Abstract
The cellular processes that regulate Bcl-2 at the posttranslational levels are as important as those that regulate bcl-2 synthesis. Previously we demonstrated that the suppression of FK506-binding protein 38 (FKBP38) contributes to the instability of Bcl-2 or leaves Bcl-2 unprotected from degradation in an unknown mechanism. Here, we studied the underlying molecular mechanism mediating this process. We first showed that Bcl-2 binding-defective mutants of FKBP38 fail to accumulate Bcl-2 protein. We demonstrated that the FKBP38-mediated Bcl-2 stability is specific as the levels of other anti-apoptotic proteins such as Bcl-XL and Mcl-1 remained unaffected. FKBP38 enhanced the Bcl-2 stability under the blockade of de novo protein synthesis, indicating it is posttranslational. We showed that the overexpression of FKBP38 attenuates reduction rate of Bcl-2, thus resulting in an increment of the intracellular Bcl-2 level, contributing to the resistance of apoptotic cell death induced by the treatment of kinetin riboside, an anticancer drug. Caspase inhibitors markedly induced the accumulation of Bcl-2. In caspase-3-activated cells, the knockdown of endogenous FKBP38 by small interfering RNA resulted in Bcl-2 down-regulation as well, which was significantly recovered by the treatment with caspase inhibitors or overexpression of FKBP38. Finally we presented that the Bcl-2 cleavage by caspase-3 is blocked when Bcl-2 binds to FKBP38 through the flexible loop. Taken together, these results suggest that FKBP38 is a key player in regulating the function of Bcl-2 by antagonizing caspase-dependent degradation through the direct interaction with the flexible loop domain of Bcl-2, which contains the caspase cleavage site.
Keywords: Apoptosis, Cancer, Proteases/Death Protease, Protein/Protein-Protein Interactions, Protein/Isomerase, Protein/Stability
Introduction
FKBP38,2 also known as FKBP8, is a member of the FK506-binding protein immunophilin family of proteins (1–3). It shares a peptidylprolyl cis-trans isomerase (4) domain that may allow the immunophilins to assist protein folding or serve as scaffold proteins to facilitate protein-protein interactions (5). In addition, FKBP38 contains a tripartite tetratricopeptide repeat (TPR) domain, calcium/calmodulin-binding motif, and a transmembrane motif (TM). The TPR domain of FKBP38 interacts with the heat shock protein 90 (HSP90) (6, 7). The structural basis of HSP90 binding by the TPR domain is defined (8), and the residues involved in the molecular interaction are well conserved in FKBP38. FKBP38 is localized predominantly at the outer membrane of the mitochondria and the endoplasmic reticulum (ER) membrane, and it was shown to be associated with the anti-apoptotic proteins Bcl-2 and Bcl-XL at these organelles, thus modulating apoptosis (2). FKBP38 is also known as an important modulator in neuronal hedgehog signaling and in controlling cell size and as an endogenous inhibitor of mTOR (9–11). The depletion of FKBP38 by small interfering RNA (siRNA) was shown to cause a loss of Bcl-2 protein or to prolong PHD2 protein stability in a transcription-independent manner (12, 13). This suggests that FKBP38 plays an important role on the stability of proteins.
Bcl-2 is the prototype member of a protein family that functions to suppress apoptosis in a variety of cell systems. The formation of heterodimers with pro-apoptotic proteins, such as Bax and Bak, provides mechanistic basis for modulating apoptotic cell death (14, 15). The dysregulation of Bcl-2 can lead to various diseases, such as autoimmunity, neurodegeneration, or cancer (16, 17). Indeed, Bcl-2 is overexpressed in various cancers, including most B cell-derived lymphoma, breast cancer, prostate cancer, and colorectal adenocarcinoma (4, 18, 19), which may be correlated with chemoresistance in cancer cells.
Bcl-2 activity is regulated by various mechanisms, including transcription, posttranslational modifications, and degradation. Cyclic AMP response element is a major positive regulatory site in the bcl-2 promoter (20). This element is required for the binding of NF-κB and CREB (cAMP-response element-binding protein), which results in the activation of bcl-2 expression in lymphoma cells (21, 22). On the other hand, increasing evidence suggests that Bcl-2 expression is also regulated at the posttranslational level through the modulation of protein stability. Various stimuli can induce the down-regulation of Bcl-2, including lipopolysaccharide, β-amyloid, chromium (VI), tumor necrosis factor-α, and kinetin riboside (23–27), through a degradation machinery, thus resulting in the promotion of apoptosis. Although it has been shown that the Bcl-2 degradation is mediated via the ubiquitin-proteasomal pathway (25, 28), earlier studies showed that Bcl-2 is identified as a caspase substrate, and cleavage of Bcl-2 appears to inactivate Bcl-2 function in cell survival pathways (29, 30), suggesting a model where caspase cleavage of Bcl-2 produces a product that can further facilitate caspase activation to ensure the execution of the cell (31).
Previously, we showed that the flexible loop of Bcl-2 is important for the molecular interaction between FKBP38 and Bcl-2 (32). We also demonstrated that the knockdown of FKBP38 by siRNA results in a decrease in the level of Bcl-2 protein, causing an apoptotic cell death (28). From these data, we suspected that FKBP38 can modulate the cleavage of Bcl-2 by forming a complex with Bcl-2, thus contributing to cell survival. In this study, to better understand the function of Bcl-2, we focused on the detailed molecular mechanism of FKBP38-mediated Bcl-2 stability. Our results showed that FKBP38 stabilizes Bcl-2 via a posttranslational mechanism, thus modulating the anti-apoptotic activity of Bcl-2. In addition, we presented that FKBP38, which binds to Bcl-2, interferes with its degradation by blocking the caspase-mediated cleavage pathway and results in accumulation of Bcl-2 and resistance to the treatments of anticancer drugs, suggesting a potential mechanism of chemoresistance in Bcl-2-overexpressed cancer cells.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagent
Human FKBP38 cDNA was cloned into the BamHI and XhoI sites of a pXJ-FLAG-S plasmid (33). The constructs for the various deletion mutants of FKBP38, including the calmodulin binding domain deletion mutant (residues Δ294–311, ΔCBD), the TPR domain deletion mutant (residues Δ150–286, ΔTPR), and the N-terminal deletion mutant, including the FK-506 binding domain (residues Δ1–152, ΔNTD), were prepared by subcloning the PCR-amplified fragment into pXJ-FLAG-S. Mutants in the TPR domain of FKBP38 (K167G/R168G, N172K, and K284G/R252G) were generated using complementary oligonucleotides with the desired mutation and were also cloned into the pXJ-FLAG-S plasmid. FKBP38 and its deletion mutants that fused to FLAG were subcloned into the XhoI and BamHI sites of pECFP-C1 (Clontech). Human Bcl-2, D34A mutant Bcl-2, transmembrane deletion mutant Bcl-2 (Bcl-2ΔTM, residues Δ207–239), and flexible loop deletion mutant Bcl-2 (Bcl-2ΔL, residues Δ35–89) were cloned into the XhoI and BamHI sites of pEYFP-C1 (Clontech). Human Mcl-1L was kindly provided by Dr. J. H. Bae (CHA University, Seongnam, Korea). All of the constructs were confirmed by DNA sequencing.
