Abstract
CDKN1C is a cyclin-dependent kinase inhibitor and is a candidate tumor suppressor gene. We previously found that the CDKN1C protein represses E2F1-driven transcription in an apparent negative feedback loop. Herein, we explore the mechanism by which CDKN1C represses transcription. We find that adenoviral-mediated overexpression of CDKN1C leads to a dramatic reduction in phosphorylation of the RNA polymerase II (pol II) C-terminal domain (CTD). RNA interference studies demonstrate that this activity is not an artifact of CDKN1C overexpression, because endogenous CDKN1C mediates an inhibition of RNA pol II CTD phosphorylation in HeLa cells upon treatment with dexamethasone. Surprisingly, we find that CDKN1C-mediated repression of RNA pol II phosphorylation is E2F1-dependent, suggesting that E2F1 may direct CDKN1C to chromatin. Chromatin immunoprecipitation assays demonstrate that CDKN1C is associated with E2F1-regulated promoters in vivo and that this association can dramatically reduce the level of RNA pol II CTD phosphorylation at both Ser-2 and Ser-5 of the C-terminal domain repeat. In addition, we show that CDKN1C interacts with both CDK7 and CDK9 (putative RNA pol II CTD kinases) and that CDKN1C blocks their ability to phosphorylate a glutathione S-transferase-CTD fusion protein in vitro. E2F1 and CDKN1C are found to form stable complexes both in vivo and in vitro. Molecular studies demonstrate that the E2F1-CDKN1C interaction is mediated by two E2F domains. A central E2F1 domain interacts directly with CDKN1C, whereas a C-terminal E2F1 domain interacts with CDKN1C via interaction with Rb. The results presented in this report highlight a novel mechanism of tumor suppression by CDKN1C.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, E2F Transcription Factor, RNA Polymerase II, Tumor Suppressor
Introduction
The mammalian cell cycle is orchestrated by the activity of the cyclin-dependent kinases (CDKs),3 and because of this they are considered important targets in cancer drug development (1–3). A key G1 target of the CDKs is pRB, the protein product of the RB tumor suppressor gene, which in turn regulates the E2F family of transcription factors. Hypophosphorylated Rb binds to and converts E2F family members from potent activators of transcription to potent repressors (4). In normal cells, Rb is phosphorylated by CDKs and then releases E2F family members. Freed E2Fs drive expression of genes essential for cell cycle progression and DNA synthesis. Excessive activation of E2F1 can also drive apoptosis, and thus, E2F1 may serve as an oncogene or tumor suppressor gene depending on context (5). In the absence of functional Rb the ability of CDK inhibitors to restrain E2F-regulated transcription is greatly diminished. Because of this central role in cell cycle control the CDK inhibitor/CDK/RB/E2F pathway is altered at some point in the vast majority of human cancers (6, 7).
Because of their central importance the CDKs are tightly regulated enzymes. There are two families of cyclin-dependent kinase inhibitors: the CDKN1 and CDKN2 proteins. The CDKN2 proteins are smaller and directly target the kinase subunits of the cyclin-CDK complexes. They are primarily active against CDK4 with some activity toward CDK6. CDKN2A (p16) is the most well known member of this family. CDKN2A is commonly lost in human cancers, and thus, it is classified as a tumor suppressor gene (8). The second group of CDK inhibitors are the CDKN1 proteins that include CDKN1A (p21, WAF1, and p21CIP1) (9, 10), CDKN1B (p27 and p27KIP1) (11, 12), and CDKN1C (p57 and p57KIP2) (13, 14). These proteins recognize the cyclin-CDK complex rather than simply the CDK subunit and generally have a broader activity spectrum. Biochemically the CDKN1 family has been shown to bind and regulate a diverse group of CDKs, including G1- and S-phase CDKs (cyclin E-CDK2, cyclin D2-CDK4, and cyclin A-CDK2), and, to lesser extent, of the mitotic cyclin B-Cdc2 (13). Structurally members of the CDKN1 family share a conserved N-terminal CDK inhibitory domain, whereas the C-terminal domains are highly divergent in structure (13, 14).
Certain CDKs can also regulate transcription directly independent of their role in cell cycle control. One important target of CDKs 7–9 is the C-terminal heptad repeat domain of RNA polymerase II (pol II) (15). The CDK7-containing CAK complex is a component of the multisubunit transcription factor TFIIH, which phosphorylates the serine 5 of the heptapeptide repeat of RNA pol II large subunit, thus promoting transcription initiation, promoter clearance, and recruitment of the capping enzyme (16, 17). CDK9 forms a complex with cyclin T, which preferentially phosphorylates Ser-2 of the heptapeptide repeat of RNA pol II large subunit promoting transcription elongation and recruitment of the 3′ RNA processing activities (18). The CDK8-cyclin C complex is also known to regulate RNA pol II activity (19).
This report is primarily concerned with CDKN1C, which we will refer to as, p57, its shorter name, in most of the figures. CDKN1C is maternally expressed, partially paternally imprinted (20), and implicated in sporadic cancers and Beckwith-Wiedemann syndrome, a familial disorder characterized by an increased risk of childhood cancer (21). Baculovirus-produced CDKN1C interacted tightly in IP/Western blots with cyclin A-CDK2, cyclin E-CDK3, and cyclin D2-CDK4, whereas its interaction with cyclin D2-CDK6 was modest and its interaction with cyclin H-CDK7 was not detected (14). Subsequent work has shown that bacterially produced CDKN1C can inhibit the kinase activity of baculovirus-produced cyclin H-CDK7 toward a cyclin A-CDK2 substrate (22). Whether CDKN1C can inhibit CDK7 and CDK9 activities toward the pol II CTD is not clear.
We recently demonstrated that E2F1 activates transcription of two members of the CDKN1 family CDKN1B (23) and CDKN1C (24). Activation of these negative cell cycle regulators appears to represent a negative feedback loop that limits excessive E2F1 activity that would otherwise drive unwanted apoptosis. During the discovery of these negative feedback pathways we happened upon the discovery of several novel interactions. Specifically, we find that CDKN1C binds directly to E2F1 and that this CDKN1C-E2F complex negatively regulates transcription by inhibiting the phosphorylation of the RNA pol II CTD. This discovery represents a novel activity for CDKN1C that may be important in its role as a tumor suppressor.
EXPERIMENTAL PROCEDURES
Mammalian Cell Lines and Expression Vectors
HeLa and MDA-MB-231cells were grown in Dulbecco's modified Eagle's media with 10% fetal bovine serum. H1299 cells were cultured in Dulbecco's modified Eagle's media supplemented with 5% fetal bovine serum. All drug treatments were performed as previously described (24, 25). pCMV6-XL5-CDK7 and pCMV6-XL5-CDK9 were purchased (OriGene Technologies, catalog numbers TC119024 and TC119344, respectively). FLAG-tagged forms of CDK7 and CDK9 were obtained using the PCR to amplify the cDNA from the purchased expression vectors. EcoRI and EcoRV sites were added to forward and reverse primers, respectively, to facilitate sub-cloning. PCR products were ligated into the p3×FLAG-myc-CMV-26 expression vector (E6401, Sigma). The primers used were; CDK7f, 5′-CGC GAA TTC AGC TCT GGA CGT GAA GTC TCG G-3′; CDK7r, 5′-CCG ATA TCA GAA AAA TTA GTT TCT TGG GCA A-3′; CDK9f, 5′-CGC GAA TTC AGC AAA GCA GTA CGA CTC GGT G-3′; and CDK9r, 5′-CCG ATA TCA TGA AGA CGC GCT CAA ACT CCG T-3′.
