Abstract
The budding yeast, Saccharomyces cerevisiae, has three cullin proteins, which act as platforms for Cullin-based E3 ubiquitin ligases. Genetic evidence indicates that Cul8, together with Mms1, Mms22, and Esc4, is involved in the repair of DNA damage that can occur during DNA replication. Cul8 is thought to form a complex with these proteins, but the composition and the function of Cul8-based E3 ubiquitin ligases remain largely uncharacterized. Herein, we report a comprehensive biochemical analysis of Cul8 complexes. Cul8 was found to form a Cul8-Mms1-Mms22-Esc4 complex under physiological conditions, with Mms1 bridging Cul8 and Mms22 and Mms22 bridging Mms1 and Esc4. Domain analysis demonstrated that the N-terminal region of Mms1 and the C-terminal region of Mms22 are required for the Mms1-Mms22 interaction, whereas the N-terminal region of Mms22 is required for the Mms22-Esc4 interaction. We also found other Cul8-Mms1-binding proteins Ctf4, Esc2, and Orc5 using yeast two-hybrid screening. Esc4 and Ctf4 bound to Mms22 directly and bound to Cul8-Mms1 in the presence of Mms22, whereas Esc2 and Orc5 interacted with both Cul8 and Mms1, independently. We found that Cul8, Mms1, and Mms22 participated in the regulation of transcriptional silencing of yeast telomeres. These results suggest that Cul8-Mms1, as part of various protein complexes, is involved in the regulation of chromatin metabolism.
Keywords: Proteases/Ubiqiuitin, Proteases/Ubiquitination, Protein/Protein-Protein Interactions, Protein/Degradation, Protein/Domains, Protein/Targeting, Protein/Turnover
Introduction
The ubiquitin/proteasome-dependent protein degradation system is a highly regulated and specific system for targeted protein degradation and is involved in virtually all of the biological processes in eukaryotes. In this system, the ubiquitin is conjugated to a substrate protein, and subsequent polyubiquitination of the substrate ultimately creates a recognition site for the 26 S proteasome. The specificity of protein degradation is determined by the recognition of specific substrates at specific times.
Protein ubiquitination involves three enzymes: E1,3 E2, and the ubiquitin ligase E3. Only E3 ligases can directly recognize specific substrates, i.e. they determine substrate specificity. There are many different types of E3 ligases. One such E3 is the cullin-based ubiquitin ligase. The Skp1-cullin-F-box protein (SCF) complex is the most studied cullin-based E3 ubiquitin ligase (1, 2). SCF consists of the RING finger protein, Rbx1, cullin, Cul1/Cdc53, the adapter protein, Skp1, and a substrate recognition subunit, which is an F-box protein. Rbx1, Cul1, and Skp1 are constitutive subunits of SCF, but the F-box protein is a variable subunit; multiple F-box proteins are present in cells and respond to different substrates.
Multiple and different cullins are found in different species, e.g. humans have six cullins, namely Cul1 through Cul5 and Cul7. Each cullin has specific adapter proteins and substrate recognition substrates, such as Elongin BC and Cul2-box proteins for Cul2, BTB (Bric-a-brac, Tramtrack, Broad complex) proteins for Cul3, Ddb1 and Cul4-box proteins for Cul4, Elongin BC and SOCS box proteins for Cul5, and Skp1 and F-box proteins for Cul7. The budding yeast, Saccharomyces cerevisiae, has three cullins: Cul1/Cdc53, Cul3, and Cul8/Rtt101. It is well established that Cul1 is part of the SCF complex, which has many functions, including cell cycle regulation. Cul3 may be involved in the degradation of the RNA polymerase subunit, Rpb1 (3). Unlike Cul1 and Cul3, Cul8 is not similar to sequences of human cullins (4). Cul8 is involved in repression of yeast transposable element transition (5). Cells that have been cul8-deleted (cul8Δ) show constitutive checkpoint activation and anaphase delay (4). A double deletion of CUL8 and RAD9—the protein product of the latter mediates the activation of the DNA damage checkpoint—suppresses checkpoint activation and anaphase delay, suggesting that spontaneous DNA damage occurs in cul8Δ cells (4).
Genome-wide genetic interaction mapping has shown that CUL8, MMS1, MMS22, and ESC4/RTT107 form an epistatic group (6, 7). Acetylation of Lys56 of histone H3 is important for damage tolerance (8), and a recent study has shown that Cul8, Mms1, and Mms22 act downstream of the Lys56 acetylation pathway (7). Deletion of CUL8, MMS1, MMS22, or ESC4 sensitizes the cells to DNA-damaging agents (9–12). Cul8, Mms1, and Mms22 have been proposed to promote sister-chromatid exchanges at stalled DNA replication forks (13). Recently, Luke and co-workers (14) showed that Cul8 forms a complex with Mms1 and that either Crt10 or Mms22 interacts with Mms1 to produce a Cul8-Mms1-Crt10 or Cul8-Mms1-Mms22 complex in vivo, suggesting that Mms1 probably acts as the adapter protein of Cul8-type complexes. Furthermore, Esc4 reportedly interacts with Mms22 and Cul8 (12, 15). However, the molecular composition(s) and function(s) of Cul8-based E3 ubiquitin ligases remain largely uncharacterized. Herein, we report that Cul8 forms at least four different protein complexes: Cul8-Mms1-Mms22-Esc4, Cul8-Mms1-Mms22-Ctf4, Cul8-Mms1-Esc2, and Cul8-Mms1-Orc5. We also show that Cul8 is involved in telomeric transcriptional silencing.
EXPERIMENTAL PROCEDURES
Yeast Techniques
Yeast strains used in this study are listed in Table 1 and were constructed using PCR-based homologous recombination. For the spot assays, the cells were grown to an A600 of 1.0 and serially diluted in 10-fold increments. Then 4 μl of each serial dilution was spotted onto the indicated medium. The serially diluted cultures were incubated at 30 °C for 2–4 days. To measure cell viability, the cells were cultured in yeast extract-peptone-dextrose (YPD) to an A600 of 1.0, and then methyl methanesulfonate (MMS) was added to the culture. The cells were cultured for an additional 1.5 h, then diluted with YPD, and plated onto YPD plates. The cells were incubated for 2 days, and the visible colony numbers were counted.
TABLE 1.