The antibodies against Bcl-2, Bcl-XL, GFP, Mcl-1, and HSP90 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), the antibody against GAPDH was obtained from Ambion (Austin, TX), and the polyclonal antibody against FKBP38 was kindly provided by Dr K. Nakayama (Kyushu University, Fukuoka, Japan). Monoclonal antibodies against the FLAG epitope and β-actin came from Sigma, and the antibodies against cleaved caspase-3, caspase-8, and caspase-9 came from Cell Signaling Technology (Danvers, MA). Cycloheximide, kinetin riboside, and chloroquine were obtained from Sigma, and the protease inhibitor complete-mini protease tablets from Roche Applied Science. Z-VAD-fmk was purchased from Santa Cruz Biotechnology, and Z-DEVD-fmk, Z-LEHD-fmk, and lactacystin was from Calbiochem. Dulbecco's modified Eagle's medium and fetal calf serum were purchased from HyClone (Logan, Utah).
Cell Culture and Transfection
Huh7, B104, HeLa, and MCF-7/caspase-3(+), provided by Dr. A. G. Porter (IMCB, Singapore) (34), were maintained in Dulbecco's modified Eagle's medium, which contained 10% fetal calf serum and 1% penicillin/streptomycin in a humidified 5% CO2 incubator at 37 °C. In a 6-well tissue culture plate (Falcon, England), cells were seeded at a density of about 2 × 105 cells per well. When the density of the cells reached 70–80%, they were transfected with plasmids using a LipofectamineTM reagent (Invitrogen). After incubation for 24 h, the transfected cells were treated as indicated for analysis.
Immunoprecipitation and Immunoblotting
Cell lysates were prepared in a lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) that contained protease inhibitors. Equal amounts of protein were immunoprecipitated using anti-FLAG and collected with Protein A/G-Sepharose beads (Santa Cruz Biotechnology) at 4 °C for 16 h. The immunoprecipitate was then washed 4 times in the cold lysis buffer described above. The bound proteins were resolved by SDS-PAGE and analyzed by Western blotting. Specific reactive bands were detected using a goat anti-rabbit or goat anti-mouse IgG conjugated to horseradish peroxidase, and the immunoreactive bands were visualized with a SUPEX Western blotting detection kit (Neuronex, Korea).
Protein Expression, Purification. and GST Pulldown Assay
FKBP38ΔTM (residues 1–313) and FKBP38NTD (residues 1–150) were subcloned into pGEX-4T-1 and expressed in Escherichia coli BL21 cells. GST-tagged proteins were purified by glutathione-Sepharose 4B resin followed by Superdex-200 size exclusion column (GE Healthcare).
Bcl-2ΔTM and Bcl-XLΔTM (residues Δ219–239 and Δ213–233, respectively) were subcloned into pSUMO and pET29b, respectively. The hexahistidine-tagged proteins were purified on Ni2+-nitrilotriacetic acid resin. SUMO was cleaved by SUMO protease 1, and free Bcl-2ΔTM and His-Bcl-XLΔTM were further purified by Superdex-200 size exclusion column.
For GST pulldown assays, aliquots of GST-tagged-FKBP38 proteins with Bcl-2ΔTM or His-Bcl-XLΔTM proteins were incubated in a TNE buffer containing 20 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40 for 16 h at 4 °C, and then protein samples were mixed with 20 μl of glutathione-Sepharose 4B resin for 30 min. After washing three times with the TNE buffer, the samples were subjected to SDS-PAGE gel analysis.
Cleavage of Bcl-2 Protein
2 μg of Bcl-2ΔTM or His-FKBP38-bound Bcl-2ΔTM were incubated with 50 ng of recombinant human active caspase-3/CPP32 (BioVision, Mountain View, CA) in a 30-μl reaction buffer containing 50 mm HEPES, pH 7.4, 100 mm NaCl, 10 mm dithiothreitol, 10% sucrose, and 0.1% CHAPS for 16 h at 37 °C. The reactions were stopped and subjected to SDS-PAGE gel analysis.
Fluorescence Resonance Energy Transfer (FRET) Assay
Cells grown on coated glass coverslips were fixed with 3.7% paraformaldehyde in phosphate-buffered saline for 30 min at 4 °C at 24 h post-transfection. A Zeiss LSM510 META confocal microscopy (Zeiss, Oberkochen, Germany) was used for FRET analysis by using the acceptor photobleaching method with modification (35). Briefly, images with the following configuration were recorded: CFP, excitation wavelength 458 nm and emission filter BP 475–525 nm; YFP, excitation wavelength 514 nm and emission filter LP560. Visualization was with an oil immersion 63× objective. Cells were bleached in the YFP channel by scanning a region of interest 10 times using the 514 argon laser line at 75% intensity. The CFP signal of the region of interest was acquired before and after the photobleaching. The YFP bleaching efficiency (BE) was calculated with the formula BE = (I5 − I6) × 100/I5, where In was the YFP intensity at the nth time point. A non-bleached region in the same size was always selected as a control. At least 10 events with BE > 90% and without bleach in the control region were selected to calculate the CFP-increasing or FRET energy transfer efficiency (EF) by using the formula EF = (I6 − I5) × 100/I6, where In was the CFP intensity at the nth time point.
RNA Interference Experiments
siRNA duplexes that target Bcl-2 (5′-AAGUACAUCCAUUAUAAGCUG-3′), FKBP38 (5′-AAGAGUGGCUGGACAUUCUGG-3′), and control (5′-AAGUCUCCAAGCGGAUCUCGU-3′) were purchased from Dharmacon (Lafayette, CO). Transfection of the siRNA duplexes at 100 nm final concentrations was carried out using a LipofectamineTM reagent. After transfection, the cells were processed for immunoblotting and immunocytochemistry as indicated.
Quantitative Reverse Transcription-PCR
The cells were transfected as described above. Cellular RNA was extracted using a RNeasy extraction kit from Qiagen (Hilden, Germany), and 2 μg of total RNA was reverse-transcribed into cDNA by ImProm-IITM reverse transcriptase (Promega, Madison, WI). The primers used in PCR were: bcl-2 forward (5′-CCCAAGCTTATGGCGCACGCTGGGAGAACG-3′) and reverse (5′-AAACTCGAGTCACTTGTGGCCCAGATAGGC-3′); β-actin forward (5′-GACCTGACTGACTACCTCATGAAGAT-3′) and reverse (5′-GTCACACTTCATGATGGAGTTGAAGG-3′).
Cell Viability Assay
Cell viability was determined using the CellTiter 96 Aqueous One Solution Reagent kit (Promega) according to the manufacturer's instructions 24 h after treatment, and absorbance at 490 nm was measured.
Statistical Analysis
All traces and immunoblots presented are representative of at least three separate experiments. All quantitative data are presented as the means ± S.E. of a minimum of three experiments. Comparisons between two groups were analyzed via a t test, and values of p < 0.05 were considered to be significant.
