Abstract
The tumor suppressor p53 is regulated by numerous post-translational modifications. Lysine methylation has recently emerged as a key post-translational modification that alters the activity of p53. Here, we describe a novel lysine methylation site in p53 that is carried out by two homologous histone methyltransferases, G9a and Glp. G9a and Glp specifically methylate p53 at Lys373, resulting mainly in dimethylation. During DNA damage, the overall level of p53 modified at Lys373me2 does not increase, despite the dramatic increase in total p53, indicating that Lys373me2 correlates with inactive p53. Further, reduction of G9a and/or Glp levels leads to a larger population of apoptotic cells. Examination of the Oncomine data base shows that G9a and Glp are overexpressed in various cancers compared with corresponding normal tissues, suggesting that they are putative oncogenes. These data reveal a new methylation site within p53 mediated by the methylases G9a and Glp and indicate that G9a is a potential inhibitory target for cancer treatment.
Keywords: Chromatin/Epigenetics, Chromosomes/Histones, Cancer Therapy, p53, Protein Methylation, Cancer, Epigenetics, Histone Methylation
Introduction
The tumor suppressor p53 is a sequence-specific transcription factor. The transcription activity is critical for its tumor suppressive function; indeed, more than half of human tumors bear a mutation in the p53 gene, largely within the DNA binding domain (1). Many mechanisms regulate transcriptional activation by p53, including nuclear localization, DNA binding, post-translational modifications (PTMs),2 and co-factor recruitment. These regulatory pathways are interdependent; for example, p53 acetylation promotes both its ability to bind to DNA and its recruitment of cofactors (2). Among all of these regulatory steps during the activation of p53, PTMs have attracted extensive attention because of their complex roles in tumorigenesis and possible clinical applications.
Lysine methylation is a relatively stable protein PTM relative to acetylation; and similar to acetylation, lysine methylation regulates diverse substrates (3). Regulatory potential is greatly increased because multiple methyl groups can be added to a single lysine residue, leading to mono-, di-, or trimethylation of a substrate protein. Indeed, within histone substrates, these various levels of methylation localize to different chromosomal regions and correlate with distinct biological outcomes. For example, histone H3 lysine 9 trimethylation (H3Lys9me3) generally localizes in the heterochromatic regions, whereas mono- and dimethylation (H3Lys9me1 and H3Lys9me2) localize within euchromatin (4).
Recently, lysine methylation has been shown to regulate the activity of p53 (3). Multiple lysines on p53 are methylated (Lys370, Lys372, and Lys382) by the methyl transferases Smyd2, Set7/9, and PR-Set7 (also known as Set8), respectively, which were previously characterized as histone methylases (5–7). These methylases specifically monomethylate their corresponding sites in p53. However, Lys370 and Lys382 dimethylation (Lys370me2 and Lys382me2) have both been detected on p53, but the cognate enzymes to these methylation sites remain unknown (8, 9).
Interestingly, similar to histones, the variable levels of p53 methylation result in different biological outcomes. For example, Lys370me1 is associated with repression of p53 function, whereas Lys370me2 is associated with p53 activation. In addition, Lys382me1 antagonizes Lys382 acetylation (Lys382ac), whereas Lys382me2 is linked to p53 response to DNA damage (7, 9). Further, as has also been observed for histone modifications, the PTMs of p53 cross-talk between one another, which greatly increases the potential complexity of p53 regulation by modification. For example, Lys372me1 suppresses Lys370me1, and Lys382me1 blocks Lys382ac (5, 7).
G9a and Glp are homologous histone methyl transferases that methylate histone H3 at Lys9 and Lys27. Gene knockouts of G9a or Glp in the mouse result in embryonic lethality (10, 11), suggesting that G9a and Glp are critical during mammalian development. However, little is known about the pathophysiological roles of G9a and Glp in oncogenesis. Here, we investigate p53 methylation mediated by G9a and Glp. Our results indicate that there is a potential role for G9a and Glp in human cancers and link p53 methylation site to tumorigenesis.
