Abstract
RecJ is a single-stranded DNA (ssDNA)-specific 5′-3′ exonuclease that plays an important role in DNA repair and recombination. To elucidate how RecJ achieves its high specificity for ssDNA, we determined the entire structures of RecJ both in a ligand-free form and in a complex with Mn2+ or Mg2+ by x-ray crystallography. The entire RecJ consists of four domains that form a molecule with an O-like structure. One of two newly identified domains had structural similarities to an oligonucleotide/oligosaccharide-binding (OB) fold. The OB fold domain alone could bind to DNA, indicating that this domain is a novel member of the OB fold superfamily. The truncated RecJ containing only the core domain exhibited much lower affinity for the ssDNA substrate compared with intact RecJ. These results support the hypothesis that these structural features allow specific binding of RecJ to ssDNA. In addition, the structure of the RecJ-Mn2+ complex suggests that the hydrolysis reaction catalyzed by RecJ proceeds through a two-metal ion mechanism.
Keywords: DNA Recombination, DNA Repair, DNase, Enzyme Mechanisms, Enzyme Structure, Protein-DNA Interaction, Protein Domains, Protein Metal Ion Interaction, OB fold, RecJ
Introduction
All of the organisms possess numerous pathways to repair DNA damages to maintain the integrity of the information contained in the genome. The single-stranded DNA (ssDNA)2 processing exonuclease RecJ has been implicated in many of these repair pathways. RecJ was originally identified as an essential gene for the RecF pathway of homologous recombination (1). RecJ produces 3′-ssDNA tails, which are required to initiate recombination from a double-stranded break (2). RecJ is also an exonuclease that mediates the excision step during mismatch repair (3, 4) and degrades abasic residues generated during base excision repair (5). Almost all eubacteria and archaea contain RecJ orthologs (6). These observations suggest that RecJ plays an important role in DNA repair.
RecJ is the only known ssDNA-specific 5′ exonuclease. It specifically hydrolyzes ssDNA in the 5′ to 3′ direction, which is dependent on metal cations (7). It processively degrades its substrate at ∼700 (8) or 1,000 (9) nucleotides/single binding event and appears to have a site that can bind seven nucleotides (9).
The crystal structure of a truncated Thermus thermophilus RecJ (ttRecJ) shows that the core domain of ttRecJ (cd-ttRecJ) (residues 40–463) consists of two domains that are interconnected by a long helix, forming a central cleft (10). Mn2+ in the active site is located on the wall of the cleft and is coordinated by conserved residues characteristic of a family of phosphoesterases that includes RecJ proteins. Although cd-ttRecJ contains the conserved residues of DHH (motifs I–IV) and DHHA1 motifs (11) (supplemental Fig. S1), the entire structure including the N- and C-terminal regions has not been determined.
To elucidate how RecJ achieves its high specificity for ssDNA, we determined the intact ttRecJ structures (666 residues) both in a ligand-free form and in a complex with Mn2+ or Mg2+ at 2.15–2.50 Å resolution. Intact ttRecJ consists of four domains that form a molecule with an O-like structure. Interestingly, one of two newly identified domains contains an oligonucleotide/oligosaccharide-binding (OB) fold. A protein fragment corresponding to this fold region bound to ssDNA. These structural and biochemical analyses explain why RecJ is a ssDNA-specific exonuclease. Furthermore, two Mn2+ ions in the active site located on the hole suggest that RecJ utilizes a two-metal ion mechanism for the exonuclease activity.
EXPERIMENTAL PROCEDURES
Materials
DNA-modifying enzymes, including restriction enzymes and LA Taq polymerase, were obtained from Takara Bio Inc. KOD-Plus polymerase was obtained from Toyobo. Yeast extract and polypeptone were obtained from Difco. The DNA oligomers were synthesized by BEX Co. [γ-32P]ATP was obtained from ICN. All of the other reagents used were of the highest available commercial grade.
Preparation of ttRecJ
Sequence analysis of the T. thermophilus HB8 genome identified one open reading frame encoding the RecJ protein. Using this sequence information, we synthesized two primers for the amplification of the target gene (ttha1167) by PCR. Amplification was performed using standard protocols, and the amplified gene fragment was ligated into a pT7Blue T-vector (Merck) by TA cloning and confirmed by sequencing. The fragment bearing the target gene from pT7Blue-ttha1167 was ligated to pET-11a (Merck) at the NdeI and BamHI sites. Escherichia coli Rosetta2(DE3)pLysS (Merck) cells transformed with the resulting plasmid were cultured at 37 °C to 4 × 108 cells/ml in 1.5 liters of LB medium containing 50 μg/ml ampicillin. The cells were then incubated for 6 h in the presence of isopropyl-β-d-thiogalactopyranoside, harvested by centrifugation, and stored at −20 °C.
