Abstract
β-Propeller proteins function in catalysis, protein-protein interaction, cell cycle regulation, and innate immunity. The galactose-binding protein (GBP) from the plasma of the horseshoe crab, Carcinoscorpius rotundicauda, is a β-propeller protein that functions in antimicrobial defense. Studies have shown that upon binding to Gram-negative bacterial lipopolysaccharide (LPS), GBP interacts with C-reactive protein (CRP) to form a pathogen-recognition complex, which helps to eliminate invading microbes. However, the molecular basis of interactions between GBP and LPS and how it interplays with CRP remain largely unknown. By homology modeling, we showed that GBP contains six β-propeller/Tectonin domains. Ligand docking indicated that Tectonin domains 6 to 1 likely contain the LPS binding sites. Protein-protein interaction studies demonstrated that Tectonin domain 4 interacts most strongly with CRP. Hydrogen-deuterium exchange mass spectrometry mapped distinct sites of GBP that interact with LPS and with CRP, consistent with in silico predictions. Furthermore, infection condition (lowered Ca2+ level) increases GBP-CRP affinity by 1000-fold. Resupplementing the system with a physiological level of Ca2+ did not reverse the protein-protein affinity to the basal state, suggesting that the infection-induced complex had undergone irreversible conformational change. We propose that GBP serves as a bridging molecule, participating in molecular interactions, GBP-LPS and GBP-CRP, to form a stable pathogen-recognition complex. The interaction interfaces in these two partners suggest that Tectonin domains can differentiate self/nonself, crucial to frontline defense against infection. In addition, GBP shares architectural and functional homologies to a human protein, hTectonin, suggesting its evolutionarily conservation for ∼500 million years, from horseshoe crab to human.
Keywords: Carbohydrate/Lectin, Immunology/Innate Immunity, Immunology/LPS, Methods/Computer Modeling, Methods/Mass Spectrometry, Methods/Surface Plasmon Resonance, Protein/Ligand Binding, Protein/Protein-Protein Interactions
Introduction
The β-propeller protein family members have diverse functions: enzyme catalysis, protein-protein interactions, and cell cycle regulation (1, 2). A subset of this family of proteins has pathogen-binding properties (1, 3–8), indicating a role in defense against microbial infection. Pathogen binding occurs through the recognition of evolutionarily conserved structures on pathogens, referred to as pathogen-associated molecular patterns (PAMPs),5 e.g. lipopolysaccharide (LPS) of Gram-negative bacteria and lipoteichoic acid (LTA) of Gram-positive bacteria. Within the subset of pathogen binding β-propeller protein family, several members are classified as having Tectonin domains (4, 5, 7–9). The Tectonin domains were first found in the Tectonins I and II proteins of the slime mold, Physarum polycephalum. The Tectonins I and II are expressed on the cell surface and are involved in the formation of a signaling complex during phagocytosis (5). Because Physarum feeds on bacteria, it has been suggested that the Tectonin domains recognize LPS in the substratum of Gram-negative bacteria (10). However, whether the Tectonin domains can directly bind to PAMPs such as LPS has not been demonstrated experimentally.
The galactose-binding protein (GBP) of the horseshoe crab, Carcinoscorpius rotundicauda, is a plasma lectin that contains Tectonin domains. It was proposed to bind PAMPs while interacting with other pattern-recognition receptors (PRRs) to form a pathogen-recognition interactome (11, 12). The C-reactive protein (CRP), an acute phase protein whose level increases rapidly and dramatically upon acute phase infection-inflammation, interacts with GBP (13). Previously, we found that interaction between GBP and CRP is induced by infection (12), likely through infection-activated serine proteases, Factor C and C2/Bf, which catalyze the assembly of the PRR-interactome (14). Because of its relative abundance in the plasma and its propensity to form an PRR-interactome, GBP is a useful model for studying the role of Tectonin domain-containing proteins in antimicrobial defense. We hypothesized that GBP plays a critical bridging role in the PRR-interactome formation. However, the molecular basis of the interactions between GBP and PAMP, and GBP and CRP is still unknown. Furthermore, it is not yet fully understood how microbial infection induces interaction between GBP and CRP (15).
Here, we examined the molecular interfaces between GBP and LPS, and GBP and CRP under normal and infection conditions. We demonstrated that of the six β-propellers or Tectonin domains of GBP, domains 6 to 1, interact with LPS, and domain 4 interacts strongly with CRP. GBP isolated from infected animals binds both LPS and CRP with dramatically increased affinities. In addition, we showed that hTectonin (15), a human tectonin domain-containing protein, shares structural and functional homology to GBP. This warrants further analysis of the structure-function of β-propeller Tectonin domains in infection and immune response. Altogether, our results define the molecular basis for GBP-LPS and GBP-CRP interactions, support a fundamental role of these interactions in boosting immune defense, and demonstrate the conservation and importance of Tectonin domain-containing proteins in innate immune response throughout evolution.
MATERIALS AND METHODS
Organisms
Horseshoe crabs were collected from the Kranji estuary, Singapore. The animals were infected intracardially with 1.2 × 106 colony-forming units of Pseudomonas aeruginosa/100 g of body weight. Before and 6 h after infection, the animals were partially bled, and cell-free plasma was obtained by centrifugation at 150 × g for 15 min at 4 °C (12). The animals were handled according to the guidelines of the National Advisory Committee for Laboratory Animal Research, Singapore.