Yeast strains
The parental strains were W303-1a: MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 and L40: MATa his3Δ 200 trp1-901 leu2-3112 ade2 LYS2::(4lexAop-HIS3) URA3::(8lexAop-lacZ)GAL.
| Strain name | Background | Genotype | Source |
|---|---|---|---|
| YLD70 | W303-1a | pep4Δ::LEU bar1Δ::TRP1 | Dr. Lucy Drury |
| YSM229 | W303-1a | pep4Δ::LEU bar1Δ::TRP1 ura3::GAL-CUL8-3FLAG(URA3) | This study |
| YSI25 | W303-1a | CUL8-18MYC(TRP) MMS22-3PK(KanMX) MMS1-3HA(LEU) | This study |
| YSI29 | W303-1a | CUL8-18MYC(TRP) MMS22-3PK(KanMX) MMS1-3HA(LEU) ESC4-5FLAG(hph) | This study |
| YSI38 | W303-1a | CUL8-18MYC(TRP) MMS22-3PK(KanMX) mms1Δ::LEU ESC4-5FLAG(hph) | This study |
| YSI34 | W303-1a | CUL8-18MYC(TRP) mms22Δ::KanMX MMS1-3HA(LEU) ESC4-5FLAG(hph) | This study |
| YTY143 | W303-1a | CUL8-18MYC(TRP) MMS22-3PK(KanMX) MMS1-3HA(LEU) | This study |
| +pRS416ADH1p-3FLAGCTF4 | |||
| YTY144 | W303-1a | CUL8-18MYC(TRP) MMS22-3PK(KanMX) MMS1-3HA(LEU) | This study |
| +pRS416ADH1p-3FLAGESC2 | |||
| YTY141 | W303-1a | CUL8-18MYC(TRP) MMS22-3PK(KanMX) MMS1-3HA(LEU) | This study |
| +pRS416ADH1p-3FLAGORC5 | |||
| YTY080 | W303-1a | CUL8-3HA(LEU) | This study |
| YSI8 | W303-1a | MMS1-3HA(LEU) | This study |
| YSI41 | W303-1a | MMS22-3HA(LEU) | This study |
| YTK86 | L40 | cul8Δ::KanMX | This study |
| YTK87 | L40 | mms1Δ::KanMX | This study |
| YTK88 | L40 | mms22Δ::KanMX | This study |
| YTY106 | W303-1a | Mms22-3HA-LEU Ctf4-5FLAG-HIS | This study |
| YSI27 | W303-1a | cul8Δ::KanMX | This study |
| YTY083 | W303-1a | ctf4Δ::URA | This study |
| YTY084 | W303-1a | cul8Δ::KanMX ctf4Δ::URA | This study |
| YTY089 | W303-1a | rad54Δ::TRP | This study |
| YTY091 | W303-1a | cul8Δ::kanMX rad54Δ::TRP | This study |
| YTY055 | W303-1a | mms1Δ::kanMX | This study |
| YTY085 | W303-1a | ctf4Δ::URA mms1Δ::kanMX | This study |
| YSI2 | W303-1a | mms22Δ::kanMX | This study |
| YTY086 | W303-1a | ctf4Δ::URA mms22Δ::kanMX | This study |
| YTY048 | W303-1a | rtt109Δ::kanMX | This study |
| YTY088 | W303-1a | ctf4Δ::URA rtt109Δ::kanMX | This study |
| YTY056 | W303-1a | esc4Δ::kanMX | This study |
| YTY087 | W303-1a | ctf4Δ::URA esc4Δ::kanMX | This study |
| YTY127 | W303-1a | cul8Δ::TRP rtt109Δ::KanMX | This study |
| YSH313 | MATa ade2 lys2 leu2 his3 ura3 GAL 10-LacZ::LYS2 gal3::HIS3 URA3-Tel | Dula and Holmes (35) | |
| YTY121 | YSH313 | cul8Δ::KanMX | This study |
| YTY122 | YSH313 | mms1Δ::KanMX | This study |
| YTY123 | YSH313 | mms22Δ::KanMX | This study |
| YTY125 | YSH313 | esc4Δ::KanMX | This study |
| YTY124 | YSH313 | esc2Δ::KanMX | This study |
| YTY126 | YSH313 | ctf4Δ::KanMX | This study |
Immunopurification and Mass Spectrometry
To purify Cul8-binding proteins, cells were cultured in 1 liter of YP-galactose to an A600 of 1.0. Then the cells were collected and suspended in lysis buffer (40 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.2% (w/v) Triton X-100, protease inhibitors) and lysed with a French press (Thermo). The extracts were clarified by centrifugation; the cleared lysates were incubated with anti-FLAG M2-agarose (Sigma). Bound proteins were eluted by incubating the beads with FLAG peptides. The eluates were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. The gels were cut into five pieces according to molecular weight. The proteins were digested with trypsin and analyzed by liquid chromatography-tandem mass spectrometry, as described (16).
Immunoprecipitation and Immunoblotting
Yeast extracts used for immunoprecipitation were prepared as follows. The cells were harvested from 25 ml of mid-log phase cultures and lysed in 50 mm HEPES-KOH, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 5% (w/v) glycerol, 0.1% (w/v) Nonidet P-40, 0.1% (w/v) Tween 20, and protease inhibitors by bead beating in a multi-bead shocker (MB400U; Yasui Kikai). For Sf21 expression, baculoviruses were prepared with the Bac-to-Bac Baculovirus expression system (Invitrogen), according to the manufacturer's instructions. Sf21 cells were incubated at 28 °C for 3 days after infection. The cells were lysed with the same buffer that was used to lyse the yeast mentioned in this section. The extracts were clarified by centrifugation. Clarified lysates were incubated at 4 °C for 3 h with protein G-Sepharose beads (GE Healthcare), to which 2 μg of antibody was bound. The beads were washed, and bound proteins were solubilized in SDS-PAGE sample buffer. For immunoblotting, the total cell lysates were prepared by precipitation with 10% trichloroacetic acid. The proteins were transferred to a polyvinylidene difluoride membrane and detected with mouse monoclonal antibodies anti-Myc 9E10 (1:1000), anti-FLAG M2 (Sigma) (1:5000), anti-HA 12CA5 (1:1000), anti-Pk (Funakoshi) (1:1000), and anti-HSV (Novagen) (1:1000), or with rabbit polyclonal antibodies anti-Myc (sc-789; Santa Cruz) (1:2500), anti-FLAG (Rockland, 600-461-383) (1:1000), anti-HA (sc-8050; Santa Cruz) (1:2500), and anti-V5 (MBL) (1:1000). Either horseradish peroxidase-conjugated anti-mouse IgG (Sigma; 1:5000) or horseradish peroxidase-conjugated protein A (GE Healthcare; 1:2500) was used as the secondary antibody. The proteins were detected with either SuperSignal West Pico Substrate or SuperSignal West Dura Substrate (Pierce).