All of the following procedures were performed at room temperature unless stated otherwise. Frozen cells (30 g) were thawed, suspended in 100 ml of buffer I (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm phenylmethanesulfonyl fluoride, and 1 mm EDTA), and disrupted by sonication on ice. The lysate was incubated at 70 °C for 15 min and centrifuged (38,000 × g) for 60 min at 4 °C. Ammonium sulfate was added to the resultant supernatant to a final concentration of 1.5 m and loaded onto a Toyopearl Ether-650M (Tosoh) column (bed volume, 20 ml) that was equilibrated with buffer II (50 mm Tris-HCl, pH 8.0, and 1 mm EDTA) containing 1.5 m ammonium sulfate. The proteins were eluted with a linear gradient of 1.5–0 m ammonium sulfate (total volume, 250 ml). The resultant supernatant was dialyzed against buffer II and loaded onto a Toyopearl SuperQ-650 M (Tosoh) column (bed volume, 10 ml) that was equilibrated with buffer II. The proteins were eluted with a linear gradient of 0–1 m NaCl (total volume, 200 ml). The fractions containing ttRecJ were collected and concentrated using a Vivaspin concentrator (molecular weight cut-off, 10,000). The concentrated solution was applied to a Superdex 200 HR 10/30 column (GE Healthcare Biosciences) that was equilibrated with buffer III (20 mm Tris-HCl, pH 7.5, and 100 mm KCl) and eluted with the same buffer using an ÄKTA explorer system (GE Healthcare Biosciences). The fractions containing ttRecJ were concentrated and stored at 4 °C. At each step, the fractions were analyzed by SDS-PAGE. The concentration of the purified protein was determined using the molar absorption coefficient at 278 nm calculated according to the formula of Kuramitsu et al. (12).
Preparation of ttRecJ Region 457–532 (ttRecJ-OB Domain)
DNA fragments expressing the ttRecJ region 457–532 were generated by PCR using T. thermophilus HB8 genome as a template. Amplification was performed using standard protocols, and the amplified gene fragment was ligated to pT7Blue vector and confirmed by sequencing. The fragment bearing the target gene from pT7Blue-ttrecj457–532 was ligated into pET-15b (Merck) at the NdeI and BamHI sites. E. coli Rosetta2(DE3)pLysS cells transformed with the resulting plasmid were cultured at 37 °C to 1 × 108 cells/ml in 1.5 liters of LB medium containing 50 μg/ml ampicillin. The cells were then incubated for 6 h in the presence of isopropyl-β-d-thiogalactopyranoside, harvested by centrifugation, and stored at −20 °C.
All of the following procedures were performed at room temperature unless stated otherwise. Frozen cells (15 g) were thawed, suspended in 100 ml of buffer IV (50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm phenylmethanesulfonyl fluoride, and 5 mm imidazole), and disrupted by sonication on ice. The lysate was centrifuged (38,000 × g) for 60 min at 4 °C. The resultant supernatant was loaded onto a His-Bind resin (Merck) column (bed volume, 5 ml) equilibrated with buffer V (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 5 mm imidazole). The proteins were eluted with a linear gradient of 0.06–1 m imidazole (total volume, 100 ml). The resultant supernatant was dialyzed against buffer VI (20 mm Tris-HCl, pH 8.0, 100 mm KCl, and 20 mm EDTA), and the His6 tag was cleaved with thrombin (Merck); thus, residues GSHM remained in the N terminus of the ttRecJ-OB domain. The solution was loaded onto a His-Bind resin column (bed volume, 5 ml) equilibrated with buffer V to remove the His6 tag. Ammonium sulfate was added to the flow-through fraction to a final concentration of 1.8 m and loaded onto a Toyopearl Butyl-650 M (Tosoh) column (bed volume, 5 ml) that was equilibrated with buffer II containing 1.8 m ammonium sulfate. The proteins were eluted with a linear gradient of 1.8–0 m ammonium sulfate (total volume, 100 ml). Fractions containing the target protein were collected and stored at 4 °C. This protein fragment was adsorbed onto a membrane of a Vivaspin centrifugal concentrator, and hence, the protein solution could not be concentrated by ultrafiltration.
Preparation of cd-ttRecJ
We constructed an expression plasmid for His10-tagged cd-ttRecJ, although we had previously prepared cd-ttRecJ (13). DNA fragments expressing the ttRecJ region 40–463 were generated by PCR using the T. thermophilus HB8 genome as a template. Amplification was performed using standard protocols, and the amplified gene fragment was ligated into pT7Blue vector and confirmed by sequencing. The fragment bearing the target gene from pT7Blue-ttrecj40–463 was ligated to pET-19b (Merck) at the NdeI and BamHI sites. E. coli Rosetta2(DE3)pLysS cells transformed with the resulting plasmid were cultured at 37 °C to 1 × 108 cells/ml in 1.5 liters of LB medium containing 50 μg/ml ampicillin. The cells were then incubated for 5 h in the presence of isopropyl-β-d-thiogalactopyranoside, harvested by centrifugation, and stored at −20 °C.
All of the following procedures were performed at room temperature unless stated otherwise. Frozen cells (6 g) were thawed, suspended in 80 ml of buffer IV, and disrupted by sonication on ice. The lysate was centrifuged (38,000 × g) for 60 min at 4 °C. The resultant supernatant was loaded onto a His-Bind resin column (bed volume, 5 ml) that was equilibrated with buffer V. The proteins were eluted with a linear gradient of 0.02–0.5 m imidazole (total volume, 100 ml). The resultant supernatant was dialyzed against buffer VII (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 1 mm EDTA), and the His10 tag was cleaved with enterokinase (Merck); thus, residues HM remained in the N terminus of cd-ttRecJ. The solution was loaded onto a His-Bind resin column (bed volume, 3 ml) equilibrated with buffer V to remove the His10 tag. Ammonium sulfate was added to the flow-through fraction to a final concentration of 1.4 m and loaded onto a Toyopearl Ether-650M column (bed volume, 5 ml) equilibrated with buffer II containing 1.4 m ammonium sulfate. The proteins were eluted with a linear gradient of 1.4–0 m ammonium sulfate (total volume, 100 ml). The fractions containing the target protein were collected and concentrated using a Vivaspin concentrator (molecular weight cut-off, 10,000). The concentrated solution was applied to a Superdex 200 HR 10/30 column that was equilibrated with buffer III and eluted with the same buffer using an ÄKTA explorer system. The fractions containing the target protein were concentrated and stored at 4 °C.
