Abstract
The genome of extraembryonic tissue, such as the placenta, is hypomethylated relative to that in somatic tissues. However, the origin and role of this hypomethylation remains unclear. The DNA methyltransferases DNMT1, -3A, and -3B are the primary mediators of the establishment and maintenance of DNA methylation in mammals. In this study, we investigated promoter methylation-mediated epigenetic down-regulation of DNMT genes as a potential regulator of global methylation levels in placental tissue. Although DNMT3A and -3B promoters lack methylation in all somatic and extraembryonic tissues tested, we found specific hypermethylation of the maintenance DNA methyltransferase (DNMT1) gene and found hypomethylation of the DNMT3L gene in full term and first trimester placental tissues. Bisulfite DNA sequencing revealed monoallelic methylation of DNMT1, with no evidence of imprinting (parent of origin effect). In vitro reporter experiments confirmed that DNMT1 promoter methylation attenuates transcriptional activity in trophoblast cells. However, global hypomethylation in the absence of DNMT1 down-regulation is apparent in non-primate placentas and in vitro derived human cytotrophoblast stem cells, suggesting that DNMT1 down-regulation is not an absolute requirement for genomic hypomethylation in all instances. These data represent the first demonstration of methylation-mediated regulation of the DNMT1 gene in any system and demonstrate that the unique epigenome of the human placenta includes down-regulation of DNMT1 with concomitant hypomethylation of the DNMT3L gene. This strongly implicates epigenetic regulation of the DNMT gene family in the establishment of the unique epigenetic profile of extraembryonic tissue in humans.
Keywords: Development Differentiation/Tissue, DNA/Methylation, DNA/Methyltransferase, Epigenetics, Gene Transcription, Extraembryonic Tissue, Placenta, Trophoblast
Introduction
DNA methylation is a major epigenetic modification regulating gene expression, silencing repetitive DNA elements, and maintaining chromosomal structures (such as centromeres) in vertebrates. Occurring exclusively at cytosine within CpG dinucleotides in mammals, DNA methylation is coordinated by a family of DNA methyltransferases (DNMTs)5 comprising DNMT1, -3A, -3B, and -3L. DNMT1 is the “maintenance” methyltransferase that ensures faithful transmission of methylation profile from maternal to daughter cells during cell division. In contrast, DNMT3A and -3B are de novo methyltransferases that show a degree of specificity in both target sequence and temporal activity (reviewed in Ref. 1). DNMT3L (DNMT3-like) lacks catalytic activity and has a very restricted gene expression profile (2). It associates directly with both DNMT3A and -3B to potentiate methyltransferase activity (3).
Formation of extraembryonic tissues, in particular the placenta, requires a coordinated series of epigenetic modifications that precisely regulate gene expression at key developmental time points (reviewed in Ref. 4). These tissues have a globally lower level of genomic 5-methylcytosine (5MeC) than somatic tissues (5–9). More recently, it has been demonstrated that the placenta also has a different repertoire of imprinted gene expression (expressed in a parent of origin specific manner) than somatic tissues (reviewed in Ref. 10) and methylation-induced silencing of tumor suppressor genes otherwise only seen in human cancers (11–14).
Mounting evidence suggests that disruption of placental development in the first trimester of pregnancy is involved in the etiology of disorders such as pre-eclampsia and intrauterine growth restriction (reviewed in Ref. 15). Despite the significance of this for pregnancy outcome and the demonstrated link between epigenetic modification and gene expression, little is known about the processes underlying the establishment of the placental epigenetic profile during early development. Given the link between DNMT enzyme activity, establishment and maintenance of the epigenome, and the potential for methylation-based regulation of DNMT gene expression, we tested for evidence of epigenetic regulation of DNMT genes (DNMT1, -3A, -3B, and -3L) by promoter methylation in human extraembryonic tissue.
EXPERIMENTAL PROCEDURES
Tissue Collection
Tissue collection for this study was approved by the Human Research Ethics Committees at the Royal Women's Hospital (03/51), Mercy Hospital for Women (R07/15), and Monash Medical Centre (07084C). For human placental tissue sampling, a core of full thickness tissue was isolated from two randomly chosen sites. Tissue sections from other mammalian placentas were processed by maceration with a scalpel blade prior to immediate DNA isolation.
First trimester cytotrophoblast cells were isolated as per Ref. 16, and purity of preparations was determined using antibodies to the trophoblast-specific cytokeratin-7 (1:100 dilution, clone OV-TL 12/30 (DakoCytomation, Glostrup, Denmark)) using immunocytochemistry as described previously (17). HIPEC65 and chorionic villus samples (CVS) cells were cultured as described previously (17).
Genomic DNA
Tissue samples were incubated overnight at 50 °C with shaking in DNA extraction buffer (100 mm NaCl, 10 mm Tris-HCl, pH 8, 25 mm EDTA, 0.5% SDS), containing 200 μg/ml Proteinase K. DNA was isolated by two rounds of phenol/chloroform extraction, followed by RNase A treatment, precipitation in absolute ethanol containing 10% sodium acetate (3 m, pH 5.2), and resuspension in 100 μl of nuclease-free water (Ambion, Austin, TX). DNA was stored at −20 °C.
Methylation Analysis
SEQUENOM MassARRAY EpiTYPER analysis was carried out as described previously (13) using primers 5′-aggaagagagGGAGAGAGGYGATATTTTGTGT (F) and 5′-cagtaatacgactcactatagggagaaggctCAAAAACTCTCACAAACCCTTAAAA (R) for DNMT1 and 5′-aggaagagagGAATGAGTTGGAAGGATTTAGGTTT (F) and 5′-cagtaatacgactcactatagggagaaggctCTAAACCCCACTCCCCACTAAAAT (R) for DNMT3L (lowercase denotes sequence tags added to facilitate downstream EpiTYPER analysis). Bisulfite DNA sequencing was performed as described previously (13). DNA samples were processed using the MethylEasy bisulfite modification kit (Human Genetic Signatures, North Ryde, Australia) according to the manufacturer's instructions. Amplification was performed on converted genomic DNA using primers directed to modified DNA. Primers for sequencing of human DNMT1 assay 1 were 5′-TAAAGTTTGTTGTATTTGGGGATTAA (F) and 5′-CAAAAACTCTCACAAACCCTTAAAA (R). For DNMT1 assay 4, primers 5′-TTTATTAGTAAGATTTTTTTGATGTTTA (F) and 5′-CCCAAATACAACAAACTTTAAATTC (R) were used. Amplification of the baboon DNMT1 promoter was carried out using human primer sequences that show 100% homology with the corresponding baboon genomic region. Marmoset amplification primers were 5′-TTAGTTTGGTTGTATAGGAAGTGGG (forward) and 5′-ACATACTTAAAATTCCTACCAAAACAC (reverse). Mouse primers were 5′-TTTTTTAAGTTTTTAGTTAATGGGTTTGT (forward) and 5′-ACAACACTACCTACTCTCAAATACC (reverse). The human DNMT3L promoter region was amplified using the same primers used for EpiTYPER analysis. Amplification conditions were as follows: 95 °C for 5 min, 56 °C for 1 min 30 s, and 72 °C for 1 min 30 s for 40 cycles, 72 °C for 7 min. Resulting amplicons were cloned into TOPO TA Cloning Vector (Invitrogen) for automated fluorescent sequencing as described (18). Data were analyzed using BiQ Analyzer software (19), and clones showing less than 80% conversion or 90% homology to the reference sequence were not included in subsequent analyses.