EXPERIMENTAL PROCEDURES
Peptides and Antibodies
Peptides were synthesized as described previously (5, 9). The amino acid numbering refers to the human p53 protein sequence, unless otherwise indicated. The p53Lys373me2 antibody was generated in rabbits using a peptide corresponding to the CSHLKSKK(me2)GQST sequence from p53. Crude rabbit antiserum was used in Western blot analysis at a dilution of 1:500 or 1:1,000.
Purification of Recombinant G9a and Glp
Full-length human G9a (amino acids 1–1211) and Glp (amino acids 1-1299) were cloned into a baculovirus vector pFastBac-HTA-FLAG. pFastBac-HTA-FLAG was generated by adding a FLAG tag into vector pFastBac-HTA (Invitrogen). Baculoviruses containing cDNA of G9a or Glp were used to infect insect Sf9 cells, as described previously (5, 9). FLAG-tagged G9a and Glp were purified from Sf9 extracts using anti-FLAG M2 antibody conjugated to agarose beads. Colloidal staining was used to confirm the purity of the proteins.
In Vitro Methylation Assay
Methylation assays were performed as described previously (5, 9). Briefly, 1 μg of peptide, 2 μg of histone H3 or GST-p53 were used as a substrate, and 1 μg of recombinant G9a or Glp was used as enzyme to perform the methylation assay. The total volume was 20 μl, and reactions were incubated at 30 °C for 30 min. Half of the reaction was used for colloidal staining and the other for fluorography. Transient transfection assays were performed using Lipofectamine 2000, as described previously (5, 9).
Small Interference RNA (siRNA) Knockdown
MCF7 cells were seeded in 6-well plates. The next day, cells were transfected with 100 nm luciferase siRNA (control), G9a siRNA1, siRNA2, Glp siRNA1 or siRNA2 using Dharmafect 1 (Dharmacon) twice in succession, with a 1-day interval between transfections. For transfections containing G9a_si1, G9a_si2, Glp_si1, and Glp_si2, a 25 nm concentration of each siRNA was mixed to a total concentration of 100 nm. 24 h after the second transfection, cells were either untreated or treated with 0.5 μm adriamycin for 36 h followed by flow cytometry analysis.
Western Blot Analysis
Cells were lysed with radioimmune precipitation assay buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate) and 1 × protease inhibitor (Roche Applied Science). Cell debris was removed by centrifugation at 13,000 rpm for 10 min at 4 °C, and 25 μg of cleared supernatant was electrophoresed using NUPAGE gels (Invitrogen).
Flow Cytometry Analysis
Flow cytometry was performed as described previously (5, 9). Briefly, cells were fixed with 70% ethanol overnight at −20 °C, treated with RNase A for 1 h at room temperature, and stained with 100 μg/ml propidium iodide for 10 min before analysis using a FACSCalibur (Becton Dickinson).
Oncomine Data Base
The use of the data base is described previously (12). Data sets with differential expression of G9a or Glp between normal and cancer tissues (p < 0.001) were selected.
RESULTS
G9a and Glp Methylate p53 at Lys373 in the C Terminus
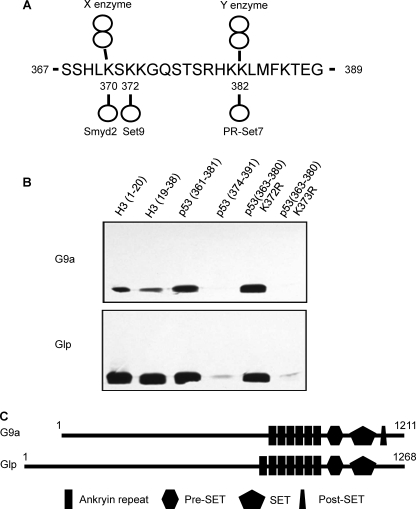
Several methylation sites in p53 (Lys370me, Lys372me, and Lys382me) were identified previously (Fig. 1A) (5–7). During our search for methylases that modify p53 in vitro (5), two additional histone methylases, G9a and Glp, modified p53. Both enzymes methylated a p53 peptide encompassing the C terminus from 361 to 381, but not a p53 peptide encompassing 374–391 (Fig. 1B). In addition, controls confirmed that G9a and Glp methylated histone H3 peptides, H3 (1–20) and H3 (19–38), consistent with previous observations that these enzymes methylate histone H3 at lysine 9 and lysine 27 (11). Notably, methylation of the p53 peptide is comparable in efficiency with the methylation of the two histone peptides (Fig. 1B). Further mapping showed that both G9a and Glp methylate p53 at Lys373 because a 361–381 peptide that was substituted from lysine to arginine at 373 (K373R) was not methylated, whereas a K372R peptide was methylated. The methylases G9a and Glp are homologous proteins, and both contain a SET domain and ankyrin repeats (Fig. 1C). Recently, ankyrin repeats in G9a have been shown to bind to histone H3 lysine 9 (H3Lys9me2), the same modification that is carried out by G9a and Glp.