RESULTS
Bcl-2 Is Accumulated by the Overexpression of FKBP38
To check whether FKBP38 is involved in mediating the stability of Bcl-2, we first looked at the effect of transfected FKBP38 on the level of Bcl-2. For this, we used Huh7 (Bcl-2−/−) cells (Fig. 1A) to exclude the effect of endogenous Bcl-2. FKBP38-fused to a FLAG epitope tag (FLAG-FKBP38) was transiently transfected into the cells along with Bcl-2 tagged with YFP (YFP-Bcl-2). A significant increase in the Bcl-2 level was observed (Fig. 1B), suggesting that the increased level of Bcl-2 is FKBP38-dependent. To confirm the effect of FKBP38 on Bcl-2, a dose-response experiment was performed in Huh7 cells. The cells were transfected with a fixed amount of Bcl-2 and varying amounts of FKBP38 as indicated (Fig. 1C), showing that the accumulation of Bcl-2 is proportional to the amount of FKBP38 transfected. To further confirm this regulatory effect, a time-course experiment for the Bcl-2 expression either alone or together with FKBP38 was performed (Fig. 1D). Huh7 cells were transfected with the plasmids expressing YFP-Bcl-2 and FLAG-FKBP38, and at different time points cell extracts were prepared and analyzed by immunoblotting. We showed that in the absence of FKBP38, no Bcl-2 was detected 48 h after transfection. However, in the presence of FKBP38, the Bcl-2 protein levels were significantly increased and well maintained up to 48 h (Fig. 1D). Taken together, our data indicate that FKBP38 plays an important role in stabilizing Bcl-2 and results in the accumulation of Bcl-2.
FIGURE 1.
Coexpression of FKBP38 augments the expression levels of Bcl-2. A, expression of Bcl-2 in HeLa and Huh7 cell lines was determined by immunoblotting using GAPDH as the internal control. B, Huh7 cells were transfected with 1 μg of plasmid pEYFP-Bcl-2 either alone or together with 1 μg of pXJ-FLAG-FKBP38 construct. Total cell extracts were incubated with anti-GFP or anti-FLAG antibodies and then analyzed by immunoblotting. C, Huh7 cells were transfected with a fixed amount of pEYFP-Bcl-2 (1 μg) and various amounts of pXJ-FLAG-FKBP38 (0, 0.1, 0.3, and 0.8 μg). D, cells were transfected with 1 μg of plasmid pEYFP-Bcl-2 either alone or together with 1 μg of pXJ-FLAG-FKBP38 construct and at different time points cell extracts were prepared and analyzed by immunoblotting.
The N-terminal Domain and TPR Domain of FKBP38 Are Important for the Stability of Bcl-2
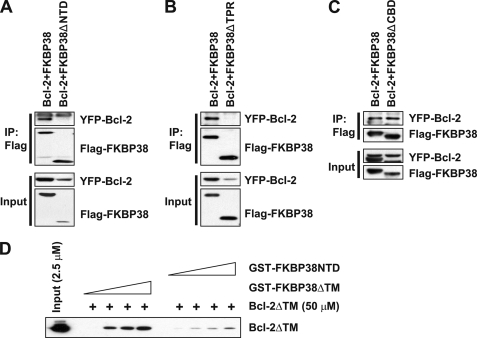
We then investigated the mechanism by which FKBP38 induces the accumulation of Bcl-2 protein. To determine whether Bcl-2 is stabilized by binding to FKBP38, we first examined three truncation mutants of FKBP38 (FKBP38ΔNTD, FKBP38ΔTPR, and FKBP38ΔCBD) to identify the binding site (supplemental Fig. S1, A and B). B104 cells were co-transfected with YFP-Bcl-2 and the FLAG-tagged FKBP deletion mutants followed by immunoprecipitation with an antibody against FLAG. Interestingly, both the N-terminal domain deletion mutant (FKBP38ΔNTD) and the TPR domain deletion mutant (FKBP38ΔTPR) failed to interact with Bcl-2 (Figs. 2, A and B). However, the Ca2+/calmodulin binding domain deletion mutant (FKBP38ΔCBD) showed little effect on the interaction with Bcl-2 (Fig. 2C). Similar results were obtained from the Huh7 cells (data not shown). To test whether the interaction between FKBP38 and Bcl-2 is direct, we performed GST pulldown assays showing that Bcl-2ΔTM binds to GST-tagged FKBP38ΔTM in a dose-dependent manner (Fig. 2D). In contrast, the binding affinity between Bcl-2ΔTM with FKBP38NTD was weak (Fig. 2D). The binding patterns observed in GST pulldown assays were similar to the immunoprecipitation patterns between Bcl-2 and FKBP38 and FKBP38NTD (supplemental Fig. S2). As described in a previous study (2), the N-terminal portion of FKBP38 that includes the FK506 binding domain is important for the binding with Bcl-2. In addition, we observed that the TPR domain has an effect on the interaction between the two proteins. To confirm the effects of the TPR domain of FKBP38 on its binding with Bcl-2, B104 cells were co-transfected with plasmids expressing YFP-Bcl-2 and either FLAG-FKBP38 or FLAG-FKBP38ΔTPR. Cell lysates were immunoprecipitated with an anti-FLAG antibody followed by immunoblotting to detect the bound YFP-Bcl-2 and the endogenous HSP90 that bind to FKBP38 through its TPR domain (6, 7). As expected, FKBP38ΔTPR showed no interaction with HSP90 and Bcl-2 (Fig. 3A), indicating that the TPR domain of FKBP38 is not only important for the interaction with HSP90 but also critical for its interaction with Bcl-2. As several residues in the TPR domain are involved in the binding with the MEEVD motif located at the C terminus of HSP90 (36–38), we examined whether those residues are also involved in the interaction with Bcl-2. We mutated the target sites (K167G/R168G, N172K, and K284G/R252G) in the TPR domain of FKBP38. Interestingly, although the mutants showed markedly reduced binding to HSP90, they did not affect its binding affinity to Bcl-2 (Fig. 3B), suggesting that Bcl-2 interacts with the TPR domain of FKBP38 through the irrelevant pathway to HSP90.
FIGURE 2.
Bcl-2 interacts with FKBP38 through its FK-506 binding and TPR domains. A, B, and C, cell lysates from B104 cells transiently overexpressing YFP-Bcl-2 with FLAG-FKBP38 full-length, FLAG-FKBP38ΔNTD (A), FLAG-FKBP38ΔTPR (B), or FLAG-FKBP38ΔCBD (C) were immunoprecipitated (IP) with anti-FLAG antibody. The cell lysates and the immune complexes were then analyzed by immunoblotting with anti-Bcl-2 or anti-FLAG antibodies. D, GST pulldown assay using GST-tagged FKBP38ΔTM and FKBP38NTD with Bcl-2ΔTM. The FKBP38 variants were applied at final concentrations of 0, 50, 100, and 200 μm, and Bcl-2 was used with a final concentration of 50 μm.
FIGURE 3.
Bcl-2 interacts with the TPR domain of FKBP38 via an HSP90-independent pathway. A, B104 cells were transfected with plasmid pEYFP-Bcl-2 either alone or together with FKBP38 constructs (pXJ-FLAG-FKBP38 and -FKBP38ΔTPR). Then cell lysates were immunoprecipitated (IP) with anti-FLAG antibody and analyzed by immunoblotting with anti-HSP90, anti-GFP, and anti-FLAG antibodies. B, in similar experiments, cells were transfected with pEYFP-Bcl-2 and pXJ-FLAG-FKBP38 wild-type or mutant constructs, FKBP38 (K167G/R168G), FKBP38 (N172K), and FKBP38 (K284G/R252G).