The novel expression vector pcDNA3-HA-E2F1–103-284 was generated by PCR using primers: 5′-CGCGGGATCCCATATGGCCGAGAGCAGTGGGC-3′ and 5′-CGCGGAATTCTCAGATCTGAAAGTTCTCCGAAGA-3′. The product of this reaction was cleaved with BamHI and EcoRI and cloned into those sites of pcDNA3-HA to form pcDNA3-HA-103–284. Previously described mammalian expression plasmids include pcDNA-FLAG-CDKN1C (24), pGL2-E2F1 (26), and pcDNA3-HA-E2F1 (27). Transient transfections for protein expression utilized the calcium phosphate method: H1299 cells growing in p100 plates and 10 μg of each CMV expression vector (or empty control to a total of 20 μg). Transfected cells were collected 48 h post transfection and were processed for Western blots and immunoprecipitation assays as described below.
A shRNAi vector targeting CDKN1C (TRCN0000039678) was obtained from Open Biosystems and was used as previously described (24). Pools of CDKN1C-deficient cells were created by transecting the CDKN1C shRNAi vector into HeLa cells with subsequent selection for transfected cells using puromycin as previously described (24).
Adenoviruses
Dr. Matthew Stewart (University of Illinois at Urbana-Champaign) provided Ad-p57 virus-expressing mouse cdkn1c (28). Dr. Haura (Moffitt Cancer Center) provided Ad-GFP, an adenoviral vector expressing the green fluorescent protein (GFP), which was used as a negative control. Ad-CDK7 was purchased from Vector Biolabs (Philadelphia, PA, catalog no. 1568). Viruses were propagated in 293T cells and titrated with the Adeno-X Rapid Titer Kit (BD Biosciences, 631028). Western blots verified viral expression of cdkn1c, CDK7, and absence of E1A expression (to ensure non-recombination).
Bacterially Expressed Proteins
GST-CTD (29), in which the C-terminal heptad repeat domain of RNA pol II is fused to the glutathione S-transferase affinity tag, was a kind gift from Dr. Arno Greenleaf (Duke University Medical Center). Bacterial expression vectors for GST-Rb 379–928, GST-E2F1–1-147, -1–358, -1–284, and -113–284 have been previously described (30–32). GST vectors for E2F1 192–437 and 300–437 are novel constructs generated from previously described mammalian expression vectors (33). The E2F expression vectors were cut with BamHI and PmeI (blunt) to excise the insert. The pGEX2T vector was prepared by cutting with EcoRI, which was then filled in with DNA pol I (Klenow fragment) to generate a blunt end, and cut with BamHI. Recombinant GST fusion proteins were expressed in BL21 Gold bacterial cells. Protein expression was induced with 0.2 mm isopropyl-1-thio-d-galactopyranoside. Cells were pelleted and resuspended in STE buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mm EDTA) supplemented with final concentrations of 5 mm dithiothreitol and 1.5% N-laurylsarcosine. The mixture was sonicated 3 times for 10 s each, mixed with 3% Triton X-100, and then cleared by centrifugation at 13,000 × g for 10 min (for GST proteins the pellet was discarded). The CDKN1C protein was expressed in bacteria using a novel vector (pET-CDKN1C) created by cutting pCMVCDKN1C (TC127869, Origene Technologies) with SacI and HindIII, and cloning into pET23b. Bacterially expressed CDKN1C was prepared as described for GST proteins, however, CDKN1C was primarily found in inclusion bodies (the pellet of the 13,000 rpm spin), and thus, CDKN1C was solubilized in 8 m urea (also containing 50 mm Tris (pH 8), 100 mm NaCl, 10 mm dithiothreitol, and 1 mm EDTA) prior to use in immunoprecipitation reactions. Twenty micrograms of crude bacterial lysates was used in immunoprecipitation reactions without purification as described below.
IP and Pull-down Assays
Mammalian cells were washed twice in phosphate-buffered saline and resuspended in lysis buffer containing 50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 5 mm EDTA, and 2% Nonidet P-40, supplemented with protease inhibitors (5 μg/ml each of antipain, aprotinin, leupeptin, and soybean trypsin inhibitor and 0.5 μg/ml pepstatin) and 0.5 mm phenylmethylsulfonyl fluoride. Protein concentrations were determined by the Bradford assay (Bio-Rad). For each IP, 50 μl of protein A/G plus-agarose beads (Santa Cruz Biotechnology, sc-2003) were incubated with 1 μg of the CDKN1C polyclonal antibody (Santa Cruz Biotechnology, sc-1040) or 1 μg of rabbit IgG (Santa Cruz Biotechnology, sc-2027) for 2 h at 4 °C. Beads were washed three times with the same protein lysis buffer, incubated with 800 μg of protein extract for 2 h at 4 °C, and washed again. Proteins bound to the beads were dissolved in loading buffer, boiled, and subjected to SDS-PAGE followed by Western blotting. IP and GST bead (G4510, Sigma) pull-down reactions on bacterially expressed proteins were carried out using 20 μg of crude bacterial lysates that were prepared as described above.
In Vitro Kinase Assay
Cell extracts were prepared from H1299 cells that had been infected by Ad-p57 or not by resuspending cells in modified TGN buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Tween 20, 0.3% Nonidet P-40, 1 mm sodium fluoride, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture from Roche Molecular Biochemicals). Cleared supernatants were immunoprecipitated with anti-CDK7 antibody (BD Pharmingen) or anti-CDK9 antibody (Cell Signaling Technology) and protein A/G-agarose, the beads were washed with TGN buffer followed by TGN buffer plus 0.5 m LiCl, and two washes with kinase buffer (20 mm HEPES, pH 7.5, 50 mm NaCl, 10 mm MgCl2, and 1 mm dithiothreitol). Finally, the immunoprecipitate was resuspended in 50 μl of kinase buffer containing 5 mm ATP and 1 μg of purified GST-CTD substrate. The kinase reaction was conducted at 30 °C for 20 min and stopped by addition of SDS-PAGE loading buffer. Proteins were separated on SDS-PAGE and transferred to nitrocellulose. The phosphorylation of GST-CTD was determined by RNA pol II phosphorylation-specific antibodies. Immunoprecipitated CDK7 was confirmed by Western blotting with anti-CDK7 monoclonal antibody.
Western Blotting
Western blots were performed as previously described (1, 25, 27). Briefly, cell lysates were normalized for total protein content (50 μg) and subjected to SDS-PAGE. Detection of proteins was accomplished using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (ECL) purchased from Amersham Biosciences. Various antibodies used included: a rabbit polyclonal antibody to GST (a kind gift from Dr. Jia Fang, Moffitt Cancer Center), a cyclin A rabbit polyclonal (provided by Dr. Jack Pledger, Moffitt Cancer Center), an E2F1 monoclonal antibody, KH20, that recognizes the E2F1 N terminus (sc-56662, Santa Cruz Biotechnology), an E2F1 monoclonal antibody, KH95, that recognizes the E2F1 C terminus (sc-251, Santa Cruz Biotechnology), a human CDKN1C polyclonal antibody (sc-1040, Santa Cruz Biotechnology), a mouse CDKN1C polyclonal antibody (sc-8298, Santa Cruz Biotechnology), an RB monoclonal antibody (OP28, Calbiochem), and an actin monoclonal antibody (A5441, Sigma). A monoclonal antibody recognizing the RNA pol II large subunit regardless of modification was supplied by Covance Research Products (8WG16). Bethyl laboratories provided antibodies against RNA pol II phosphorylated at Ser-2 (A300-654A-1) and Ser-5 (A300-655A-1).
RNase Protection Assays
RNA protection assays were performed using the Riboquant, a multiprobe template from BD Pharmingen (now discontinued), as previously described (24).