Yeast Two-hybrid Screening
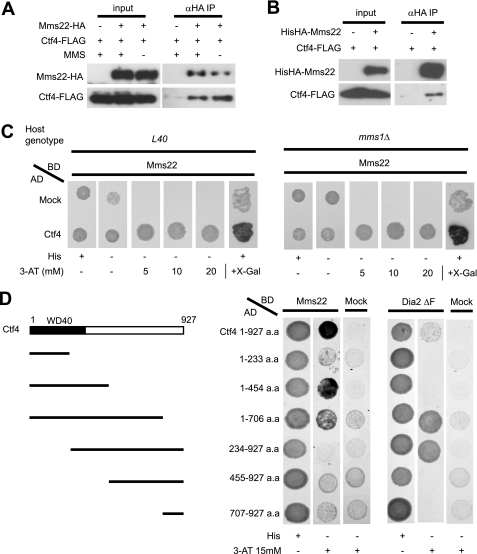
For two-hybrid screening, bait genes were cloned into pLexA, which was derived from pACT2 (Clontech), but its GAL4AD had been replaced by LexABD of pEG202 (Funakoshi). The B42-fused yeast open reading frame prey library was derived from a modified pJG4-5 (Clontech) in which GAL1p was replaced with ADH1p (17, 18). The wild type yeast host cell, L40, was transformed with pLexA-MMS1 or pLexA-MMS22 and the B42-fused yeast open reading frame prey library. Transformants were screened first for the HIS+ phenotype and then for β-galactosidase activity. Using MMS1 as bait, 13 clones of ORC5 and a single clone of ESC2 were obtained. Using MMS22 as bait, seven clones of CTF4 were isolated.
To examine protein-protein interactions, strains harboring bait and prey clones were grown to an A600 of 1.0, and 4 μl of each culture was spotted onto synthetic defined plates containing Ade and Ura. The cultures were incubated at 30 °C for 3–4 days and then photographed.
RESULTS
Cul8 Forms a Cul8-Mms1-Mms22-Esc4 Complex
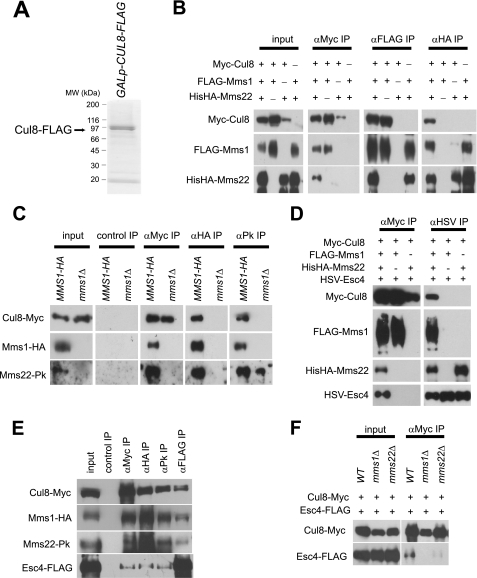
To identify Cul8-binding proteins, we generated yeast cells that could express C-terminal FLAG-tagged Cul8, with its expression under the control of the GAL1 promoter. Proteins bound to Cul8 in vivo were purified by co-immunoprecipitation with anti-FLAG, fractionated by SDS-PAGE, and stained with Coomassie Brilliant Blue (Fig. 1A). Mass spectrometric analysis of the co-precipitated proteins revealed that Mms1 and Mms22 co-immunoprecipitated with Cul8.
FIGURE 1.
Cul8-Mms1-Mms22-Esc4 complex formation. A, yeast cell extracts were prepared from the Cul8–3FLAG-expressing strain. Proteins that bound to Cul8 in vivo were isolated by immunoprecipitation with anti-FLAG, resolved by SDS-PAGE, and then stained with Coomassie Brilliant Blue. B, baculoviruses expressing Myc-Cul8, FLAG-Mms1, and His6-HA-Mms22 were co-transfected into Sf21 cells. The cells were lysed, and the proteins were immunoprecipitated with antibodies to the three tags. Immunoprecipitates were analyzed by immunoblotting. α means anti-. C, yeast strains YSI25 (containing Cul8-Myc, Mms1-HA, and Mms22-Pk) and YSI38 (containing Cul8-Myc, mms1Δ, and Mms22-Pk) were cultured in YPD. The proteins in extracts were immunoprecipitated with the antibodies used for the experiments of Fig. 1B, and immunoprecipitates were analyzed as in B. D, baculoviruses expressing Myc-Cul8, FLAG-Mms1, His6HA-Mms22, and HSV-Esc4 were co-transfected into Sf21 cells. Immunoprecipitation was performed, and the immunoprecipitates were analyzed in the same manner as the experiments of Fig. 1B. E, yeast strain YSI29, for which its endogenous Cul8, Mms1, Mms22, and Esc4 were epitope-tagged as indicated in the figure, was cultured in YPD. Immunoprecipitation was performed, and the immunoprecipitates were analyzed as for the experiments shown in Fig. 1B. F, yeast strains, YSI29 (for which its endogenous Cul8, Mms1, Mms22, and Esc4 were epitope-tagged as indicated in the figure) and YSI38 and YSI34 (which were identical to YSI29 except that MMS1 or MMS22 was deleted, respectively) were cultured in YPD. Immunoprecipitation was performed, and the immunoprecipitates were analyzed in the same manner as the experiments in B. IP, immunoprecipitation; WT, wild type.
To investigate how Cul8 binds to Mms1 and Mms22, Sf21 insect cells were co-infected with various combinations of baculoviruses that contained the genes for Myc-Cul8, HA-Mms1, and His6-FLAG-Mms22. Complexes were immunoprecipitated from cell lysates with antibodies to the epitope (Fig. 1B). Immunoblot analyses of the resulting precipitates revealed that Cul8 interacted with Mms1 and Mms22 when all three proteins were co-expressed, suggesting that they formed a trimeric complex. Additionally, Cul8 bound to Y1 in the absence of Mms22, and Mms1 bound to Mms22 in the absence of Cul8. Conversely, Cul8 did not bind to Mms22 in the absence of Mms1. Next, we examined whether the trimeric complex was formed in yeast cells under physiological conditions. Using a yeast strain in which Cul8, Mms1, and Mms22 were epitope-tagged with Myc, HA, and Pk, respectively, we found that endogenous Cul8 interacted with endogenous Mms1 and Mms22. When MMS1 was deleted, the interaction between Cul8 and Mms22 was not detected (Fig. 1C). Combined with the binding data, the simplest explanation for these observations is that in the proposed trimeric complex, Mms1 bridges Cul8 and Mms22. This finding is consistent with that of Luke and co-workers (14), who showed that an interaction between overexpressed Mms1 and Mms22 occurred subsequent to DNA damage. However, we observed binding between Mms1 and Mms22 even in the absence of DNA damage by MMS, and that MMS was not required for Mms1-Mms22 binding (supplemental Fig. S1). Therefore, we concluded that Mms1-Mms22 binding was independent of prior DNA damage, at least under the conditions we used.
