtf4 bound labeled ssDNA and dsDNA weakly (<105 m−1, data not shown). Thus, hCtf4 binds preferentially to double-stranded/single-stranded DNA junction structures but does not differentiate between a 3′- or 5′-recessed DNA end.
Using nitrocellulose filter binding assays, we examined the DNA binding properties of the purified hCtf4 derivatives containing the WD, SepB, HMG, WD + SepB, and SepB + HMG domains (to the 5′-32P-labeled 3′-recessed hairpin C DNA). As shown in Fig. 2D, the HMG region alone bound DNA more efficiently than the other truncated proteins, although the DNA binding activity of full-length hCtf4 was significantly greater than that observed with the different derivatives at all concentrations examined. These findings suggest that each hCtf4 domain contributes to the overall DNA binding property of the full-length protein. Previous studies reported that the cloned HMG domain of Xenopus Ctf4 bound DNA structures resembling Holiday junctions (19). The efficiency of this interaction, however, was not reported.
hCtf4 Interacts with Replicative DNA Polymerases
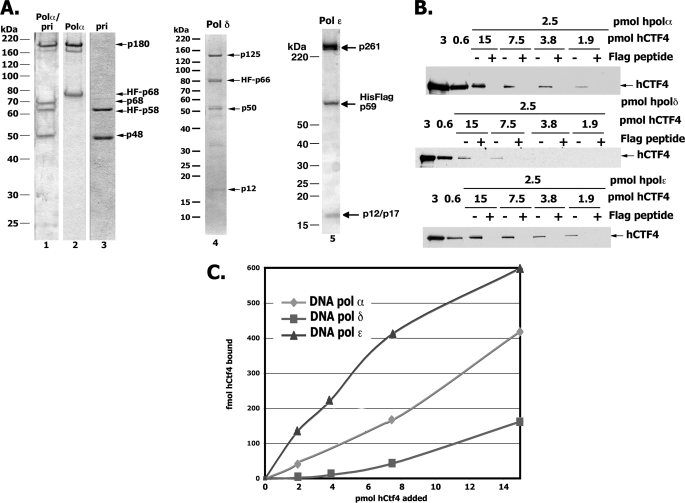
Previous reports indicated that Ctf4 interacted with Pol α of yeast, Xenopus, and humans (6, 10, 13). We examined whether interactions could be detected between purified hCtf4 and DNA polymerase holoenzymes α, δ, and ϵ (Fig. 3A). For this purpose, each polymerase, containing a FLAG tag fused to one of its noncatalytic subunits, was mixed with increasing amounts of hCtf4 and the interactions assessed by FLAG immunoprecipitation followed by Western blot analyses with anti-hCtf4 specific antibodies. As shown in Fig. 3B, hCtf4 interacted weakly with all of the replicative polymerase, albeit with different affinities. Under the conditions used, hCtf4 (7.5 pmol) bound to Pol ϵ 2.5- and 8-fold more efficiently than to Pol α/primase and Pol δ, respectively (Fig. 3C). Its interaction with the Pol α-primase complex appears to be limited to Pol α (p180/p70) because binding to the primase subcomplex (p58/p48) was not detected (data not shown).
FIGURE 3.
hCtf4 interacts with replicative Pols α, δ, and ϵ. A, Coomassie-stained SDS-10% polyacrylamide gel of purified DNA polymerases used in this study: lane 1, four-subunit Pol α-primase complex, 1.0 μg; lane 2, two-subunit Pol α, 0.6 μg; lane 3, two-subunit primase, 0.5 μg; lane 4, Pol δ, 0.8 μg; lane 5, DNA Pol ϵ, 1.5 μg. The subunits of each DNA polymerase are labeled. B, indicated levels of FLAG-tagged polymerases and untagged hCtf4 were incubated in reaction mixtures as described under “Experimental Procedures.” FLAG-antibody precipitated material was then resolved by SDS-10% PAGE followed by Western blot analyses using specific hCtf4 antibodies. “+” lanes denote reaction mixtures precipitated with FLAG antibodies in the presence of 1 mg/ml of FLAG peptide, whereas “−” lanes indicate precipitations carried out in the absence of FLAG peptide. C, quantification of results obtained in B.