Purification of GBP
The cell-free plasma was incubated overnight at 4 °C with Sepharose CL-6B (Pharmacia) preequilibrated with 10 mm Tris, 150 mm NaCl (TBS), pH 8.8, and washed with >10 column volumes until a steady base line was obtained. GBP was eluted with TBS, pH 7.4, containing 0.4 m GlcNAc (Sigma). GlcNAc was removed from the eluted protein by ultrafiltration through 3-kDa molecular weight cutoff micropore filters (Amicon). Purified GBP from the plasma of naïve and infected animals is referred as GBPn and GBPi, respectively.
Yeast Two-hybrid Assay
Co-transformations of the different bait and prey plasmids into Saccharomyces cerevisiae were performed to study protein-protein interactions. For details, see supplemental Materials and Methods.
ELISA to Test for Bacterial Ligand Binding
The GBP ligands were immobilized on ELISA plates. Their interactions with GBP were quantified (supplemental Materials and Methods).
Surface Plasmon Resonance Analysis (SPR)
Real-time biointeractions between GB and ligands (GlcNAc, LPS, ReLPS, and lipid A from Salmonella minnesota) and GBP and CRP were performed using a Biacore 2000. The purified GBP solution contained hetero-oligomers, albeit with reasonable representation of monomeric GBP. Although earlier studies (4) with hetero-oligomeric solutions of proteins from another species of horseshoe crab have utilized a Langmuir 1:1 binding equation as a standard for protein-ligand binding affinity calculations, here, we have analyzed the binding affinities for both native GBP and dithiothreitol-treated GBP (which gave more monomeric forms) and tested the SPR data by both the Langmuir 1:1 binding as well as the two-state conformational change binding and compared the binding affinity values for both fits. Details on SPR are in the supplemental Materials and Methods.
Amide Hydrogen Exchange Mass Spectrometry and Data Analysis (HDMS)
To determine the interaction interface for protein-protein and protein-ligand interaction, HDMS was performed. For details, see supplemental Materials and Methods.
Protein Homology Modeling and Docking
GBP was homology-modeled using the crystal data of tachylectin-1 (TL-1),6 (16), which shares 66.7% sequence identity with GBP (17). The three-dimensional model of the horseshoe crab CRP was prepared by homology modeling from the crystal structure of human CRP (18) and human serum amyloid protein, which share 30 and 31% sequence similarity, respectively (supplemental Fig. 1). Details on molecular modeling and docking are in the supplemental Materials and Methods.
Protein-Protein Docking
The HADDOCK 2.0 program (19, 20) was used to generate the three-dimensional models of GBP-CRP heterodimer by protein-protein docking. Peptide sequences from the HDMS analysis involved in protein dimerization with >30% solvent-accessible surface area/residue (NACCESS program) (21) were defined as active residues in the guided docking procedure, whereas amino acids within 3 Å interatomic distance from them were considered passive. Generation, refinement, and scoring of the random GBP-CRP dimer models were performed similarly. For details, see supplemental Materials and Methods.
The three-dimensional model of the GBP-CRP binding described here enables structural interpretation of the observed HDMS data on the peptide sequences involved in the protein-protein interactions. The ligand-GBP models were used for structural rationalization of the SPR data on the binding affinities of LPS components to the GBP and comparison with other sugar-binding proteins containing Tectonin domains. The observed data were compared with structural information and cross-checked by comparison with binding affinity data predicted from force field-based molecular mechanics calculations. The three-dimensional models are intended to represent working models that are sufficient for interpretation and rationalization of the experimental data.
RESULTS
Native GBP Has a Propensity to Oligomerize
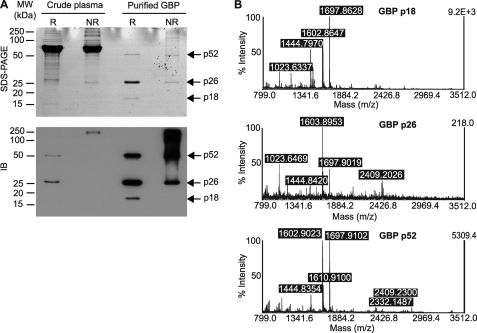
GBP was purified from the plasma, through binding to the repeating units of α-1,6-linked d-galactose and 3,6-anhydro-l-galactose on the Sepharose CL-6B (supplemental Fig. 2). On reducing SDS-PAGE, GBP showed three bands: 52 kDa (nonreducible dimer), 26 kDa (monomer), and 18 kDa (N-terminal domain) (Fig. 1A, lanes R), which were confirmed by mass spectrometry (Fig. 1B). Under nonreducing conditions, native GBP in the crude plasma was predominantly in larger oligomeric forms, although purified GBP showed substantial representation of dimeric and monomeric forms (Fig. 1A, lower panel, lanes NR).
FIGURE 1.
GBP tends to exist in oligomeric forms. A, crude plasma and purified GBP were separated by SDS-PAGE with or without reducing agent. Immunoblotting (IB) was performed with anti-GBP antibody. R, reducing condition; NR, nonreducing condition. B, matrix-assisted laser desorption ionization time-of-flight spectra identified the purified 52, 26, and 18 kDa protein bands as the dimer, monomer, and N-terminal fragment of GBP, respectively.