Crystallization, Data Collection, Molecular Replacement, Model Building, and Refinement
Crystallization conditions for ttRecJ were surveyed by the hanging drop vapor diffusion method using crystal screen kits (Crystal Screen, Crystal Screen2, Crystal Screen Cryo, Crystal Screen Lite, Natrix, Index, Salt RX, and MembFac; Hampton Research) at 293 K. The initial protein concentration was 10 mg/ml in 20 mm Tris-HCl, pH 7.5, and 100 mm KCl; 1 μl of the protein solution was mixed with the same volume of reservoir solution and equilibrated against it. After further optimization, crystals suitable for x-ray diffraction were obtained in an optimized reservoir solution containing 0.1 m lithium sulfate monohydrate, 0.1 m N-(2-acetamido) iminodiacetic acid (pH 6.5), 12% w/v polyethylene glycol 4000, and 2% (v/v) 2-propanol at 293 K.
The crystals were soaked in a cryoprotective solution containing 20% glycerol and flash-cooled in a nitrogen stream (93 K). The data were collected under cryo conditions at the Beamline BL26B2 at SPring-8. Crystals of Mn2+ and Mg2+ complexes were prepared by soaking the crystals in a reservoir solution containing 10 mm MnSO4 and 10 mm MgSO4, respectively. The diffraction images were processed using the program HKL2000 (14). Assuming that an asymmetric unit contained one subunit, the VM and Vsol values (15) were ∼2.95 Å3 Da−1 and 0.58, respectively. The regions of residues 49–292 and 326–433 of cd-ttRecJ (Protein Data Bank code 1IR6) were applied as a search model, and the program MOLREP (16) was used for molecular replacement phase determination. The initial model was built with the aid of the amino acid sequence using the program PHENIX (17) and refined with the programs Xtalview (18) and CNS (19). The success of model refinement was evaluated at each stage by the free R-factor and by inspection of stereochemical parameters with the program MolProbity web server (20). The final refinement indicated that more than 91.8% of the residues belonged to the favored regions in the Ramachandran plot.
Data for the crystals of the Mn2+ and Mg2+ complexes were collected at wavelengths of 1.00000 and 1.70000 Å at the Beamline BL26B2 at SPring-8. The diffraction images were processed using the program HKL2000. The structures were determined by molecular replacement using only the intact ttRecJ as a starting model and refined by the programs Xtalview and CNS.
The regions containing residues 109–118 and 659–666 in the ligand-free and Mn2+-bound forms and residues 109–118, 138–151, 163–170, and 659–666 in the Mg2+-bound form were disordered. The model structure of ttRecJ-Mn2+-7-mer ssDNA (5′-GAAAGAA-3′) was built up using the programs Xtalview and CNS. The figures were made using the programs CCP4 (21) and PyMOL (DeLano Scientific).
Electrophoretic Mobility Shift Assay
Custom-synthesized 21-mer ssDNA (ATGACAACTAAAGCAACACCC) and 21-mer single-stranded RNA (ssRNA) (AUGACAACUAAAGCAACACCC), obtained from BEX, were radiolabeled at their 5′-ends with [γ-32P]ATP using polynucleotide kinase. The 21-bp double-stranded DNA (dsDNA) was made by annealing 5′-32P-labeled 21-mer ssDNA with the complementary oligonucleotide. To analyze the interaction between oligo(ribo)nucleotides and intact ttRecJ, ttRecJ-OB domain or cd-ttRecJ, the reaction mixture (10 μl, pH 7.5) containing 50 mm Tris-HCl, 100 mm KCl, 20 mm EDTA, 10 nm 5′-32P-labeled oligo(ribo)nucleotide, and 0–400 nm protein was incubated at 37 °C for 10 min. A native PAGE 6× loading dye (50 mm Tris-HCl, pH 7.5, 100 mm KCl, 30% glycerol, 0.2% bromphenol blue, and 0.2% xylene cyanol (2 μl)) was added to the mixture, loaded onto a 9% (w/v) polyacrylamide gel, and electrophoresed in 1× TBE (89 mm Tris borate and 2 mm EDTA) buffer (13). The gel was dried and placed in contact with an imaging plate. The bands were visualized and analyzed using a BAS2500 image analyzer (Fuji Photo Film).
Assay for Exonuclease Activity
The reaction mixture (10 μl, pH 7.5), which contained 50 mm HEPES, 100 mm KCl, 5 mm MgCl2, 10 nm 5′-32P-labeled 21-mer ssDNA, varying concentrations of cold 21-mer ssDNA, and 2.5 nm ttRecJ or 25 nm cd-ttRecJ, was incubated at 37 °C. For each time point, the reaction was quenched by immediately adding 1 μl of 100 mm EDTA and 11 μl of phenol/chloroform. The samples were centrifuged, and an equal volume of sample buffer (5 mm EDTA, 80% deionized formamide, 10 mm NaOH, 0.1% bromphenol blue, and 0.1% xylene cyanol) was added to the supernatant. The samples were denatured at 95 °C for 3 min, loaded onto a 25% (w/v) acrylamide gel containing 8 m urea and 1 × TBE buffer, and then electrophoresed in 1 × TBE buffer. The gel was dried and placed in contact with an imaging plate. The bands were visualized and analyzed using a BAS2500 image analyzer. Three independent experiments were performed.