To check whether the Bcl-2 stability is due to its binding to FKBP38, Huh7 cells were transfected with plasmids expressing YFP-Bcl-2 and either FLAG-FKBP38ΔNTD, -FKBP38ΔTPR, or -FKBP38ΔCBD. We showed that increasing amounts of the deletion mutants (ΔNTD and ΔTPR) of FKBP38 do not lead to a significant accumulation of Bcl-2 (Figs. 4, A and B), except for FKBP38ΔCBD (Fig. 4C). Therefore, we conclude that the stabilization of Bcl-2 is attributable to the binding of Bcl-2 with FKBP38. Also, the function of FKBP38 was specific to Bcl-2, as the protein levels of Bcl-XL, which was also reported to bind to FKBP38 (2), and Mcl-1L remained relatively unchanged under co-transfection with FKBP38 (Figs. 4, D and E). Conversely, in a reciprocal experiment with increasing amounts of YFP-Bcl-2 expression vector, we failed to observe any enhanced expression of co-expressed FLAG-FKBP38 (data not shown). Together, the results suggest that the FKBP38 effect on Bcl-2 stability is specific and unidirectional.
FIGURE 4.
The stability of Bcl-2 is specifically increased by its binding to FKBP38. Huh7 cells were transfected with a fixed amount of pEYFP-Bcl-2 (1 μg) and various amounts of pXJ-FLAG-FKBP38ΔNTD (A), pXJ-FKBP38ΔTPR (B) or pXJ-FKBP38ΔCBD (C) (0, 0.1, 0.3, and 0.8 μg). The cell lysates were analyzed by immunoblotting with anti-GFP and anti-FLAG antibodies. D and E, Huh7 cells were transfected with 1 μg of plasmid pECFP- Bcl-XL or 3 μg of plasmid pcMcl-1 either alone or together with 1 μg of the abovementioned FKBP38 truncation constructs. Total cell extracts were incubated with anti-Bcl-XL and -Mcl-1 antibodies and then analyzed by immunoblotting.
FKBP38 Stabilizes Bcl-2 by a Posttranslational Mechanism
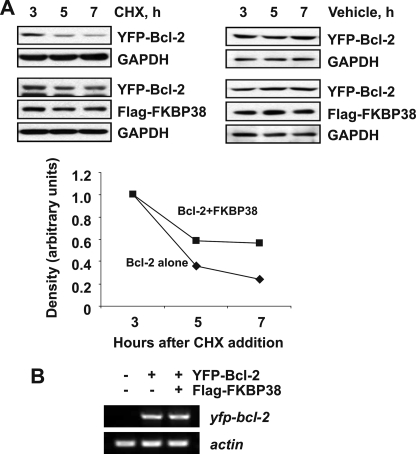
To examine whether Bcl-2 accumulation is posttranslational, Huh7 cells were transfected with plasmids expressing either YFP-Bcl-2 alone or together with FLAG-FKBP38 and incubated in the presence or absence of cycloheximide that blocks de novo protein synthesis. As shown in Fig. 5A, in the presence of cycloheximide, the overexpressed FKBP38 resulted in a decrease in Bcl-2 degradation, whereas in the absence of cycloheximide, the Bcl-2 level remained steady. The half-life of the transfected Bcl-2 protein increased from 4.5 h to more than 7 h as a consequence of FKBP38 overexpression, indicating that FKBP38 increases the stability of Bcl-2. To investigate if there is any transcriptional effect on the bcl-2 gene, the levels of bcl-2 RNA were also measured by quantitative reverse transcription-PCR (Fig. 5B). The RNA levels were similar in cells either transfected with the Bcl-2 plasmid alone or the Bcl-2 and FKBP38 plasmids. Therefore, we speculate that the stabilization effect seen on Bcl-2 is due to a posttranslation event by FKBP38.
FIGURE 5.
FKBP38 increases the stability of Bcl-2 by a posttranslational mechanism. A, Huh7 cells were transfected with pEYFP-Bcl-2 with/without pXJ-FLAG-FKBP38. 24 h after the transfection, cycloheximide (CHX) and phosphate-buffered saline as a control were added to the cultures, and the levels of Bcl-2 protein at different time points were examined by immunoblotting. Relative protein levels are calculated from band intensity as shown below the blot. B, FKBP38 overexpression does not affect Bcl-2 RNA levels. Quantitative reverse transcription-PCR was performed with cDNA obtained from Huh7 cells transfected with plasmid pEYFP-Bcl-2 either alone or together with pXJ-FLAG-FKBP38 construct.
FKBP38 Prevents the Down-regulation of Bcl-2 by Caspases
To define the mechanism that might provide a molecular basis for the FKBP38-mediated accumulation of Bcl-2, Bcl-2-overexpressed Huh7 cells were treated with lactacystin and chloroquine, which are proteasomal and lysosomal inhibitors, respectively. Both of the inhibitors failed to stabilize Bcl-2 protein (Fig. 6A), indicating the proteasomal and lysosomal degradation pathways are unlikely to be the major modes of the Bcl-2 down-regulation in Huh7 cells. As Bcl-2 was shown to be cleaved at Asp-34 by caspases during apoptosis and also by recombinant caspase-3 in vitro (29, 39), we tested the effects of caspases on the FKBP38-induced accumulation of Bcl-2. When Bcl-2-overexpressed cells were treated with the pan-caspase inhibitor, Z-VAD-fmk, the level of Bcl-2 increased (Fig. 6A), although its accumulation did not reach the levels obtained with the overexpression of FKBP38 (fourth and fifth lanes in Fig. 6B and second and third lanes in Fig. 6C). On the other hand, the Bcl-2 level in the cells co-transfected with FKBP38 and Bcl-2 in the absence of Z-VAD-fmk was slightly lower than that in its presence (fifth and eighth lanes Fig. 6B and third and fourth lanes in Fig. 6C), indicating that a lower level of caspase activity still takes place in the presence of FKBP38. To further verify that the interaction between FKBP38 and Bcl-2 lessens caspase activity against Bcl-2, cells were transfected with plasmids expressing YFP-Bcl-2 and either FLAG-FKBP38, FLAG-FKBP38ΔNTD, or FLAG-FKBP38ΔTPR and then treated with or without Z-VAD-fmk. As shown in Fig. 6C, the levels of Bcl-2 in the cells co-expressing Bcl-2 along with the deletion mutants were much lower compared with those in the cells expressing both Bcl-2 and FKBP38 in the absence of Z-VAD-fmk (third, fifth, and seventh lanes). The Bcl-2 levels in the cells transfected with deletion mutants were recovered to a varying degree by the treatment with Z-VAD-fmk, thus confirming that FKBP38 by itself can enhance the stability of Bcl-2 but cannot completely block the turnover of Bcl-2 via caspase-mediated degradation as seen in cells transfected with FLAG-FKBP38ΔNTD (Fig. 6C). The specificity of Bcl-2 accumulation to caspase inhibitors was also determined. Huh7 cells were transfected with plasmids expressing YFP-Bcl-2 alone or together with FLAG-FKBP38 and then treated with a pan-caspase inhibitor (Z-VAD-fmk), caspase-3 inhibitor (Z-DEVD-fmk), or caspase-9 inhibitor (Z-LEHD-fmk). As shown in Fig. 6D, both caspase-3 and -9 inhibitors prevented the down-regulation of Bcl-2; however, the effect of caspase-3 was greater than that of caspase-9 in the modulation of the Bcl-2 protein levels.