HPLC Measurement of 5MeC Levels
Global DNA methylation was calculated by denaturing DNA to single nucleotides and measuring the level of 5MeC relative to total cytosine levels (as described in Refs. 5 and 20). Five μg of genomic DNA was digested with an RNase mixture (Ambion/Applied Biosystems, Austin, TX), which was then removed with phenol-chloroform extraction. Genomic DNA was then precipitated in 100% ethanol and one-tenth volume of 3 m sodium acetate (pH 5.2) and washed with 70% ethanol, dried, and resuspended in 30 μl of Tris-EDTA. Between 500 ng and 3 μg of genomic DNA in 10 μl of nuclease-free water was then transferred to each of three wells of a 96-well plate. Ten μl of sodium acetate (15 mm, pH 5.3) and 0.5 μl ZnSO4 (10 mm) were then added to each well, and DNA was denatured at 100 °C for 5 min, prior to cooling to 4 °C. One μl of nuclease P1 (Sigma) and 1 μl of calf intestinal alkaline phosphatase (1 unit/μl; Invitrogen) were added to the denatured DNA, and the digestion reaction was incubated at 37 °C for a minimum of 2 h. Following digestion, 2 μl of Tris (0.8 mm, pH 7.5) was added, and the plate was incubated at 37 °C overnight. HPLC analysis was performed using the Varian Helix denaturing HPLC system (Varian Inc., Palo Alto, CA) and a Varian Pursuit 5 PFP 150 × 4.6-mm column (Varian Inc., Palo Alto, CA), with 0.1% formic acid (v/v) in water as a solute carrier and 20% (v/v) acetonitrile as the organic solvent. A detection wavelength of 260 nm was used to identify specific spectra peaks for 5MeC and cytosine, and 5MeC percentage was calculated using the equation, 5MeC percentage = 5MeC/(5MeC + C) × 100.
RNA Extraction and Real-time Reverse Transcription-PCR
Total RNA extraction was performed using the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Cells or tissue was homogenized in TRIzol, and RNA was precipitated in 100% isopropyl alcohol and resuspended in nuclease-free water (Ambion/Applied Biosystems, Austin, TX). RNA was stored at −80 °C until needed. cDNA was synthesized using the Superscript III kit (Invitrogen) as per the manufacturer's protocol. DNMT1 expression levels were analyzed by quantitative real-time PCR using the ABi 7300 sequence detection system (Applied Biosystems). DNMT1 cDNA was amplified using primers that target the exon-intron boundary at exons 25 and 26: 5′-GTGGGGGACTGTGTCTCTGT-3′ (forward) and 5′-TGAAAGCTGCATGTCCTCAC-3′ (reverse). GAPDH was used as the endogenous control and amplified using the following primers: 5′-TTCGACAGTCAGCCGCATCTT-3′ (F) and 5′-CCCAATACGACCAAATCCGTT-3′ (R). PCR was set up in 30-μl reactions (15 μl of Platinum SYBR Green mix (Invitrogen), 1 μl of each primer (300 nm final concentration), 8 μl of nuclease-free water, and 5 μl of cDNA template). DNMT1 expression levels in different samples were determined using the ΔCt method, with GAPDH as the control gene.
BeWo Syncytialization Experiment
The protocol for the syncytialization of BeWo cells was based on Ref. 21. BeWo cells were seeded in a 6-well plate, at a concentration of 1 × 105 cells/ml in 1.5 ml of Ham's F-12 culture medium. Epidermal growth factor was added 4 h postseeding, at a final concentration of 10 ng/ml. Twenty-four h after epidermal growth factor treatment, medium containing EGF was removed, and new medium containing Forskolin (100 μm final concentration) was added. Cells were collected 48 h after forskolin treatment. Ethanol (95%) was added to the non-treated cells, as a carrier control.
DNMT1 Gene Reporter Assays
A 910-bp region of the promoter/exon 1/intron 1 of the human DNMT1 gene was amplified using primers 5′-CTCTCTCTCGAGCTGTATTTGGGGATCAAAAGAGAA (F1) and 5′-CTCTCTGTCGACTGGGCCTCTATACACTGTGAGAT (R1) with Phusion Hi-Fidelity DNA polymerase (Finnzymes, Espoo, Finland). The SalI/XhoI-digested amplicon was cloned into a promoterless luciferase expression vector pGL3:basic (Promega, Madison, WI) to produce the plasmid pGL3:−318/+592. Reverse orientation cloning was also carried out to produce pGL:+592/−318. Following sequence verification, DNA was in vitro methylated using either SssI, HpaII, or HhaI methylase (New England Biolabs, Ipswich, MA) according to the manufacturer's instructions. Complete methylation was confirmed by comparative restriction enzyme digestion with BstUI, HpaII, and/or HhaI restriction (data not shown). 100 ng of pGL3:control vector (SV40 constitutive promoter), pGL3:basic (promoterless), and unmethylated or methylated DNMT1 promoter constructs were then transfected into JAR choriocarcinoma cells. Co-transfection with 10 ng of pRn (HSV-TK promoter linked to Renilla luciferase gene) was carried out in all cases to normalize for transfection efficiency.
For transfection experiments, a total of 8 × 103 JAR cells were plated 24 h prior to transfection in each well of a 96-well plate (96F Nunclon Delta white microwell). Transfection was carried out using Lipofectamine LTX reagent (Invitrogen). Luciferase and Renilla activity was measured 24 h post-transfection using the DualGLO assay system (Promega) on a FluorSTAR Optima microplate reader (BMG Labtech, Offenburg, Germany).
RESULTS
DNMT Family Genes Show Distinct Methylation Profiles in Human Placental Tissue
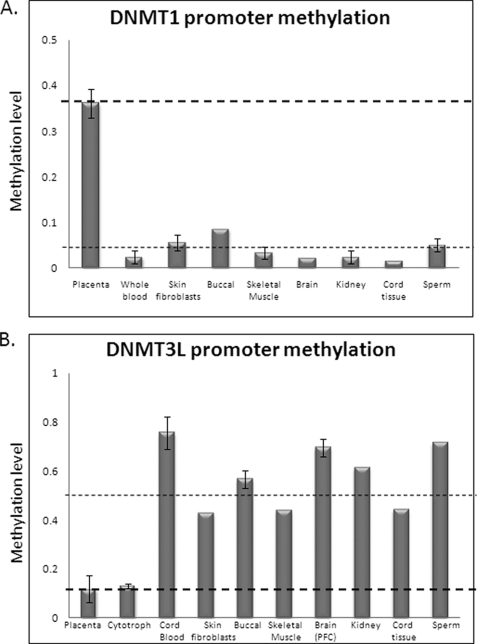
In order to examine the methylation status of DNMT genes in human placental tissue, we designed locus-specific methylation assays for DNMT1, DNMT3A, DNMT3B, and DNMT3L gene regulatory regions. Mass spectrometry-based DNA methylation profiling (SEQUENOM MassARRAY EPITYPER) revealed an absence of methylation of DNMT3A or -3B genes in all human tissues, including placenta and purified cytotrophoblasts (supplemental Fig. 1). This was confirmed by bisulfite DNA sequencing (data not shown). In contrast, the DNMT1 gene promoter was found to be methylated in full term placental tissue (mean methylation of 36.6 ± 5.9%; n = 7) relative to all somatic tissues (mean methylation of 4.4 ± 1.2% in eight tissues tested) (Fig. 1A). Interestingly, the DNMT3L gene promoter showed a reciprocal pattern of promoter methylation, with higher methylation in somatic tissues (55.5%; n = 9) than in full term placental tissue (10.42%; n = 4) (Fig. 1B). This was confirmed by bisulfite DNA sequencing (supplemental Fig. 2). The same DNMT1 and DNMT3L methylation patterns were found in first trimester placental tissue, suggesting that this level of methylation is present throughout the majority of pregnancy (supplemental Fig. 3).