In addition to its interaction with the replicative polymerases, hCtf4 also interacted with the four-subunit hGINS and two-subunit (Sld5-Psf2) complexes, as well as hMcm10. These observations were made both in vivo and in vitro (data not shown, and Ref. 13). The findings that hCtf4 can bind to proteins involved in the DNA unwinding reaction (GINS complexed to Cdc45 and Mcm2–7) and synthesis of DNA (DNA Pols) suggest that it is likely to play important roles at the replication fork.
hCtf4 Stimulates Pol α Activity
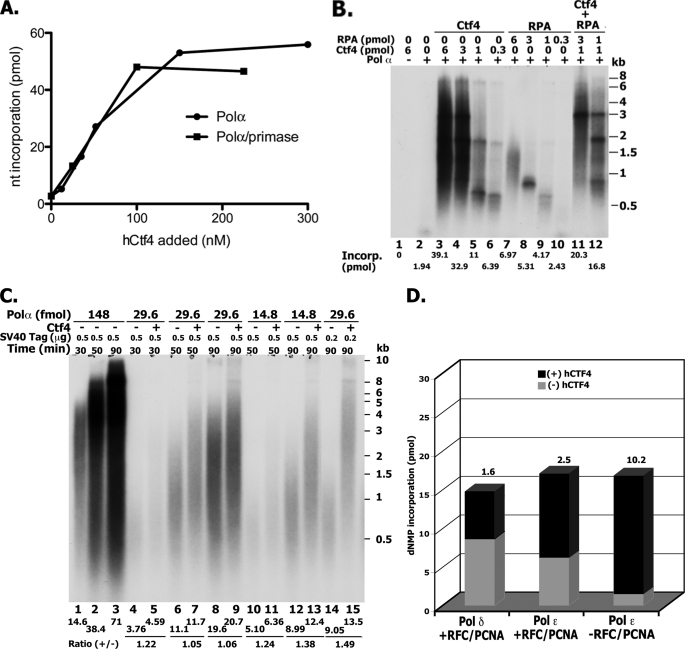
We examined the influence of hCtf4 on the enzymatic activities associated with the Pol α-primase complex. As shown in Fig. 4A, hCtf4 stimulated [α-32P]dATP incorporation (in the presence of dCTP) in the presence of the 89-mer TG40 template-primer and Pol α (either the four-subunit (Pol α-primase) or two-subunit complex). DNA synthesized in the presence of saturating levels of hCtf4 (200 nm) was 15–20-fold greater than in its absence; maximal stimulation was observed at ∼120 nm hCtf4 with ½ Vmax of 60 nm. Denaturing PAGE analyses showed that the fully elongated product (89-mer) was formed by either the four- or two-subunit Pol α complex in the presence of hCtf4 (data not presented).
FIGURE 4.
Influence of hCtf4 on Pol α. A, stimulation of Pol α polymerization activity using the 89-mer TG40 oligonucleotide as primer-template. Reactions were carried out as described under “Experimental Procedures,” hCtf4 as indicated and either the four-subunit Pol α-primase complex (4.7 fmol, ■) or two-subunit (p180-p70) Pol α complex (4.2 fmol, ●). After 30 min at 37 °C, aliquots were subjected to DEAE-cellulose paper adsorption, salt washing, and then counted. B, stimulation of Pol α activity using singly primed M13 DNA as template. Reactions containing 29.6 fmol of the p180-p70 Pol α complex and other proteins, as indicated, were carried out as described under “Experimental Procedures.” C, effect of hCtf4 on SV40 DNA replication catalyzed by Pol α. Reaction mixtures (20 μl) containing 40 mm creatine phosphate (diTris salt), pH 7.8, 4 mm ATP, 7 mm magnesium acetate, 2 mm DTT, 75 μg/ml of BSA, 15 μm [α-32P]dATP (10,500 cpm/pmol), 100 μm each of dCTP, dTTP, and dGTP, 0.75 μg of creatine phosphokinase, 150 μm each of GTP, CTP, and UTP, 30 fmol of Topo I, 26.1 fmol of pSLVD ori+ DNA, 6.25 or 2.5 pmol of SV40 T antigen monomer (as indicated), 2.2 pmol of RPA, 0.15 pmol of hDNA primase (p58-p45), 200 nm hCtf4 (where indicated), and varying levels of the p180-p70 Pol α (as indicated) were incubated at 37 °C for 30, 50, or 90 min. Aliquots were used to determine nucleotide incorporation and size of products (1% alkaline-agarose gel electrophoresis). D, influence of hCtf4 on Pol ϵ and δ. Reactions were carried out as described in A using the TG40 repeat oligonucleotide as primer-template with the following additions: reactions with the four-subunit Pol ϵ (30 fmol) or the four-subunit Pol δ (50 fmol) contained 50 mm potassium glutamate and where indicated 100 nm hCtf4, 600 fmol of hPCNA, and 45 fmol of hRFC.