GBP Binds to the Sugar Moieties of Gram-negative Bacterial LPS
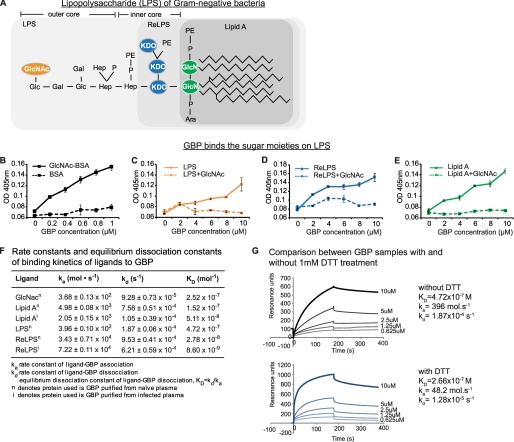
Because GBP binds to the Gal residues of Sepharose and was eluted by GlcNAc, we envisaged that GBP binds to the sugar moieties of bacterial LPS (Fig. 2A). ELISA showed that purified GBP bound GlcNAc specifically (Fig. 2B). GBP binds to immobilized LPS, but this binding was inhibited when GBP was incubated with GlcNAc prior to being added to LPS (Fig. 2C). The same was also observed with ReLPS and lipid A (Fig. 2, D and E). Likewise, the binding of GBP to Gram-positive bacterial LTA was abrogated by GlcNAc (supplemental Fig. 3). Thus, we can assume that GBP binds to LPS or LTA through their sugar moieties. To confirm this observation, we evaluated the avidity of GBP for LPS, lipid A, and GlcNAc by SPR measurements using purified GBP. Because the purified GBP more likely contains monomeric as well as oligomeric forms of the protein, we used the term “apparent KD” to refer to the potential avidity of GBP to its ligands. We found that GBP-GlcNAc, GBP-lipid A, and GBP-LPS showed similar apparent KD values of 1.52–2.52 × 10−7 m (Fig. 2F), corroborating the notion that GBP binds LPS via its GlcNAc moiety because all of these PAMPs contain GlcNAc. A SPR binding experiment for GBP-LPS under reducing conditions (1 mm dithiothreitol) also showed a similar apparent KD of 2.66 × 10−7 m, albeit with about 1 order of magnitude higher values of rate constants of the analyte-ligand association and dissociation (Fig. 2G). This indicates the presence of increased numbers of smaller and faster diffusing monomeric forms of GBP in the mass transport-limited processes on the chip surface and suggests that monomeric GBP recognizes and binds to bacterial LPS rather than its oligomeric forms.
FIGURE 2.
GBP binds sugar moieties of LPS with high affinity. A, chemical structure of LPS. GlcNAc is located on the outer core of LPS. Hep, heptose; P, phosphate; PE, phosphoethanolamine; KDO, 3-deoxy-α-d-manno-octulosonic acid; Ara, arabinose. The outermost sugar residue of each LPS truncate is colored. B–E, ELISA to measure GBP-ligand binding. The GBP ligands (LPS, ReLPS, lipid A, LTA, or GlcNAc-bovine serum albumin (BSA)) were incubated overnight in binding buffer (see supplemental Materials and Methods) on 96-well PolysorpTM microplates. The unbound sites were blocked with 1% bovine serum albumin, and serially diluted GBP (with or without preincubation with GlcNAc) was added to each well. Anti-GBP antibody was added followed by horseradish peroxidase-linked secondary antibody. The peroxidase enzyme activity was determined at 405 nm. F, SPR-derived binding constants of GBP to LPS, LPS-truncates, or GlcNAc. The apparent KD values were calculated by using BIAevaluation software version 4.1. Suffix n and i refer to naïve (uninfected) and infected experimental conditions, respectively. G, SPR analysis of GBP binding with LPS, with and without dithiothreitol (DTT) treatment.
GBP Is a Six-bladed β-Propeller Protein
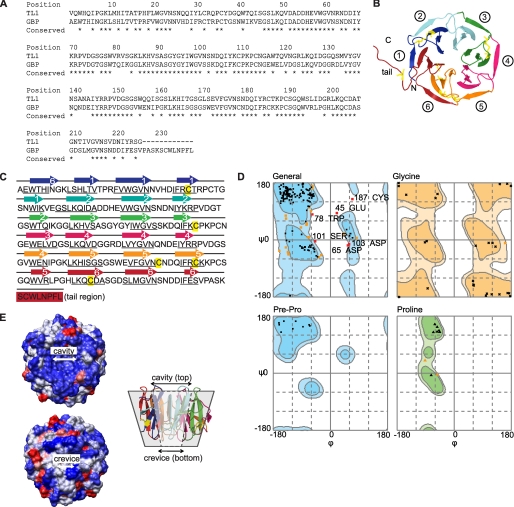
By homology modeling to TL-1, we predicted GBP to be a six-bladed β-propeller protein containing six Tectonin domains (Fig. 3, A and B). Each of the Tectonin domains is made up of four β-sheets (Fig. 3C, arrows), which is in agreement with the secondary structure predictions and the Tectonin domain classification scheme. The Ramachandran plot of the GBP structure shows only minimal number of ϕ and ψ outliers (Fig. 3D and Table 1). The surface of GBP is predominantly hydrophilic, with several scattered hydrophobic patches (Fig. 3E). The GBP forms a hexagonal torus with a larger “cavity” on the top of the central tunnel and a smaller “crevice” at the bottom.
FIGURE 3.
Three-dimensional model of GBP. A, sequence alignment of GBP to TL-1 is shown. B, homology model of GBP predicted a 6-bladed β-propeller protein, containing 8 cysteine residues (yellow). C, protein sequence of GBP shows six Tectonin domain repeats with an 8-residue tail. The modeled β-sheets are numbered accordingly. β-Strands (underlined) are predicted by PSIPRED. D, Ramachandran plot shows that the outlier residues remain close to the boundaries of the permitted ψ-ϕ values (light blue contours), indicating a reliably modeled structure. E, GBP is predominantly hydrophilic (blue) with scattered hydrophobic patches (red). The molecule folds into a toroidal structure around a funnel-shaped tunnel with a larger cavity on the top and a smaller crevice at the bottom.