FIGURE 1.
SEQUENOM MassARRAY EpiTYPER analysis of DNMT1 and DNMT3L genes in human placenta and somatic tissues. A, placenta-specific methylation of the DNMT1 promoter assessed using the DNMT1_5 assay, spanning 230 bp from −133 to +97 relative to the transcription start site. Mean methylation in somatic tissues was 4.4% (across eight different tissues), whereas full term placental tissue shows a mean methylation level of 36.6% (n = 8 placentas). B, placenta-specific hypomethylation of the DNMT3L gene assessed using the DNMT3L_1 assay spanning 371 bp, −18/+353 relative to the transcription start site. Mean methylation in term placenta and purified cytotrophoblasts was 14 and 13%, respectively, whereas the mean for other tissues was 53% (n = 8) across three analyzable CpG units.
DNMT1 Is Monoallelically Methylated in Human Placental Tissue and Cytotrophoblasts
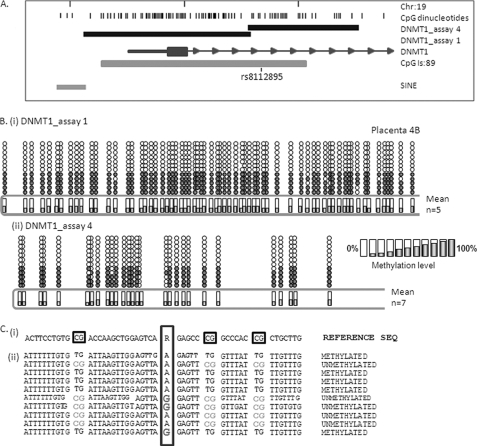
Bisulfite sequencing analysis using two independent assays spanning the entire DNMT1-associated CpG island demonstrated a pattern consistent with monoallelic methylation for the entire region spanning the promoter, exon 1, and part of intron 1 (Fig. 2) in full term human placental tissue. In each instance, all alleles show nearly complete methylation or a nearly complete lack of methylation.
FIGURE 2.
Monoallelic methylation of the human DNMT1 gene. A, location of the DNMT1 CpG island/promoter DNA methylation assays used for bisulfite sequencing. Genomic coordinates on chromosome 19 are shown, along with location of individual CpG dinucleotides (dashes) and CpG island location (gray bar) in relation to the DNMT 1 gene (exon 1 shown as a blue box, intron shown as an arrowed line) according to the University of California Santa Cruz genome browser (hg_18 assembly). The arrows denote transcriptional direction. The location of rs8112895 SNP in the DNMT1_assay_4 is also shown. B, representative DNA methylation of a single full term human placental sample for assay_1 (i) and assay_4 (ii) showing a monoallelic pattern of methylation within the human placenta. Between eight and 12 individual clones were sequenced for each sample. The circles correspond to CpG sites denoted by black dashes in A. Closed circles, methylation; open circles, lack of methylation. Mean methylation levels seen at each CpG site in multiple placental samples for each assay are shown as shaded bars between 0 and 100% (n = 4). Missing circles indicate CpG sites for which no information was obtained. C, DNMT1_4 methylation assay spanning SNP rs8112895 was used for bisulfite sequencing methylation analysis in purified first trimester cytotrophoblast cells. i, reference genomic DNA sequence surrounding rs8112895 SNP (R) and CpG dinucleotides (boxed). ii, individual clone sequences showing the presence of bisulfite-converted CG sites (TG) and unconverted CG sites (CG) in both maternal and paternal alleles from this heterozygous sample. This confirms a lack of parent of origin of the observed DNMT1 promoter methylation.
Because one of the assays used in this analysis (DNMT1_4) spans a polymorphic region of relatively high heterozygosity (rs8112895; Fig. 2), we were able to test for a parent-of-origin effect (i.e. imprinting). If this region were imprinted, paternal and maternal alleles would show distinct, reciprocal methylation patterns. This was not the case, with examples of both methylated and unmethylated maternal and paternal alleles detected in several heterozygous placental samples (Fig. 2). Therefore, the DNMT1 gene promoter shows random, monoallelic methylation in the placenta.
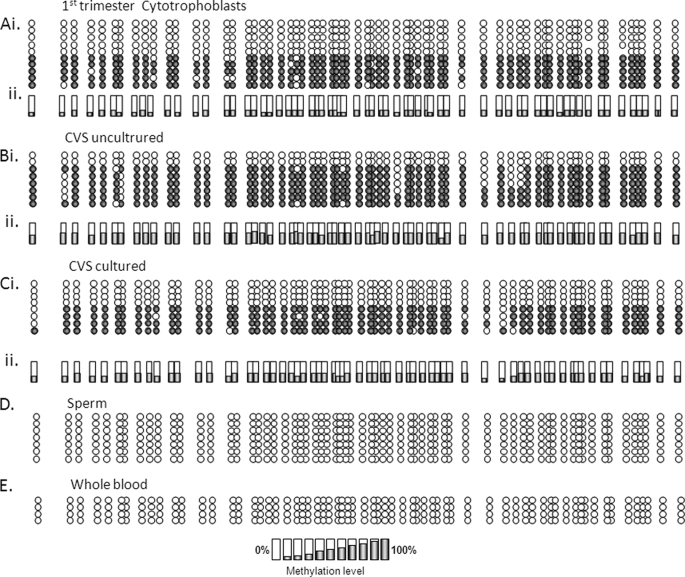
Examination of first trimester purified cytotrophoblasts or CVS (uncultured and cultured) tissue/cells confirmed monoallelic methylation in both early and late pregnancy, even in CVS-derived cells that adapt to in vitro culturing (i.e. not trophoblasts) (Fig. 3, A–C). A complete lack of DNMT1 methylation was seen in somatic (whole blood) and germ line (sperm) tissues using this approach (Fig. 3, D–E), consistent with data (see above) obtained using SEQUENOM MassARRAY EpiTYPER.
FIGURE 3.
Distribution of monoallelic methylation of the DNMT1 promoter in first trimester placental tissue and trophoblasts. Bisulfite sequencing data obtained using the DNMT1_1 assay in the following tissues: purified cytotrophoblasts (A) and uncultured (B) and cultured (C) (n = 10 passages) CVS biopsies. i, examples of data from individual samples; ii, mean methylation level for biological replicates (cytotrophoblasts, n = 2; uncultured and cultured CVS, n = 6). Shown are both sperm (D) and whole blood (E) lack of DNMT1 methylation as assessed by this methodology.