FIGURE 1.
G9 and Glp methylate p53 peptides at Lys373. A, summary of known p53 methylation sites. X and Y indicate that the enzymes for these dimethyl sites are not identified. The methylation status (mono- or di-) has been validated by Western blotting and mass spectrometry. B, fluorography of histone and p53 peptide methylation by G9a and Glp enzymes. C, schematic showing domain structure of human G9a and Glp enzymes.
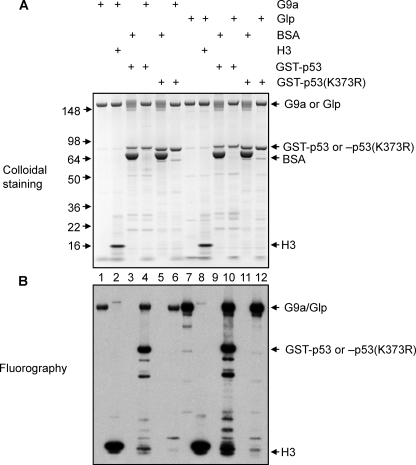
To investigate whether Lys373 is the only methylation site in p53 that is catalyzed by G9a and Glp, we prepared full-length G9a and Glp from Sf9 cells infected with baculovirus expressing these enzymes. Glutathione S-transferase (GST)-tagged full-length human p53 and p53 (K373R) were generated in bacteria as substrates. We performed in vitro methylation assays using these reagents, along with histone H3 as a positive control (Fig. 2). The enzymes automethylate in the in vitro assay as was detected in the bovine serum albumin negative control reactions (Fig. 2, lanes 1 and 7) compared with the reaction without enzyme (Fig. 2, lanes 3 and 9). GST-p53 was efficiently methylated by either enzyme; indeed, the methylation of p53 (Fig. 2, lanes 4 and 10) was comparable with the methylation of the positive control, histone H3 (Fig. 2, lanes 2 and 8). Further, Lys373 appears to be the only methylation site targeted by either G9a and Glp in vitro because GST-p53 (K373R) was not methylated by either enzyme (Fig. 2, lanes 6 and 12).
FIGURE 2.
G9a and Glp methylate full-length p53 at Lys373. A, colloidal staining of in vitro methylation of p53 by G9a or Glp. Bovine serum albumin (BSA) serves as a negative control for enzymes, and H3 serves as a positive control for substrates. 2 μg of substrate and 1 μg of enzyme were used for each reaction. The components in each reaction are indicated. B, fluorography of in vitro methylation of p53 by G9a or Glp. A and B were identical volume aliquots from the same reaction.