FIGURE 6.
FKBP38 prevents the down-regulation of Bcl-2 by caspases activation. A, Huh7 cells were transfected with pEYFP-Bcl-2. Six hours after the transfection, lactacystin (1 μm), chloroquine (20 μm), or Z-VAD-fmk (20 μm) was added to the cultures, and the protein levels were determined by immunoblotting with anti-Bcl-2 or anti-cleaved caspase-3 antibodies after 24 h. B, in similar experiments, Huh7 cells were transfected with pEYFP-Bcl-2 with/without pXJ-FLAG-FKBP38. C, Huh7 cells were transfected with plasmid encoding Bcl-2 (pEYFP-Bcl-2) either alone or together with FKBP38 constructs (pXJ-FLAG-FKBP38, pXJ-FKBP38ΔNTD, or pXJ-FKBP38ΔTPR). Six hours after the transfection, Z-VAD-fmk (20 μm) was added to the cultures, and the levels of Bcl-2 protein were determined by immunoblotting. D, Huh7 cells were transfected with plasmid pEYFP-Bcl-2 either alone or together with pXJ-FLAG-FKBP38 construct. Six hours after the transfection, Z-VAD-fmk (20 μm), Z-DEVD-fmk (50 μm), or Z-LEHD-fmk (50 μm) were treated, and the levels of Bcl-2 protein were determined by immunoblotting.
FKBP38 Protects Bcl-2 Degradation by Caspases through the Direct Interaction with the Flexible Loop Domain of Bcl-2
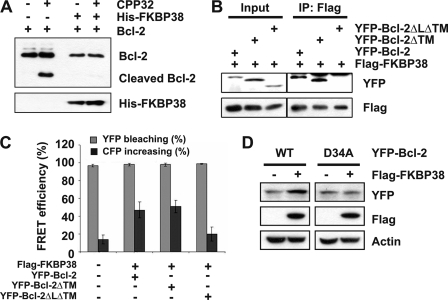
To delineate the molecular basis of the FKBP38 role on the caspase-dependent degradation of Bcl-2, we employed an in vitro caspase cleavage assay using the active form of recombinant human caspase-3/CPP32 and purified Bcl-2 and Bcl-XL. To this end, we first checked the cleavage of Bcl-2 in the absence or presence of FKBP38. As shown in Fig. 7A, Bcl-2 cleavage was not detected in the presence of FKBP38, whereas the cleaved fragment of Bcl-2 was produced in the absence of FKBP38, suggesting that the binding of FKBP38 to Bcl-2 protects Bcl-2 from degradation. Because the flexible loop of Bcl-2 contains the caspase cleavage site, we examined the role of the flexible loop of Bcl-2 in the cleavage pathway. For the study, Huh7 cells were co-transfected with FLAG-FKBP38 and the YFP-tagged Bcl-2 deletion mutants followed by immunoprecipitation with an antibody against FLAG. Our data showed that the flexible loop deletion mutant (Bcl-2ΔLΔTM) failed to interact with FKBP38 (Fig. 7B), which is consistent with in vitro observation (32). In addition, we also performed FRET experiments between the YFP-fused Bcl-2 and the loop deletion mutant of Bcl-2 and the CFP-FKBP38 (Fig. 7C), demonstrating that the loop deletion mutant shows significantly reduced FRET efficiency, confirming that the flexible loop of Bcl-2 is the primary binding to FKBP38. To further check whether the known caspase cleavage site of Bcl-2 (residue Asp-34) was mutated to Ala (mutant D34A), we tested it characteristics. As shown in Fig. 7D, a significant increase was observed in the wild-type Bcl-2 level in the presence of FKBP38 but not in D34A, suggesting that the stability of Bcl-2/D34A mutant is no longer dependent on the presence of FKBP38. To address the question of whether the protective role and resulting stability by FKBP38 is specific to Bcl-2, we also checked Bcl-XL using a GST pulldown assay. Interestingly, the results showed that FKBP38-bound Bcl-XL was barely detected as compared with FKBP38-bound Bcl-2 (supplemental Fig. S3, A and B). In addition, cleavage of Bcl-XL by caspase-3 was detected in the presence or absence of FKBP38 (supplemental Fig. S3C). Taken together, these results suggest that FKBP38 protects Bcl-2 degradation by caspases through the direct interaction with the flexible loop domain of Bcl-2, which contains the caspase cleavage site.
FIGURE 7.
FKBP38 interacts with the flexible loop of Bcl-2-contained caspase cleavage site. A, 2 μg of free Bcl-2ΔTM- or His-FKBP38-bound Bcl-2ΔTM were incubated with 50 ng of recombinant human active caspase-3/CPP32. The proteins were resolved by 12% SDS-PAGE gel and Bcl-2ΔTM, and proteolyzed fragments were detected with anti-Bcl-2 antibody. B, cell lysates from Huh7 cells transiently overexpressing FLAG-FKBP38 with YFP-Bcl-2 full-length, YFP-Bcl-2ΔTM, or YFP-Bcl-2ΔLΔTM were immunoprecipitated with anti-FLAG antibody. The cell lysates (1/20 of those used in immunoprecipitation (IP)) and the immune complexes were then analyzed by immunoblotting with anti-GFP or anti-FLAG antibodies. C, shown is FRET between Bcl-2 with FKBP38. Cells were seeded on a coverslip inside a six-well plate and co-transfected with plasmid encoding FKBP38 (pECFP-FKBP38) together with constructs (pEYFP-Bcl-2, pEYFP-Bcl-2ΔTM, or pEYFP-Bcl-2ΔLΔTM). 24 h later a FRET assay were performed as described under “Experimental Procedures.” Data represent three independent experiments. D, cell lysates from Huh7 cells transiently overexpressing FLAG-FKBP38 with YFP-Bcl-2 full-length or YFP-Bcl-2 D34A were immunoblotted with anti-FLAG, anti-GFP, and anti-actin antibodies. WT, wild type.
Finally, to find out whether FKBP38 affects general caspase activity, Huh7 cells were treated with siRNAs targeting FKBP38, and then the expression levels of processed caspase-3, -8, and -9 were monitored. We showed that the levels of cleaved caspase-3, -8, and -9 did not show much difference between cells treated and untreated with FKBP38 siRNA, suggesting that FKBP38 does not induce significant changes in general caspase activity in Huh7 cells (supplemental Fig. S4).
FKBP38 Overexpression Increased Endogenous Bcl-2 Protein Levels and Prevented Apoptotic Cell Death
As Bcl-2 was previously shown to be down-regulated under exposure to anti-cancer drug such as kinetin riboside (24), here we tested the effect of FKBP38 on kinetin riboside-induced Bcl-2 down-regulation. HeLa cells were transfected with plasmids expressing FLAG-mock or FLAG-FKBP38 and then treated with kinetin riboside in a dose-dependent manner. The overexpression of FKBP38 resulted in a reduced rate of Bcl-2 down-regulation compared with that of the mock transfection (Fig. 8A). We examined cell viability under identical conditions. As shown in Fig. 8B, FKBP38 also reduced kinetin riboside-induced cell toxicity. These results suggest that the overexpressed FKBP38 in HeLa cells is involved in the modulation of endogenous Bcl-2.