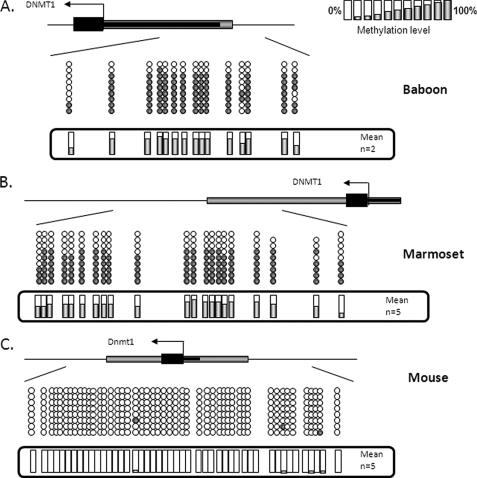
Monoallelic DNMT1 Methylation Is Primate-specific
Using bisulfite DNA sequencing, we found evidence for monoallelic DNMT1 gene methylation in baboon (full term (Fig. 4A) and first trimester (not shown)) and marmoset (full term) placental tissue (Fig. 4B) but no evidence for DNMT1 gene methylation in mouse (Fig. 4C), cow, or guinea pig placental tissue (data not shown). Therefore, placental methylation of the DNMT1 gene in mammals appears to be primate-specific.
FIGURE 4.
DNMT1 promoter methylation is restricted to primates. Bisulfite DNA sequencing confirmed promoter methylation of the DNMT1 gene in full term baboon (A) and marmoset (B) placental tissue, with no evidence for methylation in mouse (C), bovine, or guinea pig placental tissue (data not shown).
DNMT1 Methylation Is Reduced in All Choriocarcinoma (CCA)-derived Cell Lines
We have previously demonstrated hypermethylation of several tumor suppressor and vitamin D regulatory genes in CCA cell lines and speculated that this plays a role in the generation of related gestational tumors (12, 13, 17). Interestingly, when DNMT1 methylation was examined in JAR, BeWo, and JEG-3 CCA cell lines, we found an almost complete loss of methylation (supplemental Fig. 4); however, this was not associated with an increase in gene expression (Fig. 5A). In contrast, DNMT3L methylation was increased in JEG-3 cells (mean of 33 and 40% for MassARRAY and bisulfite sequencing, respectively) and decreased in JAR cells (mean 6.5%) relative to first trimester cytotrophoblasts (mean 13.1% using MassARRAY analysis) (supplemental Figs. 2 and 5). The differential changes in DNMT3L methylation in CCA cell lines relative to purified cytotrophoblasts reveals a previously uncharacterized level of epigenetic variation within these cell lines.
FIGURE 5.
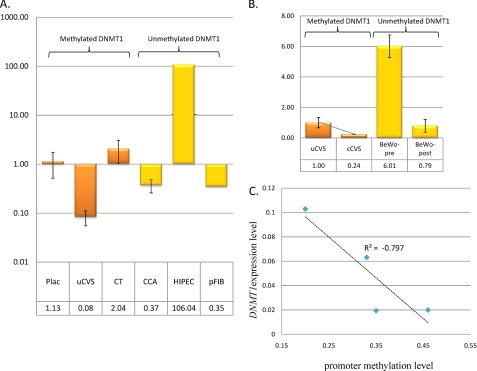
Widely variable DNMT1 expression in placental tissue and derived cells. A, DNMT1 expression levels in methylated samples (orange bars) versus non-methylated (yellow bars) relative to the mean full term placenta DNMT1 expression level. Data are shown for full term placenta (Plac; n = 4), uncultured first trimester CVS tissue (uCVS; n = 2), purified first trimester cytotrophoblasts (CT; n = 2), choriocarcinoma cell lines (CCA; n = 3), SV40-immortalized trophoblast cell line HIPEC65, and placental fibroblasts (pFIB; n = 1). No obvious correlation between gene expression and promoter methylation was apparent. B, decreasing DNMT1 expression was seen in following culturing of CVS tissue for 10 passages (cCVS; n = 2) and with syncitialization of BeWo cell lines (BeWo-post; n = 2). Expression level is relative to the mean of uncultured CVS tissue. The colored bars denote methylation status as per A. C, correlation between DNMT1 expression level and methylation status in CVS tissue (pre- and post-culturing). Culturing for 10 passages results in a decrease in DNMT1 expression, whereas promoter methylation is increased. The correlation co-efficient is −0.797, suggesting an inverse relationship between methylation and expression levels in this system.
DNMT1 Promoter Methylation Directly Attenuates Gene Transcription
Previous studies have examined regulation of the DNMT1 gene promoter, but to date, none has examined the effect of DNA methylation on the currently annotated major promoter of this gene (22–24). In order to gain insight into the likely consequences of placenta-specific methylation, we carried out in vitro reporter analysis using a 910-bp amplified region of the human DNMT1 promoter (containing 56 CpG sites) that spans several highly conserved sequence regions in vertebrates known to confer constitutive promoter activity (22) (Fig. 6). The amplified region also includes the most proximal repetitive SINE element upstream of the start site of transcription. Such sequences occupy the majority of DNA within the 5 kb upstream of the DNMT1 transcription start site.
FIGURE 6.
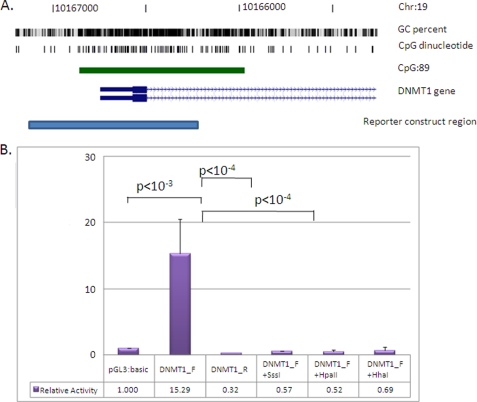
Methylation of the DNMT1 promoter directly attenuates gene transcriptional level. A, position of the cloned DNMT1 promoter region (pale blue bar; −536/+374 relative to the transcription start site). Shown are genome coordinates, GC percentage, location of individual CpG dinucleotides, CpG island (green bar), and DNMT1 gene (exons shown as blue boxes, intron as an arrowed line) according to the University of California Santa Cruz genome browser (hg_18 assembly). The arrows denote transcriptional direction. B, luciferase reporter constructs were transfected into human JAR cells with or without prior in vitro methylation with SssI, HpaII, or HhaI methylases (n = 8 for each). A 15-fold increase in promoter activity was detected for the unmethylated promoter region relative to the promoterless vector. This activity was abolished with in vitro methylation prior to transfection (SssI, HpaII, and HhaI methylases). Luminescence values were normalized to Renilla luciferase to correct for transfection efficiency, and all data are displayed relative to the mean basal promoter activity of the pGL3:basic vector. The error bars denote 95% confidence intervals. Statistical analysis for comparing between transfected groups was carried out using a two-tailed Student's t test with unequal variance.
Choriocarcinoma-derived JAR cells were transfected with two different DNMT1 reporter constructs ((+)- and (−)-orientation), with or without in vitro methylation with SssI, HhaI, or HpaII methylases. SssI methylase acts at all CpGs, HpaII methylase methylates only the CpG within the sequence 5′-CCGG, and HhaI methylates CpG within 5′-GCGC. Transfection of the (+)-orientation reporter construct confirmed the basal activity of the 910-bp cloned fragment with a greater than 15-fold increase in promoter activity relative to the promoterless control vector (n = 16) (Fig. 6). This is consistent with previous data from a colorectal cancer-derived cell line (25). Promoter activity was negligible in the (−)-orientation, demonstrating a directionality of DNMT1 promoter activity (Fig. 6). In vitro methylation of the reporter construct with SssI, HpaII, or HhaI methylases prior to transfection abolished promoter activity (p < 0.0001), suggesting that only limited methylation of this region is required to abolish transcription.