G9a and Glp Dimethylate, and May Monomethylate, p53 in Vitro
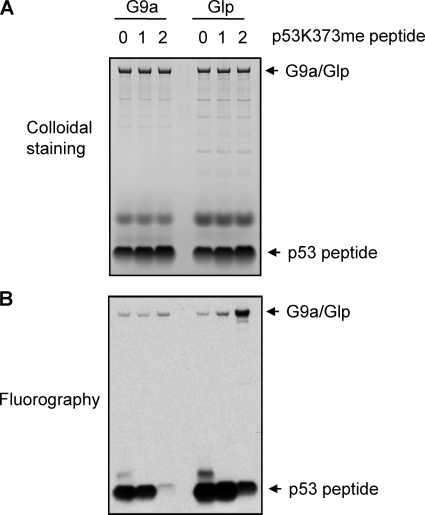
As mentioned above, the status of methylation can correlate with different biological outcomes. For example, in p53, Lys370me1 is linked to repression of p53 activity, whereas Lys370me2 is linked to p53 activation (5, 9). Methylation of p53 at Lys382 also results in different functional consequences, in that Lys382me1 represses p53 activity, whereas Lys282me2 is involved in the DNA damage response mediated by p53 (7, 8). For this reason, we investigated the methylation status of p53Lys373 by G9a and Glp. We performed in vitro methylation reactions using p53 peptide (363–381) as substrate, either unmodified (Lys373me0) or chemically monomethylated (Lys373me1) or dimethylated (Lys373me2) (Fig. 3). Both G9a and Glp methylated p53Lys373me0 and Lys373me1, and Glp weakly methylated Lys373me2. These results suggest that G9a and Glp mainly dimethylate and possibly monomethylate p53 at Lys373. Similarly, G9a and Glp mainly dimethylate the histone H3 Lys9 (10, 11).
FIGURE 3.
p53Lys373 methylation status by G9a and Glp. A, colloidal staining of in vitro methylation by G9a and Glp of peptides representing p53 (363–380) incorporating Lys373me0, me1, and me2. 1 μg of substrate and 1 μg of enzyme were used for each reaction. The components for each reaction are indicated. B, fluorography of in vitro methylation of p53 peptides by G9a or Glp. A and B were identical volume aliquots from the same reaction.
G9a and Glp Dimethylate p53 at Lys373 in Cells
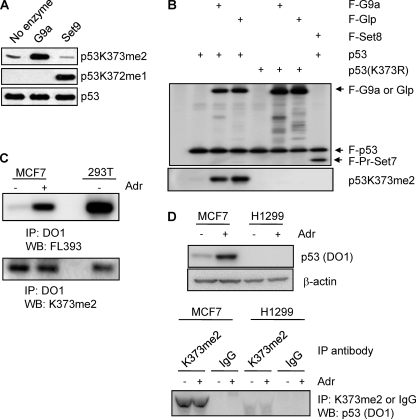
To investigate the roles of G9a and Glp-mediated dimethylation of p53 at Lys373 in cells, we generated a rabbit polyclonal antibody using a p53 peptide dimethylated at Lys373 (Lys373me2). Dot-blot analysis showed that this antibody (p53Lys373me2) recognized the p53 peptide p53Lys373me2 with a much higher affinity than other p53 methylation sites (supplemental Fig. S1). In addition, the p53Lys373me2 antibody specifically recognized p53 methylation carried out by recombinant G9a by not by Set9, a Lys372me1 methylase (6) (Fig. 4A). We then used this antibody to examine whether G9a and Glp can specifically methylate p53Lys373 in cells. For this purpose, vectors containing FLAG-tagged p53 or p53 (K373R) cDNA were co-transfected with FLAG-tagged G9a or Glp into 293T cells (Fig. 4B). FLAG-p53 was immunoprecipitated using FLAG resin and eluted using FLAG peptides, followed by Western blotting using the Lys373me2-specific antibody. We observed a strong Lys373me2 signal when the p53 expression vector was co-transfected with G9a or Glp expression vector compared with empty vector (Fig. 4B, left side). Importantly, there was no Lys373me2 signal when p53 (K373R) vector was used (Fig. 4B, right side). PR-Set7 (Set8), a methylase that monomethylates p53 at Lys382 (7), did not methylate p53 at Lys373. These data demonstrate that p53 is dimethylated by G9a and Glp in cells.
FIGURE 4.