TABLE 1.
List of φ-ψ outliers in the GBP model
| No. | Residue | ψ | φ | Score |
|---|---|---|---|---|
| 1 | Glu-45 | 84.6 | 20.2 | 0 |
| 2 | Asp-65 | −17.3 | 54.9 | 0.0003 |
| 3 | Trp-78 | 80.4 | −56.8 | 0.0001 |
| 4 | Ser-101 | −4.4 | −47.9 | 0.0003 |
| 5 | Asp103 | −13.2 | 58.8 | 0.0003 |
| 6 | Asp-131 | −34.5 | −161.1 | 0.0005 |
| 7 | Cys-187 | 128.5 | 69.3 | 0.0003 |
Number of residues in favored region (∼98.0% expected): 190 (86.8%).
Number of residues in allowed region (∼2.0% expected): 23 (10.5%).
Number of residues in outlier region: 6 (2.7%).
The TL-1 template contains 99.1% residues in favoured region and only 1 residue in the outlier region (Cys-187).
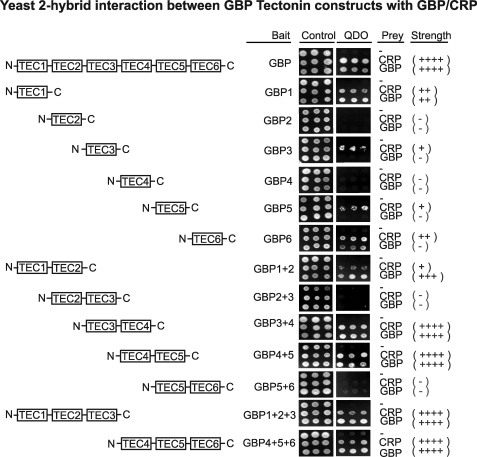
Specific Tectonin Domains of GBP Bind CRP Preferentially
Based on the homology-modeled GBP structure, we subcloned the six Tectonin domains individually, in duos (domains 1–2, 2–3, 3–4, 4–5, 5–6), and in trios (domains 1–2–3, 4–5–6). Yeast two-hybrid analysis showed that each domain appears to interact differentially with GBP or CRP (Fig. 4). The GBP Tectonin domains 3–4 and 4–5 may interact more efficiently with CRP as suggested by the comparatively faster growth of the co-transformed yeast cells on the quadruple dropout plate.
FIGURE 4.
Yeast two-hybrid analysis shows that specific Tectonin domains of GBP interact with CRP. Single, double, and triple GBP Tectonin domain subclones were tested for their interaction with CRP and with full-length GBP. The pGBKT7 vector containing GBP Tectonins domains/full-length GBP and pGADT7 vector containing CRP/full-length GBP were co-transformed and spotted on SD-Leu-Trp plates (double-dropout control) and SD-His-Ade-Leu-Trp (quadruple-dropout). The strength of interaction is indicated as +/−.
Saccharides and Lipid A Dock to Similar Sites in GBP
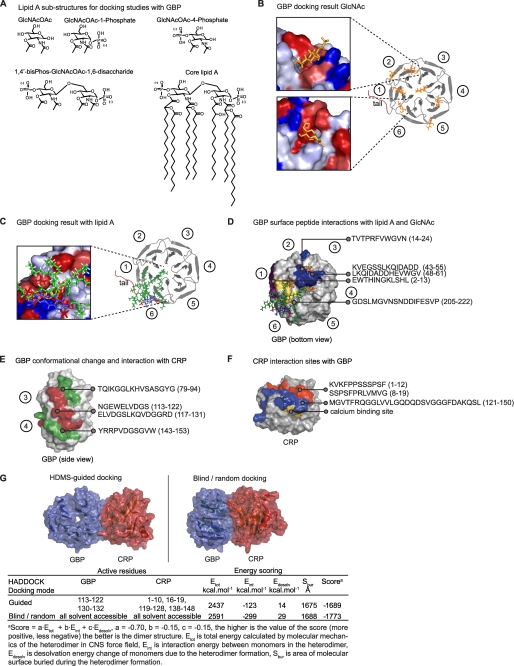
To define the binding sites on GBP that interact with LPS and saccharides, we utilized the three-dimensional model structure of GBP for docking studies. Proteins containing β-propeller repeats such as TL-2 are known to undergo protein-sugar interactions via the backbone atoms of the conserved binding site residues, which are flanked by adjacent β-sheet blades of the Tectonin domains (3, 24, 25). Because GBP has high affinity for GlcNAc and other sugar moieties of the LPS (see Fig. 2), it is reasonable to expect that the sugar binding sites are also localized between the adjacent β-sheet blades.
Computational docking predicted that from among a set of monosaccharides and monosaccharide N-acetylamines (Gal, GalNAc, Gln, GlcNAc, KDO, heptose), Gal binds GBP with the highest affinity (Table 2). The highest affinity binding site for GlcNAc (Fig. 5A) is located between the Tectonin domains 1 and 6 (Fig. 5B). Additional possible binding sites were predicted in clefts between other adjacent β-propellers (Fig. 5B) resembling those in TL-2 (3), as well as in the central cavity similar to the predicted binding site in TL-1 (26).
TABLE 2.