ChIP Assay
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (34) with minor adaptations. After viral infection, cells were cross-linked with 1% formaldehyde for 10 min at ambient temperature. The reaction was stopped by adding glycine to a final concentration of 0.125 m. Cells were washed twice with cold phosphate-buffered saline, scraped, and lysed for 5 min on ice in cold lysis buffer (10 mm Tris-HCl, pH 8.1, 10 mm EDTA, 0.5 mm EGTA, 0.25% Triton X-100, 0.5% Nonidet P-40, and protease and phosphatase inhibitors) at a density of 4 × 106 cells/ml. The nuclei were washed for 10 min in cold salt wash buffer (10 mm Tris-HCl, pH 8.1, 1 mm EDTA, 0.5 mm EGTA, 200 mm NaCl and protease and phosphatase inhibitors) and resuspended subsequently in 600 μl of sonication buffer (10 mm Tris-HCl, pH 8.1, 1 mm EDTA, 0.5 mm EGTA, 1% SDS, and protease and phosphatase inhibitors). The chromatin was sheared using a Biorupter XL water bath sonicator to lengths ranging from 200 to 1000 bp. After preclearing the sheared chromatin with preblocked protein A/G beads, equal amounts of chromatin were immunoprecipitated overnight at 4 °C with antibodies in a buffer containing 0.15% SDS and 0.22% Triton X-100. The immunoprecipitated chromatin was collected on protein A/G beads. From the non-bound material of the rabbit IgG immune precipitation 10% was collected as input sample. The bead-bound chromatin was washed for 5 min each sequentially in low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), high salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 500 mm NaCl), LiCl 250 buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1) and twice with TE10:1 (10 mm Tris-HCl, pH 8.1, 1 mm EDTA). DNA was eluted twice with TE10:1 elution buffer containing 1% SDS, the samples were vortexed for 1 min, and then incubated at 65 °C for 15 min and ambient temperature for 5 min. The two elution fractions for each sample were collected from the beads and pooled. Cross-linking was reversed by adding 20 μl 5 m NaCl and incubating the samples for 4 h at 65 °C. To purify the DNA, samples were treated with DNase-free RNase (30 min, 37 °C), followed by proteinase K treatment (1 h at 45 °C in 40 mm Tris, 10 μm EDTA). The DNA was then purified using a Qiagen PCR Purification Kit. The manufacturer's protocol was altered by leaving the PE wash on the column for 5 min before spinning and the 50 μl elution buffer for 2 min. Of each sample, except input, 3 μl was used for real-time PCR. For the input sample 3 μl of a 1:10 dilution was used.
Antibodies used for immunoprecipitation were RNA pol II 8WG16 monoclonal antibody (Covance, MMS-126R), phospho RNA pol II Ser-2 (Bethyl Laboratories Inc., A300-654A-1), phospho RNA pol II Ser-5 (Bethyl Laboratories Inc., A300-655A-1), and normal rabbit IgG polyclonal antibody (Millipore, 12-370). Real-time PCR was performed using Bio-Rad iQ SYBR Green Supermix on a MyiQ Single Color Real-time PCR detection system. The following primers were used: E2F3 forward, 5′-gacttggaaactccgactgc-3′; E2F3 reverse, 5′-catctctcgctcctgctct-3′; cylin A2 forward, 5′-cagccttcggacagcctcgc-3′; and cyclin A2 reverse, 5′-caaactggctggggcgggag-3′. Human myoglobin intron primer mix was obtained from Diagenode (catalog no. pp-1006-500).
RESULTS
Expression of CDKN1C Leads to a Decrease in the Phosphorylation State of the RNA pol II C-terminal Domain
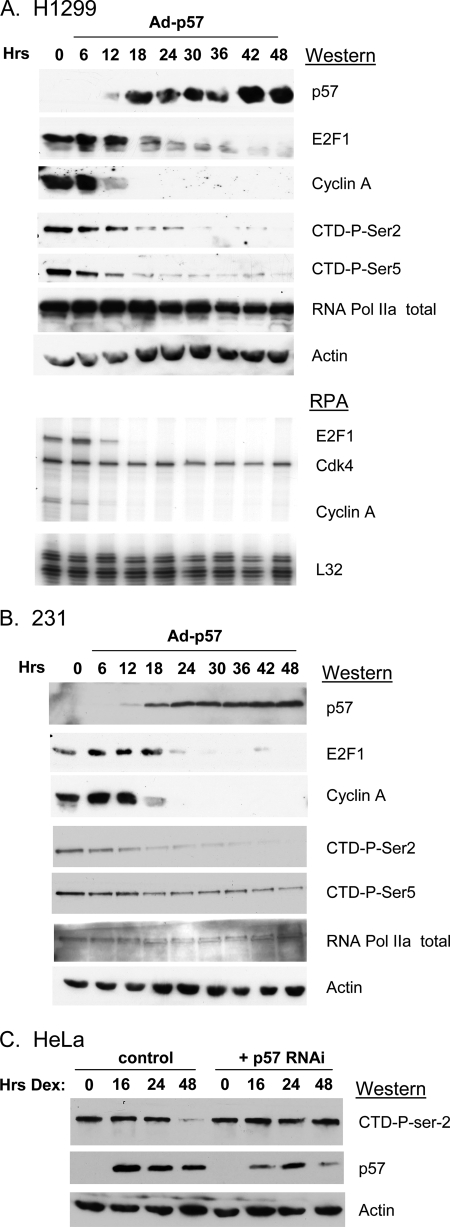
We previously demonstrated that CDKN1C can inhibit E2F1-driven transcription in a feedback loop (24). To better understand the mechanism of CDKN1C feedback we examined the effects of adenovirus-mediated CDKN1C expression on the protein and mRNA levels of various E2F-regulated genes (not shown). Surprisingly, we observed dramatic reductions in the total phosphorylation state of the RNA pol II CTD repeat domain following CDKN1C expression. Fig. 1 highlights these results in NCI-H1299 (lung) and MDA-MB-231 (breast) cancer cells. In each case CDKN1C expression leads to a dramatic decrease in the expression of the E2F-regulated transcripts, E2F1 and cyclin A, at both the protein and mRNA levels. More examples of CDKN1C-mediated down-regulation of E2F-regulated transcripts can be seen in previously published work (24). CDKN1C expression also resulted in a significant to moderate decreases in the phosphorylation state of both Ser-2 and Ser-5 of the CTD. There was little change in the expression of total RNA pol II or actin in response to CDKN1C expression in either cell line.
FIGURE 1.
Expression of CDKN1C leads to a decrease in the phosphorylation state of the RNA pol II C-terminal domain. A and B, the indicated cell lines were infected with an adenovirus expressing mouse cdkn1c, Ad-p57 (28). Cells were harvested at the indicated times post-infection and were subjected to Western blotting or RNase protection assays using the indicated antibodies or RNase protection assay probes, as indicated (described under “Experimental Procedures”). The viral multiplicity of infection was 200 to 1. Ad-GFP was used as a negative control and had no effect (not shown). C, HeLa cells (first four lanes) were treated with 100 nm dexamethasone, and extracts were prepared and subjected to Western blotting at the indicated times after treatment. In the second four lanes HeLa cells were transfected with an shRNAi vector targeting CDKN1C and subjected to selection and propagation in puromycin prior to treatment with dexamethasone. Similar results were obtained in multiple experiments.