G9a and Glp methylate p53 in mammalian cells. A, testing of polyclonal antibody recognizing p53Lys373me2. Full-length p53 was unmethylated or methylated by either recombinant G9a or Set9 in vitro. Methylation products were subjected to Western blot analysis with antibodies specific to p53Lys373me2, p53Lys372me1, and total p53. B, Western blot analysis of 293T cells transfected with vectors as indicated using FLAG antibody (upper panel) and p53Lys373me2 antibody (lower panel). F, FLAG-tagged; K373R, lysine to arginine substitute at Lys373. C, Western blot analysis of MCF7 cells untreated or treated with adriamycin for 8 h. 293T cell lysate was used as a control. Immunoprecipitates (IP) using DO1 were subjected to Western blot (WB) analysis using rabbit polyclonal antibody, FL393 (upper panel), and Lys373me2 antibody (lower panel). D, upper panels, Western blot analysis of MCF7 and H1299 cells untreated or treated with adriamycin (Adr) for 8 h using p53 (DO1) and β-actin antibodies. Lower panels, same amount of cell lysates from MCF7 or H1299 was immunoprecipitated with p53Lys373me2 or IgG (negative control) antibody followed by Western blot analysis using p53 (DO1) antibody.
To assess the p53Lys373me2 endogenously, MCF7 cells were treated with adriamycin, a DNA-damaging agent, for 8 h. MCF7 cells express wild-type p53 and thus can be used to examine endogenous p53 signaling upon DNA damage. A p53 mouse monoclonal antibody, DO1, was used to immunoprecipitate total p53 followed by Western blotting with rabbit polyclonal antibody, FL393, and with the p53Lys373me2 antibody. After an 8-h adriamycin treatment, the level of total p53 was greatly induced (Fig. 4C, upper left). Surprisingly, the Lys373me2 levels remained essentially unchanged (Fig. 4C, lower left), suggesting that Lys373me2 occurs on inactive p53 and not on activated p53. Consistent with the signal occurring on inactive p53, we also detected the Lys373me2 signal on p53 in 293T cells (Fig. 4C, right), in which endogenous p53 is inactivated by a virus protein, SV40 large T antigen.
To demonstrate clearly the specificity of p53Lys373me2 antibody in cells, we performed the reverse immunoprecipitation in MCF7 and H1299 cell lysates with p53Lys373me2 antibody or rabbit IgG followed by Western blot analysis with DO1 antibody (Fig. 4D). H1299 is a p53-null lung cancer cell line that we used as a negative control. We detected p53Lys373me2 signal only in MCF7 cells with immunoprecipitation using p53Lys373me2 antibody, but not immunoprecipitation with IgG. Additionally, we did not detect p53Lys373me2 in H1299 cell lysate. These data show that G9a and Glp dimethylate p53 at Lys373 in cells.
Reduction of the Levels of G9a and Glp Increases Cell Apoptosis
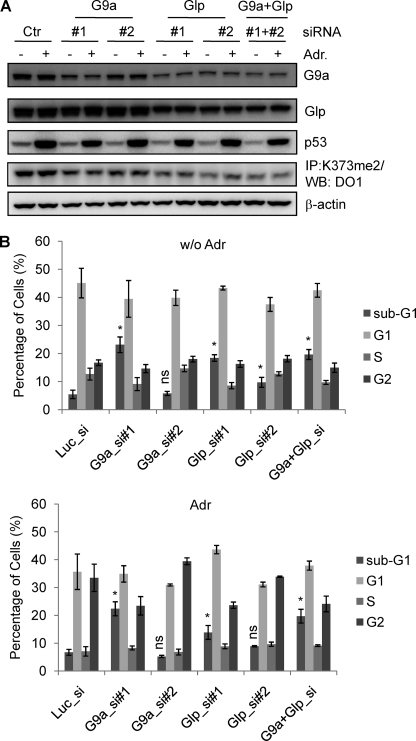
Because Lys373me2 may be linked to the repressed state of p53, we hypothesized that Lys373me2 by G9a and Glp may help to maintain p53 in an inactive form. This hypothesis suggests that reduction of G9a and Glp levels will result in hyperactivation of p53. To test this prediction, we utilized siRNAs that target luciferase (control), G9a, and/or Glp in MCF7 cells (Fig. 5A and supplemental Fig. S2). Two siRNAs for G9a (G9a_si1 and si2) and two siRNAs for Glp (Glp_si1 and si2) were used to reduce enzyme levels and to control for “off-target” effects. After