Widely Variable DNMT1 Expression Levels in Human Placenta and Derived Cells
In order to ascertain the effects of the observed DNMT1 promoter methylation level in placental tissue and derived cells, we carried out quantitative reverse transcription-PCR using primers designed to interrogate all annotated transcripts arising from the most distal DNMT1 transcription start site associated with the methylated promoter in the placenta. We found clear evidence for DNMT1 expression in full term placenta (with monoallelic methylation) at lower levels than that detected in fetal brain (46%) but higher than that in fetal liver and (127%) and several adult tissues (data not shown). Widely divergent levels of expression were found in primary first trimester CVS tissue (mean 14-fold decrease relative to mean of full term tissue) and purified cytotrophoblasts (mean 2-fold increase), both of which show similar levels of DNMT1 promoter methylation. We also found lower levels of expression in three independent CCA cell lines (mean 2.7-fold decrease) and placental fibroblasts (mean 3.2-fold decrease), each of which lack promoter methylation, whereas the SV40-transformed trophoblast line HIPEC65 (26) (also unmethylated) showed a major increase in expression (106-fold relative to full-term placenta) (Fig. 5A).
This complicated pattern of expression prompted us to examine the effects of DNMT1 expression and methylation in two cell culture systems under controlled conditions, with assessment of both DNA methylation and gene expression status pre- and postculturing. Primary CVS tissue cultured for 10 passages showed a clear decrease in DNMT1 expression relative to primary tissue (4.2-fold reduction) (Fig. 5B) concomitant with an increase in promoter methylation (mean methylation 0.26 in P0 versus 0.4 at P10). This occurred in the absence of any decrease in proliferative capacity. Correlation between methylation and expression level was −0.797 (Fig. 5C), supporting a direct relationship between DNMT1 methylation and expression in this system. Conversely, differentiation of BeWo trophoblast cells to syncitia (associated with a decrease in proliferation) was also associated with a decrease in DNMT1 expression in the absence of any change in promoter methylation status (Fig. 5B). In this case, expression level appears to be directly linked to proliferative status.
Global Hypomethylation of Extraembryonic Tissue in the Absence of DNMT1 Promoter Methylation
Although all placentas share a basic common function in modulating exchange between maternal and fetal circulations, the human placenta is unique in structure and function (27). In order to determine the extent to which DNMT1 promoter methylation predicts or is associated with global methylation levels across species, we measured 5MeC content by HPLC in a range of different human and animal placental tissues. We also included in vitro-derived trophoblast stem cells obtained following differentiation and selection of hCG secreting cells from human embryonic stem cells in culture (28).
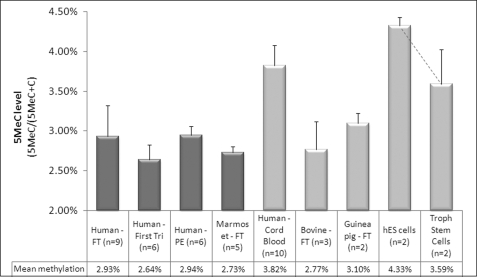
Human cord blood (lacking DNMT1 methylation) showed a mean global 5MeC content of 3.82%, consistent with previous reports for peripheral blood (5). Mean global 5MeC levels for full term and first trimester human placental tissue (with DNMT1 methylation) combined were variable (ranging from 2.63 to 3.62%) with a mean level of 2.93% (95% confidence interval, 2.54–3.32%), also in accord with previously published data (5). No significant change in global methylation was seen in placentas obtained from pre-eclamptic pregnancies (Fig. 7). Full term marmoset tissue (with DNMT1 promoter methylation) showed global methylation levels similar to that seen in human placental tissue (range 2.64–2.93%). However, despite lacking DNMT1 methylation, bovine and guinea pig full term placental tissue has a similar mean global 5MeC level (2.77 and 3.1% methylated cytosine, respectively), demonstrating that global hypomethylation can occur in the absence of DNMT1 promoter methylation. This is further supported by our demonstration that human embryonic stem cells with a mean global 5MeC level of 4.33% undergo substantial global demethylation associated with differentiation into hCG-secreting cytotrophoblast stem cells (mean methylation of 3.59%) (Fig. 7), despite the absence of concomitant DNMT1 promoter methylation in either cell population (data not shown).
FIGURE 7.
Methylation status of DNMT1 does not predict global 5MeC levels in extraembryonic tissue. Global 5MeC content was measured in a variety of placental and umbilical cord blood cells. Human full term (FT), first trimester, full term pre-eclamptic (PE), and full term marmoset tissue (with DNMT1 promoter methylation) show mean global 5MeC levels of <3% (range 2.64–2.93%). Despite lacking methylation of the equivalent DNMT1 gene promoter, bovine and guinea pig FT placental tissues show a similar mean global 5MeC level (2.77 and 3.1% methylation, respectively), indicating that DNMT1 promoter methylation is not an absolute prerequisite for reduced genomic methylation in extraembryonic tissue. Human cord blood (also lacking DNMT1 promoter methylation) shows a mean global 5MeC content of 3.82%, consistent with previous reports in peripheral blood (5). Human embryonic stem cells showed the highest global 5MeC level of all tissues/cells examined with a mean of 4.33%. Differentiation of these cells into cytotrophoblast stem cells was associated with a reduction in global methylation levels (mean 3.59%) in the absence of concomitant DNMT1 promoter methylation (data not shown). However, the small sample numbers resulted in a lack of significance when tested using a paired Student's t test. The dark bars correspond to samples with DNMT1 promoter methylation, whereas light bars identify samples lacking DNMT1 promoter methylation.
DISCUSSION
DNMT Enzymes in Early Mammalian Development
The primary aim of this study was to investigate the potential role of promoter methylation of the DNMT family of genes in the establishment of the unique epigenetic profile of the placenta, including global DNA hypomethylation. We found no evidence for methylation-associated down-regulation of the DNMT3A or -3B genes in the placenta or any human tissue but found clear evidence for placenta-specific monoallelic methylation of the DNMT1 gene and hypomethylation of the DNMT3L gene. This supports recent data obtained by whole genome profiling of DNA methylation in full term placental tissue (29). In vitro reporter analysis demonstrated the sensitivity of this region to silencing by methylation, with similar attenuation of transcriptional activity by SssI (all CpG sites), HpaII (9 of 56 CpG sites), and HhaI (13 of 56 CpG sites) methylases.
DNMT1 acts as a “maintenance” methyltransferase, playing an essential role in the faithful propagation of the genomic methylation profile from mother to daughter cells following cell division (30). Elegant work by Kato et al. (31) has demonstrated the specificity of other members of the DNMT family in the de novo establishment of methylation, particularly at imprinting-associated differentially methylated regions, interspersed repeats (including LINE1 elements), and satellite repeats in mice. Although Dnmt3a mainly methylates imprinted regions and short interspersed (SINEB1) elements, both Dnmt3a and Dnmt3b are involved in establishing gene-specific (non-imprinted) and long interspersed repeat (IAP and LINE) methylation. Interestingly, DNMT3L appears to be the only member of this family universally required for appropriate levels of de novo methylation at all sequence families, highlighting the general importance of this protein in de novo methylation establishment (31).