The percentage of undegraded ssDNA was plotted for each incubation period. Because the plot showed a linear region, the initial rate of the reaction could be obtained. In this assay, the enzyme was mixed with a large excess of the substrate. Assuming that the enzyme and substrate reacted according to a scheme similar to the Michaelis-Menten mechanism, the second step was considered the rate-limiting step when the enzyme-bound intermediate was in the steady state. The initial rate was plotted against substrate concentration. The data were fitted to the Michaelis-Menten equation, and the kinetic constant was calculated using the Igor Pro 3.14 software (WaveMetrics). The kcat and Km values were determined from nonlinear regression analysis.
RESULTS AND DISCUSSION
Overall Structure of ttRecJ
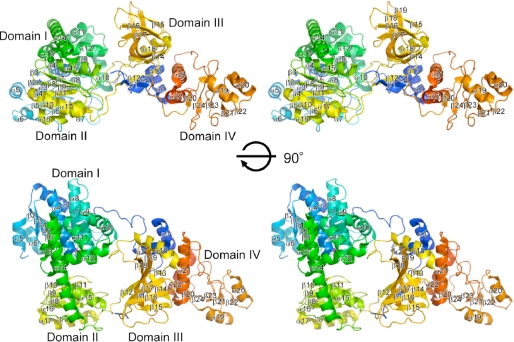
We determined the crystal structures of the intact ttRecJ by molecular replacement using cd-ttRecJ as an initial search model. Data collection and refinement statistics are shown in Table 1. The ttRecJ structure can be divided primarily into four domains (Fig. 1). Domains I (residues 47–291) and II (residues 323–425) are interconnected by a long helix (α14; residues 292–322) to form an active center, as identified previously for the structure of cd-ttRecJ (10). Domain III comprises the N-terminal region (residues 1–46) and ∼110 residues (residues 426–535). Domain IV comprises the C-terminal region of ∼120 residues (residues 536–658). This is the first report of the crystal structure of the full-length RecJ, and it is the first time that domains III and IV were structurally determined.
TABLE 1.
Data collection and refinement statistics
| ttRecJ (free) |
ttRecJ-Mn2+ (complex) |
ttRecJ-Mg2+ (complex) |
||
|---|---|---|---|---|
| Space group P43212 | Space group P43212 | Space group P43212 | Space group P43212 | |
| Data collection | ||||
| Cell dimensions | ||||
| a, b, c (Å) | 83.0, 83.0, 249.2 | 83.1, 83.1, 249.9 | 83.2, 83.2, 250.3 | 83.4, 83.4, 251.1 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.70000c | 1.00000 |
| Resolution (Å)a | 50–2.50 (1.90–2.50) | 50–2.30 (1.73–2.30) | 50–2.65 (2.74–2.65) | 50–2.15 (1.76–2.15) |
| Rmerge (%)a | 7.0 (30.2) | 7.8 (33.9) | 32.2 (6.6) | 6.4 (27.6) |
| Average I/σ(I)a | 45.4 (6.2) | 50.6 (9.0) | 38.5 (5.0) | 47.2 (3.3) |
| Completeness (%)a | 100 (99.9) | 100 (100) | 99.9 (99.5) | 98.5 (86.0) |
| Redundancya | 13.1 (9.9) | 13.4 (12.9) | 7.9 (5.1) | 17.2 (3.9) |
| Refinement | ||||
| Resolution (Å) | 42.7–2.50 | 41.6–2.30 | 43.0–2.15 | |
| No. reflections | 30,698 | 39,390 | 47,494 | |
| Rwork/Rfree | 0.230/0.282 | 0.232/0.281 | 0.235/0.279 | |
| No. atoms | ||||
| Protein | 5,020 | 5,020 | 4,773 | |
| Ion | – | 2 | 1 | |
| Water | 180 | 293 | 290 | |
| B-factors | ||||
| Protein | 42.0 | 36.3 | 33.8 | |
| Ion | 31.5 (Mn1), 65.2 (Mn2) | 15.3 | ||
| Water | 36.4 | 37.5 | 36.3 | |
| Root mean square deviations | ||||
| Bond lengths (Å) | 0.011 | 0.011 | 0.011 | |
| Bond angles (°) | 1.7 | 1.6 | 1.7 | |
| Ramachandran plot | ||||
| Favored regions (%)b | 91.8 (591/644) | 93.2 (600/644) | 93.3 (567/608) | |
| Allowed regions (%)b | 5.2 (34/644) | 4.6 (30/644) | 3.7 (23/608) | |
| Disallowed regions (%)b | 3.0 (19/644) | 2.2 (14/644) | 3.0 (18/608) | |
a The values in parentheses are for the outermost shell.
b The value was calculated using MolProbity (20). The values in parentheses are the numbers of residues.
c The diffraction data collected at 1.70000 Å was used to ascertain the presence of Mn2+ by use of an anomalous difference Fourier map.
FIGURE 1.