a 48-h siRNA treatment, MCF7 cells were subjected to adriamycin treatment for 36 h (reduction of RNA and protein shown in supplemental Fig. S2 and Fig. 5A, respectively), followed by flow cytometry and analysis of DNA content. In most cases, knockdown of G9a and/or Glp dramatically increased the sub-G1 (apoptotic) population (Fig. 5B), and where the difference was not significant (i.e. G9a_si2), the siRNA treatment was not as effective. It is noteworthy that the increase in the sub-G1 population correlated with a decrease in the p53Lys373me2 signal (Fig. 5A). This inverse correlation strongly suggests that knockdown of G9a and/or Glp releases p53 from repression mediated by Lys373me2 and leads to an increase in the proportion of apoptotic cells. Interestingly, the combination of G9a and Glp siRNA knockdown did not further increase the percentage of apoptotic population, indicating that G9a and Glp are not playing redundant roles in this process. This observation is reminiscent of the H3Lys9 dimethylation by G9a and Glp, in which disruption of either G9a or Glp dramatically decreases H3Lys9me2 signal. Therefore, G9a and Glp are playing cooperative roles rather than simply redundant roles, which is consistent with previous observations that these two proteins exist in a complex and affect the activity of each other (11). Taken together, the data suggest that G9a and Glp repress apoptosis, consistent with the hypothesis that p53Lys373me2 is negatively regulating p53.
FIGURE 5.
G9a and Glp knockdown increases apoptosis in MCF7 cells. A, Western blot (WB) analysis with MCF7 cell lysates mock-transfected or transfected with G9a and/or Glp siRNAs followed by adriamycin (Adr) treatment. Antibodies were G9a, Glp, p53, and β-actin as indicated on the right side. To detect Lys373me2 signal, same amount of cell lysates were immunoprecipitated (IP) with p53Lys373me2 antibody followed by Western blot analysis using p53 (DO1) antibody. B, flow cytometry analysis of MCF7 cells transfected with luciferase (control), G9a (G9a_si1 and si2), Glp (Glp_si1 and si2), and both G9 and Glp siRNAs followed by 36-h adriamycin treatment. Propidium iodide was used to stain the DNA for flow cytometry. Shown are the average percentage of cells from three or four independent repeats, and only the statistical analyses of sub-G1 are shown; *, p < 0.05 (compared with Luc_si); ns, not significant. Error bars, S.E.
G9a Is Overexpressed in Various Cancers
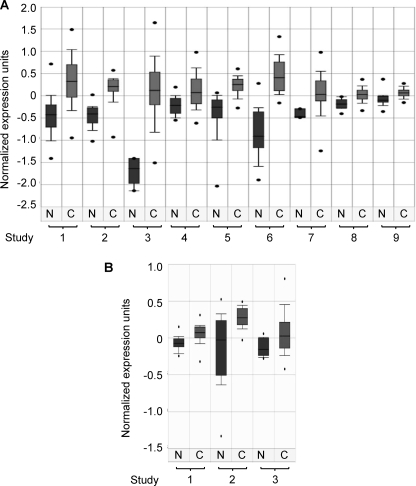
Because G9a- and Glp-mediated Lys373me2 appears to repress p53 activity, the enzymes are potential oncogenes and, if overexpressed, could suppress p53 activity leading to cancer. To test this possibility, we investigated the expression levels of G9a and Glp in nine different types of cancers and their corresponding normal tissues, using the public gene expression data set in the Oncomine data base (12). Strikingly, G9a was up-regulated in all nine cancer types compared with the normal tissues (Fig. 6A). Glp is also overexpressed in brain tumors, multiple myeloma, and head and neck cancers (Fig. 6B). Of note, these data sets result from nine independent studies for G9a and three studies for Glp. Therefore, these data strongly suggest that overexpression of G9a and/or Glp promotes tumor cell growth.
FIGURE 6.