The stimulation of Pol α activity by hCtf4 was also observed using singly primed M13 DNA as the template-primer. As shown, hCtf4 stimulated Pol α-catalyzed DNA synthesis ∼20-fold (Fig. 4B, compare lanes 2 and 3) and the two- and four-subunit Pol α complexes behaved identically (Fig. 4A and data not shown). Maximal stimulation was observed with ∼350 nm hCtf4 and a ½ Vmax was obtained at ∼80 nm. Because RPA can stimulate Pol α activity, its effects on the replication reaction were compared with hCtf4 (Fig. 4B, lanes 7–10). In this case, the DNA template was fully covered by RPA based on the previously determined binding capacity of one molecule of RPA for a single-stranded region of ∼20 nucleotides (21). As shown, although RPA stimulated the polymerase activity, both the extent of the stimulation as well as size of the products formed were substantially lower than that detected with hCtf4. When both hCtf4 and RPA were added together, the level of DNA synthesized was slightly more than the sum observed with each alone (Fig. 4B, lanes 11 and 12). However, the size of DNA chains produced was increased significantly. These findings suggest that hCtf4, which binds to both primer-template ends and Pol α, may facilitate and/or stabilize the interaction of the polymerase with primer ends, resulting in enhanced processivity. We conjecture that RPA may facilitate the translocation of Pol α by reducing structural impediments such as hairpin structures in the M13 template. In support of this notion, in the presence of hCtf4 an increased level of RPA (Fig. 4B, compare lanes 11 and 12) resulted in products with less visible “stop sites.”
We also examined whether hCtf4 stimulated the SV40 DNA replication reaction (Fig. 4C). The SV40 DNA replication system is a well studied replication system that uses the replication proteins of the host and a single viral-encoded protein, T antigen, which binds to the SV40 origin and unwinds the duplex (22). Our experiments were carried out in the presence of SV40 T antigen, DNA containing the SV40 core origin (pSLVD, 8 kbp), cloned DNA primase complex (p58-p45), RPA, and variable levels of the two-subunit Pol α complex (p180-p70) in the presence or absence of hCtf4. Under these conditions, DNA synthesis is catalyzed only by Pol α (the monopolymerase reaction) (23). As shown, relatively high levels of Pol α (148 fmol) supported extensive DNA synthesis that increased with time (Fig. 4C, lanes 1–3). At this level of Pol α, hCtf4 addition had no discernable effect on either the amount or size of DNA products synthesized (data not shown). At low levels of Pol α and T antigen, the presence of hCtf4 increased DNA synthesis marginally (up to ∼50%, compare lanes 14 and 15) but increased the mean length of DNA chains synthesized significantly, from 0.75 kb to more than 2 kb (compare lanes 14 and 15).
The possibility that the effects of hCtf4 were due to the stimulation of the SV40 T antigen helicase and/or DNA primase activities were examined. No increase in either of these activities was detected (data not shown). Furthermore, we noted that hCtf4 did not stimulate SV40 replication in reactions in which the monopolymerase system was supplemented with Pol δ, RFC, and PCNA (reconstituted dipolymerase system). This may be due to the marked increase in DNA synthesis and size of chains produced by the dipolymerase system compared with the monopolymerase reaction. Together, the above results suggest that hCtf4 can increase the activity of Pol α in a coupled unwinding-replication system.
hCtf4 Stimulates Pol ϵ but Pol δ Poorly
We examined the influence of hCtf4 on the polymerase activities of Pol δ and Pol ϵ. In the presence of the 89-mer TG40 oligonucleotide primer-template, the four-subunit hPol ϵ complex catalyzed significant levels of DNA synthesis in the absence of RFC, PCNA, and a DNA-binding protein. The addition of hCtf4 to these reactions increased DNA synthesis ∼10-fold compared with that observed in its absence (Fig. 4D). Under the conditions used, maximal stimulation occurred with 300 nm hCtf4 and a ½ Vmax of ∼60 nm. Supplementing reactions with RFC and PCNA further increased the activity of Pol ϵ 4-fold and the addition of hCtf4 and RFC + PCNA stimulated DNA synthesis ∼2.5-fold over that observed without hCtf4. The addition of either RPA or Escherichia coli SSB had no effect on the level of DNA synthesized. Similar experiments were carried out with hPol δ. In contrast to Pol ϵ, under the conditions used, Pol δ catalyzed detectable levels of DNA synthesis only in the presence of RFC and PCNA; hCtf4 addition stimulated DNA synthesis ∼1.6-fold (Fig. 4D). In the absence of RFC and PCNA, DNA synthesis was not detected even in the presence of hCtf4. These findings indicated that both Pol ϵ and δ were marginally stimulated by hCtf4 in the presence of the clamp and clamp loader, whereas in their absence hCtf4 only stimulated Pol ϵ. To evaluate the influence of hCtf4 on DNA synthesis catalyzed by Pol δ and Pol ϵ on longer and more natural DNA templates, we examined its effect on the elongation of singly primed ϕX174 and M13 DNAs (data not shown). DNA synthesis on these templates by Pol δ and ϵ requires the action of both RFC and PCNA as well as the presence of a DNA-binding protein (either E. coli SSB or RPA). Under these conditions, significant enhancement of either Pol δ or ϵ activity by hCtf4 was not detected.
The findings described in Fig. 4 indicate that hCtf4 stimulates DNA synthesis catalyzed by Pol α and Pol ϵ. The stimulatory effects of hCtf4 on Pol ϵ activity were more pronounced in the absence of RFC and PCNA.