Esc4 has been identified as an Mms22-binding protein by two-hybrid screening (15), and Esc4 also co-immunoprecipitates with Cul8 (12). We co-expressed Myc-Cul8, FLAG-Mms1, His6-HA-Mms22, and HSV-Esc4 in Sf21 insect cells in various combinations and looked for an interaction between Esc4 and the other proteins by co-immunoprecipitation (Fig. 1D). Esc4 interacted with Cul8, Mms1, and Mms22 when all four proteins were co-expressed. In the absence of Mms1, Esc4 bound to Mms22, but not to Cul8; moreover, Esc4 did not bind to Mms1 or Cul8 in the absence of Mms22. Next, we examined whether the Cul8-Mms1-Mms22-Esc4 complex could form in yeast cells (Fig. 1E). In an extract of yeast strain that carried epitope-tagged Cul8-Myc, Mms1-HA, Mms22-Pk, and Esc4-FLAG, we observed that Esc4 interacted with Cul8, Mms1, and Mms22. However, only some of the Esc4 present formed a complex with Cul8-Mms1-Mms22. Therefore, Cul8-Mms1-Mms22 and Esc4 might have partially overlapping functions, which is supported by the recent observation that cul8Δ and esc4Δ have additive effects on MMS sensitivity (14). The interaction between Cul8 and Esc4 was not detected in mms1Δ cells and was greatly reduced in mms22Δ cells (Fig. 1F). Therefore, Esc4 interacted with the Cul8-Mms1-Mms22 complex via Mms22. Because weak interaction between Cul8 and Esc4 was observed in the absence of Mms22, we assume that there is an unidentified Mms22-like protein present in vivo that can also bridge the interaction between Esc4 and Cul8-Mms1.
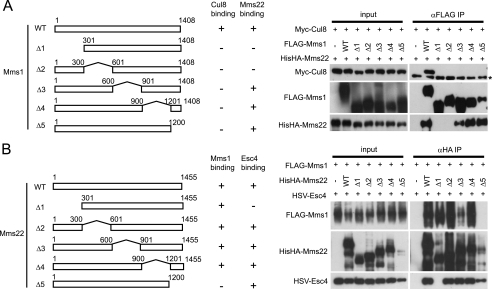
Mms1 and Mms22 Regions Involved in Complex Formation
Next, we attempted to identify regions of Mms1 and Mms22 responsible for their interactions with their binding partners of the Cul8-Mms1-Mms22-Esc4 complex. First, we generated genes for Mms1 deletion mutants that lacked 300 sequential amino acid residues at different locations within the protein (Fig. 2A, left panel) and co-expressed them with Cul8 and Mms22 in Sf21 cells. We examined the ability of the Mms1 deletion mutants to interact with Cul8 and Mms22 by immunoprecipitation and immunoblot analysis (Fig. 2A, right panel). Although full-length Mms1 bound to Cul8, all of the deletion mutants lost the ability to bind to Cul8, suggesting that an intact Mms1 structure was required for interaction with Cul8. Conversely, full-length Mms1 and its deletion mutants Δ601–900, Δ901–1200, and Δ1201–1408 bound Mms22, implying that the N-terminal region of Mms1 (residues 1–600) was required for binding to Mms22. Next, a series of Mms22 deletion mutants was expressed in Sf21 cells (Fig. 2B). Because the Mms22 deletion mutants showed different expression levels, cell extracts (excluding that of Δ1201–1455) that contained similar amounts of the Mms22 mutants were mixed with cell lysates that had expressed Mms1 and Esc4. After incubation at 4 °C for 1 h, the lysates were subjected to immunoprecipitation using anti-HA antibody. Immunoblot analysis of the resulting precipitates revealed that full-length Mms22, Δ1–300, Δ301–600, and Δ901–1200 efficiently bound Mms1. However, the interaction between Mms22 and Mms1 was reduced when residues 601–900 were deleted and not detected when residues 1201–1455 of Mms22 were deleted. Furthermore, Δ1–300 almost completely lost the ability to bind to Esc4. These results suggest that the N- and C-terminal regions of Mms22 were required for interaction with Esc4 and Mms1, respectively. Although Δ1201–1455 was expressed at low levels, it is unlikely that this mutant had a global folding defect because it interacted with Esc4.
FIGURE 2.
Mms1 and Mms22 regions involved in complex formation. A, identification of the Cul8- and Mms22-binding domains in Mms1. Baculoviruses expressing Myc-Cul8, His6HA-Mms22, and full-length or truncated mutants of FLAG-Mms1 were co-transfected into Sf21 cells. The cells were lysed, and the tagged proteins were immunoprecipitated and then analyzed by immunoblotting. The left panel demonstrates schematic diagrams of Mms1 deletion mutants. The asterisk indicates a cross-reacting band. B, identification of the Mms1- and Esc4-binding domains in Mms22. Baculoviruses expressing His6HA-Mms22, HSV-Esc4, and full-length or truncated mutants of FLAG-Mms1 were transfected separately into Sf21 cells. The cells were lysed, and the lysates were mixed to equalize the amounts of the Mms22 proteins in the mixtures. The proteins were then immunoprecipitated from the mixtures and analyzed by immunoblotting. The left panel demonstrates schematic diagrams of Mms22 deletion mutants. IP, immunoprecipitation; WT, wild type.
Identification of a New Complex Containing Cul8 Using Yeast Two-hybrid Screening
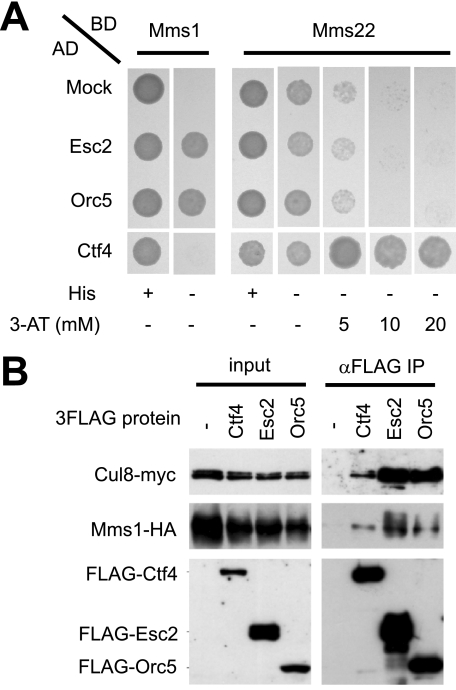
To further investigate the function of the Cul8 complex, we performed yeast two-hybrid screening using either Mms1 or Mms22 as bait. Use of Mms1 as the bait resulted in the isolation of Esc2 (a members of the RENi family of SUMO-like domain proteins) (19–22) and Orc5 (a subunit of the origin recognition complex, or ORC) (23–27). When Mms22 was the bait, we identified Ctf4, a replication fork-associated protein (28–31). We found that Esc2 and Orc5 interacted with Mms1 but not with Mms22 and that Ctf4 interacted with Mms22 but not with Mms1 (Fig. 3A and supplemental Fig. S2A). To investigate how Esc2, Orc5, or Ctf4 interacted with Cul8-Mms1, we expressed FLAG-tagged Esc2, Orc5, or Ctf4 under the control of the ADH1 promoter in yeast that also expressed both Cul8-Myc and Mms1-HA. The cell lysates were subjected to immunoprecipitation with anti-FLAG. Immunoblot analysis of the resulting precipitates showed that all FLAG-tagged proteins interacted with Cul8-Myc and Mms1-HA (Fig. 3B). We did not notice any differences in binding between these proteins in the absence or presence of MMS, suggesting that these interactions were not dependent on DNA damage (data not shown). Because Esc2, Orc5, and Ctf4 were isolated as proteins bound to the Cul8 complex, we tested the possibility that they were substrates for the complex. These proteins were relatively stable, however, and did not accumulate substantially in cul8Δ cells or upon treatment of the cells with the proteasome inhibitor, MG132 (data not shown). Although we cannot rule out the possibility that ubiquitination of these proteins occurred locally and transiently, we presume that they were not substantially degraded by the Cul8 complex.