To determine if the observed effect of CDKN1C expression on CTD phosphorylation was an artifact of overexpression, we turned to a well described model in which endogenous CDKN1C is induced by dexamethasone treatment of HeLa cells (35). Fig. 1C demonstrates that overnight treatment of HeLa cells with dexamethasone results in a dramatic up-regulation of CDKN1C, due to the presence of a glucocorticoid response element in its promoter (35), with sustained CDKN1C expression for 48 h. By 48 h CTD-Ser-2 phosphorylation has significantly decreased, showing that CDKN1C can regulate CTD phosphorylation. To determine if the decreased CTD-Ser-2 phosphorylation was dependent upon the up-regulation of CDKN1C, HeLa cells were transfected with an shRNAi expression vector targeting CDKN1C (p57 RNAi), and transfected clones were selected by exposure to puromycin. As expected, pools of puromycin-resistant cells reveal that the RNAi significantly reduced the level of dexamethasone-induced CDKN1C and attenuated the down-regulation of Ser-2 phosphorylation in response to treatment. Observing this activity in HeLa cells, which lacks functional Rb family members due to expression of the E7 viral oncoprotein, also suggests that this CDKN1C activity is not dependent upon its well known ability to regulate Rb phosphorylation.
CDKN1C Interacts with RNA pol II and Its CTD Kinases
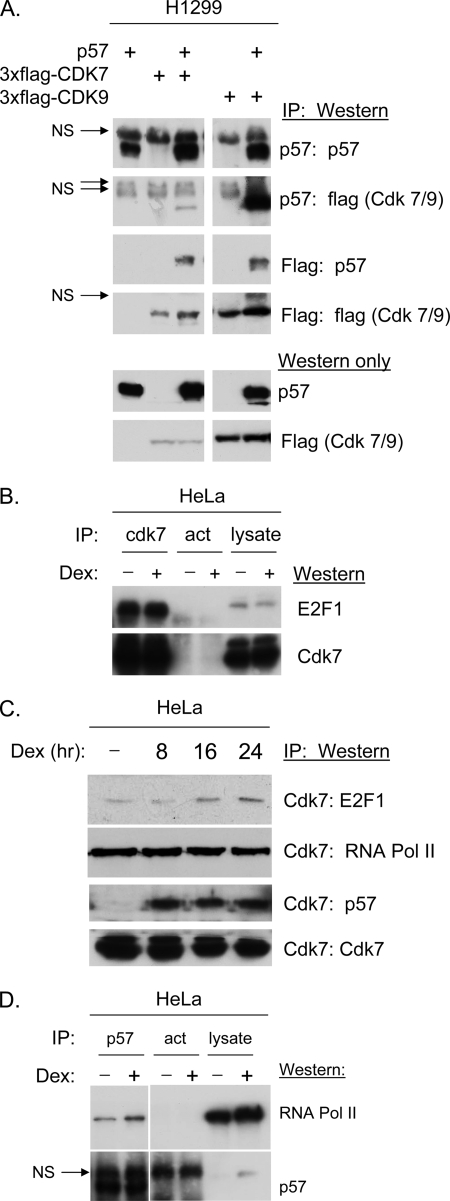
The primary kinases thought to be responsible for the phosphorylation of the pol II CTD are CDK 7 and CDK9 (2, 15). Previous work has specifically demonstrated that CDKN1C can coimmunoprecipitate cyclins D1, E, and A as well as CDK2 and CDK4 (13). To determine if CDKs 7 and 9 might also associate with CDKN1C plasmids expressing FLAG-tagged CDK 7, FLAG-tagged CDK9 and/or CDKN1C were co-transfected into H1299 cells, and extracts were subjected to IP/Western blotting. Fig. 2A reveals that both CDK7 and CDK9 are efficiently co-immunoprecipitated with CDKN1C antibody. The fact that the CDK9 band is much stronger in CDKN1C IPs than the CDK7 band may be explained by its higher expression level revealed by the Western blots at the bottom of Fig. 2 and not necessarily by higher affinity. Identical results were obtained when FLAG antibodies were used to co-immunoprecipitate CDKN1C (see middle panels of Fig. 2A).
FIGURE 2.
CDKN1C interacts with CDK7 and 9 in vivo. A, H1299 cells were co-transfected with the indicated expression vectors or empty vector. IP/Western blots were performed using CDKN1C antibody sc-1040 (Santa Cruz Biotechnology) for both IP and Western blot. The FLAG antibody F-1804 from Sigma was used for detection of tagged CDK7 and CDK9 by Western blot. An anti-FLAG antibody conjugated to agarose (A-2220, Sigma) was used for FLAG-IPs. B–D, HeLa cells extracts (with and without prior dexamethasone (Dex) treatment as indicated) were subjected to IP/Western blots using the indicated combinations of antibodies. Results reveal that endogenous CDK 7associates with both E2F1 and CDKN1C and that CDKN1C interacts with RNA pol II in vivo.
Additional IP/Western blots were performed to verify that endogenous CDKN1C interacts with endogenous CTD kinases. Previous work has shown an interaction between the E2F1 activation domain and CDK7 (36). Fig. 2B reveals that a CDK7 antibody efficiently immunoprecipitates E2F1 from HeLa cells regardless of dexamethasone treatment, as expected (36). Fig. 2C reveals that CDK7 antibody also efficiently immunoprecipitates RNA pol II and CDKN1C from HeLa cells. RNA pol II interacted with CDK7 regardless of dexamethasone treatment, whereas the interaction with CDKN1C was dependent upon dexamethasone treatment. Although we were unable to detect interactions using the CDK9 antibody, in our experiments, the results with CDK7 clearly indicate that CDKN1C stably interacts with endogenous CDK7 and interacts with E2F1 and RNA pol II. This observation suggests the possibility that CDKN1C/RNA polymerase complexes may exist in vivo. To test this hypothesis CDKN1C antibody was used in IP/Western experiments. Fig. 2D reveals CDKN1C antibody co-immunoprecipitates RNA pol II.
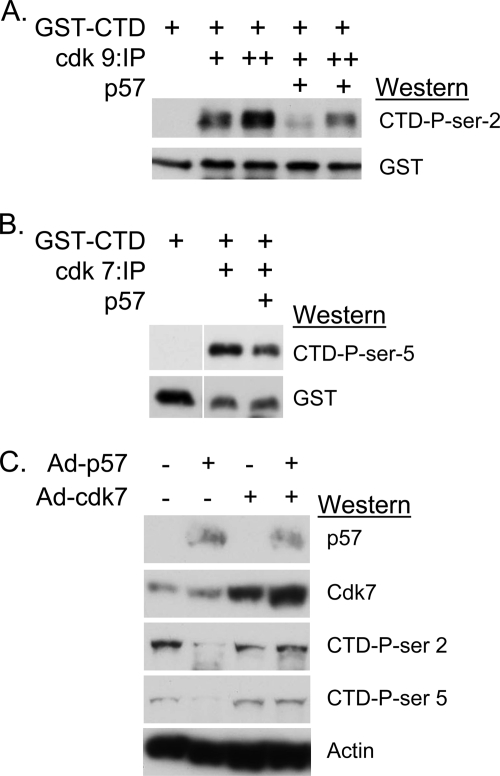
While CDKN1C has been shown to inhibit the activity of CDK7 toward a cyclin A-CDK2 substrate in vitro (22), it is not clear if CDKN1C can biochemically block the activity of CDKs toward the CTD. To test this, active CDKs were immunoprecipitated from either H1299 cells or H1299 cells overexpressing CDKN1C and subjected to in vitro kinase assays using a bacterially produced GST-RNA pol II CTD fusion protein (29) as substrate. Phosphorylation of GST-CTD was detected by Western blot using the appropriate anti-phospho-Ser antibodies. Fig. 3 (A and B) demonstrates that immunopurified CDKs 7 and 9 both efficiently phosphorylate GST-CTD when purified from control H1299 cells, but this activity was significantly reduced in extracts of cells overexpressing CDKN1C. A rescue experiment, shown in Fig. 3C, was performed as an additional test to demonstrate that CDKN1C functionally inhibits the activity of cdk7 toward RNA pol II. H1299 cells were infected with Ad-p57 to express cdkn1c, with a CDK7-expressing adenovirus, with a control virus or combinations. Results reveal that cdkn1c expression blocks CTD phosphorylation at both Ser-2 and Ser-5 in H1299, as expected. Although overexpression of CDK7 did not affect the baseline level of CTD phosphorylation; its overexpression restored CTD phosphorylation in the presence of exogenous cdkn1c. These results support the hypothesis that CDKN1C can block the ability of cyclin-dependent kinases to phosphorylate the RNA pol II CTD.