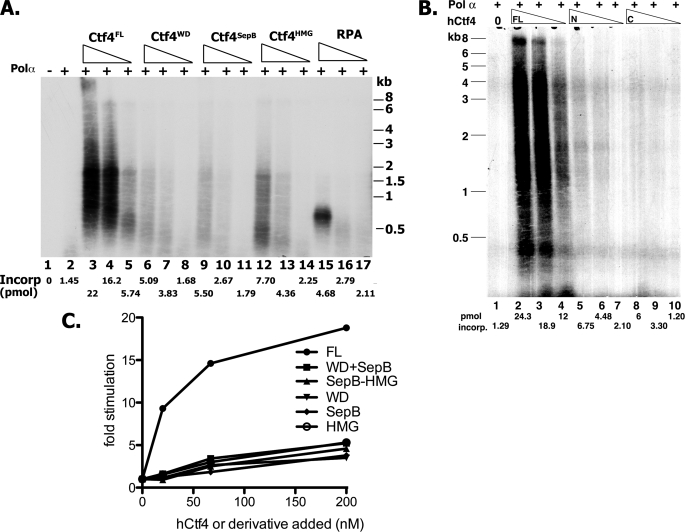
Influence of hCtf4 Domains on Pol α-catalyzed DNA Synthesis
It was previously shown that in human cells both full-length hCtf4 and an N-terminal truncated derivative (aa 330–1129) interacted with the p180 subunit of Pol α, as well as Mcm10, whereas the WD40 domain (aa 1–336) and the C terminus HMG region (aa 984–1129) did not (13). We investigated whether truncated proteins, each containing one of these domains, like full-length hCtf4, stimulated the Pol α polymerase activity using the singly primed M13 assay. None of these truncated preparations stimulated Pol α activity as effectively as full-length hCtf4 (Fig. 5A). Derivatives containing either the WD and SepB domains alone were ∼5–10-fold less active than full-length hCtf4 (compare lanes 6–11 to 3–5), whereas the HMG domain was slightly more active (∼20–30% of the activity detected with full-length hCtf4) (compare lanes 12–14 to 3–5). Derivatives containing two domains (WD + SepB and SepB + HMG) were as active as the truncated proteins with only one domain (Fig. 5, B and C). These findings suggest that the full-length protein is required for maximal stimulation of Pol α activity. The quantitative effects of hCtf4 reported above have been observed with different preparations of both hCtf4 (N-terminal FLAG-tagged and untagged) and DNA polymerase.
FIGURE 5.
Influence of various hCtf4 derivatives on the activity of Pol α. A, effect of individual hCtf4 domains and hRPA. Reaction mixtures, as described in the legend to Fig. 4B, contained 15.2 fmol of p180-p70 Pol α complex and 200, 67, and 20 nm of either full-length (FL) hCtf4, or the indicated truncated derivatives and hRPA. After 45 min at 37 °C, aliquots were used to measure nucleotide incorporation and subjected to alkaline-agarose gel electrophoresis. B, effects of the WD-SepB and SepB-HMG derivatives. Reaction mixtures, as described in A, with 200, 67, and 20 nm of the indicated hCtf4 derivatives were used. C, quantification of total dNMP incorporation in A and B.
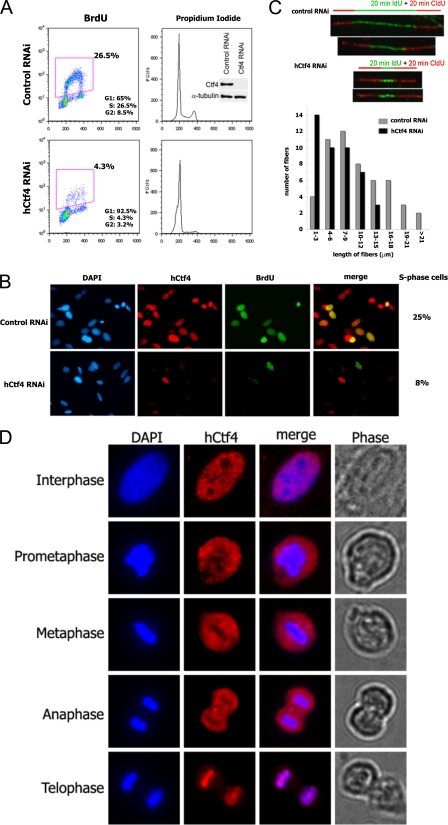
hCtf4 Is Required for S-phase Progression
To evaluate the function of hCtf4 in S-phase, asynchronous HeLa cells were transfected with siRNA to deplete hCtf4. This treatment reduced the level of hCtf4 >90% based on Western blot analyses (Fig. 6A, inset) and resulted in a 6-fold decrease in BrdUrd incorporation into cells (4.31% in hCtf4 siRNA-treated versus 26.5% in control siRNA-treated cells). Furthermore, propidium iodide staining of cells showed that, unlike control siRNA-treated cells, hCtf4-depleted cells were arrested predominantly at the G1/S phase demonstrating that hCtf4 is required for S-phase completion (Fig. 6A). Cells depleted of hCtf4 remained in the G1/S phase for at least 48 h post-transfection, whereas control cells progressed through the cell cycle (data not shown). Similar results were obtained with four siRNAs that targeted different regions of hCtf4 mRNA.