FIGURE 3.
Identification of Esc2, Orc5, and Ctf4 as components of a Cul8 complex. A, two-hybrid experiments. The yeast strain, L40, was transformed with bait (BD) plasmids and prey (AD) plasmids. Yeast strains were plated onto synthetic defined plates, with or without histidine, or various amount of 3-amino-1,2,4-triazole (3-AT). Because LexA-Mms22, to some extent, self-activated HIS3, at least 10 mm of 3-AT was required to suppress the background level of expression. B, co-immunoprecipitation of the Cul8 complex with Ctf4, Esc2, or Orc5. FLAG-Ctf4, FLAG-Esc2, or FLAG-Orc5 was expressed from a single-copy plasmid under the control of the ADH1 promoter. The FLAG-tagged proteins were immunoprecipitated with anti-FLAG, and their possible co-immunoprecipitation with Cul8 and Mms1 was examined by immunoblotting with anti-HA or anti-Myc, respectively. IP, immunoprecipitation.
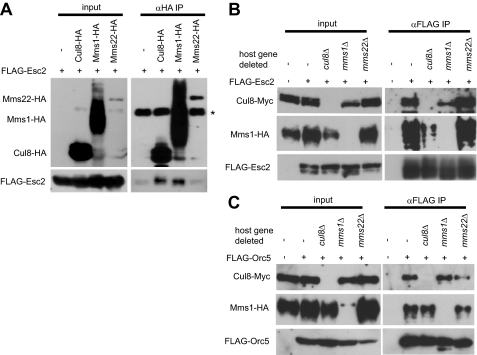
Cul8-Mms1-Esc2 and Cul8-Mms1-Orc5 Complex Formation
To identify which subunits of the Cul8 complex bind to Esc2, we expressed FLAG-Esc2 under the control of the ADH1 promoter in yeast that also expressed Cul8-HA, Mms1-HA, or Mms22-HA. The cell lysates were subjected to immunoprecipitation with anti-HA. Immunoblot analysis of the resulting precipitates showed that both Cul8 and Mms1 interacted with Esc2, whereas Mms22 did not (Fig. 4A). These findings are consistent with the results of our two-hybrid experiments, which revealed an interaction between Mms1 and Esc2 but not between Mms22 and Esc2. The results also imply that although Esc2 can interact with Cul8-Mms1, it cannot do so if Mms22 is bound to Cul8-Mms1. By immunoprecipitation experiments, it was found that Esc2 interacted with Mms1 in cul8Δ extracts and with Cul8 in mms1Δ extracts (Fig. 4B). Furthermore, Orc5 interacted with Mms1 in cul8Δ extracts and with Cul8 in mms1Δ extracts (Fig. 4C). The results of two-hybrid experiments are consistent with those of immunoprecipitation; Esc2 and Orc5 interacted with Cul8 in mms1Δ cells and with Mms1 in cul8Δ cells (supplemental Fig. S2B). Together, these results indicate that both Esc2 and Orc5 bound independently to Cul8 and Mms1.
FIGURE 4.
Interaction of Cul8-Mms1 with Esc2 or Orc5. A, Esc2 immunoprecipitated with Cul8 and Mms1 but not with Mms22. FLAG-Esc2 was expressed from a single-copy plasmid under the control of the ADH1 promoter. Endogenous Cul8-HA, Mms1-HA, or Mms22-HA was immunoprecipitated with anti-HA, and then any FLAG-Esc2 that had co-immunoprecipitated was identified by immunoblotting with anti-FLAG. The asterisk indicates a cross-reacting band. B, co-immunoprecipitation of Cul8-Myc or Mms1-HA with Esc2. FLAG-Esc2 was expressed from a single-copy plasmid under the control of the ADH1 promoter. FLAG-Esc2 was immunoprecipitated with anti-FLAG, and then endogenous Cul8-Myc and Mms1-HA that had co-immunoprecipitated were identified by immunoblotting. C, co-immunoprecipitation of Cul8-Myc or Mms1-HA with Orc5. The experiments were performed as in C, except that Orc5-FLAG was used instead of Esc2-FLAG. IP, immunoprecipitation.