G9 and Glp are up-regulated in many types of cancer. A, boxplot of G9a expression in normal and cancerous tissues. N, normal tissue; C, cancerous tissue. The listed nine independent studies include: 1, nontumor liver versus hepatocellular carcinoma, p = 6.3e-24; 2, normal bladder versus superficial transitional cell carcinoma and invasive transitional cell carcinoma, p = 6.7e-8; 3, normal bone marrow versus B cell acute lymphoblastic leukemia, p = 3.9e-7; 4, adjacent normal colon mucosa versus primary colon carcinoma, p = 2e-6; 5, normal skin and benign nevus versus cutaneous melanoma, p = 4e-6; 6, normal adjacent prostate, normal adult prostate, normal pubertal prostate versus prostate carcinoma, p = 7e-6; 7, normal ovary versus ovarian serous adenocarcinoma, p = 1e-5; 8, normal lung versus lung adenocarcinoma, p = 6.6e-5; 9, normal B cells versus B cell chronic lymphocytic leukemia, p = 7.5e-5. B, boxplot of Glp expression in normal tissues and cancerous tissues. The listed three independent studies include: 1, normal brain versus astrocytoma, p = 1.4E-4; 2, normal bone marrow versus smoldering multiple myeloma, p = 5.1E-4; 3, normal versus head and neck cancer, p = 6.1E-4.
DISCUSSION
The tumor suppressor p53 is heavily decorated with a variety of PTMs, which are centrally involved in regulation of p53 activity (1). Most of these modification sites reside within the N-terminal transactivation and C-terminal regulatory domains. However, these two regions contain many fewer inactivating mutations occurring in human cancer compared with the central DNA binding domain (1); this relationship leads to provocative questions about the role and overall importance of these modifications in p53 biology (13). In particular, regarding lysine modifications, mouse genetic models expressing p53 with seven (p537KR: lysines 367, 369, 370, 378, 379, 383, and 384 in mouse) or six (p536KR: lysines 367, 369, 370, 378, 379, and 383 in mouse) lysine-to-arginine mutations at its C terminus display only minor effects in either tumorigenesis or the DNA damage response (13–15). Lys373 (Lys370 in mouse), the site described in this study, is one of these lysine sites. One simple explanation for the modest defects and low incidence of mutations found at these modification sites is that redundancies exist between the sites. For example, p300/CBP acetylates p53 at numerous lysines (120, 164, 370, 372, 373, 381, 382, and 386 in human). p53 with substitutions at individual lysine residues behaves similarly to wild-type p53. However, substitution of all eight substrate lysines to arginine (8KR) leads to a strong defect in p53 activity (16). This result also potentially explains the observations that p537KR and p536KR mice are normal (14, 15).
However, in contrast to acetylation, all identified p53 methylases are site-specific, i.e. each of Smyd2, Set7/9, PR-Set7 (Set8), G9a, and Glp methylates a single site on p53. Therefore, the lack of mutation(s) in these lysine sites cannot simply be explained by redundancies among these sites. Thus, a key question is whether these sites regulate p53 activity in growth and cancer pathways. One explanation is that the methylases engage in cross-talk, to cooperate or antagonize each other's activity. Indeed, positive-acting Lys372me1 by Set7/9 inhibits negative-acting Lys370me1 mediated by Smyd2 (5). This coordination between sites and enzymes could lead to altered p53 activity beyond the effect of altering a single modification site. Another explanation is that the same lysine residue could be positively (acetylation) or negatively (methylation) modified. Therefore, mutation from lysine to arginine not only blocks positive modification, acetylation, but also prevents negative modifications such as methylation. The ultimate outcome of lysine-to-arginine mutation is the combinatorial effect of the loss of these positive and negative modifications.
Compared with acetylation, which seems to act only positively to up-regulate p53 activity, a subset of the methylation sites behaves negatively, to maintain p53 in an inactive state. These negatively acting sites include Lys370me1, Lys382me1, and, in this current study based on increased apoptosis in the reduced enzyme conditions, Lys373me2. Hence, these methylations are all potentially oncogenic because high levels of the enzymes, leading to constitutive methylation, would lead to underactive p53.