FIGURE 6.
Depletion of hCtf4 inhibits DNA replication in vivo. HeLa cells were transfected with hCtf4 RNA interference (RNAi) or control nonspecific RNAi for 48 h as described under “Experimental Procedures.” A, flow cytometry analyses of BrdUrd incorporation, DNA content, and cell cycle distribution of cells following siRNA treatments. The cell cycle distribution of HeLa cells after siRNA transfections are indicated. Whole cell extracts (30 μg) were analyzed for the depletion of hCtf4 following treatment with the siRNAs using hCtf4-specific antibodies. Anti-α-tubulin was used as a loading control (inset). B, indirect immunofluorescence of HeLa cells treated with either hCtf4 RNAi or control siRNA. Cells were stained with anti-hCtf4 antibody (red, center left panels) and BrdUrd (green, center right panels). DNA was counterstained with DAPI (left panels). Merged images of hCtf4 and BrdUrd staining are shown in the right panels. The percentage of S-phase cells, determined by counting BrdUrd positive cells in the control cells (25%) and hCtf4 RNAi-treated cells (8%), is shown. C, examination of the movement of replication fork in HeLa cells treated with control siRNA or hCtf4 siRNA. Cells were incubated for 20-min with IdU followed by a 20-min pulse incubation with CldU. In the upper panel, individual replication units were visualized by immunofluorescence of the incorporated halogenated nucleotides in isolated DNA fibers, as described under “Experimental Procedures.” The number of fibers for each specified length originating from new origin initiations (IdU incorporation) in hCtf4 RNAi-treated HeLa cells were compared with control siRNA-treated cells (lower panel). The data were derived from one of three independent experiments in which >100 fibers were analyzed per experiment. D, subcellular localization of hCtf4 during the cell cycle. Immunofluorescence staining showing the distribution of hCtf4 (red) in interphase, prometaphase, metaphase, anaphase, and telophase. HeLa cells were stained with hCtf4 antibodies and DNA was counterstained with DAPI (blue). Merged images show superimposed hCtf4 (red) and DAPI (blue) signals. Morphology of cells at different stages of cell cycle is shown in phase contrast images in the right panels.
Immunofluoresence staining of asynchronous cells depleted of hCtf4 also showed a significant decrease of hCtf4 expression and number of BrdUrd positive cells (Fig. 6B), in keeping with the immunoblot results shown in Fig. 6A. Notably, there was a population of siRNA-treated cells that exhibited low levels of hCtf4 yet were stained positive for BrdUrd. This suggests that DNA replication may require low levels of hCtf4.
We measured the influence of hCtf4 on the rate of DNA fork progression by DNA fiber analyses. This technique utilizes immunofluorescence microscopy to visualize the rate of fork progression. HeLa cells were incubated for 20 min with IdU and then with CldU for 20 min, followed by treatment with specific antibodies and fluorescent tags. After cell lysis and DNA spreading, fibers containing both green IdU fluorescent label flanked on each side with the red CldU fluorescent label represent DNA molecules that were replicated from origins (green label) and subsequently elongated bidirectionally (red label).
Fig. 6C presents representative DNA fibers isolated from wild-type and hCtf4-depleted cells. The green IdU label, indicating DNA synthesis from replication origins, was shorter in cells partially depleted of hCtf4 than from fibers isolated from siRNA control cells. Furthermore, the combined length of red CldU- and green IdU-stained fibers were shorter in hCtf4-depleted cells than in control cells, indicating that decreased levels of hCtf4 resulted in slower fork movement. Based on the measurements of 100 individual DNA fibers isolated from each cell sample, the majority of the green-labeled IdU fibers from hCtf4-depleted cells were 1–3 μm in length, whereas those from control cells were 7–9 μm in length (Fig. 6C). Quantification of the doubly labeled fibers indicated that the average rate of fork progression in hCtf4-depleted cells was 1.5-fold slower than that observed in control cells.