Overall structure of ttRecJ. Stereo view of a ribbon model of the overall structure of ttRecJ complexed with Mn2+. The color scheme for each segment is the same as that in supplemental Fig. S1.
A DALI search for domain IV did not identify any structures with a Z-score >4 and a root mean square deviation of <3.0 Å, suggesting that this fold was novel. It is unlikely that this domain participates in DNA binding or catalytic activity. This domain may be involved in interactions with other proteins. However, it should be noted that the ssDNA-binding protein (SSB) recognized a region within the core domains of RecJ (8).
Domain III Includes a Novel OB Fold Domain
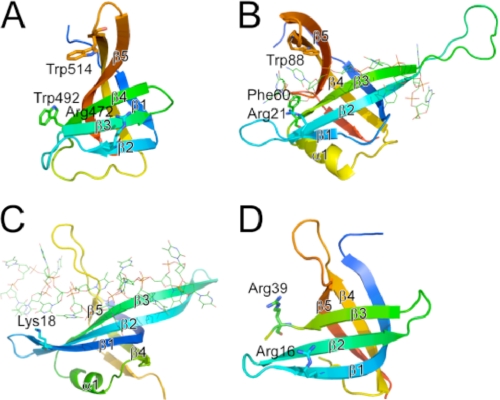
Unexpectedly, the structure of the full-length ttRecJ revealed significant structural similarities between a region in domain III (residues 457–532; Fig. 2A) and the OB fold domains of other proteins (Fig. 2, B–D). This region (the ttRecJ-OB domain) containing five β-strands (β15–β19) is similar to a typical OB fold domain consisting of a five-stranded β-sheet coiled to form a closed β-barrel (22). The DALI search for the ttRecJ-OB domain revealed significant matches, a Z-score of ∼8 and a root mean square deviation of <3.0 Å, with many SSBs (Fig. 2B) and PriB proteins (Fig. 2C), primosomal DNA replication proteins. In the common architecture of the OB fold, the β-sheet was capped by an α-helix located between the third and fourth strands (22); however, the ttRecJ-OB domain lacks this helix. An OB fold member that lacks this helix is also included in the N-terminal region of Deinococcus radiodurans RecO (23) (Fig. 2D).
FIGURE 2.
Structural comparison between ttRecJ-OB domain (A) and the OB fold domains of other proteins (B–D). A, region with residues 457–532 of ttRecJ (Protein Data Bank code 2ZXP). B–D, OB fold domains of E. coli SSB (residues 1–112; Protein Data Bank code 1EYG) (B), E. coli PriB (residues 1–102; Protein Data Bank code 2CCZ) (C), and D. radiodurans RecO (residues 2–79; Protein Data Bank code 1W3S) (D). OB folds displayed are colored ranging from blue at the N terminus to red at the C terminus. Displayed residues are assumed to interact with ssDNA. The secondary structure elements that form the OB fold are represented by β1–β5 and α1 according to the literature (22). β1–β5 in (A) and (B) correspond to β15–β19 and β2–β6 in the entire structure of ttRecJ and E. coli SSB, respectively. Residues considered important for binding to ssDNA are shown by sticks. ssDNA is shown by lines.
Some proteins containing OB folds have been shown to bind nucleic acids and prefer ssDNA over dsDNA (24). Structural and mutational studies of other OB fold proteins revealed that binding to nucleic acids was mediated by the following three major structural regions: the β1–β2 loop, the C-terminal part of β3, and the β4–β5 loop (25). The first region provides positively charged residues to form hydrogen bonds with the phosphate groups of the substrate; the remaining two provide one conserved aromatic side chain that stacks with the bases. In the ttRecJ structure, the residues in these three regions might be Arg472, Trp492, and Trp514, respectively (Fig. 2A). These findings also support the hypothesis that the ttRecJ-OB domain is involved in binding to ssDNA. Thus, considering the low sequence similarities of the RecJ-OB domains to other OB fold domains, we propose that the RecJ-OB domain is a novel member of the OB fold superfamily and is involved in binding to ssDNA.
To verify whether the ttRecJ-OB domain can bind to DNA, we prepared the ttRecJ-OB domain as a protein fragment and investigated its DNA binding ability by the electrophoretic mobility shift assay. The ttRecJ-OB domain could bind to both ssDNA and dsDNA (supplemental Fig. S2B). The intact ttRecJ could also bind to ssDNA, but not efficiently to dsDNA or ssRNA (supplemental Fig. S2A). The complexes between the ttRecJ-OB domain and DNA formed smeared bands on the gel, suggesting that the binding affinity of this domain with ssDNA was weaker than that of intact ttRecJ. We then prepared cd-ttRecJ lacking domains III and IV and measured DNA binding and exonuclease activity. The cd-ttRecJ exhibited ssDNA-specific 5′-3′ exonuclease activity, but a mobility shift was not observed (supplemental Fig. S2C) under the same conditions as used those for the intact RecJ (supplemental Fig. S2A). The kcat and Km values of the intact ttRecJ for 21-mer ssDNA were 0.0047 s−1 and 0.037 μm, whereas those of cd-ttRecJ were 0.0073 s−1 and 20 μm, respectively (Table 2). Compared with intact ttRecJ, cd-ttRecJ exhibited much lower affinity (∼540-fold) for the ssDNA substrate. These results therefore support the possibility that the ttRecJ-OB domain is involved in binding to DNA. However, it is also indicated that the ttRecJ-OB domain alone is insufficient for conferring ssDNA specificity on ttRecJ.