DNMTs and the Placental Epigenome
Genome-wide hypomethylation, coupled with gene-specific hypermethylation of tumor suppressor genes, is a common feature of human cancers. Hypomethylation appears to be an early feature of the tumorigenic process and is present in benign lesions (32, 33–35). Interestingly, the placenta parallels human cancers in both the overall decreased level of genomic DNA methylation and the specific hypermethylation of several tumor suppressor genes (5, 6, 11–14). In this regard, it is unique among non-malignant human tissues. In quantitative terms, human full term placenta shows a reduction in global DNA methylation of ∼20% relative to the brain, liver, and peripheral blood lymphocytes (5, 6). This is largely due to reductions in methylation levels at specific repetitive DNA elements that comprise the majority of mammalian genomic DNA (9, 36, 37). Despite this general global hypomethylation, it is clear that some DNA methylation (locus-specific and/or repeat-based) is important for correct placental development. Inhibition of DNA methylation by 5-azacytidine treatment disrupts trophoblast invasive and migratory potential in vitro (38) and proper placental development in vivo (39).
Monoallelic Methylation of Genes in the Absence of Imprinting
In the current study, we utilized a polymorphic sequence downstream of the transcription start site of the DNMT1 gene in combination with bisulfite sequencing methylation analysis to show conclusively the presence of monoallelic methylation of the DNMT1 gene in the absence of a parent-of-origin effect. Although we cannot exclude the possibility of a mixed cell population consisting of a combination of cells with full methylation of both alleles and another population lacking DNMT1 methylation, the demonstration of monoallelic methylation in purified cell populations (e.g. CK7+ cytotrophoblasts) does not support a “mixed cell population” model of DNMT1 methylation in the placenta.
Monoallelic gene expression, in the absence of classical parent-of-origin imprinting has, has recently been described for several genes in different human tissues (40). A similar phenomenon has been described for several tumor suppressor genes in placenta, although conflicting data related to a parent-of-origin effect (i.e. imprinting) have been reported (11, 13, 14, 17). This is probably due to the differing methodologies used to test for a parent-of-origin effect.
Placental Pathologies and DNA Methylation
In this study, we showed consistent hypomethylation of the DNMT1 gene in three independent CCA cell lines, relative to first trimester cytotrophoblasts or placental tissue. It is speculated that higher DNMT1 expression is involved in the etiology of these tumors. No consistent changes were detected at the DNMT3L promoter with increasing methylation in JEG-3 cells and decreasing levels in JAR cells. These data are in marked contrast to the increasing levels of methylation we have previously reported for several tumor suppressor genes in CCA cell lines, highlighting the complexity of locus-specific methylation changes that accompany choriocarcinoma development.
Increasing DNMT gene activity, such as that predicted to occur in CCA-derived trophoblast cells, independent of promoter methylation, has been described in several human cancers (e.g. see Refs. 41–47). We speculate that increasing DNMT1 activity facilitates the aberrant tumor suppressor methylation associated with cancer development. The efficacy of DNMT1 inhibition in restoring tumor suppressor activity, promoting cell cycle arrest, and inhibiting tumor growth (reviewed in Ref. 48) supports this hypothesis.
DNMT1 Expression Is Regulated by Multiple Mechanisms in Extraembryonic Tissue
Expression analysis revealed a complex pattern of DNMT1 regulation in placental tissue and derived cells, likely to involve promoter methylation and other additional levels of regulation. No evidence was found for a direct association of DNMT1 methylation with overall gene expression level, with widely variable expression in different populations of cells (e.g. cytotrophoblasts and CVS tissues), even where similar overall levels of promoter methylation were apparent. Similarly, no direct evidence for a clear association of expression levels with proliferative status was found, supporting previously reported data (49, 50). However, under controlled conditions in culture, we observed both a clear negative correlation of DNMT1 promoter methylation with gene expression level in CVS-derived tissue and a correlation between expression and proliferation in a model of trophoblast differentiation.
These data support a complex interplay of regulatory elements in DNMT1 expression in the placenta that encompasses a “basal” promoter activity associated with the maintenance of DNA methylation during cell division in addition to cell cycle-independent regulation of gene activity that may play an as yet uncharacterized, functional role. This is further supported by previous data demonstrating a link between DNMT1 transcriptional activity and specific cell signaling pathways. For example, DNMT1 has been shown to respond to both proto-oncogene and tumor suppressor pathways (22). The APC tumor suppressor protein inhibits DNMT1 expression in HT-29 colorectal cancer cells (25), and it has been postulated that the observed up-regulation of DNMT1 seen in several cancers may be related to tumor suppressor silencing (including the APC gene). We have previously reported methylation-mediated APC down-regulation in human placenta (13) and have identified placenta-specific silencing of several tumor suppressor genes, raising the intriguing possibility of a link between these two systems in placental development and function. The placenta is also a known endocrine organ, producing several hormones, including estrogen, previously shown to promote expression of another DNA methyltransferase, DNMT3B, in human endometrial cells (51).
An alternative explanation for the observed complexity of DNMT1 regulation in the placenta may lie in the propensity for the placenta to utilize cryptic promoter sequences as alternative regulatory elements for gene expression. This is a common feature of placenta-specific gene expression, often associated with the changed epigenetic state of repetitive elements (52, 53). Thus, methylation of the DNMT1 promoter may be associated with a switch to other regulatory elements in this tissue, possibly in response to specific environmental cues during pregnancy. The upstream region of DNMT1 is highly enriched for repetitive DNAs; however, these are primarily short and long interspersed nuclear elements (SINEs and LINEs), neither of which have yet been shown to function as cryptic promoters in this tissue.
Future examination of placenta-specific profiles of DNMT transcription and enzyme activity in response to specific environmental cues should provide valuable insights into the role of the DNMT1 promoter methylation that we have identified in human placental tissue.
DNMT1 Methylation Is Not Essential for Global Methylation in Extraembryonic Tissue
Analysis of global DNA methylation levels by HPLC has revealed consistently lower levels of methylation, albeit with some individual variability, in full term placental and CVS tissue (5, 9, 36, 54). Using the same approach, we found similar levels of global methylation at between 2.73 and 3.1% in human, marmoset, guinea pig, and bovine full term placental tissue.
Previous studies in mice have suggested that the inner cell mass of the blastocyst undergoes more extensive de novo methylation than the trophectoderm (prior to implantation), leading to an overall hypomethylation in extraembryonic lineages (8, 55, 56). At present, it is unclear whether this reduced level of de novo methylation results from lower DNMT gene activity. Our data showing a lack of Dnmt1 promoter methylation in non-primates and a reduction in global methylation levels in hESC-derived cytotrophoblast stem cells in the absence of DNMT1 methylation support a DNMT1-independent mechanism for reduced global DNA methylation in the early stages of extraembryonic tissue development. This would suggest an as yet unidentified role for DNMT1 promoter methylation in primate placental development. Alternatively, our data could equally be explained by the existence of distinct mechanisms leading to decreasing global methylation in different systems.
Interestingly, recent data have implicated a role for silencing (or down-regulation) of DNMT1 in the invasive and migratory potential of some cancer cell lines. Specifically, inhibition of DNMT1 activity in prostate cancer-derived PC3 cells causes growth retardation while enhancing invasiveness and migratory capacity (57). This is also supported by data demonstrating a similar effect on proliferation and invasion in some pancreatic and gastric cancer cell lines following 5-azacytidine (a DNMT1 inhibitor) treatment (58, 59). It is tempting to speculate that the methylation-associated DNMT1 down-regulation described in this study may play an important role in the migratory and invasive properties of primate placentation. Future experiments, examining the effect of overexpression of DNMT1 in primary trophoblast cells, will enable this to be examined directly.