Relevant to this issue is the fact, mentioned above, that the p53 gene is mutated in only half of human tumors. It remains unclear how tumor formation escapes p53-mediated surveillance mechanisms in the other half of cancers apparently lacking p53 gene mutations. Our results and observations from other studies suggest that inappropriate levels of PTMs, due to overexpression of the cognate enzymes, may contribute to the inactivation of p53. One example is LSD1, a histone and p53 demethylase, which is overexpressed in neuroblastoma (17) and is associated with the relapse of prostate cancer (18). Our previous findings show that LSD1 removes a positive-acting p53 methylation at Lys372me2 (9), thus, high levels of LSD1 could be oncogenic because of constitutive repression of p53.
In a similar vein, our current observations indicate that p53Lys373me2 is negative acting and that the cognate methylases G9a and Glp are up-regulated in many types of cancers. These findings, both for LSD1 removing an activating methylation and for G9a/Glp adding a repressing methylation, strongly suggest that enzymes that negatively regulate p53 activity can act as oncogenes when expressed at aberrantly high levels. These results suggest that new therapies are possible via inhibition of these enzymes, which in turn would restore the activity of p53. This concept is particularly intriguing in tumor/cancers that bear a wild-type p53 gene. Future efforts will be needed to address the precise roles of p53 lysine modifications in tumor pathways.
Supplementary Material
Acknowledgment
We thank Dr. Nan Roche for critically reading the manuscript.
This work was supported, in whole or in part, by the NCI, National Institutes of Health Intramural Research Program and Center for Cancer Research (J. H.). This work was also supported by NCI, National Institutes of Health Grant CA 078831 (to S. L. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1 and S2.
- PTM
- post-translational modification
- Glp
- G9a-like protein
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- LSD1
- lysine specific demethylase 1.
REFERENCES
- 1.Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 2.Prives C., Manley J. L. (2001) Cell 107, 815–818 [DOI] [PubMed] [Google Scholar]
- 3.Huang J., Berger S. L. (2008) Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 4.Schotta G., Lachner M., Sarma Lys., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. (2004) Genes Dev. 18, 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Lysubicek S., Opravil S., Jenuwein T., Berger S. L. (2006) Nature 444, 629–632 [DOI] [PubMed] [Google Scholar]
- 6.Chuikov S., Lysurash J., Lys., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McLysinney Lys., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 7.Shi X., Lysachirskaia I., Yamaguchi H., West L. E., Wen H., Wang E. W., Dutta S., Appella E., Gozani O. (2007) Mol. Cell 27, 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lysachirskaia I., Shi X., Yamaguchi H., Tanoue Lys., Wen H., Wang E. W., Appella E., Gozani O. (2008) J. Biol. Chem. 283, 34660–34666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J., Sengupta R., Espejo A. B., Lee M. G., Dorsey J. A., Richter M., Opravil S., Shiekhattar R., Bedford M. T., Jenuwein T., Berger S. L. (2007) Nature 449, 105–108 [DOI] [PubMed] [Google Scholar]
- 10.Tachibana M., Sugimoto Lys., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Lysato H., Shinkai Y. (2002) Genes Dev. 16, 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Lysodama T., Hamakubo T., Shinkai Y. (2005) Genes Dev. 19, 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes D. R., Yu J., Shanker Lys., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo F., Wahl G. M. (2006) Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 14.Lysrummel Lys., A., Lee C. J., Toledo F., Wahl G. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng L., Lin T., Uranishi H., Gu W., Xu Y. (2005) Mol. Cell. Biol. 25, 5389–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y., Zhao W., Chen Y., Zhao Y., Gu W. (2008) Cell 133, 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulte J. H., Lim S., Schramm A., Friedrichs N., Lysoster J., Versteeg R., Ora I., Pajtler Lys., Lyslein-Hitpass L., Lysuhfittig-Lysulle S., Metzger E., Schüle R., Eggert A., Buettner R., Lysirfel J. (2009) Cancer Res. 69, 2065–2071 [DOI] [PubMed] [Google Scholar]
- 18.Lysahl P., Gullotti L., Heukamp L. C., Wolf S., Friedrichs N., Vorreuther R., Solleder G., Bastian P. J., Ellinger J., Metzger E., Schüle R., Buettner R. (2006) Cancer Res. 66, 11341–11347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.