TABLE 2.
Kinetic constants for 5′-3′ exonuclease activity of ttRecJ and cd-ttRecJ
| Protein | Kma | kcata | kcat/Km |
|---|---|---|---|
| μm | s−1 | m−1s−1 | |
| ttRecJ | 0.037 ± 0.0054 | 0.0047 ± 0.00025 | 130,000 ± 13,000 |
| cd-ttRecJ | 20 ± 2.8 | 0.0073 ± 0.00071 | 370 ± 14 |
a The assays were performed at pH 7.5 as described under “Experimental Procedures.” Km and kcat were determined by nonlinear regression analysis of the assay data.
Because of the presence of domain III, the active site containing the metal center is located within the hole (supplemental Fig. S3). Near the active site, the hole between the three domains is too narrow (11 Å wide) for binding to dsDNA. Therefore, a ring-shaped structure is thought to ensure tight RecJ-ssDNA interaction by encircling the ssDNA.
Metal Ion-binding Sites and the Catalytic Mechanism
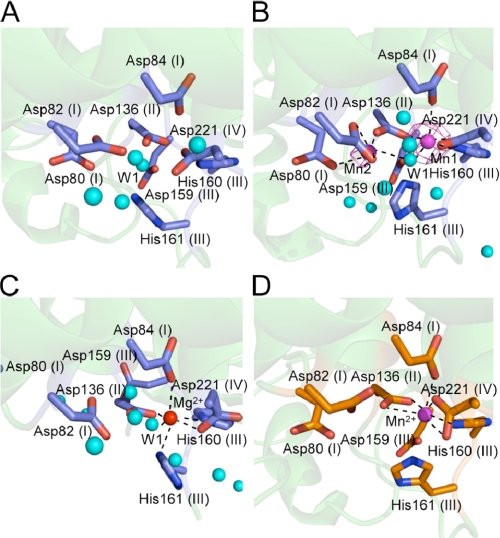
In the active site of the ttRecJ-Mn2+ complex, we identified electron density corresponding to two Mn2+ ions near motifs I–IV in the hole using an anomalous difference Fourier map (Fig. 3B). Although Mn1 was coordinated by four oxygen atoms and one nitrogen atom, Mn2 was coordinated by five oxygen atoms. Mn1 was located at the same positions as Mn2+ in cd-ttRecJ (Figs. 3D and 4) and a single Mg2+ ion in the Mg2+ complex (Fig. 3C). The density of Mn2 was much weaker than that of Mn1, and the B-factor of Mn2 (65.2) was higher than that of Mn1 (31.5), suggesting that Mn2 had a lower occupancy than Mn1.
FIGURE 3.
Metal ion-binding site. A, intact ttRecJ ligand-free form. B, Mn2+ ion-binding sites in intact ttRecJ. Electron density (magenta) of the Mn2+ ions derived from an anomalous difference Fourier map (λ = 1.70000 Å) is superimposed on the model. The position of the Mn2+ ion is shown by a magenta sphere. Water molecules are shown by cyan spheres. C, Mg2+ ion-binding site in intact ttRecJ. The position of the Mg2+ ion is shown by a red sphere. D, Mn2+ ion-binding site in cd-ttRecJ (Protein Data Bank code 1IR6). The position of the Mn2+ ion is shown by a magenta sphere. Wat1 (W1) is assumed to be the nucleophilic water. The motif names are indicated in parentheses. The dashed lines show the coordination with the metal ion.
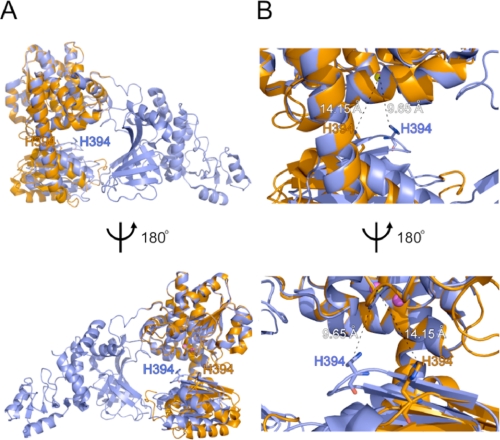
FIGURE 4.
Superimposition of the intact structure and cd-ttRecJ. A, superimposition of the overall intact ttRecJ (slate blue) and overall cd-ttRecJ (orange) structures. The figures were produced using the LSQKAB program in CCP4 (21). Mn2+ ions in the intact structure are shown as magenta spheres. The Mn2+ ion in cd-ttRecJ is shown as a yellow sphere. His394 in the DHHA1 motif is shown in stick form. B, an enlarged view of the area around His394. The dashed line shows the shortest distance between an atom of His394 and Mn1 or Mn2+.