Computed binding energies for top scoring saccharides and lipid A poses docked to GBP
| Ligand | Einta |
|---|---|
| kcal·mol−1 | |
| Galactose (Gal) | −58.8 |
| Glucose (Glu) | −51.3 |
| Glucosamine (Gln) | −45.2 |
| N-Acetylglucosamine (GlcNAc) | −35.0 |
| 3-Deoxy-α-d-manno-octulosonic acid (KDO) | −52.5 |
| 2-N-Acetyl-3-O-acetylglucosamine (GlcNAcOAc) | −66.1 |
| GlcNAcOAc-1-phosphate | −65.7 |
| GlcNAcOAc-4-phosphate | −63.3 |
| 1,4′-Bisphosphate-GlcNAcOAc-1,6-disaccharide | −140.0 |
| Core lipid A | −126.0 |
a Eint is the sum of electrostatic and Van der Waals ligand-receptor binding energy contributions as defined in the AMBER99 force field (41).
FIGURE 5.
Docking and identification of GBP surfaces that bind GlcNAc, lipid A, and/or CRP. A, GlcNAc and lipid A structures used for docking to GBP are shown. B, GlcNAc (orange) was docked to GBP, and binding energies were quantified. Circled numbers correspond to the Tectonin domains. Inset, GlcNAc were docked to the clefts (hydrophobic, red; hydrophilic, blue) between the propeller blades. C, lipid A (fatty acid chains, green; glucosamine, blue; phosphates, red) was docked to GBP. The lipid A molecule overlapped one of the GlcNAc binding sites. D–F, HDMS experiments are shown. D, GBP interaction sites with GlcNAc (blue)/lipid A (yellow). Peptides showing change in deuterium uptake were mapped onto the surface of GBP. GBPi showed an additional peptide (2–13) binding to lipid A (purple). Top docking results (GlcNAc, orange; lipid A, green-blue-red) are included for comparison. E, GBP interaction sites with CRP. Peptides involved in deuterium uptake (red, decrease; green, increase). F, CRP peptides that bind GBP. Surfaces in blue and red both showed decreased deuterium incorporation. The calcium (yellow) binding site on CRP is in close proximity and overlapping with the colored surfaces.
2-N-Acetyl-3-O-acetyl-β-d-glucosamine (GlcNAcOAc), the principal component of the disaccharide head group of lipid A, was predicted to show an enhanced affinity (Eint) toward GBP (Fig. 5B and Table 2). Phosphate groups at position 1 or 4 of the GlcNAcOAc did not significantly affect the binding affinity with GBP. However, significant affinity toward GBP was predicted for the polar disaccharide head group (1,4′-bisphospho-β-(1,6)-2,2′-N-acetyl-3,3′-O-acetyl-d-glucosamine disaccharide) of the lipid A (Fig. 5A), which was higher than the sum of the Eint of its components (GlcNAcOAc-1-Phos, GlcNAcOAc-4-Phos). The core lipid A (head group together with its fatty acid chains) showed binding affinity similar to that of the head group itself. Molecular docking predicted that the lipid A head group binding site coincides with the GlcNAc binding site (Fig. 5C, zoomed view; GlcNAc is overlapped by the head group of the larger lipid A molecule).
GBP Interacts with LPS and CRP via Distinct Interaction Surfaces
Because each of the Tectonin domains folds into structurally similar β-propellers, we asked how these domains differentiate self from nonself molecules. We used computational docking to predict, and HDMS (27, 28) to validate and map, the lipid A and CRP binding sites on the Tectonin domains (Fig. 5, supplemental Fig. 4, and supplemental Tables 1 and 2). HDMS of the GBPn-lipid A complex showed GBP peptide 205–222 with decreased deuterium uptake, suggesting a lipid A binding site within domains 6 to 1 (Fig. 5D, yellow surface), in agreement with docking predictions (Fig. 5C). Interestingly, GBPi showed an additional lipid A-binding peptide (24, 25, 29–36), supporting our postulate of an infection-related conformational change. Furthermore, this peptide contains an LPS-binding motif, HINGK, resembling the pattern of lipid A-binding residues, BHB(P)HB (B, basic; H, hydrophobic; P, polar) (25).
In the presence of CRP, the GBP peptides (113–122 and 117–131) corresponding to the outermost β-strand of the fourth Tectonin domain showed decreased deuterium exchange (supplemental Fig. 4), suggesting that it contains the GBP-CRP interaction site (Fig. 5E, red surface). The adjacent peptides, 79–94 and 143–153 (Fig. 5E, green surfaces), showed increased deuterium exchange, indicating greater solvent accessibility. This is likely due to induced conformational changes upon interaction (red surface). Conversely, in the CRP-GBP interaction, three peptides forming a continuous surface patch in the CRP molecule (1–12, 8–19, red, and 121–150, blue; Fig. 5F) showed decreased deuterium exchange (supplemental Fig. 4), indicating interaction sites with GBP. Peptide 121–150 harbors residues (Asp136, Gln137, Asp138, Gln148; Fig. 5F, yellow surface), known to be crucial for calcium binding, thus explaining why CRP binds GBP only at low Ca2+ levels (12, 14). Incidentally, hypocalcaemia prevails in infection-inflammation (12, 34).
Following from HDMS-identified GBP-CRP interaction surfaces, a guided docking run showed a nonsymmetric heterodimer model with a higher score (higher stability) than that generated by random docking (supplemental Fig. 5). This confirms the preference of specific Tectonin domains participating in the GBP-CRP interaction. Taken together, the interaction domains indicated by yeast two-hybrid assay and the HDMS-identified interaction interfaces between CRP and GBP are in general agreement with the in silico docking. This lends credence to the modeled structures of these two proteins. Nevertheless, further experimental evidence from x-ray crystallographic structures of the CRP and GBP, individually as well as co-crystals, would be needed to support the proposed model structures.