FIGURE 8.
Overexpression of FKBP38 prevents kinetin riboside-induced Bcl-2 down-regulation and apoptotic cell death. A, HeLa cells were transfected with or without pXJ-FLAG-FKBP38. Kinetin riboside (KR) at the indicated concentrations was added 24 h after transfection, and cell extracts were prepared for immunoblotting 48 h after transfection. B, HeLa cells were transfected with or without pXJ-FLAG-FKBP38. Twenty-four hours after the transfection, kinetin riboside (30 μm) was added to the culture. Cell viability was analyzed after 24 h by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) conversion assay. Results are the mean ± S.E. values from three separate experiments. **, p < 0.01.
FKBP38-dependent stabilization of Bcl-2 was further examined using siRNAs, demonstrating that the endogenous FKBP38 and Bcl-2 protein levels were significantly reduced by the treatment of siRNA targeting FKBP38 in MCF-7 cells expressing caspase-3 (MCF-7/caspase-3(+)) (Figs. 9, A and B), whereas the level of Bcl-XL remained little changed (Fig. 9A). Moreover, Bcl-2 siRNA did not affect the levels of FKBP38 protein or mRNA (data not shown). To check the effects of various inhibitors on the down-regulation of Bcl-2 upon depletion of FKBP38, MCF-7/caspase-3(+) cells transfected with FKBP38 specific siRNA were treated with lactacystin, chloroquine, or Z-VAD-fmk. The cells exposed to Z-VAD-fmk showed a minor increase of the Bcl-2 levels as compared with vehicle treatment (Fig. 9C). To further verify this finding, FLAG-FKBP38 was re-transfected into the cells pretransfected with FKBP38 siRNA. As shown in Figs. 9, D and E, the increased level of Bcl-2 was observed in FLAG-FKBP38-oeverexpressing cells with control siRNA and FKBP38 siRNA. To address the physiological context of the effect of FKBP38 on the Bcl-2 stability, MCF-7/caspase-3(+) cells, which were transfected with control siRNA and FKBP38 siRNA, were treated with etoposide. Etoposide-treated cells resulted in increases in caspase-3 and -8 activities, although the level of increase is less with caspase-8. Also, we showed that the Bcl-2 level decreases compared with the untreated cells, whereas the level of Bcl-XL remained unchanged (Fig. 9F). We demonstrated that in the absence of FKBP38 knockdown, Bcl-2 exhibits a relatively minor down-regulation in response to etoposide, whereas in the presence of FKBP38 knockdown Bcl-2 shows a significant down-regulation. In addition, the activities of caspase-3, -8, and -9 in etoposide-untreated cells showed little change in both FKBP38 siRNA and control siRNA treated cells (Fig. 9F), and similar results were obtained in FKBP38-knockdown HeLa cells treated with kinetin riboside and etoposide (Fig. 9G), suggesting that Bcl-2 stability after apoptotic stimuli is mainly mediated by FKBP38, without affecting other Bcl-2 family proteins or inherent caspase activity and not limited to a particular cell type. Taken together, our results provide evidence that FKBP38 is a key chaperone in Bcl-2 stability and plays an important role in the maintenance of Bcl-2 protein level in cells.
FIGURE 9.
FKBP38 siRNA induces the down-regulation of Bcl-2 in MCF-7/caspase-3(+) cells. A and B, MCF-7/caspase-3(+) cells were transfected with either control or FKBP38 siRNA (siFKBP38), and after 72 h, the effects of siRNA were confirmed by immunoblotting and immunocytochemistry. A, representative immunoblots after transfection with siRNA against FKBP38 are shown. B, typical images after transfection with FKBP38 siRNA are shown. C, the cells were transfected with either control or FKBP38 siRNA, and after 24 h, lactacystin (1 μm), chloroquine (20 μm), or Z-VAD-fmk (20 μm) was added to the cultures, and the levels of Bcl-2 and FKBP38 proteins were determined by immunoblotting after 48 h. D and E, reduction of Bcl-2 by FKBP38 siRNA was restored by FKBP38 overexpression. The cells were transfected with either control or FKBP38 siRNA and, after 48 h, transfected with or without pXJ-FLAG-FKBP38 again. Forty-eight hours after transfection, cell extracts were prepared for immunoblotting (D). Ctrl, control. Bcl-2 protein levels are shown below the blot (E). F, the cells were transfected with either control or FKBP38 siRNA, and after 48 h, etoposide (50 μm) was added to the cultures, and the levels of proteins as indicated were determined by immunoblotting after 24 h. G, similarly, HeLa cells were transfected with either control or FKBP38 siRNA, and after 48 h, kinetin riboside (KR, 50 μm) or etoposide (Etopo, 50 μm) was added to the cultures, and the levels of proteins as indicated were determined by immunoblotting after 10 or 20 h, respectively.
DISCUSSION
Overexpression of Bcl-2 is linked to resistance to chemotherapy and a poor prognosis in cancer (40). However, the molecular mechanism for the Bcl-2-mediated resistance to chemotherapeutic agents currently remains elusive.
Although function of Bcl-2 is tightly regulated through its molecular interaction with proapoptotic partners, its antiapoptotic activity is also regulated through posttranslational modifications such as ubiquitination, phosphorylation, and caspase-mediated cleavage in response to a variety of external stimuli (23, 25, 28, 29, 39, 41, 42). Here, we show that FKBP38, a noncanonical FKBP family member, contributes to the accumulation and stabilization of Bcl-2 by hindering its degradation through a caspase-dependent mechanism. Our results demonstrated that the level of overexpressed Bcl-2 remains very low at 24 h in Huh7 cells that retain their caspase-3 activity under normal conditions (Figs. 1 and 6A). However, its level remains significantly high after cells were treated with Z-VAD-fmk, a pan-caspase inhibitor (Fig. 6). In addition, we showed that proteasomal and lysosomal degradation pathways are unlikely to be major modes of Bcl-2 instability in Huh7 cells (Figs. 6, A and B), suggesting that Bcl-2 stability is mainly controlled by caspase activation in Huh7 cells.
In a recent study showing a caspase-mediated Bcl-2 cleavage, the cleavage was observed only after 12 h of drug treatment, whereas the proteasome-mediated cleavage was observed at earlier time periods (<12 h) in human non-small lung cancer H-460 cells (41). Interestingly, it was shown that resistance to cisplatin is mediated by Bcl-2 accumulation at early time periods and also by preventing the proteasomal degradation of Bcl-2, not by caspase-mediated cleavage (42). These data suggest that the drug resistance mechanism via Bcl-2 accumulation is complex and may also depend on modes of Bcl-2 degradation. Indeed, Bcl-2-overexpressing cells did not show any protection against caspase-3 activation or poly(ADP-ribose) polymerase cleavage at later time points, which coincides with the caspase-mediated cleavage of Bcl-2 (30), suggesting that the inhibition of caspase-mediated Bcl-2 cleavage may be a key late event for ensuring Bcl-2 stability and may consequently contribute to resistance to chemotherapy.