Mn1 was coordinated with the side chain of Asp84 (2.38 Å, motif I), Asp136 (2.09 Å, motif II), His160 (2.27 Å, motif III), Asp221 (2.04 Å, motif IV), and one water molecule (2.52 Å, Wat1). Mn2 was coordinated with the side chains of Asp80 (2.25 Å, motif I), Asp82 (2.30 Å and 2.54 Å, motif I), Asp136 (1.92 Å, motif II), and the same water molecule (2.54 Å, Wat1). The water molecule (Wat1) coordinated by the metal ions was located at the same position as in the ligand-free form (Fig. 3A). The Mg2+ ion was coordinated in a nearly perfect octahedral geometry by four oxygen atoms and two nitrogen atoms. Coordination of the Mg2+ ion was similar to that of Mn1, except for an additional coordination with His161 (motif III) (Fig. 3C). This determined structure did not contain a second Mg2+ ion. Even in the Mn2+ complex, the affinity of Mn2 was considered to be weaker than that of Mn1. The binding of a nucleotide or ssDNA may stabilize the second metal ion, similar to what is observed for E. coli DNA polymerase I (26).
With regard to the catalytic mechanism, it is significant that the same water molecule is coordinated by two Mn2+ ions. In Saccharomyces cerevisiae cytosolic exopolyphosphatase, belonging to another DHH protein family, two metal cations are also coordinated by such a geometry in its active site (27). Therefore, we concluded that ttRecJ possessing two metal ions was an active form. The water molecule (Wat1) near the two Mn2+ ions is a probable nucleophile candidate for initiating an attack on the scissile phosphodiester bond of ssDNA, because Wat1 is located at the surface of the cleft with access to ssDNA. Coordination such as this is consistent with a two-metal ion mechanism of DNA cleavage, as suggested previously for other nucleases (28, 29).
Considering the spatial arrangement, Mn1 might be located to the 3′-side of the cleavage site and Mn2 to the 5′-side (supplemental Fig. S4). In other nucleases, metal ions are known to act as ligands for oxygen atoms of DNA phosphate groups. Mn2 might interact with the 5′-terminal nucleotide of the substrate and also with the mononucleotide, a product generated by cleavage of the phosphodiester bond.
Structural Difference between Intact ttRecJ and cd-ttRecJ
By comparing domains I and II between the intact and cd-ttRecJ forms, the movement of the loop containing the DHHA1 motif could be observed (Fig. 4). This loop was placed closer to the catalytic residues in the intact ttRecJ than in cd-ttRecJ. This might have been induced by an interaction between the N-terminal and middle regions in domain III outside the core domain. The distance between His394 in the DHHA1 motif and the metal ion (Mg2+ or Mn1) in the intact ttRecJ (10 Å) was shorter than that (Mn2+) in cd-ttRecJ (14 Å) but was too far for coordination. An E. coli RecJ H429A mutant showed markedly decreased activity, suggesting the functional importance of this residue (30). We hypothesize that this highly conserved His residue is involved in proper binding with ssDNA. Difference in the loop containing this His residue may be another reason for the differences in the Km values.
Model for the ttRecJ-ssDNA Complex
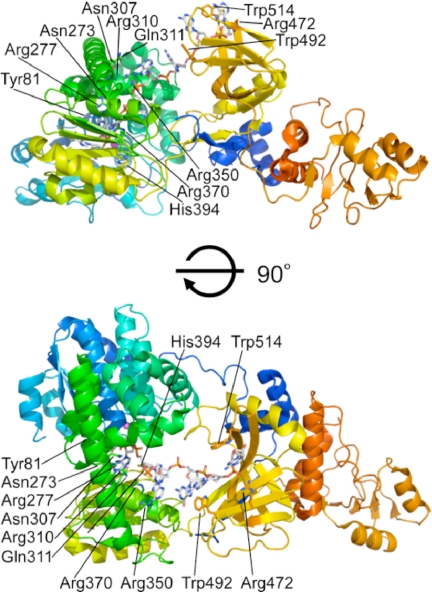
The ttRecJ-OB domain extended from the catalytic core domain (domains I and II) and was situated over the cleft (Fig. 1). Based on the results of this study and previous experimental data, it can be assumed that ssDNA enters into the active site within the hole from the side where the OB domain is located. The hole is more open on this side than at the other. Furthermore, highly conserved residues in domains I and II, Arg110 (disordered in the intact ttRecJ structures), Asn273, Arg277, Asn307, Arg310, Gln311, Arg350, and Arg370 (supplemental Fig. S1) were located on the side of the OB domain (Fig. 5). Among them, Asn273, Arg277, and Arg370 were considered responsible for binding to ssDNA (6, 30).
FIGURE 5.
A model structure of ttRecJ-ssDNA complex. The side chains of the possible residues for binding to ssDNA and ssDNA are shown as stick forms. Mn2+ ions are shown as magenta spheres. An ssDNA (7-mer) was built into the active site as described under “Experimental Procedures.”
If this is the case, then the ttRecJ-OB domain is probably situated at the entrance to the active site. Thus, it can be postulated that ssDNA enters the hole and passes under the bridge (α14 helix) between domains I and II. Based on these arguments and structures of the SSB protein-ssDNA complex, a model of the ttRecJ-ssDNA complex was constructed (Fig. 5). In this model, the 5′-terminal residue is located at an interior wall, which contains the catalytic site, and its base may be stacked with the side chain of Tyr81, which is highly conserved (supplemental Figs. S1 and S4). Consequently a dNMP (a product) is released from the gap between domains I and II. This model can also explain why RecJ has a site that can bind seven nucleotides (9) as well as high processivity (8, 9).