Changes in the Nuclear Localization of hCtf4 during Mitosis
The subcellular localization of hCtf4 was monitored during the cell cycle. Although the level of hCtf4 did not change during the cell cycle (data not shown), immunostaining analyses revealed dramatic changes in nuclear localization of hCtf4 (Fig. 6D). In these experiments, DAPI staining was used to visualize the level of chromosomal condensation, whereas phase contrast was employed to follow cell morphology during the cell cycle. At interphase (the period of active replication) when chromatin is not condensed and the nuclear envelope is intact, hCtf4 was localized to the nucleus but widely dispersed within the nucleoplasm. In prometaphase, when the nuclear envelope has dissolved and cells assume a rounder shape and chromosomes start to condense, hCtf4 remained dispersed but was not associated with the condensed chromosomes. During metaphase and anaphase, when chromosomes line up on the equatorial plate, and chromosomes begin to move to the opposite poles of the cell, but the cell has not divided into two daughter cells, the condensed chromatin was still devoid of hCtf4. At the end of anaphase, however, when chromosomes have been pulled apart and are localized in two daughter cells, hCtf4 was dramatically associated with the separated chromatids. Thus, during mitosis, hCtf4 is displaced from mitotic chromatin but rapidly re-associates with the daughter chromatids at telophase. The significance of these changes is described in more detail below.
DISCUSSION
In this report, we describe the isolation of hCtf4 and characterization of some of its biochemical and biological properties. Based on its hydrodynamic properties, hCtf4 (126 kDa monomer) appears to be a homodimeric protein. hCtf4 is a member of a conserved family of proteins, including the budding yeast and human Ctf4, SepB of Aspergillus nidulans and S. pombe Mcl1, which are required for multiple chromosome dynamics (8, 11, 24). Members of this family contain either two or three recognizable motifs, including an N-terminal WD40 domain (of variable repeats) followed by three highly conserved SepB boxes. Higher eukaryotes include a C-terminal HMG AT-hook DNA-binding domain that is missing in lower eukaryotes. SepB, Mcl1, and hCtf4 all possess five to seven degenerate WD40 repeats (β-propeller domains), whereas ScCtf4 contains only a single WD40 repeat. However, all family members harbor a Sep domain (∼300 aa) within their central region. Protein structure algorithms predict that the SepB domain can form a four-bladed β-propeller (24). Thus, hCtf4, the A. nidulans SepB, and Mcl1 family members contain multiple β-propeller domains likely to mediate interactions with a number of proteins. In keeping with this notion, interactions between hCtf4, Mcm10, GINS, Pol α, and other polymerases, as reported here, have been detected. We also noted that the SepB domain (but not the WD40 domain) contributes to the dimerization of hCtf4. All hCtf4 derivatives described here, containing the SepB domain (full-length hCtf4, SepB alone, WD40 + SepB, and SepB + HMG), were dimeric in structure, whereas derivatives lacking this domain were monomers.
We examined the DNA binding properties of full-length hCtf4 and derivatives containing one or two contiguous domains. Full-length hCtf4 bound to the 94-mer hairpin C DNA containing two relatively short dAT regions. DNA binding (shown by a mobility shift and nitrocellulose assays) was observed only at high molar ratios of hCtf4:DNA (>10) (Fig. 2), suggesting that hCtf4 binds to DNA weakly. Under the conditions described in Fig. 2, primer-template DNA structures (containing either 3′- or 5′-recessed ends) were bound preferentially, whereas duplex or single-stranded DNA was not. These properties suggest that hCtf4 could help recruit and/or stabilize replication proteins, such as polymerases, to primer-template junctions.
In addition to its identification in budding yeast by genetic screens for mutants that affected chromosome transmission fidelity (7, 9), Ctf4 was also isolated as a Pol α-binding protein by affinity chromatography (10). More recent results indicate that Ctf4, in conjunction with Mcm10, plays a role in the loading of Pol α onto the activated replisome complex (13). The availability of highly purified hCtf4 and replicative polymerases prompted us to examine their interactions. Interactions between hCtf4 and hPol ϵ (four-subunit complex) and hPol α were detected, whereas weak interaction with Pol δ (four-subunit complex) was also observed. At stoichiometric levels, hCtf4 bound to Pol ϵ more efficiently than to Pol α (∼3-fold) and to Pol δ poorly (∼10-fold). These interactions appear functional because low levels of hCtf4 markedly stimulated the polymerase activities of both Pol α and Pol ϵ but marginally affected Pol δ activity. We failed to detect any physical or functional interactions between hCtf4 and the purified primase complex (p58-p48). The polymerase activities of the four- and two-subunit Pol α complexes were stimulated identically by hCtf4 suggesting that the primase subunits play no role in this activation.
All truncated hCtf4 derivatives examined stimulated the Pol α-catalyzed elongation of single-primed M13 DNA less effectively than the full-length protein. Zhu et al. (13) reported that in vivo both full-length hCtf4 and the truncated derivative containing both the SepB and HMG domains interacted with the p180 subunit of Pol α and Mcm10, whereas the WD40 domain alone did not. In budding yeast, Ctf4 lacking the WD40 domain supported Pol α binding to the RPC in vivo (6). Although these in vivo findings as well as the in vitro observations reported here reveal that Pol α is an important target of full-length Ctf4, it is unclear how these functional interactions affect replication.