FIGURE 3.
CDKN1C can inhibit CDK7 and CDK9 kinase activity toward the RNA pol II CTD. A, CDK9 was immunopurified from extracts of control H1299 cells or H1299 cells infected with Ad-p57 and subjected to kinase assays using bacterially produced GST-CTD as substrate. A single “+” sign indicates that 250 μg of cell extract was used for IP, whereas “++” indicates 500 μg was used. Following 30-min incubations, reactions were subjected to SDS-PAGE and Western blotting using the indicated antibodies. The Western blot signal using phospho-serine specific antibodies indicated the relative level of kinase activity toward CTD. B, CDK7 was immunoprecipitated from 30 μg of cell extract, and the kinase reaction was carried out for 20 min. C, H1299 cells were infected for 48 h with the indicated combination of viral vectors to express cdkn1c (p57) or CDK7. Overexpression of CDK7 rescues CTD-Ser-2 phosphorylation in the presence of excess cdkn1c. A similar viral vector for CDK9 was not available.
CDKN1C Associates with E2F-regulated Promoters
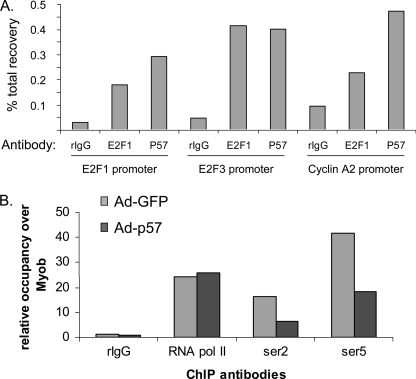
The CDKN1C relative CDKN1A has been shown to associate with E2F1 on the Wnt4 promoter curtailing Wnt4 transcription (37). To determine if CDKN1C might have a similar activity a ChIP assay was employed. Quantitative ChIP assays were performed using E2F1 and CDKN1C antibodies on the E2F1, E2F3, and cyclin A promoters. Control antibodies in these experiments included rabbit IgG, as a negative control, and an antibody against acetylated histone H3, as a positive control (not shown). ChIP signal for each promoter was normalized to the total input DNA. Fig. 4A reveals that both the E2F1 and CDKN1C antibodies pull down significantly more of each promoter than does the control antibody. Furthermore, E2F and CDKN1C antibodies did not immunoprecipitate the second intron of the myoglobin gene locus, which served as a negative control (not shown).
FIGURE 4.
ChIP assays indicate that E2F1 and CDKN1C coexist in complexes on the E2F1, E2F3, and cyclin A2 promoters. A, H1299 cells were treated with either Ad-GFP or Ad-p57 for 48 h as indicated. Quantitative ChIP assays were performed on the E2F1, E2F3, and cyclin A2 promoters using antibodies as indicated on the x-axis. “IgG” represents a negative control (Santa Cruz Biotechnology, sc-1027). E2F1 represents E2F1 antibody (Santa Cruz Biotechnology, sc-193). “P57” represents CDKN1C antibody (Santa Cruz Biotechnology, sc-1040). Results were normalized to % total recovery from the input chromatin. B, quantitative ChIP assays on the E2F3 promoter were performed using antibodies against various RNA pol II phospho-isoforms. Additionally, in this experiment results were normalized to relative occupancy over the occupancy of the myoglobin B intron.
Antibodies against the various phosphorylated isoforms of RNA pol II were used in ChIP assays to determine whether association of CDKN1C would affect the phosphorylation state of the RNA pol II associated with the E2F3 promoter. Fig. 4B reveals that infection of H1299 cells with Ad-p57 dramatically reduces the phosphorylation state of RNA pol II associated with the E2F3 promoter. CDKN1C expression does not affect the total amount of RNA pol II associated with the promoter. Although this result may have been predicted, based on the effect of CDKN1C expression on total RNA pol II phosphorylation as observed in Western blots, it is important to establish that promoter DNA-associated pol II is subject to CDKN1C regulation.
E2F1 Is Required for CDKN1C to Block the Phosphorylation of RNA pol II
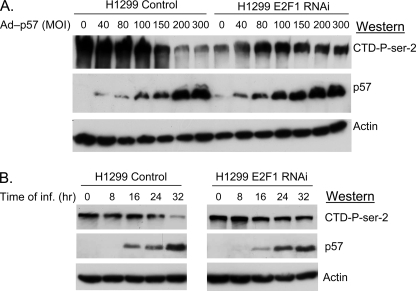
We next utilized a well characterized E2F1-deficient H1299 cell line to determine if the activity of CDKN1C toward RNA pol II was dependent on E2F1 (1). In this experiment, shown in Fig. 5A, control H1299 cells (stably harboring a non-targeted shRNA vector) or a previously characterized (1) E2F1-deficient H1299 (expressing an E2F1 shRNAi) were infected with various doses of Ad-p57. In the control cells increasing the dose of virus increased the amount of CDKN1C expressed and decreased the phosphorylation state of RNA pol II CTD-Ser-2. In contrast, CDKN1C expression did not decrease the pol II phosphorylation in the E2F1-deficient cells. This result indicates that E2F1 may be required to bring CDKN1C to DNA and thereby to RNA pol II and its kinases. The apparent decrease in basal RNA pol II phosphorylation in the E2F1-deficient cells (time 0) was not reproducible. Fig. 5B shows a similar experiment in which control and E2F1-deficient H1299 cells are infected with a fixed concentration of virus and are harvested at various times after infection.
FIGURE 5.
E2F1 is required for CDKN1C to block the phosphorylation of RNA pol II. A, H1299 cells stably expressing a non-targeted shRNAi vector (H1299) and E2F1-deficient H1299 cells stably expressing an E2F1 shRNAi (1) were infected with the indicated multiplicity of the Ad-57 virus. Cells were harvested after 32 h, and extracts were subjected to Western blots as indicated. B, H1299 cells were infected with a constant multiplicity of infection of 200 to1 with Ad-p57 virus and harvested at various times, as indicated.
E2F1 and CDKN1C Form Stable Complexes
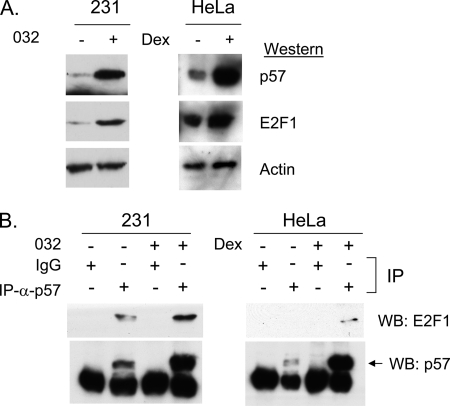
Several cell cycle regulatory proteins, such as 14-3-3 tau (38), MDM2 (39, 40), and ARF (41–43), are known to bind E2F1 directly. In fact, a direct interaction between E2F1 and CDKN1C relative CDKN1A has been proposed (37). To determine if CDKN1C and E2F1 might interact we examined HeLa cells and MDA-MB-231 cells using IP/Western blots. To increase the sensitivity of the assay the HeLa cells were treated with 100 nm dexamethasone to increase endogenous CDKN1C levels (35), and 231 cells were treated with SNS-032 (formerly BMS-387032) (44). SNS-032 is a small molecule CDK inhibitor that has been shown to dramatically up-regulate E2F1 and CDKN1C expression in 231 cells (24). The Western blot of Fig. 6A reveals that SNS-032 treatment of MDA-MB-231 cells induces CDKN1C protein, and addition of dexamethasone to HeLa cells induced CDKN1C. Fig. 6B reveals that antibodies against CDKN1C efficiently co-immunoprecipitate E2F1 in both cell lines; this supports the hypothesis that endogenous E2F1 and CDKN1C interact.