Cul8-Mms1-Mms22-Ctf4 Complex Formation
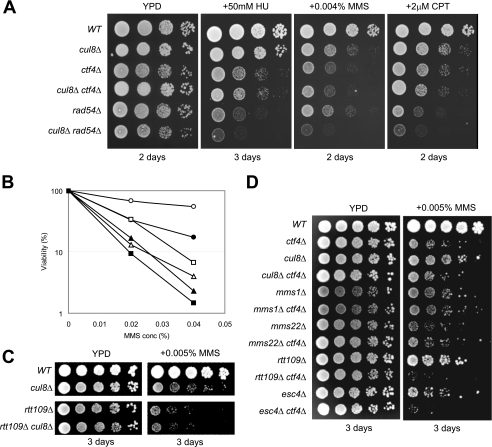
Next, we investigated the interaction between the Cul8 complex and Ctf4. Unlike Esc2 and Orc5, Mms22 clearly interacted with endogenous Ctf4 (Fig. 5A). Furthermore, Mms22 and Ctf4 co-immunoprecipitated when overexpressed in Sf21 cells, suggesting that these proteins directly bound to each other (Fig. 5B). The two-hybrid assay also showed that Mms22 and Ctf4 interacted even in the absence of Mms1 (Fig. 5C). Recent studies have shown that the F-box protein, Dia2, binds to Ctf4 via an N-terminal TPR motif (32, 33). To determine the Ctf4 sequences needed to bind Mms22 and Dia2, we performed two-hybrid experiments using truncated mutants of Ctf4 (Fig. 5D). For Mms22 to interact with Ctf4, the N-terminal 454 residues of the latter, which include the WD40 motif, were found to be necessary. Conversely, Dia2 ΔF did not require all of the first 454 residues of Ctf4 for binding but rather needed residues 234–706. These results suggest that the two proteins recognized different parts of Ctf4. It has been reported that ctf4Δ cells are sensitive to DNA-damaging agents (34). If Ctf4 is part of a Cul8 complex and plays a role in the suppression of drug sensitivity, it might be expected that the presence of Cul8 and Ctf4 has a nonadditive effect on drug sensitivity. The double mutant cul8Δctf4Δ was equally sensitive to hydroxyurea, MMS, and camptothecin, compared with ctf4Δ (Fig. 6, A and B), suggesting that Cul8 and Ctf4 are part of the same pathway. In comparison with cul8Δ, ctf4Δ was much more sensitive to MMS, indicating that Ctf4 has an additional function that does not involve a Cul8 complex. As a control, the ctf4Δrad54Δ double mutant had an additive effect on drug sensitivity, suggesting that Ctf4 and Rad54 function in separate pathways. The cul8Δ, mms1Δ, and mms22Δ mutations had nonadditive effects on ctf4Δ, indicating that they function as a Cul8-Mms1-Mms22-Ctf4 complex (Fig. 6D). The histone acetyltransferase, Rtt109, which is responsible for histone H3 Lys56 acetylation, has been found to act upstream of Cul8 (7). As expected, rtt109Δcul8Δ had a nonadditive effect with respect to MMS sensitivity (Fig. 6C). The rtt109Δ mutant was much more sensitive to MMS than was cul8Δ, indicating that Rtt109 has a function unrelated to that of the Cul8 complex. The rtt109Δctf4Δ double mutant had an additive effect, suggesting that the functions of these proteins partially overlap (Fig. 6D). Recent reports showed that, in terms of MMS sensitivity, esc4Δcul8Δ produced an additive effect (14). As expected, esc4Δctf4Δ had an additive effect on MMS sensitivity (Fig. 6D), suggesting that Ctf4 and Esc4 assemble into the Cul8-Mms1-Mms22 complex separately and that the Cul8-Mms1-Mms22-Ctf4 and Cul8-Mms1-Mms22-Esc4 complexes function differently when suppressing MMS sensitivity.
FIGURE 5.
Interaction between Ctf4 and Mms22. A and B, co-immunoprecipitation of Ctf4 and Mms22. A, endogenous Mms22-HA was immunoprecipitated from yeast extracts, and endogenous Ctf4-FLAG that had co-immunoprecipitated was detected by immunoblotting with anti-FLAG. B, recombinant His6HA-Mms22 and Ctf4-FLAG were expressed in Sf21 insect cells. Co-immunoprecipitation of Ctf4 with Mms22 was assessed. C, two-hybrid interactions between Mms22 and Ctf4. The L40 host strain and mms1Δ were transformed with bait (BD) plasmids and prey (AD) plasmids. Yeast strains were plated onto synthetic defined plates, with or without histidine, and with or without various amounts of 3-amino-1,2,4-triazole (3-AT) or histidine and 5-bromo-4-chloro-3-indolyl β-galactoside (X-Gal). D, identification of the Mms22- and Dia2-binding domain in Ctf4. The host strain L40 was transformed with bait (BD) plasmids and various prey (AD) plasmids containing Ctf4 deletion mutants. (The Dia2 ΔF construct is missing the F-box sequence, which increased the strength of the two-hybrid interaction (32).) The yeast strains were plated onto synthetic defined plates, with or without histidine, and with or without 15 mm of 3-AT. IP, immunoprecipitation.
FIGURE 6.
Interaction between Ctf4 and Cul8. A, yeast strains, with their genotypes indicated to the left of the figure, were grown to an A600 of 1.0 and serially diluted in 10-fold increments, and then 4 μl of the serial dilutions was spotted onto YPD plates, containing 50 mm hydroxyurea, 0.004% MMS, or 2 μm camptothecin. The plates were incubated at 30 °C for the indicated number of days. B, yeast strains were cultured in YPD, then treated with or without MMS for 1.5 h. Open circles, wild type (WT); closed circles, cul8Δ; open triangles, ctf4Δ; closed triangles, cul8Δctf4Δ; open squares, rad54Δ; closed squares, cul8Δrad54Δ. The cells were plated onto YPD plates. Two days later, the colony numbers were counted. C and D, the spot assay was performed in the same manner as the experiments of Fig. 6A.
Involvement of Cul8 Complex in Telomeric Silencing
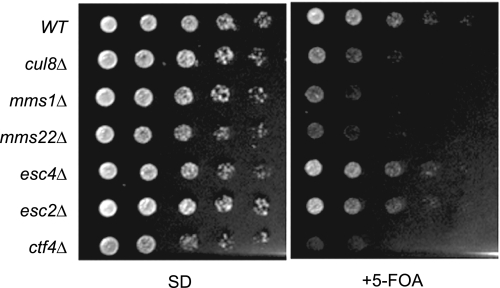
Esc4, Esc2, Orc5, and Ctf4 are involved in gene silencing (19, 22, 24, 25). Therefore, we tested whether Cul8 is also involved in gene silencing. Using a yeast strain in which URA3 was integrated into the chromosome near the telomeric region (35), we assayed for telomeric gene silencing by measuring cell growth on plates containing 5-fluoroorotic acid, which counterselects cells expressing URA3 gene (Fig. 7). The wild type, esc4Δ, and esc2Δ cells grew on 5-fluoroorotic acid plates, whereas cul8Δ, mms1Δ, mms22Δ, or ctf4Δ grew very slowly. These results indicated that the Cul8 complex was involved in telomeric gene silencing.
FIGURE 7.
Involvement of Cul8-Mms1-Mms22-Ctf4 in telomeric gene silencing. All of the strains contained URA3 at the telomeric end of chromosome VII. Yeast strains, with their genotypes indicated to the left of the figure, were grown to an A600 of 1.0 and serially diluted in 10-fold increments, and then 4 μl of the serial dilutions was spotted onto synthetic defined plates (SD) with or without 1 μg/ml of 5-fluoroorotic acid (+5-FOA). WT, wild type.