In addition, hCtf4 markedly stimulated the activities of Pol ϵ. These experiments were carried out using an oligonucleotide-primed template that Pol ϵ elongated efficiently in the absence of PCNA and RFC. In contrast, Pol δ was inactive with this template in the absence of the clamp and clamp loader. In their presence, elongation of the oligonucleotide primer-template by Pol ϵ was stimulated ∼4-fold and the addition of hCtf4 increased this activity 2-fold. Reactions with Pol δ plus RFC and PCNA were also increased marginally by hCtf4. Elongation of singly primed M13 (or ϕx) by Pol δ and ϵ are totally dependent on the presence of RFC, PCNA, and a DNA-binding protein (SSB or RPA). In their presence, hCtf4 did not affect the polymerase activities of Pol δ and ϵ. The reasons for the discrepancy between oligonucleotide and longer natural M13 DNA templates are presently unclear. We conjecture that the selective binding of hCtf4 at primer-template junctions may compete with the DNA-mediated interactions between PCNA and Pol δ and ϵ. Additional factors might be required for hCtf4 activity in the presence of RFC/PCNA and on longer natural templates like singly primed M13. The hCtf4-mediated stimulation of hPol ϵ, which was significantly effective on oligonucleotides template-primers in the absence of PCNA/RFC, was reduced by their presence. These findings suggest that hCtf4, like the clamp-clamp loader complex, increases the association of Pol α and ϵ with primer ends. Mcm10, which markedly stimulates the activity of Pol α in vitro, was previously shown to act in this manner (25).
Depletion of hCtf4 in HeLa cells markedly reduced DNA replication and halted cell cycle progression by arresting cells at the G1/S phase. A small percentage of cells, presumably containing low levels of hCtf4, carried out limited DNA replication. The rate of fork progression of such cells, measured by DNA fiber analyses, was 70% of that observed in control cells. Recent studies in human cells revealed that the formation of the putative replicative helicase, minimally containing a complex of Cdc45, Mcm2–7, and GINS, required hCtf4 (as well as RecQL4 and Mcm10) (15). These findings suggest that hCtf4 may play an important role in replisome formation as well as fork progression.
Recent studies from Labib and co-workers (6) demonstrated that the association of Pol α with the RPC in budding yeast is dependent on Ctf4, which is a component of the RPC. Although ScCtf4 is not essential in budding yeast, it is essential in fission yeast and higher eukaryotes (Refs. 11, 13, and 14, and this study). In budding yeast, as in higher eukaryotes, Pol α was destabilized and the chromosomal distribution of the Mcm2–7 complex and Pol ϵ was widely dispersed upon depletion of Ctf4 (14). These findings suggest that Ctf4 plays an important role connecting the replicative helicase and replicative polymerases. Based on the properties of hCtf4 described here and by others, we propose a model in which hCtf4 plays an important role at the replication fork (Fig. 7). In this model, we suggest that through its interactions with the polymerases and GINS, hCtf4 influences the activities of the polymerases at the fork and contributes to the regulation of fork progression. We posit that the interaction of hCtf4 with GINS connects the action of the polymerases with the CMG-helicase activity and coordinates the movement of both the helicase and rate of polymerization. The interactions between hCtf4 and polymerases (particularly Pol ϵ on the leading strand and Pol α on the lagging strand) coordinate leading and lagging strand synthesis. Due to its dimeric structure, hCtf4 may interact with multiple polymerase simultaneously. These considerations prompted us to examine whether full-length hCtf4 could bind two different polymerase at the same time. In these experiments various combinations of two different polymerase were incubated with hCtf4, followed by selective immunoprecipitation of a specific polymerase. Although hCtf4 was immunoprecipitated in all experiments, we failed to detect the presence of two different polymerases (data not shown). It may be that such interactions require other replication fork components as well as DNA.
FIGURE 7.
Model for hCtf4 role at the replication fork. See text for detail.
Ctf4 and its family of proteins play critical roles in chromatin remodeling, chromosomal partition, and sister chromatid cohesion. Ctf4 has been identified as a cohesion establishment factor that in conjunction with other RPC-associated proteins (including Ctf18, Ctf7, etc.) support sister chromatid pairing during S phase. It is likely that this property is related to its important role in maintaining fork progression and DNA replication.
We examined the subcellular localization of hCtf4 during the normal HeLa cell cycle. Although no significant alterations in the hCtf4 level were noted, the nuclear distribution of hCtf4 changed markedly. In interphase, when chromosomes are associated with proteins involved in replication, transcription, recombination, and DNA damage repair, hCtf4 and chromosomes were co-distributed widely within the nucleus. As cells progressed through the mitotic cycle, the association of hCtf4 with chromosomes was lost, specifically between prometaphase and metaphase. By late anaphase and early telophase, after chromosome segregation, hCtf4 was quantitatively associated with segregated and condensed chromosomes. The underlying mechanism contributing to this dramatic shift is unknown. Recently, Yanagida (26) proposed that chromosome condensation, catalyzed by the condensin complex in conjunction with DNA topoisomerase II as well as separase (which removes cohesin complexes from chromosomes), results in the removal of proteins during mitosis that might inhibit or reduce the fidelity of chromosome segregation. Our data suggest that hCtf4 is likely to be among the proteins removed from chromatin during mitosis and thus not required during this stage of the cell cycle. However, the re-association of hCtf4 with chromatin during telophase and the beginning of a new cell cycle is in keeping with its critical role in subsequent chromosome dynamics.