FKBP38 is abundantly expressed in various cancer cell lines and tumor tissues (12) (supplemental Fig. S5), including breast, colon, liver, lung, lymph node, prostate, and stomach cancers. FKBP38 is involved in the regulation of many cellular processes, including apoptosis, proliferation, development, and growth. A recent study demonstrated that FKBP38 has an anti-tumor effect through the induction of anti-invasive syndecan 1 and the suppression of the pro-invasive MMP9 (43). On the other hand, in response to growth factors or amino acid availability, Rheb directly interacts with FKBP38 and prevents the interaction of FKBP38 with mTOR, which allows the activation of mTOR constitutively and leads to cell proliferation (9, 44). In addition, the inhibition of mTOR by rapamycin or starvation inhibits angiogenesis and endothelial cell proliferation in vitro and in vivo (45, 46), suggesting FKBP38 may play an important role in proliferation of cancer cells. In this study, we similarly showed that overexpression of FKBP38 results in accumulation of exogenous and endogenous Bcl-2 proteins (Figs. 1, 8A, and 9) and reduces the rate of Bcl-2 degradation (Fig. 5A), thus contributing to protection of the apoptotic cell death induced by chemotherapeutic compound (Fig. 8B). We have also demonstrated that suppression of FKBP38 by siRNA decreases cellular levels of Bcl-2 protein (Figs. 9, A and B), thus resulting in the apoptotic cell death (12), which is in agreement with a previous report (2), suggesting that FKBP38 is an important regulator in promoting growth of cancer cells through accumulation of Bcl-2 protein, although it is probably dependent on the cellular context.
FKBP38 was shown to be involved in modulating stability of PHD2, which is a member of novel family of oxygen-, ferrous iron-, and 2-oxoglutarate-dependent prolyl-4-hydroxylase domain-containing enzymes (13). It was also shown that FKBP38 directly binds via its TPR domain to the S4 subunit of the 19 S proteasome and results in an increase in the proteasomal activity in the membrane fraction (47). These data suggest that FKBP38 may be involved in the degradation processes of its binding proteins in the membrane. In our study reported here, we presented that overexpressed FKBP38 forms a complex with Bcl-2 (Figs. 2 and 4) and increases the Bcl-2 stability in both Huh7 (Figs. 2, 4, and 5) and HeLa cells (data not shown). This confirms that the molecular interaction between Bcl-2 and FKBP38 is critical in enhancing the stability of Bcl-2. In fact, FKBP38 has multiple binding partners, including Bcl-2, Bcl-XL, Rheb, mTOR, and PHD2 (2, 9, 13). Therefore, we also tested the effect of FKBP38 on the stability of Bcl-XL. However, its level remained unchanged with overexpression of FKBP38 (Fig. 4D), suggesting that FKBP38 has a specific function on the stability of Bcl-2.
Previous studies showed that Bcl-2 is cleaved by caspase-3 and caspase-9 activation and its anti-apoptotic function is inactivated and promotes apoptosis (29, 39). Here, we showed that the overexpression of FKBP38 interferes with down-regulation of Bcl-2 in the caspase-mediated pathways, including caspase-3 and caspase-9, and Bcl-2 binding-defective mutants (ΔNTD and ΔTPR) of FKBP38 are unable to protect the caspase-mediated Bcl-2 degradation (Fig. 6). The structure of Bcl-2 reveals the presence of a long flexible loop between Bcl-2 homology region 4 (BH4) and BH3 (48), which contains a caspase cleavage site (29). Here we demonstrated that the loop containing the caspase cleavage site in Bcl-2 (residue 34) is protected by FKBP38 (Fig. 7C), indicating the antagonizing role of FKBP38 against Bcl-2 cleavage by caspase-3 through the direct binding (Fig. 7A). Based on these results, we hypothesize that FKBP38 binding to Bcl-2 restricts the caspase-mediated Bcl-2 degradation in the flexible loop, contributing to the stability of Bcl-2, and might shut off a positive feedback loop for executing apoptosis (31).
Is the protective role of FKBP38 specific to Bcl-2 but not to other anti-apoptotic Bcl-2 family proteins? The three-dimensional structures of Bcl-2 and Bcl-XL are very similar, and both contain the flexible loops between Bcl-2 homology region 3 (BH3) and BH4 regions (49). It is noted that their amino acid sequences in the flexible loops are quite divergent. The flexible loop of Bcl-XL contains more charged residues, especially acidic residues, contributing to a considerably lower pI value (pI = 4.86) compared with that of Bcl-2 (pI = 6.8). Another interesting feature is that unlike Bcl-XL, Bcl-2 becomes phosphorylated in response to a variety of external stimuli. It was previously shown that the primary phosphorylation sites are located in the flexible loop of Bcl-2 (residues Thr-56, Ser-70, Ser-87) (32). Interestingly, these phosphorylation sites are always followed by proline residues (49). As FKBP38 exhibits a peptidylprolyl cis-trans isomerase activity that is induced upon binding calcium-calmodulin (50), it is possible that the inducible peptidylprolyl cis-trans isomerase of FKBP38 preferentially recognizes the proline-containing sequences of Bcl-2 and modulates the folding process of Bcl-2 or protein-protein interaction. This could be a molecular signature in providing specificity to Bcl-2 by FKBP38. However, the detailed molecular mechanism awaits a further study. In conclusion, here we presented a regulatory role of FKBP38 on the caspase-mediated Bcl-2 down-regulation, which is inhibited by the overexpression of FKBP38 and thereby maintains cell survival function of Bcl-2 in cancer cells.
Supplementary Material
Acknowledgment
We are grateful to Jeff Tai for technical assistance.
This work was supported by A*STAR-Biomedical Research Council of Singapore Grant 04/1/22/12/362.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S5.