DISCUSSION
Cul8/Rtt101 is a cullin protein reportedly involved in the regulation of DNA replication subsequent to DNA damage (4, 5, 11). Herein, we show that Cul8 can physically interact with several proteins, many of which have been reported to be involved in DNA replication and gene silencing. Our results indicate that, for Cul8 to be functional, it must be part of a multi-protein complex (supplemental Fig. S3). We also established the order of complex formation using immunoprecipitation to examine the ability or inability of a given protein to interact with Cul8 when the other proteins of the complex were absent in vivo. The possibility that gene deletion indirectly affected the order of complex formation cannot be completely excluded; however, the binding data obtained with recombinant proteins from insect cells favor the simplest explanation that if a component of the complex is required for an interaction between other components, then the first component bridges the interaction between the other components.
Cul8-Mms1-Mms22-Esc4 Complex
By immunoprecipitating either the endogenous yeast proteins or the recombinant proteins of the Sf21 insect system, we found that Cul8, Mms1, Mms22, and Esc4 formed a complex. We also found that in the Cul8-Mms1-Mms22-Esc4 complex, Mms1 bridged Cul8 and Mms22 and Mms22 bridged Mms1 and Esc4. It was recently reported that Mms1 binds Cul8 directly and mediates the binding of either Mms22 or Crt10 (14). One conflict between that study and ours is that in the former, Mms22 bound to Mms1 only in the presence of MMS, whereas we observed that Mms1 and Mms22 interacted even when DNA had not been damaged. Although the reason for this discrepancy is not clear, it may have arisen because we examined the interaction of chromosomally expressed endogenous proteins, whereas the other study examined interactions with overexpressed plasmid-derived Mms22. Further experimentation will be required to understand the regulation of the interaction. Because Esc4 binding to Cul8-Mms1 is dependent on Mms22 and its interaction with Mms22 is independent of Cul8 and Mms1 in Sf21 cells, we concluded that Esc4 bound to Mms22 directly. In yeast extracts, however, we observed that a small amount of Cul8 co-immunoprecipitated with Esc4 in the absence of Mms22. We do not understand the significance of this interaction, but there might be an unidentified Mms22-like protein in vivo that also can bridge Esc4 and Mms1. Our results also show that only a fraction of the Cul8-Mms1-Mms22 complex bound to Esc4. This observation is consistent with a recent report that the effects of cul8Δ and esc4Δ are additive with respect to DNA-damaging agents (14). Presumably, Cul8-Mms1-Mms22 and Esc4 have both overlapping and independent functions. Using domain analysis, we found that the N-terminal region of Mms1 was required for binding to Mms22. The sequence of this region is highly conserved between Mms1 and human Ddb1. Furthermore, the comparable region in Ddb1 has been implicated in binding to human Ddb2 (14). These results suggest that Mms1 and Ddb1 are structurally related. Conversely, we showed that the N-terminal region of Mms22 is required for binding to Mms1. We noticed that the sequences of the Cul8-Mms1-binding proteins, Esc2 and Orc5, are weakly similar in this region. Additionally, mutations in the Esc2 sequence within this region weakened the Cul8-Mms1-Esc2 interaction.4 Therefore, this region might serve as a Cul8-Mms1-binding motif, as does the F-box sequence for Skp1.
Cul8-Mms1-Esc2 and Cul8-Mms1-Orc5 Complex
Using yeast two-hybrid screening, we isolated Esc2, Orc5, and Ctf4 bound to Cul8-Mms1. Esc2 and Orc5 also bound to Cul8-Mms1 in the absence of Mms22. We concluded that Mms22, Esc2, and Orc5 bind to Mms1 independently to form Cul8-Mms1-Mms22, Cul8-Mms1-Esc2, and Cul8-Mms1-Orc5 complexes, respectively. Unlike Mms22, which binds to Mms1 but not to Cul8, Esc2 and Orc5 bind to both Cul8 and Mms1. By analogy with the composition of the SCF complex, we hypothesize that Mms1 acts as an adapter protein for Cul8, as Skp1 does for Cul1. As noted above, Esc2 and Orc5 bind to both Cul8 and Mms1. Their abilities to bind to both Cul8 and Mms1 contrasts with the binding of the F-box proteins, which bind to Skp1 but not to Cul1. It has been reported, however, that both mammalian Cul2 and Cul5 use Elongin BC as adapter proteins but use different substrate recognition proteins (Cul2 box and Cul5 box proteins, respectively) (36). The Cul2 box consists of the BC box, which binds to elongins B and C, and the VHL box. The Cul5 box consists of the BC and SOCS boxes. Thus, these substrate recognition proteins presumably interact with adapter proteins and with cullin to determine the specificity of the complex. Perhaps Esc2 and Orc5 bind to the Cul8-Mms1 complex in a similar manner. To determine the true function of these subunits, we need to identify substrates for the Cul8 complex and determine which subunit(s) they bind. Orc5 is a subunit of the ORC, which recognizes and binds to the replication origin sequence (23). Thus, the interaction between Cul8-Mms1 and Orc5 implies that Cul8 may be involved in regulation of DNA replication origins. We do not know whether Orc5 binds to Cul8-Mms1 as a monomer or as part of an ORC. If it binds as a monomer, then Orc5 might have specific and different functions.
Cul8-Mms1-Mms22-Ctf4 Complex
Unlike Esc2 and Orc5, Ctf4 binds to Mms22, just as Esc4 does. Ctf4 is a relatively stable protein in vivo, and in our studies its stability was unaltered whether Cul8 was present or absent (data not shown). Thus, we assumed that most of the Ctf4 present was not ubiquitinated by the Cul8 complex. Genetic analysis also showed that the effects of cul8Δ and ctf4Δ on drug sensitivity were not additive, suggesting that the Cul8 complex and Ctf4 are part of the same pathway. Given the physical interaction found for the Cul8 complex and Ctf4, we speculated that Ctf4 and Esc4 might act as loading proteins for the Cul8 complex at DNA damage sites. Ctf4 is a component of the replisome, and thus its participation would be an ideal way to load the Cul8 complex at a stalled replication fork. The Cul8 complex would be required to ubiquitinate and degrade those proteins that might otherwise prevent DNA repair or allow replication to resume. In addition to Ctf4, Esc4 might also help load the Cul8 complex onto a DNA damage site independently of Ctf4. Esc4 has six BRCT motifs, which make up a phosphoprotein-binding motif that is common to many proteins involved in DNA repair (37). Both Cul8 and Esc4 have been reported to bind to chromatin, depending on DNA damage, and Esc4 is required for Cul8 chromatin binding (12). Thus, we speculate that Esc4 binds to a damaged DNA site along with many phosphoproteins (e.g. histone H2A) and by doing so recruits Cul8 to the site.