FIGURE 6.
E2F1 and CDKN1C form stable complexes. A, MDA-MB-231 cells or HeLa cells were treated with 500 nm SNS-032 (48 h) or 100 nm dexamethasone (24 h). Western blotting confirms induction of endogenous CDKN1C and continued expression of E2F1. B, MDA-MB-231 cells or HeLa cells were treated as in A, and extracts were used in IP/Western blots using antibody to CDKN1C (or control IgG) for the IP step and E2F1 and CDKN1C antibodies for the Western step.
Multiple E2F1 Regions Contribute to the Interaction with CDKN1C
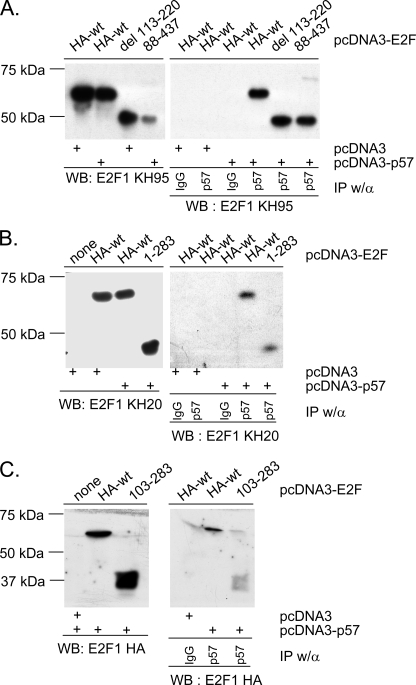
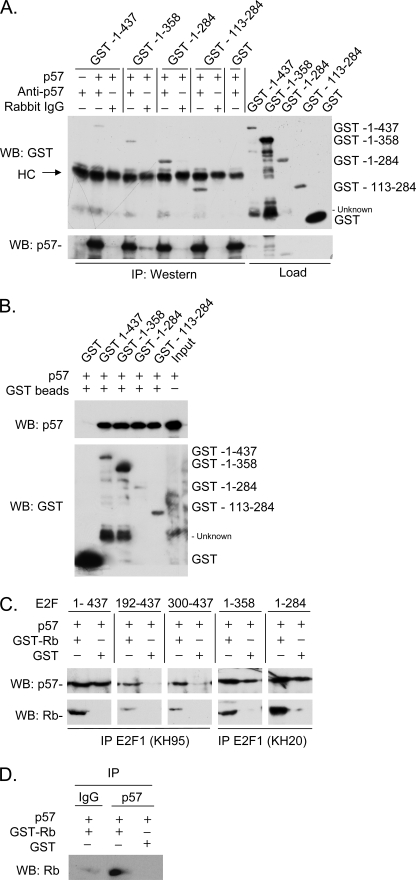
We investigated the CDKN1C-E2F1 interaction using a panel of E2F1 derivatives to map the interacting domains. For in vivo studies a CDKN1C expression vector was transfected into H1299 cells together with wild-type E2F1, one of the E2F1 derivatives highlighted in supplemental Fig. S1 or with an empty vector. The co-expressed proteins were then immunoprecipitated with CDKN1C antibody and IP pellets Western blotted to detect the various E2F1 derivatives (different E2F antibodies were used depending on the epitopes missing in the constructs, see supplemental Fig. S1). Fig. 7A reveals that the N-terminal 220 amino acids of E2F1 appear dispensable for the CDKN1C interaction. In a similar fashion, Fig. 7B demonstrates that the C-terminal 143 amino acids are not required. This analysis indicates that the central region of E2F is key to its interaction with CDKN1C. A novel construct was created containing amino acids 103–283 to determine if this region was sufficient for interaction with CDKN1C. Fig. 7C reveals that this construct is sufficient to bind CDKN1C weakly in vivo. Because this region of E2F1 is known to interact with other factors, it seemed important to determine if the interaction was direct. Thus, bacterially expressed CDKN1C was mixed with bacterially expressed E2F1 and several E2F1 mutants. Fig. 8 reveals that the central region of E2F1 is sufficient to mediate a direct interaction with CDKN1C in the absence of other mammalian proteins. Interestingly, GST-E2F1 constructs such as 192–437 and 300–473 that lack the central region but contain the C-terminal Rb-interaction domain, can also interact with CDKN1C; however, this interaction is dependent upon the presence of added Rb protein (see Fig. 8, C and D) suggesting a trimolecular complex in which Rb bridges interaction between the E2F C-terminal region and CDKN1C.
FIGURE 7.
A central E2F1 domain contributes to the interaction with CDKN1C. The indicated mammalian E2F1 expression vectors described in Fig. 3 were transfected into H1299 cells with either pcDNA3-CDKN1C or pcDNA3 (indicated at the bottom). Left panels represent Western blots that show protein expression. Right panels represent IP/Western blots in which either nonspecific IgG or CDKN1C-specific rabbit polyclonal were used, as indicated. Three different antibodies were used to detect E2Fs. KH95 recognizes the E2F C terminus and was used in panel A. KH20 recognizes the E2F N terminus and was used in panel B. An HA epitope antibody was used to detect E2F1 103–283 in panel C.
FIGURE 8.
The CDKN1C/E2F1 interaction is direct. A, crude lysates containing bacterially expressed GST-E2F1 fusions or CDKN1C (indicated with a “+” if present) were mixed and binding allowed to occur in IP buffer. Protein mixtures were immunoprecipitated by CDKN1C antibody (Anti-CDKN1C) or a negative control (Rabbit IgG) antibody. GST-E2F1 fusions were detected in immunoprecipitates by Western blotting with a rabbit GST antibody, and CDKN1C was detected using a CDKN1C antibody. B, similar to A except that GST beads were used to pull down the various GST-E2F1-CDKN1C complexes. The last lane represents the crude CDKN1C lysate and highlights an unknown protein present in some crude extracts that cross-reacts with our GST antibody. GST served as a negative control and does not pull down detectable CDKN1C. C, similar to A, except that purified GST (as a negative control) or GST-Rb was included in the binding reaction to determine if Rb would affect the interaction. Appropriate monoclonal antibodies to E2F1 were used. D, similar to C, except E2F1 was not present and the immunoprecipitation was carried out using control or CDKN1C antibody. HC stands for the immunoglobulin heavy chain.
DISCUSSION
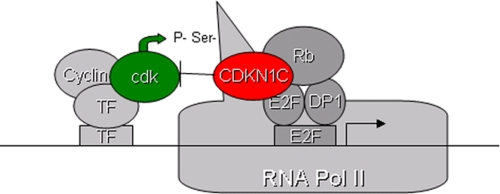
The work described in this report identifies a novel mechanism by which the tumor suppressor CDKN1C regulates transcription (shown in Fig. 9). The most novel component of this model is that CDKN1C contributes to transcriptional repression by blocking the activity of cyclin-dependent kinases toward RNA pol II. Three lines of evidence support this conclusion. First, CDKN1C overexpression blocks CTD phosphorylation in multiple cell lines (Fig. 1). This activity is not exclusive to exogenous CDKN1C, because blocking the expression of CDKN1C with RNAi prevents the notable decrease in the pol II CTD phosphorylation that occurs in dexamethasone-treated HeLa cells (Fig. 1). Because HeLa cells express the E7 oncogene, it is also safe to conclude that the activity of CDKN1C in HeLa cells is not an indirect consequence of regulating Rb phosphorylation. In addition Fig. 3 provides biochemical evidence that this activity is direct, because CDKN1C can block the activity of CDKs toward a GST-CTD substrate in vitro. Second, stable associations between CDKN1C and CDK7, CDK9, and RNA pol II are observable in IP/Western experiments (Fig. 2). Finally, CDN1C is shown to associate with three well characterized E2F-regulated promoters by ChIP assay, and the association of CDKN1C results in a decrease in phosphorylated RNA pol II at the E2F3 promoter (Fig. 4).