- FKBP38
- FK506-binding protein 38
- TPR
- tetratricopeptide repeat
- HSP90
- heat shock protein 90
- siRNA
- small interfering RNA
- FRET
- fluorescence resonance energy transfer
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TM
- transmembrane motif
- GFP
- green fluorescent protein
- Z-
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- GST
- glutathione S-transferase
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Pedersen K. M., Finsen B., Celis J. E., Jensen N. A. (1999) Electrophoresis 20, 249–255 [DOI] [PubMed] [Google Scholar]
- 2.Shirane M., Nakayama K. I. (2003) Nat. Cell Biol. 5, 28–37 [DOI] [PubMed] [Google Scholar]
- 3.Snyder S. H., Lai M. M., Burnett P. E. (1998) Neuron 21, 283–294 [DOI] [PubMed] [Google Scholar]
- 4.Buolamwini J. K. (1999) Curr. Opin. Chem. Biol. 3, 500–509 [DOI] [PubMed] [Google Scholar]
- 5.Fischer G., Aumüller T. (2003) Rev. Physiol. Biochem. Pharmacol. 148, 105–150 [DOI] [PubMed] [Google Scholar]
- 6.Edlich F., Erdmann F., Jarczowski F., Moutty M. C., Weiwad M., Fischer G. (2007) J. Biol. Chem. 282, 15341–15348 [DOI] [PubMed] [Google Scholar]
- 7.Okamoto T., Nishimura Y., Ichimura T., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y. (2006) EMBO J. 25, 5015–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 9.Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 10.Bulgakov O. V., Eggenschwiler J. T., Hong D. H., Anderson K. V., Li T. (2004) Development 131, 2149–2159 [DOI] [PubMed] [Google Scholar]
- 11.Rosner M., Hofer K., Kubista M., Hengstschläger M. (2003) Oncogene 22, 4786–4798 [DOI] [PubMed] [Google Scholar]
- 12.Kang C. B., Feng L., Chia J., Yoon H. S. (2005) Biochem. Biophys. Res. Commun. 337, 30–38 [DOI] [PubMed] [Google Scholar]
- 13.Barth S., Nesper J., Hasgall P. A., Wirthner R., Nytko K. J., Edlich F., Katschinski D. M., Stiehl D. P., Wenger R. H., Camenisch G. (2007) Mol. Cell. Biol. 27, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cory S., Adams J. M. (2002) Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 15.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 16.Baliga B. C., Kumar S. (2002) Hematol. Oncol. 20, 63–74 [DOI] [PubMed] [Google Scholar]
- 17.Raff M. C., Whitmore A. V., Finn J. T. (2002) Science 296, 868–871 [DOI] [PubMed] [Google Scholar]
- 18.Amundson S. A., Myers T. G., Scudiero D., Kitada S., Reed J. C., Fornace A. J., Jr. (2000) Cancer Res. 60, 6101–6110 [PubMed] [Google Scholar]
- 19.Osford S. M., Dallman C. L., Johnson P. W., Ganesan A., Packham G. (2004) Curr. Med. Chem. 11, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 20.Wilson B. E., Mochon E., Boxer L. M. (1996) Mol. Cell. Biol. 16, 5546–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heckman C. A., Mehew J. W., Boxer L. M. (2002) Oncogene 21, 3898–3908 [DOI] [PubMed] [Google Scholar]
- 22.Xiang H., Wang J., Boxer L. M. (2006) Mol. Cell. Biol. 26, 8599–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azad N., Vallyathan V., Wang L., Tantishaiyakul V., Stehlik C., Leonard S. S., Rojanasakul Y. (2006) J. Biol. Chem. 281, 34124–34134 [DOI] [PubMed] [Google Scholar]
- 24.Choi B. H., Kim W., Wang Q. C., Kim D. C., Tan S. N., Yong J. W., Kim K. T., Yoon H. S. (2008) Cancer Lett. 261, 37–45 [DOI] [PubMed] [Google Scholar]
- 25.Dimmeler S., Breitschopf K., Haendeler J., Zeiher A. M. (1999) J. Exp. Med. 189, 1815–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haendeler J., Zeiher A. M., Dimmeler S. (1996) Eur. J. Pharmacol. 317, 407–411 [DOI] [PubMed] [Google Scholar]
- 27.Paradis E., Douillard H., Koutroumanis M., Goodyer C., LeBlanc A. (1996) J. Neurosci. 16, 7533–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breitschopf K., Haendeler J., Malchow P., Zeiher A. M., Dimmeler S. (2000) Mol. Cell. Biol. 20, 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng E. H., Kirsch D. G., Clem R. J., Ravi R., Kastan M. B., Bedi A., Ueno K., Hardwick J. M. (1997) Science 278, 1966–1968 [DOI] [PubMed] [Google Scholar]
- 30.Grandgirard D., Studer E., Monney L., Belser T., Fellay I., Borner C., Michel M. R. (1998) EMBO J. 17, 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirsch D. G., Doseff A., Chau B. N., Lim D. S., de Souza-Pinto N. C., Hansford R., Kastan M. B., Lazebnik Y. A., Hardwick J. M. (1999) J. Biol. Chem. 274, 21155–21161 [DOI] [PubMed] [Google Scholar]
- 32.Kang C. B., Tai J., Chia J., Yoon H. S. (2005) FEBS Lett. 579, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 33.Manser E., Huang H. Y., Loo T. H., Chen X. Q., Dong J. M., Leung T., Lim L. (1997) Mol. Cell. Biol. 17, 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanc C., Deveraux Q. L., Krajewski S., Jänicke R. U., Porter A. G., Reed J. C., Jaggi R., Marti A. (2000) Cancer Res. 60, 4386–4390 [PubMed] [Google Scholar]
- 35.Karpova T. S., Baumann C. T., He L., Wu X., Grammer A., Lipsky P., Hager G. L., McNally J. G. (2003) J. Microsc. 209, 56–70 [DOI] [PubMed] [Google Scholar]
- 36.Carrello A., Ingley E., Minchin R. F., Tsai S., Ratajczak T. (1999) J. Biol. Chem. 274, 2682–2689 [DOI] [PubMed] [Google Scholar]
- 37.Cliff M. J., Harris R., Barford D., Ladbury J. E., Williams M. A. (2006) Structure 14, 415–426 [DOI] [PubMed] [Google Scholar]
- 38.Ramsey A. J., Russell L. C., Whitt S. R., Chinkers M. (2000) J. Biol. Chem. 275, 17857–17862 [DOI] [PubMed] [Google Scholar]
- 39.Chen M., Guerrero A. D., Huang L., Shabier Z., Pan M., Tan T. H., Wang J. (2007) J. Biol. Chem. 282, 33888–33895 [DOI] [PubMed] [Google Scholar]
- 40.Reed J. C. (1995) Curr. Opin. Oncol. 7, 541–546 [DOI] [PubMed] [Google Scholar]
- 41.Ling Y. H., Liebes L., Ng B., Buckley M., Elliott P. J., Adams J., Jiang J. D., Muggia F. M., Perez-Soler R. (2002) Mol. Cancer Ther. 1, 841–849 [PubMed] [Google Scholar]
- 42.Chanvorachote P., Nimmannit U., Stehlik C., Wang L., Jiang B. H., Ongpipatanakul B., Rojanasakul Y. (2006) Cancer Res. 66, 6353–6360 [DOI] [PubMed] [Google Scholar]
- 43.Fong S., Mounkes L., Liu Y., Maibaum M., Alonzo E., Desprez P. Y., Thor A. D., Kashani-Sabet M., Debs R. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14253–14258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 45.Humar R., Kiefer F. N., Berns H., Resink T. J., Battegay E. J. (2002) FASEB J. 16, 771–780 [DOI] [PubMed] [Google Scholar]
- 46.Guba M., von Breitenbuch P., Steinbauer M., Koehl G., Flegel S., Hornung M., Bruns C. J., Zuelke C., Farkas S., Anthuber M., Jauch K. W., Geissler E. K. (2002) Nat. Med. 8, 128–135 [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa T., Shirane M., Iemura S., Natsume T., Nakayama K. I. (2007) Genes Cells 12, 709–719 [DOI] [PubMed] [Google Scholar]
- 48.Petros A. M., Medek A., Nettesheim D. G., Kim D. H., Yoon H. S., Swift K., Matayoshi E. D., Oltersdorf T., Fesik S. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muchmore S. W., Sattler M., Liang H., Meadows R. P., Harlan J. E., Yoon H. S., Nettesheim D., Chang B. S., Thompson C. B., Wong S. L., Ng S. L., Fesik S. W. (1996) Nature 381, 335–341 [DOI] [PubMed] [Google Scholar]
- 50.Edlich F., Weiwad M., Erdmann F., Fanghänel J., Jarczowski F., Rahfeld J. U., Fischer G. (2005) EMBO J. 24, 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.