The Involvement of the Cul8 Complex in Telomeric Gene Silencing
Cul8-binding proteins are sometimes involved in gene silencing (19, 22, 24, 25). In our study, we showed that telomeric silencing was diminished by deletion of CUL8, MMS1, MMS22, or CTF4. Our genetic data strongly suggest that the Cul8-Mms1-Mms22-Ctf4 complex is responsible for telomeric silencing. Interestingly, it has been reported that, for fission yeast, pcul4, a possible functional homolog of Cul8, participates in gene silencing (38). Thus, the silencing function of the Cul8 complex would be conserved across species. It was recently reported that regulation of acetylation of the histone H3 Lys56 residue is required for efficient telomeric silencing (39, 40). Ctf4 is also involved in telomeric silencing (25). Cul8 might be loaded onto telomeric chromatin via interaction with Ctf4 and might be involved in telomeric silencing by modulating chromatin structure. It would then assist in silencing ubiquitinating inhibitor proteins, such as histone demethylase. Alternatively, Cul8 might regulate epigenetic transmission of silenced chromatin upon DNA replication.
In summary, we report that Cul8 of the budding yeast, S. cerevisiae, forms a variety of complexes and is involved in DNA damage responses and gene silencing. Crt10, a Cul8-Mms1-binding protein, is known to be involved in the transcription of ribonucleotide reductase genes (14). It may be that Cul8 participates in a variety of biological processes by forming different protein complexes. The substrates of the Cul8 complex are unknown, however, and thus identification of the substrates will increase our understanding of how the Cul8 complex functions.
Supplementary Material
Acknowledgments
We are grateful to Drs. Lucy Drury, John Diffley, Tsutomu Kishi, and Takashi Hishida for reagents and yeast strains. We thank Dr. Kunio Nakatsukasa for critically reading the manuscript.
This work was supported in part by grants from the Ministry of Education, Science, Sports, and Culture of Japan, the Sumitomo Foundation, the Uehara Foundation, and the Saibou Kagaku Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
T. Yamaguchi and T. Kamura, unpublished results.
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin ligase
- SCF
- Skp1-cullin-F-box protein
- YPD
- yeast extract-peptone-dextrose
- MMS
- methyl methanesulfonate
- HA
- hemagglutinin
- Pk
- small epitope (Pk) present on the P and V proteins of the paramyxovirus of simian virus
- HSV
- Herpes simplex virus of glycoprotein D.
REFERENCES
- 1.Willems A. R., Schwab M., Tyers M. (2004) Biochim. Biophys. Acta 1695, 133–170 [DOI] [PubMed] [Google Scholar]
- 2.Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 3.Ribar B., Prakash L., Prakash S. (2007) Mol. Cell. Biol. 27, 3211–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel J. J., McCarville J. F., Xiong Y. (2003) J. Biol. Chem. 278, 22828–22837 [DOI] [PubMed] [Google Scholar]
- 5.Scholes D. T., Banerjee M., Bowen B., Curcio M. J. (2001) Genetics 159, 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., Boeke J. D. (2006) Cell 124, 1069–1081 [DOI] [PubMed] [Google Scholar]
- 7.Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 8.Ozdemir A., Masumoto H., Fitzjohn P., Verreault A., Logie C. (2006) Cell Cycle 5, 2602–2608 [DOI] [PubMed] [Google Scholar]
- 9.Baldwin E. L., Berger A. C., Corbett A. H., Osheroff N. (2005) Nucleic Acids Res. 33, 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dovey C. L., Russell P. (2007) Genetics 177, 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luke B., Versini G., Jaquenoud M., Zaidi I. W., Kurz T., Pintard L., Pasero P., Peter M. (2006) Curr. Biol. 16, 786–792 [DOI] [PubMed] [Google Scholar]
- 12.Roberts T. M., Zaidi I. W., Vaisica J. A., Peter M., Brown G. W. (2008) Mol. Biol. Cell 19, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duro E., Vaisica J. A., Brown G. W., Rouse J. (2008) DNA Repair 7, 811–818 [DOI] [PubMed] [Google Scholar]
- 14.Zaidi I. W., Rabut G., Poveda A., Scheel H., Malmström J., Ulrich H., Hofmann K., Pasero P., Peter M., Luke B. (2008) EMBO Rep. 9, 1034–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin J. K., Bashkirov V. I., Heyer W. D., Romesberg F. E. (2006) DNA Repair 5, 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. (2004) Nat. Cell Biol. 6, 1135–1141 [DOI] [PubMed] [Google Scholar]
- 17.Kishi T., Ikeda A., Koyama N., Fukada J., Nagao R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14497–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishi T., Ikeda A., Nagao R., Koyama N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17418–17423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrulis E. D., Zappulla D. C., Alexieva-Botcheva K., Evangelista C., Sternglanz R. (2004) Genetics 166, 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuperus G., Shore D. (2002) Genetics 162, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novatchkova M., Bachmair A., Eisenhaber B., Eisenhaber F. (2005) BMC Bioinformatics 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohya T., Arai H., Kubota Y., Shinagawa H., Hishida T. (2008) Genetics 180, 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell S. P., Dutta A. (2002) Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 24.Foss M., McNally F. J., Laurenson P., Rine J. (1993) Science 262, 1838–1844 [DOI] [PubMed] [Google Scholar]
- 25.Suter B., Tong A., Chang M., Yu L., Brown G. W., Boone C., Rine J. (2004) Genetics 167, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mankouri H. W., Ngo H. P., Hickson I. D. (2009) Mol. Biol. Cell 20, 1683–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., Branzei D. (2009) Mol. Biol. Cell 20, 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 29.Hanna J. S., Kroll E. S., Lundblad V., Spencer F. A. (2001) Mol. Cell. Biol. 21, 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miles J., Formosa T. (1992) Mol. Cell. Biol. 12, 5724–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K. P., Shirahige K., Uhlmann F. (2006) Mol. Cell 23, 787–799 [DOI] [PubMed] [Google Scholar]
- 32.Mimura S., Komata M., Kishi T., Shirahige K., Kamura T. (2009) EMBO J. 28, 3693–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morohashi H., Maculins T., Labib K. (2009) Curr. Biol. 19, 1943–1949 [DOI] [PubMed] [Google Scholar]
- 34.Ogiwara H., Ui A., Lai M. S., Enomoto T., Seki M. (2007) Biochem. Biophys. Res. Commun. 354, 222–226 [DOI] [PubMed] [Google Scholar]
- 35.Dula M. L., Holmes S. G. (2000) Genetics 156, 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I. (2004) Genes Dev. 18, 3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover J. N., Williams R. S., Lee M. S. (2004) Trends Biochem. Sci 29, 579–585 [DOI] [PubMed] [Google Scholar]
- 38.Jia S., Kobayashi R., Grewal S. I. (2005) Nat. Cell Biol. 7, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 39.Xu F., Zhang Q., Zhang K., Xie W., Grunstein M. (2007) Mol. Cell 27, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang B., Miller A., Kirchmaier A. L. (2008) Mol. Biol. Cell 19, 4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.