FIGURE 9.
Schematic summarizing the novel tumor suppressor mechanism of CDKN1C.
The ability of CDKN1C to block CTD phosphorylation is dependent upon E2F1, suggesting that E2F1 recruits CDKN1C to DNA as shown in the model in Fig. 9. However, it is formally possible that E2F1 serves as a scaffold that brings various proteins together independent of its DNA interaction. This activity appears to be E2F1-specific, because it is lost in E2F1-deficient cells (Fig. 5) and we are unable to detect a physical interaction between CDKN1C and other E2Fs (data not shown) under conditions where the CDKN1C-E2F1 interactions are easily detected (Figs. 6 and 7). Rb contributes to the stability of this complex by bridging an interaction between the E2F1 C terminus and CDKN1C, but there is a direct physical interaction between E2F1 and Rb mediated by the central E2F1 domain that will be better defined in future work. This interaction would occur even in Rb-deficient cells possibly providing a backup mechanism by which CDKN1C can restrain the G1 transition even in the absence of Rb (such as in HeLa cells). This unexpected E2F1 activity may contribute to its tumor suppressor activity in certain contexts (5, 45, 46). The physical interaction between CDKN1C and E2F1 was unexpected; however, careful examination of the literature suggests that it is not unprecedented among the CDKN1 family. An Rb-independent mechanism of E2F feedback has been proposed for CDKN1A (47). It was shown as early as 1996 that CDKN1A could inhibit an E2F-driven promoter and that it could be found stably associated with E2F complexes, which at the time was presumed to involve a complex, including E2F/Rb/cyclin/CDK2 with the association with E2F mediated by the cyclin/CDK2 bridge (48). It has been shown that GST-CDKN1A can bind to E2F1 and DP1, and that CDKN1A can inhibit transcription of a Gal4-E2F fusion in reporter assays in vivo and in a general transcription assay in vitro (47). A recent study has used ChIP assays to demonstrate that CDKN1A is associated with the Wnt4 promoter (37). We found that the association of E2F1 with CDKN1C is much easier to detect than its association with either CDKN1A or -B, but have not fully investigated the relative importance of the various E2F1 CDKN1 family interactions.
In this model we speculate that growth-regulated promoters are driven, in part, by DNA-bound transcription factors that recruit kinases that phosphorylate the CTD. Although transcription factor-mediated recruitment is not an essential component of this model, there are many likely candidates to mediate this hypothetical recruitment. For example, CDK9 has been shown to associate directly or indirectly with B-Myb (49), RB (50), RelA (51), AR (52), and c-Myc (53). In addition, CDK7 has been shown to associate with the AR (52), p53 (54, 55), and E2F1 itself (36). Of all these candidates c-Myc is likely the most relevant, because c-Myc and E2F are known to co-regulate promoters (56) and the ability of C-Myc to recruit CDK9 for the purpose of transcriptional activation is established (57, 58). Future work to examine specific examples of E2F-regulated promoters will establish more details of this element of the model.
The biological context of this novel biochemical mechanism is not yet fully understood. Certainly this mechanism provides a backup system to provide for cell growth control in situations where the tumor suppressor Rb is inactivated (such as in HeLa cells, which were extensively studied in this report). Significant genetic evidence suggests that inactivation of Rb alone is insufficient to drive tumorigenesis in most tissues (reviewed in Ref. 59). These observations support the idea that additional failsafe cell cycle control mechanisms must be in place in most tissues. It is also noteworthy that CDKN1C is extremely responsive to treatment with glucocorticoids (see above as well as Refs. 35, 60) in cells expressing the appropriate receptors; thus, this novel mechanism may be very important in the biological response to glucocorticoids. Finally, given that CDKN1C is up-regulated in response to multiple small molecules (24, 61, 62), including glucocorticoids (35, 60), it is possible that CDKN1C reactivation could have clinical applications even in cancers that have lost Rb function. Future work will examine these ideas.
Supplementary Material
Acknowledgments
We thank numerous colleagues for gifts of essential reagents and materials (see “Experimental Procedures”). We thank Dr. Kenneth Wright for technical advice.
This work was supported, in whole or in part, by National Institutes of Health Grants CA90489 and CA119997 from NCI. This work was also supported by the Florida Department of Health (Grant 08BB-05) and by the Moffitt Research Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CDK
- cyclin-dependent kinase
- pol
- polymerase
- IP
- immunoprecipitation
- ChIP
- chromatin immunoprecipitation
- CTD
- C-terminal domain
- HA
- hemagglutinin
- CMV
- cytomegalovirus
- shRNAi
- short hairpin RNA interference
- Ad
- adenovirus
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- Rb
- retinoblastoma.
REFERENCES
- 1.Ma Y., Cress W. D., Haura E. B. (2003) Mol. Cancer Ther. 2, 73–81 [PubMed] [Google Scholar]
- 2.Shapiro G. I. (2006) J. Clin. Oncol. 24, 1770–1783 [DOI] [PubMed] [Google Scholar]
- 3.Corsino P., Horenstein N., Ostrov D., Rowe T., Law M., Barrett A., Aslanidi G., Cress W. D., Law B. (2009) J. Biol. Chem. 284, 29945–29955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cress W. D., Seto E. (2000) J. Cell. Physiol. 184, 1–16 [DOI] [PubMed] [Google Scholar]
- 5.Johnson D. G., Degregori J. (2006) Curr. Mol. Med. 6, 731–738 [DOI] [PubMed] [Google Scholar]
- 6.Nevins J. R. (2001) Hum. Mol. Genet. 10, 699–703 [DOI] [PubMed] [Google Scholar]
- 7.Sherr C. J., McCormick F. (2002) Cancer Cell 2, 103–112 [DOI] [PubMed] [Google Scholar]
- 8.Ruas M., Peters G. (1998) Biochim. Biophys. Acta 1378, F115–F177 [DOI] [PubMed] [Google Scholar]
- 9.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 11.Polyak K., Lee M. H., Erdjument-Bromage H., Koff A., Roberts J. M., Tempst P., Massagué J. (1994) Cell 78, 59–66 [DOI] [PubMed] [Google Scholar]
- 12.Toyoshima H., Hunter T. (1994) Cell 78, 67–74 [DOI] [PubMed] [Google Scholar]
- 13.Lee M. H., Reynisdóttir I., Massagué J. (1995) Genes Dev. 9, 639–649 [DOI] [PubMed] [Google Scholar]
- 14.Matsuoka S., Edwards M. C., Bai C., Parker S., Zhang P., Baldini A., Harper J. W., Elledge S. J. (1995) Genes Dev. 9, 650–662 [DOI] [PubMed] [Google Scholar]