Our results suggest that a stable complex of ttRecJ with dsDNA cannot be formed even if the ttRecJ-OB domain binds to the dsDNA or dsDNA region in branched DNA forms. In addition, the affinity of the ttRecJ-OB domain for ssDNA was weaker than that of the intact protein (supplemental Fig. S2, A and B), and the cd-ttRecJ lacking this domain still exhibited ssDNA-specific 5′-3′ exonuclease activity. Therefore, this domain might play a role in stabilizing the substrate complex by enhancing binding to ssDNA. The C-terminal regions of all RecJ homologs display region similarities to the OB domain (supplemental Fig. S1), which supports the validity of the functional importance of this domain.
Multi-domain nucleases with an OB fold domain are known in the field of ribonucleases, such as RNase E (31), RNase II (32), and Rrp44 (33). To our knowledge, however, there is no other nuclease with an O-like structure accompanied by an OB fold domain, indicating the unique structural feature of RecJ. Recently, it was reported that MRT-1 nuclease, a dual domain protein required for telomerase function and DNA cross-link repair, contains an OB fold domain (34). It could be believed that more nucleases containing an OB fold domain will be found. Our data provide structural framework for understanding the role of oligonucleotide binding by the OB fold domain in those nucleases.
Supplementary Material
Acknowledgments
We are grateful to Dr. Masaki Yamamoto, Yuka Nonaka, and Toshi Arima for help with the x-ray data collections. We are grateful to Hirohumi Ohmori for DNA sequencing.
This work was supported in part by Grants-in-Aid for Scientific Research 20570131 (to R. M.) and 19770083 (to N. N.) from the Ministry of Education, Science, Sports and Culture of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 2ZXO, 2ZXP, and 2ZXR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ss
- single-stranded
- cd-ttRecJ
- core domain of ttRecJ
- ds
- double-stranded
- OB
- oligonucleotide/oligosaccharide-binding
- SSB
- ssDNA-binding protein
- ttRecJ
- T. thermophilus RecJ
- Wat
- water molecule.
REFERENCES
- 1.Lovett S. T., Clark A. J. (1984) J. Bacteriol. 157, 190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handa N., Morimatsu K., Lovett S. T., Kowalczykowski S. C. (2009) Genes Dev. 23, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdett V., Baitinger C., Viswanathan M., Lovett S. T., Modrich P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan M., Burdett V., Baitinger C., Modrich P., Lovett S. T. (2001) J. Biol. Chem. 276, 31053–31058 [DOI] [PubMed] [Google Scholar]
- 5.Dianov G., Sedgwick B., Daly G., Olsson M., Lovett S., Lindahl T. (1994) Nucleic Acids Res. 22, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajman L. A., Lovett S. T. (2000) J. Bacteriol. 182, 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovett S. T., Kolodner R. D. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma R., Rao D. N. (2009) J. Mol. Biol. 385, 1375–1396 [DOI] [PubMed] [Google Scholar]
- 9.Han E. S., Cooper D. L., Persky N. S., Sutera V. A., Jr., Whitaker R. D., Montello M. L., Lovett S. T. (2006) Nucleic Acids Res. 34, 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamagata A., Kakuta Y., Masui R., Fukuyama K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5908–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L., Koonin E. V. (1998) Trends Biochem. Sci. 23, 17–19 [DOI] [PubMed] [Google Scholar]
- 12.Kuramitsu S., Hiromi K., Hayashi H., Morino Y., Kagamiyama H. (1990) Biochemistry 29, 5469–5476 [DOI] [PubMed] [Google Scholar]
- 13.Yamagata A., Masui R., Kakuta Y., Kuramitsu S., Fukuyama K. (2001) Nucleic Acids Res. 29, 4617–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 16.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 17.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 18.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 19.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 20.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 22.Murzin A. G. (1993) EMBO J. 12, 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leiros I., Timmins J., Hall D. R., McSweeney S. (2005) EMBO J. 24, 906–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theobald D. L., Mitton-Fry R. M., Wuttke D. S. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochkarev A., Bochkareva E. (2004) Curr. Opin. Struct. Biol. 14, 36–42 [DOI] [PubMed] [Google Scholar]
- 26.Beese L. S., Steitz T. A. (1991) EMBO J. 10, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ugochukwu E., Lovering A. L., Mather O. C., Young T. W., White S. A. (2007) J. Mol. Biol. 371, 1007–1021 [DOI] [PubMed] [Google Scholar]
- 28.Kovall R. A., Matthews B. W. (1999) Curr. Opin. Chem. Biol. 3, 578–583 [DOI] [PubMed] [Google Scholar]
- 29.Nowotny M., Gaidamakov S. A., Crouch R. J., Yang W. (2005) Cell 121, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 30.Sutera V. A., Jr., Han E. S., Rajman L. A., Lovett S. T. (1999) J. Bacteriol. 181, 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert M., Edge R. E., Lario P., Cook M. A., Strynadka N. C., Mackie G. A., McIntosh L. P. (2004) J. Mol. Biol. 341, 37–54 [DOI] [PubMed] [Google Scholar]
- 32.Zuo Y., Vincent H. A., Zhang J., Wang Y., Deutscher M. P., Malhotra A. (2006) Mol. Cell 24, 149–156 [DOI] [PubMed] [Google Scholar]
- 33.Lorentzen E., Basquin J., Tomecki R., Dziembowski A., Conti E. (2008) Mol. Cell 29, 717–728 [DOI] [PubMed] [Google Scholar]
- 34.Meier B., Barber L. J., Liu Y., Shtessel L., Boulton S. J., Gartner A., Ahmed S. (2009) EMBO J. 28, 3549–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.