A Role for DNMT3L in Human Placental Function?
In mice, Dnmt3L is expressed at high levels in the chorion, containing a multipotent trophoblast stem cell population. Disruption of Dnmt3L results in multiple defects in the developing placenta, including poor formation of the labyrinth and spongiotrophoblast layer, excess trophoblast giant cells, and insufficient attachment of the chorion to the ectoplacental cone. This is associated with an arrest of proliferation of the extraembryonic tissue (60, 61), suggesting that Dnmt3L-mediated de novo methylation is critical for proper placental development in mice. Our demonstration of placenta-specific hypomethylation of the DNMT3L gene supports a role in human placental development, although this will require further examination.
Conclusions
It is generally believed that the majority of CpG island-containing genes remain unmethylated throughout development. However, the reason for this is unclear, given the obvious potential for methylation of such sequences to play a significant role in regulation of gene expression. The unique profile of placental promoter methylation observed in this and other studies suggests that many such genes are in fact methylated in a highly tissue-specific manner. Large scale epigenome projects currently under way will provide valuable insights into the extent of such methylation in highly specialized cell types during development.
The continuing identification of examples of placenta-specific promoter methylation provides valuable insights into pathways likely to be epigenetically regulated during development of this enigmatic tissue. Uncovering the mechanisms leading to the establishment of such markings, their timing, and sensitivity to environmental and genetic disruption has the potential to reveal insights into the evolution of different placental functions and may reveal novel risk pathways associated with adverse pregnancy outcomes and tumor development.
Supplementary Material
Acknowledgments
We thank Sarah Healy, Tina Viano, and Nicole Brooks for help with collection of placental tissue for this study. We also thank Prof. Lois Salamonson (Prince Henry's Institute of Medical Research, Australia) and Prof. Samuel Breit (University of New South Wales, Australia) for CCA cell lines and Drs. Patrick Western and Craig Smith for mouse placental tissue. We especially thank Prof. Harry Moore (Centre for Stem Cell Biology, Sheffield, UK) for trophoblast stem cells and hESC DNA, Prof. Anne-Maree Hennessey and Dr. Neroli Sunderland (Royal Prince Alfred Hospital) for baboon placenta and CVS tissue, and Monash Animal Services for marmoset tissue. We also thank Mark Pertile for CVS samples and Dr. Thierry Fournier and Prof. Danielle Evain-Brion (INSERM, Paris, France) for HIPEC-65 cells.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- DNMT
- DNA methyltransferase
- 5MeC
- 5-methylcytosine
- CCA
- choriocarcinoma
- CVS
- chorionic villous sampling
- HPLC
- high pressure liquid chromatography
- SINE
- short interspersed nuclear element
- LINE
- long interspersed nuclear element.
REFERENCES
- 1.Bestor T. H. (2000) Hum. Mol. Genet. 9, 2395–2402 [DOI] [PubMed] [Google Scholar]
- 2.Aapola U., Kawasaki K., Scott H. S., Ollila J., Vihinen M., Heino M., Shintani A., Kawasaki K., Minoshima S., Krohn K., Antonarakis S. E., Shimizu N., Kudoh J., Peterson P. (2000) Genomics 65, 293–298 [DOI] [PubMed] [Google Scholar]
- 3.Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. (2004) J. Biol. Chem. 279, 27816–27823 [DOI] [PubMed] [Google Scholar]
- 4.Hemberger M. (2007) Cell. Mol. Life Sci. 64, 2422–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. (2004) Ann. Hum. Genet. 68, 196–204 [DOI] [PubMed] [Google Scholar]
- 6.Gama-Sosa M. A., Midgett R. M., Slagel V. A., Githens S., Kuo K. C., Gehrke C. W., Ehrlich M. (1983) Biochim. Biophys. Acta 740, 212–219 [DOI] [PubMed] [Google Scholar]
- 7.Okahara G., Matsubara S., Oda T., Sugimoto J., Jinno Y., Kanaya F. (2004) Genomics 84, 982–990 [DOI] [PubMed] [Google Scholar]
- 8.Santos F., Hendrich B., Reik W., Dean W. (2002) Dev. Biol. 241, 172–182 [DOI] [PubMed] [Google Scholar]
- 9.Tsien F., Fiala E. S., Youn B., Long T. I., Laird P. W., Weissbecker K., Ehrlich M. (2002) Cytogenet. Genome Res. 98, 13–21 [DOI] [PubMed] [Google Scholar]
- 10.Wagschal A., Feil R. (2006) Cytogenet. Genome Res. 113, 90–98 [DOI] [PubMed] [Google Scholar]
- 11.Guilleret I., Osterheld M. C., Braunschweig R., Gastineau V., Taillens S., Benhattar J. (2009) Epigenetics 4, 62–68 [DOI] [PubMed] [Google Scholar]
- 12.Novakovic B., Rakyan V., Ng H. K., Manuelpillai U., Dewi C., Wong N. C., Morley R., Down T., Beck S., Craig J. M., Saffery R. (2008) Mol. Hum. Reprod. 14, 547–554 [DOI] [PubMed] [Google Scholar]
- 13.Wong N. C., Novakovic B., Weinrich B., Dewi C., Andronikos R., Sibson M., Macrae F., Morley R., Pertile M. D., Craig J. M., Saffery R. (2008) Cancer Lett. 268, 56–62 [DOI] [PubMed] [Google Scholar]
- 14.Chiu R. W., Chim S. S., Wong I. H., Wong C. S., Lee W. S., To K. F., Tong J. H., Yuen R. K., Shum A. S., Chan J. K., Chan L. Y., Yuen J. W., Tong Y. K., Weier J. F., Ferlatte C., Leung T. N., Lau T. K., Lo K. W., Lo Y. M. (2007) Am. J. Pathol. 170, 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maccani M. A., Marsit C. J. (2009) Am. J. Reprod. Immunol. 62, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapia A., Salamonsen L. A., Manuelpillai U., Dimitriadis E. (2008) Hum. Reprod. 23, 1724–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novakovic B., Sibson M., Ng H. K., Manuelpillai U., Rakyan V., Down T., Beck S., Fournier T., Evain-Brion D., Dimitriadis E., Craig J. M., Morley R., Saffery R. (2009) J. Biol. Chem. 284, 14838–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong N. C., Wong L. H., Quach J. M., Canham P., Craig J. M., Song J. Z., Clark S. J., Choo K. H. (2006) PLoS Genet. 2, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. (2005) Bioinformatics 21, 4067–4068 [DOI] [PubMed] [Google Scholar]
- 20.Gama-Sosa M. A., Slagel V. A., Trewyn R. W., Oxenhandler R., Kuo K. C., Gehrke C. W., Ehrlich M. (1983) Nucleic Acids Res. 11, 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Nasiry S., Spitz B., Hanssens M., Luyten C., Pijnenborg R. (2006) Hum. Reprod. 21, 193–201 [DOI] [PubMed] [Google Scholar]