- 15.Phatnani H. P., Greenleaf A. L. (2006) Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 16.Shiekhattar R., Mermelstein F., Fisher R. P., Drapkin R., Dynlacht B., Wessling H. C., Morgan D. O., Reinberg D. (1995) Nature 374, 283–287 [DOI] [PubMed] [Google Scholar]
- 17.Roy R., Adamczewski J. P., Seroz T., Vermeulen W., Tassan J. P., Schaeffer L., Nigg E. A., Hoeijmakers J. H., Egly J. M. (1994) Cell 79, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 18.Shim E. Y., Walker A. K., Shi Y., Blackwell T. K. (2002) Genes Dev. 16, 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickert P., Seghezzi W., Shanahan F., Cho H., Lees E. (1996) Oncogene 12, 2631–2640 [PubMed] [Google Scholar]
- 20.Matsuoka S., Thompson J. S., Edwards M. C., Bartletta J. M., Grundy P., Kalikin L. M., Harper J. W., Elledge S. J., Feinberg A. P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3026–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatada I., Ohashi H., Fukushima Y., Kaneko Y., Inoue M., Komoto Y., Okada A., Ohishi S., Nabetani A., Morisaki H., Nakayama M., Niikawa N., Mukai T. (1996) Nat. Genet. 14, 171–173 [DOI] [PubMed] [Google Scholar]
- 22.Kaldis P., Russo A. A., Chou H. S., Pavletich N. P., Solomon M. J. (1998) Mol. Biol. Cell 9, 2545–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C., Hou X., Mohapatra S., Ma Y., Cress W. D., Pledger W. J., Chen J. (2005) J. Biol. Chem. 280, 12339–12343 [DOI] [PubMed] [Google Scholar]
- 24.Ma Y., Cress W. D. (2007) Oncogene 26, 3532–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y., Freeman S. N., Cress W. D. (2004) Cancer Biol. Ther. 3, 1262–1269 [DOI] [PubMed] [Google Scholar]
- 26.Johnson D. G., Ohtani K., Nevins J. R. (1994) Genes Dev. 8, 1514–1525 [DOI] [PubMed] [Google Scholar]
- 27.Ma Y., Yuan J., Huang M., Jove R., Cress W. D. (2003) J. Biol. Chem. 278, 16770–16776 [DOI] [PubMed] [Google Scholar]
- 28.Stewart M. C., Kadlcek R. M., Robbins P. D., MacLeod J. N., Ballock R. T. (2004) J. Bone Miner. Res. 19, 123–132 [DOI] [PubMed] [Google Scholar]
- 29.Morris D. P., Lee J. M., Sterner D. E., Brickey W. J., Greenleaf A. L. (1997) Methods 12, 264–275 [DOI] [PubMed] [Google Scholar]
- 30.Cress W. D., Johnson D. G., Nevins J. R. (1993) Mol. Cell. Biol. 13, 6314–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cress W. D., Nevins J. R. (1994) J. Virol. 68, 4213–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cress W. D., Nevins J. R. (1996) Mol. Cell. Biol. 16, 2119–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C., Rauscher F. J., 3rd, Cress W. D., Chen J. (2007) J. Biol. Chem. 282, 29902–29909 [DOI] [PubMed] [Google Scholar]
- 34.Gyory I., Wu J., Fejér G., Seto E., Wright K. L. (2004) Nat. Immunol. 5, 299–308 [DOI] [PubMed] [Google Scholar]
- 35.Samuelsson M. K., Pazirandeh A., Davani B., Okret S. (1999) Mol. Endocrinol. 13, 1811–1822 [DOI] [PubMed] [Google Scholar]
- 36.Vandel L., Kouzarides T. (1999) EMBO J. 18, 4280–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devgan V., Mammucari C., Millar S. E., Brisken C., Dotto G. P. (2005) Genes Dev. 19, 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B., Liu K., Lin F. T., Lin W. C. (2004) J. Biol. Chem. 279, 54140–54152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., Wang H., Li M., Rayburn E. R., Agrawal S., Zhang R. (2005) Oncogene 24, 7238–7247 [DOI] [PubMed] [Google Scholar]
- 40.Martin K., Trouche D., Hagemeier C., Sørensen T. S., La Thangue N. B., Kouzarides T. (1995) Nature 375, 691–694 [DOI] [PubMed] [Google Scholar]
- 41.Martelli F., Hamilton T., Silver D. P., Sharpless N. E., Bardeesy N., Rokas M., DePinho R. A., Livingston D. M., Grossman S. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason S. L., Loughran O., La Thangue N. B. (2002) Oncogene 21, 4220–4230 [DOI] [PubMed] [Google Scholar]
- 43.Eymin B., Karayan L., Séité P., Brambilla C., Brambilla E., Larsen C. J., Gazzéri S. (2001) Oncogene 20, 1033–1041 [DOI] [PubMed] [Google Scholar]
- 44.Misra R. N., Xiao H. Y., Kim K. S., Lu S., Han W. C., Barbosa S. A., Hunt J. T., Rawlins D. B., Shan W., Ahmed S. Z., Qian L., Chen B. C., Zhao R., Bednarz M. S., Kellar K. A., Mulheron J. G., Batorsky R., Roongta U., Kamath A., Marathe P., Ranadive S. A., Sack J. S., Tokarski J. S., Pavletich N. P., Lee F. Y., Webster K. R., Kimball S. D. (2004) J. Med. Chem. 47, 1719–1728 [DOI] [PubMed] [Google Scholar]
- 45.DeGregori J., Johnson D. G. (2006) Curr. Mol. Med. 6, 739–748 [DOI] [PubMed] [Google Scholar]
- 46.Johnson D. G. (2000) Mol. Carcinog. 27, 151–157 [DOI] [PubMed] [Google Scholar]
- 47.Delavaine L., La Thangue N. B. (1999) Oncogene 18, 5381–5392 [DOI] [PubMed] [Google Scholar]
- 48.Afshari C. A., Nichols M. A., Xiong Y., Mudryj M. (1996) Cell Growth Differ. 7, 979–988 [PubMed] [Google Scholar]
- 49.De Falco G., Bagella L., Claudio P. P., De Luca A., Fu Y., Calabretta B., Sala A., Giordano A. (2000) Oncogene 19, 373–379 [DOI] [PubMed] [Google Scholar]
- 50.Simone C., Bagella L., Bellan C., Giordano A. (2002) Oncogene 21, 4158–4165 [DOI] [PubMed] [Google Scholar]
- 51.Amini S., Clavo A., Nadraga Y., Giordano A., Khalili K., Sawaya B. E. (2002) Oncogene 21, 5797–5803 [DOI] [PubMed] [Google Scholar]
- 52.Lee D. K., Duan H. O., Chang C. (2000) J. Biol. Chem. 275, 9308–9313 [DOI] [PubMed] [Google Scholar]
- 53.Kanazawa S., Soucek L., Evan G., Okamoto T., Peterlin B. M. (2003) Oncogene 22, 5707–5711 [DOI] [PubMed] [Google Scholar]
- 54.Schneider E., Montenarh M., Wagner P. (1998) Oncogene 17, 2733–2741 [DOI] [PubMed] [Google Scholar]
- 55.Ko L. J., Shieh S. Y., Chen X., Jayaraman L., Tamai K., Taya Y., Prives C., Pan Z. Q. (1997) Mol. Cell. Biol. 17, 7220–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leung J. Y., Ehmann G. L., Giangrande P. H., Nevins J. R. (2008) Oncogene 27, 4172–4179 [DOI] [PubMed] [Google Scholar]
- 57.Cowling V. H., Cole M. D. (2007) Mol. Cell. Biol. 27, 2059–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eberhardy S. R., Farnham P. J. (2001) J. Biol. Chem. 276, 48562–48571 [DOI] [PubMed] [Google Scholar]
- 59.Chen H. Z., Tsai S. Y., Leone G. (2009) Nat. Rev. Cancer 9, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alheim K., Corness J., Samuelsson M. K., Bladh L. G., Murata T., Nilsson T., Okret S. (2003) J. Mol. Endocrinol. 30, 359–368 [DOI] [PubMed] [Google Scholar]
- 61.Yang X., Karuturi R. K., Sun F., Aau M., Yu K., Shao R., Miller L. D., Tan P. B., Yu Q. (2009) PLoS ONE 4, e5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng H., Wu M., Botnen J. H. (2009) J. Nutr. 139, 1613–1618 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.