Infection Conditions Increase the Affinity of GBP-LPS and GBP-CRP
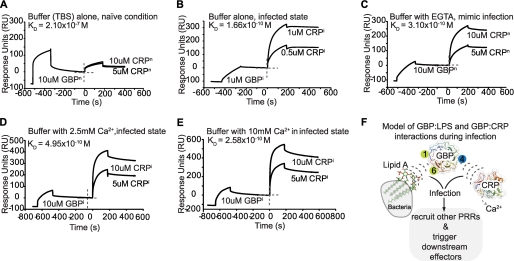
GBP interacts with CRP only during infection, suggesting that certain infection conditions prime them for interaction (12). We found that infection resulted in increased affinity of GBPi for ReLPS (ReLPSi, apparent KD of 8.60 × 10−9 m) and lipid A (lipid Ai, apparent KD of 5.11 × 10−8 m) (Fig. 2F). Next, we characterized the affinities between GBPn and CRPn, and GBPi and CRPi. The purified CRPn or CRPi was injected separately over the GBPn or GBPi that were preimmobilized on the lipid A surfaces of the Biacore chip. Fig. 6, A and B, show that GBPn-CRPn interacted with an apparent KD of 2.10 × 10−7 m, whereas GBPi-CRPi interacted with an apparent KD of 1.66 × 10−10 m, indicating that infection caused a 1000-fold increase in affinity between GBP and CRP. Such a dramatic increase in affinity probably resulted from protein conformational changes that take place during a microbial infection.
FIGURE 6.
Infection increases the affinity of LPS and CRP to GBP. A–C, SPR analysis of GBP, which was first bound to immobilized lipid A, followed by CRP. GBPn-CRPn showed apparent KD of 2.10 × 10−7 m, whereas GBPi-CRPi showed 1000-fold increased affinity (apparent KD of 1.66 × 10−10 m). Depletion of calcium resulted in a 1000-fold increase in affinity (apparent KD of 3.10 × 10−10 m) of GBPn-CRPn, similar to that of GBPi-CRPi. D and E, supplementing with 2.5 and 10 mm Ca2+ did not return the binding affinity of GBPi-CRPi to the basal state. F, proposed model of interaction and formation of the core pathogen-recognition complex. The GBP Tectonin domains 1 and 6 (green circles) bind lipid A of LPS, which are displayed on the Gram-negative bacterium (gray), whereas Tectonin domain 4 (blue circle) interacts with CRP, as determined by SPR, yeast two-hybrid and HDMS experiments. The pathogen-recognition interactome recruits other PRRs such as carcinolectins, CL5 (12), to further stabilize and form the antimicrobial complex to drive downstream effectors and complement activation pathways.
It has been documented that in an acute phase infection, the bacterial invaders usurp calcium ions (12, 34–37) from the host. To investigate whether Ca2+ plays a role in the GBP-CRP interaction, we measured the affinity between GBPn and CRPn in the presence of EGTA, which depletes Ca2+, thus mimicking a possible infection condition. The resulting apparent KD was 3.1 × 10−10 m, a 1000-fold increase in affinity (Fig. 6C), similar to that between GBPi and CRPi. However, supplementing the infected proteins with a physiological level of 2.5 mm Ca2+, or even higher, at 10 mm Ca2+ did not revert the affinity between GBPi and CRPi to basal level (Fig. 6, D and E). Because Ca2+ did not affect the GBP-LPS interaction (supplemental Fig. 6), these results indicate that infection causes irreversible conformational changes to the PRRs (GBP-CRP interaction), which likely recruit other proteins (14) to form the pathogen-recognition interactome (Fig. 6F and supplemental Fig. 7).
Antiendotoxic Potentials of GBP and CRP
Because GBP and CRP bind LPS, we tested their antiendotoxic potentials. First, we confirmed that the purified GBP and CRP were pyrogen-free. Then, we showed that when reacted with increasing doses of LPS (0.5–2 enzyme units), the proteins bound and probably disrupted the LPS micelles to increase the overall endotoxicity (supplemental Fig. 8).
Human Tectonin, a Domain Architectural Homolog of GBP
Although protein sequence BLAST against the human genome did not reveal any homologs of GBP, SMART analysis (9, 40) yielded three hypothetical proteins, Q7Z6L1, Q15040, and O95714, which contain Tectonin domains (15). The QZ7L1 was found to interact with human ficolin (15), the homolog of horseshoe crab carcinolectin 5. We have shown earlier that during infection, GBP interacts with carcinolectin 5 (12). Yeast 2-hybrid screening with hTectonin as bait against the human leukocyte cDNA library showed potential interaction partners such as neutrophil cytosol factor 1, Src-like adaptor 2, and ubiquitin-specific-processing protease (supplemental Fig. 9 and supplemental Table 3), all of which are immunoregulatory proteins.
DISCUSSION
By using both experimental and in silico approaches, we have shown that GBP, a representative Tectonin protein, with 6 β-propeller/Tectonin repeats, can distinguish host from bacteria, thus conferring self (GBP-CRP)/nonself (GBP-LPS) molecular interactions. Consistent with reports that individual β-propeller domains can self-assemble into larger multipropeller structures, the purified GBP molecules seemed to present as a mixture of monomers, dimers, and larger polymers (Fig. 1). Its propensity to homo-oligomerize may provide a supramolecular structure of Tectonin domains that contributes a stable bridge for the host-pathogen network.