- 22.Bigey P., Ramchandani S., Theberge J., Araujo F. D., Szyf M. (2000) Gene 242, 407–418 [DOI] [PubMed] [Google Scholar]
- 23.Slack A., Cervoni N., Pinard M., Szyf M. (1999) Eur. J. Biochem. 264, 191–199 [DOI] [PubMed] [Google Scholar]
- 24.Slack A., Pinard M., Araujo F. D., Szyf M. (2001) Gene 268, 87–96 [DOI] [PubMed] [Google Scholar]
- 25.Campbell P. M., Szyf M. (2003) Carcinogenesis 24, 17–24 [DOI] [PubMed] [Google Scholar]
- 26.Pavan L., Tarrade A., Hermouet A., Delouis C., Titeux M., Vidaud M., Thérond P., Evain-Brion D., Fournier T. (2003) Carcinogenesis 24, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 27.Carter A. M. (2007) Placenta 28, Suppl. A, S41–S47 [DOI] [PubMed] [Google Scholar]
- 28.Harun R., Ruban L., Matin M., Draper J., Jenkins N. M., Liew G. C., Andrews P. W., Li T. C., Laird S. M., Moore H. D. (2006) Hum. Reprod. 21, 1349–1358 [DOI] [PubMed] [Google Scholar]
- 29.Rakyan V. K., Down T. A., Thorne N. P., Flicek P., Kulesha E., Gräf S., Tomazou E. M., Bäckdahl L., Johnson N., Herberth M., Howe K. L., Jackson D. K., Miretti M. M., Fiegler H., Marioni J. C., Birney E., Hubbard T. J., Carter N. P., Tavaré S., Beck S. (2008) Genome Res 18, 1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann A., Goyal R., Jeltsch A. (2004) J. Biol. Chem. 279, 48350–48359 [DOI] [PubMed] [Google Scholar]
- 31.Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. (2007) Hum. Mol. Genet. 16, 2272–2280 [DOI] [PubMed] [Google Scholar]
- 32.Feinberg A. P., Gehrke C. W., Kuo K. C., Ehrlich M. (1988) Cancer Res. 48, 1159–1161 [PubMed] [Google Scholar]
- 33.Chalitchagorn K., Shuangshoti S., Hourpai N., Kongruttanachok N., Tangkijvanich P., Thong-ngam D., Voravud N., Sriuranpong V., Mutirangura A. (2004) Oncogene 23, 8841–8846 [DOI] [PubMed] [Google Scholar]
- 34.Jackson K., Yu M. C., Arakawa K., Fiala E., Youn B., Fiegl H., Müller-Holzner E., Widschwendter M., Ehrlich M. (2004) Cancer Biol. Ther. 3, 1225–1231 [DOI] [PubMed] [Google Scholar]
- 35.Widschwendter M., Jiang G., Woods C., Müller H. M., Fiegl H., Goebel G., Marth C., Müller-Holzner E., Zeimet A. G., Laird P. W., Ehrlich M. (2004) Cancer Res. 64, 4472–4480 [DOI] [PubMed] [Google Scholar]
- 36.Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. (1983) Nucleic Acids Res. 11, 3087–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cotton A. M., Avila L., Penaherrera M. S., Affleck J. G., Robinson W. P., Brown C. J. (2009) Hum. Mol. Genet. 18, 3544–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahnama F., Shafiei F., Gluckman P. D., Mitchell M. D., Lobie P. E. (2006) Endocrinology 147, 5275–5283 [DOI] [PubMed] [Google Scholar]
- 39.Serman L., Vlahović M., Sijan M., Bulić-Jakus F., Serman A., Sincić N., Matijević R., Jurić-Lekić G., Katusić A. (2007) Placenta 28, 803–811 [DOI] [PubMed] [Google Scholar]
- 40.Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V. V., Schupf N., Vilain E., Morris M., Haghighi F., Tycko B. (2008) Nat. Genet. 40, 904–908 [DOI] [PubMed] [Google Scholar]
- 41.Arai E., Kanai Y., Ushijima S., Fujimoto H., Mukai K., Hirohashi S. (2006) Int. J. Cancer 119, 288–296 [DOI] [PubMed] [Google Scholar]
- 42.Etoh T., Kanai Y., Ushijima S., Nakagawa T., Nakanishi Y., Sasako M., Kitano S., Hirohashi S. (2004) Am. J. Pathol. 164, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan H., Zhao Z. J., Cheng J., Su X. W., Wu Q. X., Shan Y. F. (2009) World J. Gastroenterol. 15, 2020–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin R. K., Hsu H. S., Chang J. W., Chen C. Y., Chen J. T., Wang Y. C. (2007) Lung Cancer 55, 205–213 [DOI] [PubMed] [Google Scholar]
- 45.Morey Kinney S. R., Smiraglia D. J., James S. R., Moser M. T., Foster B. A., Karpf A. R. (2008) Mol. Cancer Res. 6, 1365–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng D. F., Kanai Y., Sawada M., Ushijima S., Hiraoka N., Kitazawa S., Hirohashi S. (2006) Carcinogenesis 27, 1160–1168 [DOI] [PubMed] [Google Scholar]
- 47.Vallböhmer D., Brabender J., Yang D., Schneider P. M., Metzger R., Danenberg K. D., Hölscher A. H., Danenberg P. V. (2006) Clin. Lung Cancer 8, 39–44 [DOI] [PubMed] [Google Scholar]
- 48.Brueckner B., Kuck D., Lyko F. (2007) Cancer J. 13, 17–22 [DOI] [PubMed] [Google Scholar]
- 49.Robertson K. D., Uzvolgyi E., Liang G., Talmadge C., Sumegi J., Gonzales F. A., Jones P. A. (1999) Nucleic Acids Res. 27, 2291–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spada F., Haemmer A., Kuch D., Rothbauer U., Schermelleh L., Kremmer E., Carell T., Längst G., Leonhardt H. (2007) J. Cell Biol. 176, 565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui M., Wen Z., Yang Z., Chen J., Wang F. (2009) Mol. Biol. Rep. 36, 2201–2207 [DOI] [PubMed] [Google Scholar]
- 52.Medstrand P., van de Lagemaat L. N., Dunn C. A., Landry J. R., Svenback D., Mager D. L. (2005) Cytogenet. Genome Res. 110, 342–352 [DOI] [PubMed] [Google Scholar]
- 53.Reiss D., Zhang Y., Mager D. L. (2007) Nucleic Acids Res. 35, 4743–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraga M. F., Uriol E., Borja Diego L., Berdasco M., Esteller M., Cañal M. J., Rodríguez R. (2002) Electrophoresis 23, 1677–1681 [DOI] [PubMed] [Google Scholar]
- 55.Chapman V., Forrester L., Sanford J., Hastie N., Rossant J. (1984) Nature 307, 284–286 [DOI] [PubMed] [Google Scholar]
- 56.Sanford J., Forrester L., Chapman V., Chandley A., Hastie N. (1984) Nucleic Acids Res. 12, 2823–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yaqinuddin A., Qureshi S. A., Qazi R., Farooq S., Abbas F. (2009) J. Urol. 182, 756–761 [DOI] [PubMed] [Google Scholar]
- 58.Saikawa Y., Kubota T., Maeda S., Otani Y., Kumai K., Kitajima M. (2004) Oncol. Rep. 12, 527–531 [PubMed] [Google Scholar]
- 59.Sato N., Maehara N., Su G. H., Goggins M. (2003) J. Natl. Cancer Inst. 95, 327–330 [DOI] [PubMed] [Google Scholar]
- 60.Arima T., Hata K., Tanaka S., Kusumi M., Li E., Kato K., Shiota K., Sasaki H., Wake N. (2006) Dev. Biol. 297, 361–373 [DOI] [PubMed] [Google Scholar]
- 61.Hata K., Okano M., Lei H., Li E. (2002) Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.