Footnotes
- RPC
- replisome progression complex
- Pol
- polymerase
- AND-1
- acidic nucleoplasmic DNA-binding protein 1
- BrdUrd
- bromodeoxyuridine
- Ctf4
- chromosome transmission fidelity 4
- PBS
- phosphate-buffered saline
- DTT
- dithiothreitol
- BSA
- bovine serum albumin
- siRNA
- small interfering RNA
- HMG
- high mobility group
- DAPI
- 4′,6-diamidino-2-phenylindole
- dsDNA
- double-stranded DNA
- ssDNA
- single-stranded DNA
- PCNA
- proliferating cell nuclear antigen
- aa
- amino acid
- SSB
- single-stranded DNA-binding protein
- h
- human
- MES
- 4-morpholineethanesulfonic acid
- IdU
- iododeoxyuridine
- CldU
- chlorodeoxyuridine.
REFERENCES
- 1.Sclafani R. A., Holzen T. M. (2007) Annu. Rev. Genet. 41, 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Hatten R. A., Tutter A. V., Holway A. H., Khederian A. M., Walter J. C., Michael W. M. (2002) J. Cell Biol. 159, 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wohlschlegel J. A., Dhar S. K., Prokhorova T. A., Dutta A., Walter J. C. (2002) Mol. Cell 9, 233–240 [DOI] [PubMed] [Google Scholar]
- 4.Schlesinger M. B., Formosa T. (2000) Genetics 155, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 6.Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna J. S., Kroll E. S., Lundblad V., Spencer F. A. (2001) Mol. Cell. Biol. 21, 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouprina N., Kroll E., Bannikov V., Bliskovsky V., Gizatullin R., Kirillov A., Shestopalov B., Zakharyev V., Hieter P., Spencer F., et al. (1992) Mol. Cell. Biol. 12, 5736–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouprina N., Tsouladze A., Koryabin M., Hieter P., Spencer F., Larionov V. (1993) Yeast 9, 11–19 [DOI] [PubMed] [Google Scholar]
- 10.Miles J., Formosa T. (1992) Mol. Cell. Biol. 12, 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams D. R., McIntosh J. R. (2002) Eukaryot. Cell 1, 758–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka H., Kubota Y., Tsujimura T., Kumano M., Masai H., Takisawa H. (2009) Genes Cells 14, 949–963 [DOI] [PubMed] [Google Scholar]
- 13.Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., Dutta A. (2007) Genes Dev. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka H., Katou Y., Yagura M., Saitoh K., Itoh T., Araki H., Bando M., Shirahige K. (2009) Genes Cells 14, 807–820 [DOI] [PubMed] [Google Scholar]
- 15.Im J. S., Ki S. H., Farina A., Jung D. S., Hurwitz J., Lee J. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bermudez V. P., Maniwa Y., Tappin I., Ozato K., Yokomori K., Hurwitz J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10237–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson D. A., Pombo A. (1998) J. Cell Biol. 140, 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 19.Köhler A., Schmidt-Zachmann M. S., Franke W. W. (1997) J. Cell Sci. 110, 1051–1062 [DOI] [PubMed] [Google Scholar]
- 20.Kim C., Wold M. S. (1995) Biochemistry 34, 2058–2064 [DOI] [PubMed] [Google Scholar]
- 21.Blackwell L. J., Borowiec J. A. (1994) Mol. Cell. Biol. 14, 3993–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waga S., Bauer G., Stillman B. (1994) J. Biol. Chem. 269, 10923–10934 [PubMed] [Google Scholar]
- 23.Eki T., Matsumoto T., Murakami Y., Hurwitz J. (1992) J. Biol. Chem. 267, 7284–7294 [PubMed] [Google Scholar]
- 24.Harris S. D., Hamer J. E. (1995) EMBO J. 14, 5244–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fien K., Cho Y. S., Lee J. K., Raychaudhuri S., Tappin I., Hurwitz J. (2004) J. Biol. Chem. 279, 16144–16153 [DOI] [PubMed] [Google Scholar]
- 26.Yanagida M. (2009) Nat. Rev. Mol. Cell Biol. 10, 489–496 [DOI] [PubMed] [Google Scholar]
- 27.Bermudez V. P., MacNeill S. A., Tappin I., Hurwitz J. (2002) J. Biol. Chem. 277, 36853–36862 [DOI] [PubMed] [Google Scholar]