Real-time biointeraction analysis showed that GBP binds strongly to LPS, ReLPS, and LA, most likely through the GlcNAc sugar moiety. Calculations of the SPR data using either the Langmuir 1:1 binding and the two-state conformation change binding of both the native GBP solution, and the dithiothreitol-treated GBP (containing more GBP monomers) showed closely similar binding affinities of 3.32 × 10−7 m and 3.78 × 10−7 m, respectively. The GBP-LPS interaction seems to show a slower association (ka) and dissociation (kd) rate compared with GBP-GlcNAc, suggesting that GBP interacts with multiple sugar moieties of LPS. Interestingly, GBP binds ReLPS with a 10-fold greater affinity compared with the full-length LPS, suggesting that other sugar moieties, e.g. glucose and 2-keto-3-deoxyoctonate in LPS (Fig. 2A), are also available for GBP to bind to.
The three-dimensional model of GBP (Fig. 3B) served as a basis for us to explore the interactions between GBP and its interacting protein partners or bacterial ligands via computational docking and simulations and provides a platform to map experimental results visually to gain a structural perspective on the molecular interactions taking place. Molecular docking of GlcNAc to the GBP model predicted that GlcNAc has the highest binding affinity for GBP, occurring between Tectonin domains 6 and 1. The lipid A structure binds GBP through its glucosamine residues instead of the fatty acid chains. Therefore, GBP recognizes and preferentially binds the glucosamine disaccharide head group of the lipid A, consistent with the observation that lipid A and GlcNAc share similar binding sites in GBP.
Yeast two-hybrid analyses of the GBP subclones suggested that three contiguous Tectonin domains are sufficient to interact as strongly as the full-length GBP with itself and with CRP. Furthermore, at least two consecutive Tectonin domains are needed for consistent interactions between GBP and CRP. HDMS showed the GBP-lipid A and GBP-CRP interfaces to be consistent with docking predictions. HDMS also confirmed that lipid A preferentially binds to the cleft between domains 6 and 1, which interfaces the β-propeller folds, whereas CRP binds at domain 4. This is consistent with our earlier observations through SPR analyses that the GBP peptides synthesized from domains 6 to 1 bind to LPS (15).
We observed that infection caused a 10-fold increase in binding affinity between GBPi and LPS, with a slower release (kd rate) of LPS from GBPi, suggesting that after initial recognition and binding to the sugar moieties, the adjacent chemical groups of the LPS molecule enhances the anchorage of GBP onto the bacterium. The effect of infection is clearly demonstrated with a 1000-fold increase in affinity between GBPi and CRPi. Furthermore, the chelation of Ca2+ seemed to mimic the state of infection by producing binding affinity similar to that in an infected condition. Fluctuations in cation levels during infection were reported (36–39) to affect protein-protein interactions and consequently, regulate the immune response. However, Ng et al. (12) showed that plasma factors other than Ca2+ may also enhance the interaction of GBP and CRP. This led us to postulate that although Ca2+ depletion seems to represent the state of infection, the conformational change in these plasma PRRs is irreversible and that their binding to PAMPs would likely enable them to recruit other PRRs (14) to form the pathogen-recognition interactome (Fig. 6F and supplemental Fig. 7), which triggers downstream effectors for opsonization by macrophages.
The physiological implication of GBP was indicated by its endotoxic potential, where its interaction with LPS increased the endotoxicity. It is likely that GBP disrupts the LPS micelles, which exposes/unmasks the endotoxic potency of LPS. We suggest that in vivo, GBP and CRP bind to the bacterial surface and break down the LPS on the outer membrane of the invading bacteria. This exposure may possibly lead to the recruitment and activation of other host factors to mount a more efficient antimicrobial response. In conclusion, we have demonstrated the structural and functional basis of the Tectonin domain-containing proteins in antimicrobial defense. The apparently similar Tectonin domains of GBP can differentiate self from nonself. The horseshoe crab and the human are separated by ∼500 million years of evolutionary distance, yet the remarkable conservation in the architecture and function of Tectonin domain-containing proteins suggests their critical role in frontline defense against microbes.
Supplementary Material
Acknowledgments
We thank Zhang Jing for the ficolin clone, Michelle Mok (Proteins and Proteomics Centre) for help with mass spectrometry, and Dr. S. Kawabata (Kyushu University) for the three-dimensional atomic coordinates of TL-1.
This work was supported by grants from the Singapore-MIT Alliance, Computational and Systems Biology and the AcRF (T208B3109, MoE).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–9, Tables 1–3, “Materials and Methods,” and additional references.
H.-G. Beisel, S.-I. Kawabata, W. Bode, and S. Iwanaga, unpublished data.
- PAMP
- pathogen-associated molecular pattern
- CRP
- C-reactive protein
- ELISA
- enzyme-linked immunosorbent assay
- GBP
- galactose-binding protein
- GlcNAcOAc
- 2-N-acetyl-3-O-acetyl-β-d-glucosamine
- HDMS
- hydrogen deuterium exchange mass spectrometry
- LPS
- lipopolysaccharide
- LTA
- lipoteichoic acid
- PRR
- pattern-recognition receptor
- ReLPS
- LPS from Salmonella minnesota Re 595
- SPR
- surface plasmon resonance
- TL-1
- tachylectin-1
- TL-2
- tachylectin-2.
REFERENCES
- 1.Fülöp V., Jones D. T. (1999) Curr. Opin. Struct. Biol. 9, 715–721 [DOI] [PubMed] [Google Scholar]
- 2.Jawad Z., Paoli M. (2002) Structure 10, 447–454 [DOI] [PubMed] [Google Scholar]
- 3.Beisel H. G., Kawabata S., Iwanaga S., Huber R., Bode W. (1999) EMBO J. 18, 2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S. C., Yen C. H., Yeh M. S., Huang C. J., Liu T. Y. (2001) J. Biol. Chem. 276, 9631–9639 [DOI] [PubMed] [Google Scholar]
- 5.Huh C. G., Aldrich J., Mottahedeh J., Kwon H., Johnson C., Marsh R. (1998) J. Biol. Chem. 273, 6565–6574 [DOI] [PubMed] [Google Scholar]
- 6.Rakotobe D., Violot S., Hong S. S., Gouet P., Boulanger P. (2008) Virol. J. 5, 32 (abstr.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito T., Kawabata S., Hirata M., Iwanaga S. (1995) J. Biol. Chem. 270, 14493–14499 [DOI] [PubMed] [Google Scholar]
- 8.Schröder H. C., Ushijima H., Krasko A., Gamulin V., Thakur N. L., Diehl-Seifert B., Müller I. M., Müller W. E. (2003) J. Biol. Chem. 278, 32810–32817 [DOI] [PubMed] [Google Scholar]
- 9.Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwanaga S. (2002) Curr. Opin. Immunol. 14, 87–95 [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Ng P. M., Ho B., Ding J. L. (2007) Eur. J. Immunol. 37, 3477–3488 [DOI] [PubMed] [Google Scholar]
- 12.Ng P. M., Le Saux A., Lee C. M., Tan N. S., Lu J., Thiel S., Ho B., Ding J. L. (2007) EMBO J. 26, 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng P. M., Jin Z., Tan S. S., Ho B., Ding J. L. (2004) J. Endotoxin Res. 10, 163–174 [DOI] [PubMed] [Google Scholar]
- 14.Le Saux A., Ng P. M., Koh J. J., Low D. H., Leong G. E., Ho B., Ding J. L. (2008) J. Mol. Biol. 377, 902–913 [DOI] [PubMed] [Google Scholar]
- 15.Low D. H. P., Ang Z. W., Yuan Q., Frecer V., Ho B., Chen J., Ding J. L. (2009) PLoS ONE 4, e6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deleted in proof
- 17.Petsko G. A., Dagmar R. (2004) Protein Structure and Function, New Science Press Ltd., Sinauer Associates, Sunderland, MA [Google Scholar]
- 18.Ramadan M. A., Shrive A. K., Holden D., Myles D. A., Volanakis J. E., DeLucas L. J., Greenhough T. J. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 992–1001 [DOI] [PubMed] [Google Scholar]
- 19.de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 20.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 21.Hubbard S., Thornton J. (1996) NACCESS, version 2.1.1 [Google Scholar]
- 22.Deleted in proof
- 23.Deleted in proof
- 24.Wimmerova M., Mitchell E., Sanchez J. F., Gautier C., Imberty A. (2003) J. Biol. Chem. 278, 27059–27067 [DOI] [PubMed] [Google Scholar]
- 25.Cioci G., Mitchell E. P., Chazalet V., Debray H., Oscarson S., Lahmann M., Gautier C., Breton C., Perez S., Imberty A. (2006) J. Mol. Biol. 357, 1575–1591 [DOI] [PubMed] [Google Scholar]
- 26.Yadid I., Tawfik D. S. (2007) J. Mol. Biol. 365, 10–17 [DOI] [PubMed] [Google Scholar]
- 27.Hoofnagle A. N., Resing K. A., Ahn N. G. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 1–25 [DOI] [PubMed] [Google Scholar]
- 28.Mandell J. G., Falick A. M., Komives E. A. (1998) Anal. Chem. 70, 3987–3995 [DOI] [PubMed] [Google Scholar]
- 29.Kawabata S., Iwanaga S. (1999) Dev. Comp. Immunol. 23, 391–400 [DOI] [PubMed] [Google Scholar]
- 30.Daiyasu H., Saino H., Tomoto H., Mizutani M., Sakata K., Toh H. (2008) J. Biochem. 144, 467–475 [DOI] [PubMed] [Google Scholar]
- 31.Naberezhnykh G. A., Gorbach V. I., Likhatskaya G. N., Davidova V. N., Solov'eva T. F. (2008) Biochemistry (Mosc.) 73, 432–441 [DOI] [PubMed] [Google Scholar]
- 32.Vilar S., Cozza G., Moro S. (2008) Curr. Top. Med. Chem. 8, 1555–1572 [DOI] [PubMed] [Google Scholar]
- 33.Frecer V., Ho B., Ding J. L. (2000) Eur. J. Biochem. 267, 837–852 [DOI] [PubMed] [Google Scholar]
- 34.Zhang J., Koh J., Lu J., Thiel S., Leong B. S., Sethi S., He C. Y., Ho B., Ding J. L. (2009) PLoS Pathog. 5, e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aslam S. N., Newman M. A., Erbs G., Morrissey K. L., Chinchilla D., Boller T., Jensen T. T., De Castro C., Ierano T., Molinaro A., Jackson R. W., Knight M. R., Cooper R. M. (2008) Curr. Biol. 18, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 36.Blackwell J. M., Searle S., Goswami T., Miller E. N. (2000) Microbes Infect. 2, 317–321 [DOI] [PubMed] [Google Scholar]
- 37.Maguire M. E. (2006) Front. Biosci. 11, 3149–3163 [DOI] [PubMed] [Google Scholar]
- 38.Ong S. T., Ho J. Z., Ho B., Ding J. L. (2006) Immunobiology 211, 295–314 [DOI] [PubMed] [Google Scholar]
- 39.Papp-Wallace K. M., Maguire M. E. (2006) Annu. Rev. Microbiol. 60, 187–209 [DOI] [PubMed] [Google Scholar]
- 40.Letunic I., Copley R. R., Pils B., Pinkert S., Schultz J., Bork P. (2006) Nucleic Acids Res. 34, D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case D. A., Cheatham T. E., III, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 16, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.